To the Editor: Gastric cancer (GC) is one of the most commonly diagnosed malignancies worldwide.[1,2] Recent studies show that an aberrant gastric microbiota contributes to the onset and progression of GC.[3] A distinct cluster of bacteria consisting of Peptostreptococcus, Streptococcus, and Parvimonas among others is associated with gastric atrophy and intestinal metaplasia (IM).[4] However, the gastric microbiome is highly dynamic and influenced by several factors, including diet, xenobiotics, proton pump inhibitors, physiological changes, and host genetics.
The present study included data of 60 patients (mean age 55 ± 13 years; 58.3% male) that presented at the First Affiliated Hospital of Nanjing Medical University from February 2017 to March 2018, including 17 with chronic gastritis (CG), 13 patients with IM, and 30 individuals with GC. This study was approved by the First Affiliated Hospital of Nanjing Medical University's Ethics Committee (No. 2017-SRFA-180) and all participants provided written informed consent before participation. Gastric biopsy samples were obtained from the gastric antrum and corpus of CG patients, and targeted biopsies were obtained from patients with IM or GC.
The gut bacteria community structure was examined by sequencing the V3-V4 region of 16S rRNAs in gastric tissue samples on the Illumina MiSeq platform according to the manufacturer's instructions (Illumina technologies, USA). The microbiota community structure was analyzed with the Quantitative Insights Into Microbial Ecology (QIIME) pipeline using high-quality sequences. The taxonomy of the representative optical transform units (OTUs) was determined using the UCLUST classifier with Greenegenes (13_8 release, Lawrence Berkeley National Laboratory, CA, USA) as a reference dataset. The taxa with significantly different relative abundance between groups were identified using linear discriminant analysis effect size (LEfSe) (P < 0.05), and Wilcoxon test was used to identify significantly different OTUs (P < 0.05). Leave-one-out cross-validation (LOOCV) was performed on the random forest model (R 3.4.1, random forest 4.6–12 package, Datagurn, New Zealand). One-way analysis of variance was used to compare three groups. Continuous variables were compared between two groups using the Wilcoxon rank-sum test, and Fisher exact test was used for categorical variables. All statistical analyses were performed using SPSS v.20.0 software (SPSS, Chicago, IL, USA).
The microbiota structure of the CG, IM, and GC groups were compared in terms of alpha diversity and beta diversity. The Chao Index and diversity were both significantly elevated in the GC group compared to the others (all P < 0.05) [Figure 1A and 1B], whereas no significant difference was seen in the bacterial diversity between CG and IM groups. Beta diversity was calculated using principal co-ordinates analysis. The space between the three groups indicated that the microbiota composition of GC patients was significantly different from that of CG or IM patients [Figure 1C].
Figure 1.
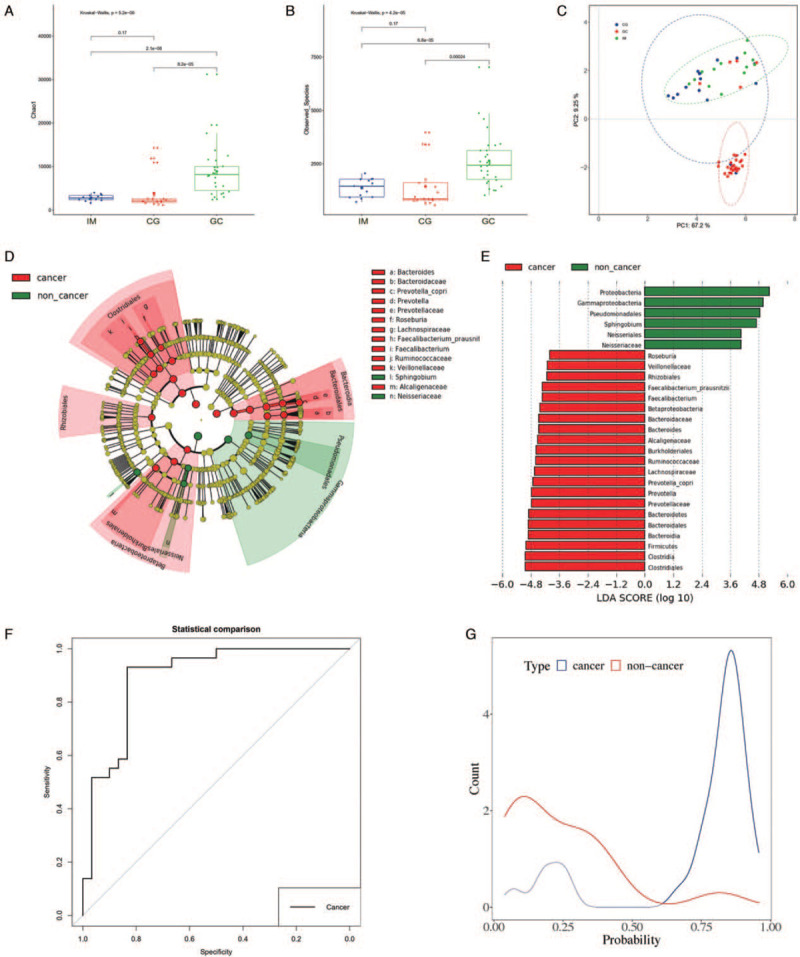
Alteration of gastric microbiota in gastric cancer. (A) Alpha diversity was estimated by the Chao Index. (B) Alpha diversity was estimated by Shannon Index. (C) Beta diversity was estimated by PCoA. (D) Cladogram representation of the gastric microbiota taxa associated with non-GC and GC. (E) Association of specific microbiota taxa with non-GC and GC by LDA effect size. (F) ROC analysis for LOOCV model. (G) Density figure for LOOCV model. CG: Chronic gastritis; GC: Gastric cancer; IM: Intestinal metaplasia; LDA: Linear discriminant analysis; LOOCV: Leave-one-out cross-validation; PCoA: Principle coordination analysis; ROC: Receiver operating characteristic.
The CG and IM groups were included into the non-GC group for further analyses. LEfSe analysis identified 21 and 6 OTUs that were significantly altered in the GC and non-GC groups respectively [Figure 1D and 1E]. To illustrate the diagnostic value of the gastric microbiome biomarker for GC, a LOOCV model was used to differentiate GC samples from non-GC samples. The fivefold cross-validation identified a 19 OTU-biomarker set (Barnesiellaceae, Phascolarctobacterium, Bacteroides uniformis, Clostridium, Trabulsiella, Lachnospira, Roseburia, Prevotella copri, Butyricimonas, Deinococci, Prevotella, Deinococcales, Thermi., Prevotellaceae, Alcaligenaceae, Dialister, Ruminococcus, Sutterella, and Bifidobacteriaceae). Predicted values higher than 0.55 indicated a high risk for GC, while values lower than 0.55 were indicative of a low risk [Figure 1F]. The LOOCV model achieved an area under the curve value of 89.3% in the discovery phase [Figure 1G], sensitivity of 83.33%, specificity 90%, false-positive rate 10%, and false negative rate 16.67%.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (USA, http://huttenhower.sph.harvard.edu/galaxy/) was used to predict the gastric microbiota function. The abundance of 29 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was significantly enriched in the GC samples, and most were associated with metabolism, such as primary bile acid biosynthesis, secondary bile acid biosynthesis, sphingolipid metabolism, biosynthesis of ansamycins, butirosin and neomycin biosynthesis, cyanoamino acid metabolism, drug metabolism, flavone and flavonol biosynthesis, fructose and mannose metabolism, glycosaminoglycan degradation, glycosphingolipid biosynthesis, pentose and glucuronate interconversions, steroid hormone biosynthesis, and nucleotide metabolism (all P < 0.05). Interestingly, the abundance of folate biosynthesis and gastric acid secretion pathways were significantly decreased in the GC samples (P < 0.05).
LEfSe analysis has been used previously to identify GC-associated specific microbial taxa and construct the diagnostic model using microbial dysbiosis index.[5] Another study used a sparse compositional correlation (SparCC) algorithm to perform a correlation analysis, which is known for its robustness of compositional effects that are influenced by the diversity and sparsity of correlation in human microbiome data sets.[6] From our biomarker set, Clostridium spp. and Prevotella spp. were also reported in the LEfSe analysis,[5] while B. uniformis and P copri have been reported in the study using SparCC algorithm.[6] LOOCV has the following advantages over the two aforementioned approaches: (1) every OTU data point is tested separately to avoid the influence of grouping, (2) almost all data points (n–1 OTUs) are used in the training model to decrease bias, and (3) no random factors are used to ensure reproducibility. However, LOOCV has the disadvantages of high cost and the time required for computation. Nevertheless, all three models are highly efficient and it is critical to select the correct computation method.
The signaling pathways of GC-associated microbiota are largely unknown. According to our KEGG analysis, primary and secondary bile acid biosynthesis were enriched in the GC group. The bile acid receptor Takeda-G protein receptor-5 is overexpressed in GS cells and promotes epithelial-mesenchymal transition,[7] and is also associated with decreased survival in patients with gastric adenocarcinomas.[8] Interestingly, a positive correlation was found between bile acid concentration and the grade of atrophy/IM in Heliobacter pylori (H. pylori)-positive but not H. pylori-negative patients.[9] This suggests that bile acid may play an important role in H. pylori-related gastritis and GC.
In conclusion, gastric microbiota species are useful diagnostic biomarkers of GC, although they need to be validated in a much larger sample. Furthermore, identification of key bacterial species and signaling pathways associated with GC development lends more insights into the underlying mechanisms.
Conflicts of interest
None.
Footnotes
How to cite this article: Dang YN, Dong Y, Mu YZ, Yan J, Lu M, Zhu YL, Zhang GX. Identification of gastric microbiota biomarker for gastric cancer. Chin Med J 2020;133:2765–2767. doi: 10.1097/CM9.0000000000001081
Yi-Ni Dang and Yu Dong contributed equally to this work.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Chen XZ, Huang CZ, Hu WX, Liu Y, Yao XQ. Gastric cancer screening by combined determination of serum Helicobacter pylori antibody and pepsinogen concentrations: ABC method for gastric cancer screening. Chin Med J 2018; 131:1232–1239. doi: 10.4103/0366-6999.231512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 2011; 140:210–220. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sung JJY, Coker OO, Chu E, Szeto CH, Luk STY, Lau HCH, et al. Gastric microbes associated with gastric inflammation, atrophy and intestinal metaplasia 1 year after Helicobacter pylori eradication. Gut 2020; 69:1572–1580. doi: 10.1136/gutjnl-2019-319826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018; 67:226–236. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019; 40:336–348. doi: 10.1016/j.ebiom.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carino A, Graziosi L, D’Amore C, Cipriani S, Marchianò S, Marino E, et al. The bile acid receptor GPBAR1 (TGR5) is expressed in human gastric cancers and promotes epithelial-mesenchymal transition in gastric cancer cell lines. Oncotarget 2016; 7:61021–61035. doi: 10.18632/oncotarget.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao WB, Tian W, Hong J, Li D, Tavares R, Noble L, et al. Expression of bile acid receptor TGR5 in gastric adenocarcinoma. Am J Physiol Gastrointest Liver Physiol 2013; 304:G322–G327. doi: 10.1152/ajpgi.00263.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatsugami M, Ito M, Tanaka S, Yoshihara M, Matsui H, Haruma K, et al. Bile acid promotes intestinal metaplasia and gastric carcinogenesis. Cancer Epidemiol Biomarkers Prev 2012; 21:2101–2107. doi: 10.1158/1055-9965.EPI-12-0730. [DOI] [PubMed] [Google Scholar]


