ABSTRACT
The choroid plexus (ChP) is a secretory tissue that produces cerebrospinal fluid (CSF) secreted into the ventricular system. It is a monolayer of secretory, multiciliated epithelial cells derived from neuroepithelial progenitors and overlying a stroma of mesenchymal cells of mesodermal origin. Zfp423, which encodes a Kruppel-type zinc-finger transcription factor essential for cerebellar development and mutated in rare cases of cerebellar vermis hypoplasia/Joubert syndrome and other ciliopathies, is expressed in the hindbrain roof plate, from which the IV ventricle ChP arises, and, later, in mesenchymal cells, which give rise to the stroma and leptomeninges. Mouse Zfp423 mutants display a marked reduction of the hindbrain ChP (hChP), which: (1) fails to express established markers of its secretory function and genes implicated in its development and maintenance (Lmx1a and Otx2); (2) shows a perturbed expression of signaling pathways previously unexplored in hChP patterning (Wnt3); and (3) displays a lack of multiciliated epithelial cells and a profound dysregulation of master genes of multiciliogenesis (Gmnc). Our results propose that Zfp423 is a master gene and one of the earliest known determinants of hChP development.
KEY WORDS: Choroid plexus, Patterning, Development, Hindbrain, Wnt, BMP, Cilium, Multiciliated cells, Multiciliated epithelium, Microvilli, Zfp423, ZNF423, Joubert syndrome, JS19, Hydrocephalus
Summary: The zinc-finger protein 423, a transcription factor and protein-protein interaction hub, is identified as a master gene in hindbrain choroid plexus patterning and multiciliogenesis.
INTRODUCTION
The choroid plexus (ChP) is a secretory structure consisting of an epithelial cell layer vascularized by fenestrated blood vessels. The ChP produces cerebrospinal fluid (CSF) and releases it into all ventricles of the vertebrate brain, from which the CSF flows into the central canal of the spinal cord and into the subarachnoid space (Damkier et al., 2013; Lehtinen and Walsh, 2011). The CSF plays an essential role in the CNS (Lehtinen et al., 2013) and contains multiple signals, including growth factors necessary for neural development (Parada et al., 2006; Zappaterra et al., 2007). Conversely, CSF overproduction, obstructed flow or inadequate resorption lead to hydrocephalus (Damkier et al., 2013). Defects in the development and maintenance of ChP can lead to hydrocephalus and ventriculomegaly (Liu et al., 2005; Spassky and Meunier, 2017). Thus, understanding the mechanisms underlying the establishment of the ChP is important for the diagnosis and prevention of hydrocephalus.
The ChP develops on the dorsal aspect of the neural tube at three locations: fourth, third and lateral ventricles in the rhombencephalon, diencephalon and telencephalon, respectively. Morphologically and functionally, it is conserved from lower vertebrates to humans (Brocklehurst, 1979). The hindbrain ChP (hChP) of the fourth ventricle appears first, followed by the appearance of the telencephalic ChP (tChP) in the lateral ventricles, and finally by the diencephalic ChP in the third ventricle (Currle et al., 2005; reviewed by Lehtinen et al., 2013). The secretory role of the ChP reflects its structure that consists of a monolayer of cuboidal epithelial cells, which undergo a series of morphological transformations, molecularly and temporally regulated during development (Awatramani et al., 2003). ChP epithelial cells, a specialized form of ependyma, derive from neuroepithelial progenitors. At first, they are pseudostratified, then become columnar, and eventually monostratified and cuboidal (Sturrock, 1979; Tennyson and Pappas, 1964); they are characterized by the presence of tufts of cilia and microvilli. Transition through these distinct stages is accompanied by structural changes such as epithelial polarization and the formation of numerous microvilli and several cilia along the apical domain of the cell membrane (Sturrock, 1979). Whereas mature ChP epithelia are histologically and ultrastructurally indistinguishable, irrespective of location, tChP and hChP are heterogeneous with regard to their transcriptional profiles and positional identities, and they are also functionally distinct, secreting two region-specific varieties of CSF (Lun et al., 2015).
Hindbrain ChP specification occurs between embryonic day 8.5 (E8.5) and E9.5 (Thomas and Dziadek, 1993), 2-3 days before hChP morphology unveils itself. The hChP epithelium derives from neuroectodermal roof-plate progenitors positive for the bone morphogenetic protein (BMP) family ligand GDF7 (Currle et al., 2005), whereas the stromal component comprises mesenchymal cells of mesodermal origin (Wilting and Christ, 1989). The adoption of ChP cell fate is established early in development and requires the expression of Notch signaling targets Hes1, Hes3 and Hes5 (Imayoshi et al., 2008). The LIM-homeobox protein Lmx1a is expressed in the hindbrain rhombic lip (Chizhikov et al., 2010). In Lmx1a−/− (Dreher) mice, the hindbrain roof plate does not form, leading to a failure of hChP development (Millonig et al., 2000). Likewise, conditional Otx2 deletion in E9 embryos impairs the development of all ChPs, whereas its abrogation at a later stage (E15) affects only the hChP (Johansson et al., 2013). The role played at later stages (E12.5-E14.0) by the lower rhombic lip (LRL) in hChP progenitor proliferation has been elegantly described: early-born, roof plate-derived hChP epithelial cells differentiate, exit the cell cycle and start releasing the mitogen sonic hedgehog (SHH), which promotes the expansion of a mitotic progenitor pool located in the transition zone, adjacent to the LRL (Huang et al., 2009b). Conversely, the role played in hChP development by the upper rhombic lip (URL), if any, has not been entirely clarified. More broadly, the earliest molecular mechanisms leading to hChP formation are incompletely characterized.
The Zfp423 gene and its human homolog ZNF423 encode a 30 zinc-finger transcription factor involved in key developmental pathways. Zfp423 acts both as a transcription factor and as a scaffold for multiple protein-protein interactions with signaling molecules involved in early patterning, cell fate specification, neurogenesis and various aspects of differentiation (Casoni et al., 2017; Hata et al., 2000; Hong and Hamilton, 2016; Huang et al., 2009a; Masserdotti et al., 2010; Massimino et al., 2018; Tsai and Reed, 1998). Moreover, Zfp423 has been implicated in the DNA damage response in vitro (Chaki et al., 2012; Ku et al., 2003) and in vivo (Casoni et al., 2017). Although all homozygous mutants described to date exhibit severe cerebellar malformations (Alcaraz et al., 2006; Casoni et al., 2017; Cheng et al., 2007; Warming et al., 2006), two allelic mutant lines described by our group, each of which has deletion of a distinct functional domain, have revealed domain-specific functions in cerebellar ventricular zone neurogenesis (Casoni et al., 2017). Zfp423, which is required for proper cerebellar granule cell development, also plays an important role in ciliary development/function in mitotic monociliated neurogenic progenitors of the external granular layer (Hong and Hamilton, 2016). In humans, ZNF423 mutations have been linked to cerebellar vermis hypoplasia, Joubert syndrome 19 and nephronophthisis 14 (Chaki et al., 2012). In addition, an allelic series of human mutations has been carefully characterized in the mouse by CRISPR/Cas9 knock/in (Deshpande et al., 2020). In this article, we report the expression of the Zfp423 protein in the roof plate and developing hChP. Moreover, by analyzing a C-terminal Zfp423 mutant (Δ28-30) (Casoni et al., 2017), which displays a near-complete deletion of this structure, we examine the roles played by the homonymous protein at the earliest stages of hChP morphogenesis, and its contribution to the development of mesenchymal and epithelial components of this tissue.
RESULTS
Zfp423 mutation causes hChP hypoplasia and downregulation of functional markers
In order to study the role of Zfp423 in ChP development, we analyzed a previously described mouse line (Casoni et al., 2017) that produces a C-terminally deleted protein. In this line, called Zfp423Δ28-30, nucleotides 3861-4108 of the ORF have been removed, causing a frameshift mutation that produces a stop codon 158 bp into exon 8. The truncated protein loses 91 C-terminal amino acids, including zinc-finger 28-30.
The analysis of Nissl-stained P18 cerebellar frontal sections revealed the presence of an elaborate hChP in the fourth ventricle in wild-type embryos, which was completely absent in the mutant (Fig. 1A,A′), except for a very small segment of ChP tissue at the very lateral edge of the ventricle (Fig. 1A′, inset). This observation prompted further studies of hChP development. Parasagittal sections were analyzed at earlier ontogenetic stages. At E14.5, mutants displayed an enlarged fourth ventricle and a rudimentary hChP primordium (Fig. 1B′, red arrowhead) when compared with wild-type littermates (Fig. 1B, white arrowhead). However, in parasagittal sections of the telencephalon, the primordium of the tChP shows no significant alterations (Fig. 1C,C′, white arrowheads), suggesting that Zfp423 specifically affects hChP development in the fourth ventricle.
Fig. 1.
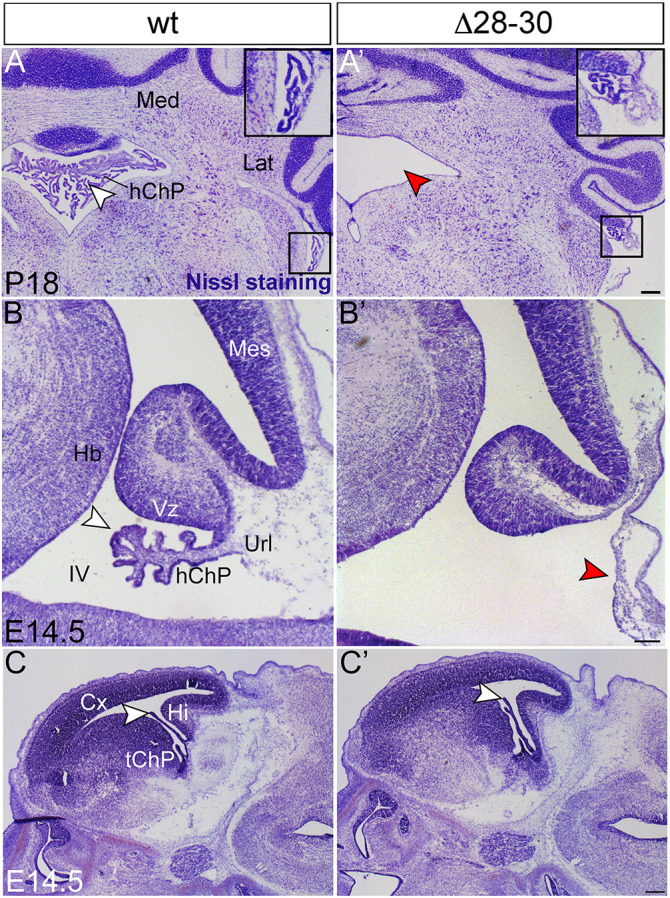
Profound malformation of the mutant hChP. (A,A′) Nissl staining of P18 mice frontal sections reveals the near-complete absence of the hindbrain ChP (hChP) in the fourth ventricle of the Zfp423 mutant (red arrowhead in A′) compared with the wild type (white arrowhead in A). A small lateral segment of the hChP is visible in wild type and mutants (insets). (B,B′) Nissl staining of E14.5 parasagittal sections shows a severe hChP hypoplasia in the mutant compared with wild type, with a flattened surface (red arrowhead in B′) compared with the wild type (white arrowhead in B). (C,C′) Conversely, the mutant tChP shows a more-normal morphology compared with the wild type (white arrowhead in C,C′). Med, medial cerebellar nucleus; Lat, lateral nucleus; Cx, cortex; hChP, hindbrain ChP; tChP, telencephalic ChP; Hb, ventral hindbrain; Hi, hippocampus; IV, fourth ventricle; Mes, mesencephalon; Url, rhombic lip; Vz, ventricular zone. Scale bars: 200 μm in A; 100 μm in B; 400 μm in C.
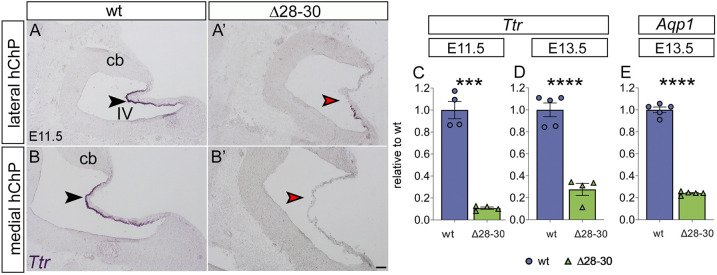
Transthyretin (Ttr) is one of the earliest functional markers of the ChP: it is a protein that transports the thyroid hormone thyroxine (T4) and retinol-binding protein bound to retinol (Power et al., 2000; Raz and Goodman, 1969) from ChP epithelial cells into the CSF. In situ hybridization was used to analyze Ttr expression in the E11.5 hChP. Our results indicate that the expression of the Ttr gene is dramatically reduced in the mutant (Fig. 2A′,B′, red arrowheads) compared with controls (Fig. 2A,B, black arrowheads). Moreover, using quantitative RT-PCR (RT-qPCR) conducted on embryonic hindbrain mRNA (Fig. 2C-E), we analyzed mRNA levels of Ttr and aquaporin 1 (Aqp1), which encodes a water transporter expressed at high levels in the ChP. Our results confirmed a sharp downregulation of Ttr in mutants at E11.5 (***P<0.0005, Fig. 2C) and E13.5 (***P<0.0001, Fig. 2D). Similarly, Aqp1 mRNA levels were sharply decreased in mutants at E13.5 (****P<0.0001, Fig. 2E). Thus, the Zfp423 mutation affects both hChP morphology and function starting at early stages of hChP embryonic development.
Fig. 2.
Functional markers of the hChP are downregulated in the Zfp423-mutant hChP during development. (A-B′) In situ hybridization of parasagittal E11.5 sections, lateral and medial, carried out with a Ttr riboprobe. Ttr is expressed both in lateral and in medial parasagittal hChP sections (black arrowheads in A, B). In the mutant, Ttr is downregulated and the expression is patchy in lateral sections of the mutant hChP epithelium (red arrowhead in A′), whereas it is completely absent in medial parasagittal sections (red arrowhead in B′). (C-E) RT-qPCR reveals a sharp downregulation of Ttr in mutants at E11.5 (C, n=4/genotype) and at E13.5 (D, n=6/wild type, n=5/Δ28-30 mutant). The Aqp1 gene is also downregulated in mutants at E13.5 (E, n=5/genotype). Data are mean±s.e.m. ****P<0.0001, ***P<0.0005; Welch's unequal variances t-test. cb, cerebellum; IV, fourth ventricle. Scale bar: 100 μm.
ZFP423 is expressed in early epithelial progenitors of the prospective hChP
To define the possible role of Zfp423 in hChP development, we first investigated the precise distribution of the corresponding protein in the hChP. To determine whether Zfp423 is expressed at the earliest stages of hindbrain and hChP development, we performed LacZ staining on heterozygous embryos derived from a gene trap line containing an integration of the β-galactosidase gene within intron 1 of the Zfp423 gene (Wurst et al., 1995). Our results indicated that Zfp423 is expressed starting at the neurula stage (E7.5. Fig. 3A) and remains expressed at E8.5 (Fig. 3B) and E9.5 (Fig. 3C) in the hindbrain. Furthermore, we immunostained sagittal sections of the hChP at E10.5 and E12.5 (Fig. 3D and E, respectively) with a previously characterized anti Zfp423 polyclonal antibody (Casoni et al., 2017; Hong and Hamilton, 2016; Shao et al., 2016). At E10.5, Zfp423 is expressed exclusively in the hChP epithelium cells facing the ventricular lumen (arrowheads in Fig. 3D), which contains proliferating basal progenitors. At E12.5, Zfp423 continues to be expressed in the proliferating hChP epithelium, labeled by the S-phase marker EdU (arrows in in Fig. 3E), but is excluded from mitotically quiescent ChP cells (arrowheads in Fig. 3E). These results indicated that Zfp423 is robustly expressed at the earliest stages of roof plate and dorsal hindbrain patterning.
Fig. 3.
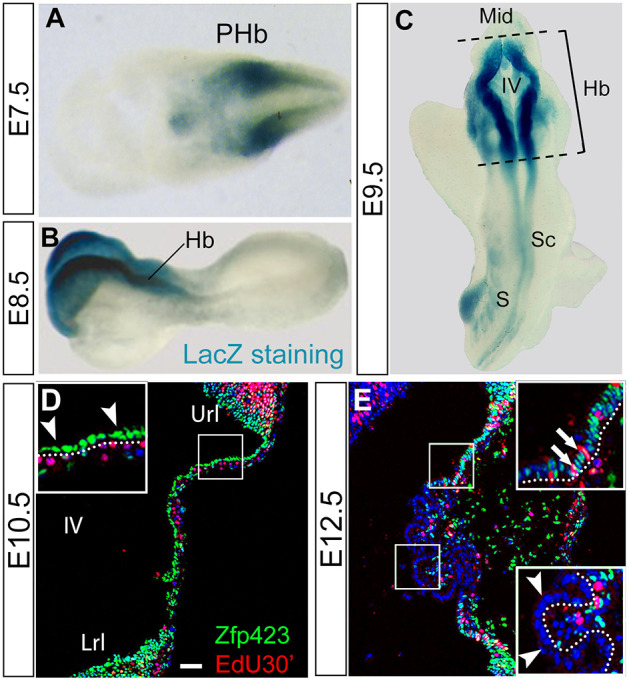
ZFP423 is expressed in ectodermal and mesodermal derivatives of the developing hChP. (A-C) Embryos from a Zfp423 β-galactosidase gene trap line (see text) were analyzed by LacZ staining, showing that Zfp423 starts to be expressed at E7.5 in the neuroectoderm, and at E8.5 and E9.5 is strongly expressed in the hindbrain. (D,E) Immunostaining of parasagittal cerebellar sections. At E10.5 (D), Zfp423 is present in the progenitors of the hChP epithelium (arrowheads) adjacent to the IV ventricle. At E12.5 (E), Zfp423 is expressed exclusively in the proliferating epithelial cells labeled by EdU (arrows), whereas it is completely absent in the mitotically quiescent cells (arrowheads). PHb, presumptive hindbrain; Hb, hindbrain; Mid, midbrain; Sc, spinal cord; S, somites; IV, fourth ventricle; Url, upper rhombic lip; Lrl, lower rhombic lip. Scale bar: 75 μm in A; 125 μm in B; 175 μm in C; 50 μm in D,E.
Because results previously obtained in apical progenitors of the cerebellar ventricular zone (Casoni et al., 2017) indicated an accumulation of DNA damage and a delay in cell cycle progression between G2 and M phases, we analyzed the number of cells labeled by the DNA damage marker γH2AX in the wild-type and mutant hChP, and found a slight, yet significant, increase in the Δ28-30. Likewise, by calculating the ratio of dotted (G2 phase) to full (M phase) phosphohistone H3 signal, we determined that cell cycle progression from G2 to M is moderately delayed in mutant hChP progenitors (Fig. S1A-D). Despite these findings, we did not observe any increase in programmed cell death at E10.5, as shown by activated caspase 3 immunostaining (Fig. S1E-G′), or at E12.5, as shown by TUNEL staining (Fig. S6).
Early development of the hChP stroma is only mildly affected in Zfp423 mutants
To begin dissecting the cascade of cell-cell interactions leading to hChP malformation in Zfp423 mutants, we used immunofluorescence to analyze markers of the hChP stroma. To this end, we immunostained the hChP for: (1) platelet endothelial cell adhesion molecule 1 (PECAM1), to visualize endothelial cells; (2) desmin, for pericytes, which regulate blood-CSF barrier permeability (Armulik et al., 2010; Daneman et al., 2010); (3) collagen type IV for the lamina densa of the basement membrane secreted by epithelial and endothelial cells; and (4) Akap12 and Igf2 for prospective leptomeninges. Our results revealed that: (1) capillaries are present at E11.5 in the hChP stroma in wild-type and mutant embryos alike, albeit discontinuous in the latter (Fig. 4A,A′, white arrowheads); (2) desmin+ pericytes are maintained in mutant embryos at E12.5 (Fig. 4B,B′); (3) collagen type IV is present in the basal membrane of the mutant hChP at E14.5 (Fig. 4C,C′); and (4) meningeal markers are either mildly downregulated or unchanged in the mutant at early stages, as shown by RT-qPCR (Fig. 4D). These findings suggest that the blood supply to the hChP is relatively preserved in Zfp423 mutants at early stages of development, and that the basement membrane is conserved, albeit reduced in its extension.
Fig. 4.
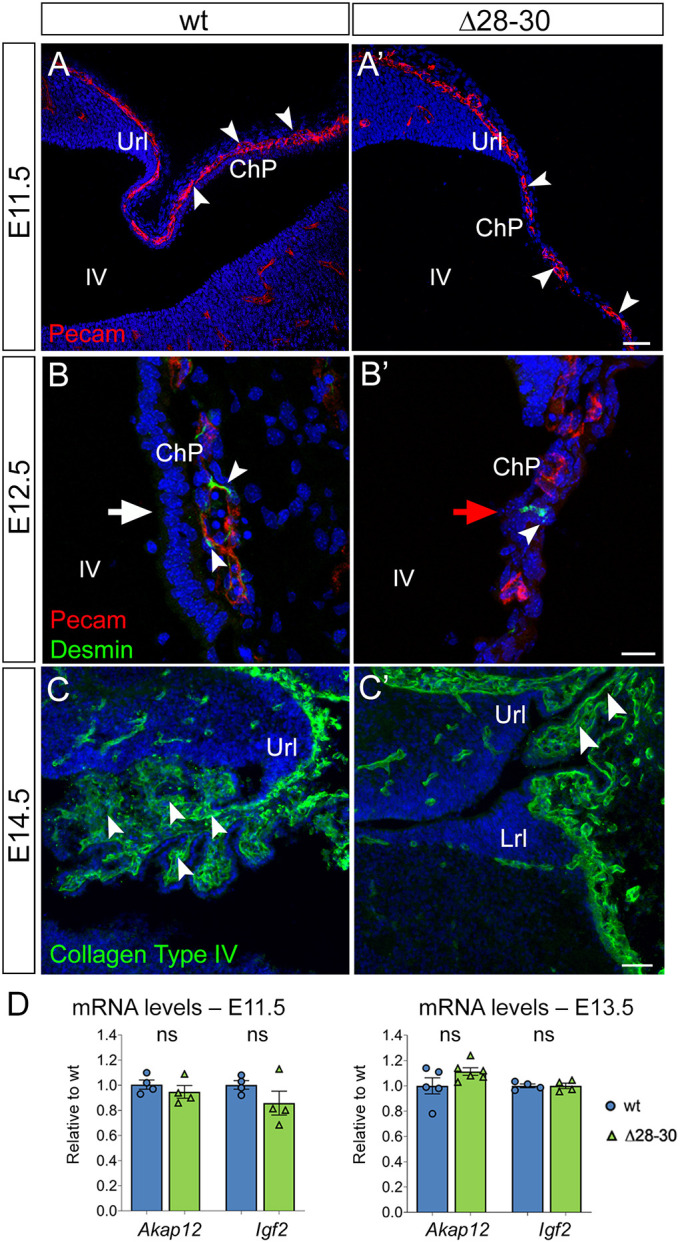
Vascular and pericyte markers are expressed in the mutant hChP. (A-C′) Parasagittal cerebellar sections harvested at different developmental stages and immunostained for different markers of the hChP stroma, vascular tree and basement membrane. (A,A′) Pecam-positive capillaries are present in the mutant hChP (white arrowheads in A and A′) at E11.5, albeit reduced in size. (B,B′) E12.5 desmin-positive pericytes are detected in mutant embryos. Epithelial nuclei are stratified in B (white arrow) and lacking in B′ (red arrow). White arrowheads indicate desmin-positive pericytes. (C,C′) Collagen type IV is present in the basal membrane of the mutant hChP at E14.5. White arrowheads indicate collagen type IV-positive basement membrane. (D) RT-qPCR reveals slight downregulation or unchanged expression of Akap12 and Igf2 in the mutant hindbrain at E11.5 and E13.5. ns, not significantly different. n=4/genotype at E11.5; n=5/wild type at E13.5 and n=6/Δ28-30 mutant for Akap12; n=4/genotype for Igf2 at E13.5. Results are mean±s.e.m. Welch's unequal variances t-test was used to test for significance. Url, upper rhombic lip; Lrl, lower rhombic lip; ChP, ChP; IV, fourth ventricle. Scale bars: 50 μm in A,A′,C,C′; 20 μm in B,B′.
Changes in microvilli and tight junctions
As mentioned previously, ChP epithelial cells exhibit microvilli and cilia on their apical plasma membrane. Microvilli expand the luminal surface of the epithelial cell to increase the capacity of the ChP to secrete CSF. Ezrin is expressed in the ChP (Gimeno et al., 2004) and concentrates in the cytoskeleton of apical microvilli (Viswanatha et al., 2014). An ezrin antibody was used to decorate hChP microvilli on E12.5 parasagittal sections. Although the apical margin is strongly positive in the wild-type hChP epithelium (Fig. 5A,A′, white arrowheads), ezrin signal is weaker, patchy and poorly polarized in the mutant epithelium, and it is also present in the basal domain of some hChP cells (Fig. 5B,B′, red arrowheads), indicating a possible defect in cell polarity (see Movies 1 and 2). Conversely, in the developing tChP (Fig. S2A,B′), ezrin is expressed at high levels and apically located in both wild-type and mutant epithelia (white arrowheads). These results further confirm that the observed cytoarchitectural changes are highly specific to the hChP.
Fig. 5.
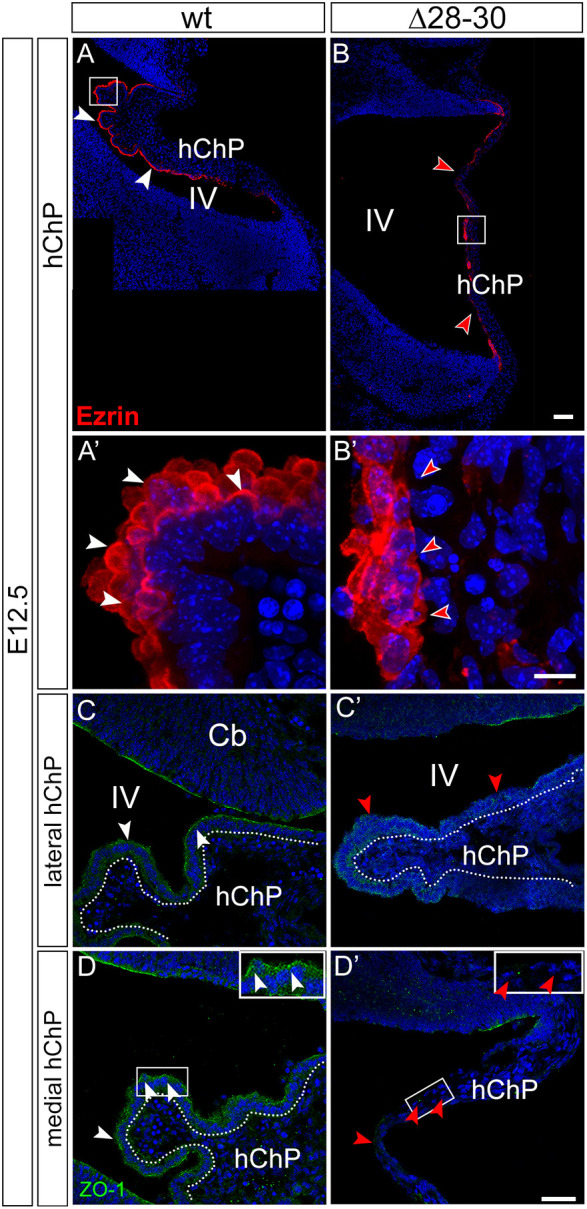
Expression of epithelial markers is downregulated in the hindbrain ChP of Zfp423 mutants. (A-B′) Parasagittal cerebellar sections at E12.5 immunostained for the apical microvilli marker ezrin. The apical margin is strongly positive in the wild-type hChP epithelium (A,A′, white arrowheads); however, ezrin signal is weaker, patchy and poorly polarized in the mutant epithelium (B,B′, red arrowheads), extending to the basal domain of some hChP cells (B′, red arrowheads). (C-D′) The signal of the tight junction marker ZO-1 is strong and continuous in the wild-type hChP epithelium (white arrowheads in C-D and inset in D); however, ZO-1 signal is weaker and discontinuous in mutant lateral sections (red arrowheads in C′), and is virtually absent from the mutant hChP epithelium in medial sections (red arrowheads in D′ and inset). Dotted lines in C-D highlight the columnar hChP epithelium, not visible in D′. Cb, cerebellum; hChP, hindbrain ChP; IV, fourth ventricle. Scale bars: 100 μm in A,B; 10 μm in A′,B′; 40 μm in C-D′.
Next, to visualize the tight junctions that interconnect ChP epithelial cells at their apical pole, we performed immunofluorescence with a zonula occludens 1 (ZO-1)-specific antibody. This marker is downregulated in both medial and lateral sections of the Zfp423 mutant hChP at E12.5 (Fig. 5C′,D′, red arrowheads) compared with the wild type (Fig. 5C,D, white arrowheads), suggesting that the apico-basal segregation of membrane proteins, normally maintained by tight junctions, may be compromised, possibly leading to a leakage of the blood-CSF barrier.
Impaired generation of multiciliated cells in the mutant hChP
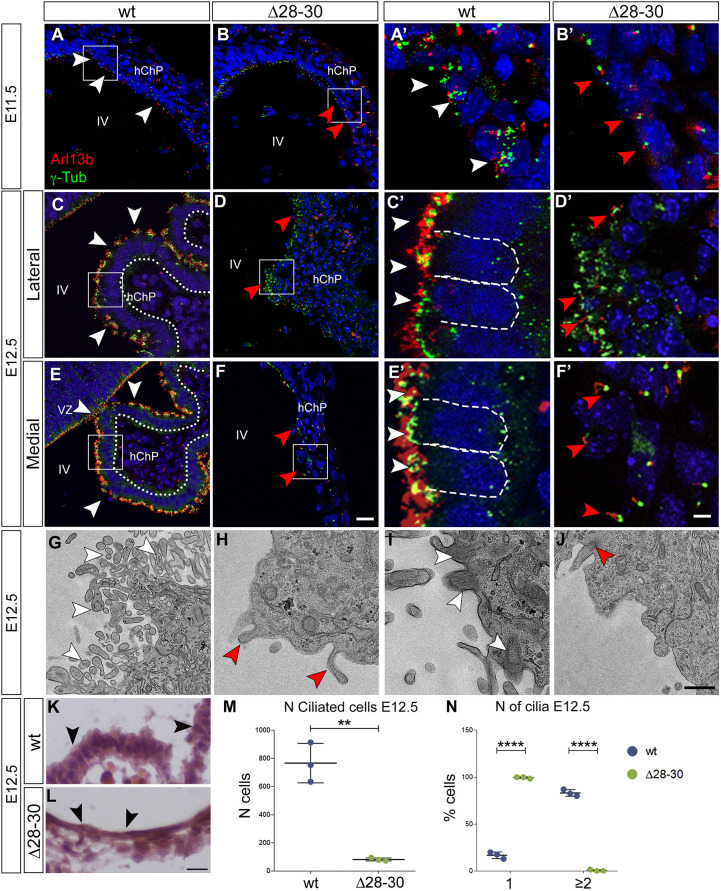
Specialized epithelial cells of the wild-type hChP are multiciliated, exhibiting apical tufts of motile cilia (9+2 microtubules) ranging in number from 4 to 20/cell. These cilia express Arl13b in the axoneme, while γ-tubulin labels the basal body (reviewed by Beisson and Wright, 2003; e.g. Casoni et al., 2017). Immunostaining with Arl13b and γ-tubulin antibodies was used to characterize ciliogenesis in parasagittal hChP sections. At E11.5, although the wild-type hChP contains some multiciliated epithelial cells, and basal body duplication is evident (Fig. 6A,A′, white arrowheads), only monociliated cells are visible in the mutant hChP (Fig. 6B,B′, red arrowheads). Both lateral and medial parasagittal sections of wild-type E12.5 embryos display a columnar hChP epithelium characterized by numerous tufts of Arl13b+ cilia with multiple γ-tubulin+ basal bodies (Fig. 6C,E, insets in C′,E′, white arrowheads; Fig. 6K, black arrowheads). Conversely, in mutant littermates the lateral sections display a smaller, squamous and disorganized hChP epithelium with poorly aligned γ-tubulin dots and absent, or very rare, Arl13b+ axonemes (Fig. 6D, inset in D′ red arrowheads). More strikingly, medial sections contain a monostratified and squamous (Fig. 6L, black arrowheads) hChP epithelium, and only sparse monociliated cells are present (Fig. 6F, inset in F′, red arrowheads). However, the analysis of the tChP in the lateral ventricles revealed that wild-type and mutant cells alike display a columnar and multiciliated epithelium positive for Arl13b+ and γ-tubulin (Fig. S8), again suggesting that Zfp423 is only required for the specification of the hChP ciliated epithelium and that alternative regulatory molecules may be in place in the embryonic lateral ventricles.
Fig. 6.
The Zfp423 mutant ChP fails to develop a multiciliated epithelium. (A-F′) Parasagittal cerebellar sections of the hChP were obtained at different developmental stages and stained for Arl13b and γ-tubulin, to visualize cilia and basal bodies, respectively. At E11.5, wild-type hChP epithelial cells display numerous γ-tubulin-positive basal bodies and several Arl13b-positive cilia (A,A′, white arrowheads); conversely, the mutant hChP contains mono-ciliated cells (B,B′, red arrowheads). At this stage, both mutant and wild-type hChP display a cuboidal epithelium (red and white arrowheads, respectively). At E12.5, the wild-type epithelium is multiciliated with several γ-tubulin-positive and Arl13b-positive dots on the apical domain of the epithelium, which are columnar in the wild type (C,C′,E,E′, white arrowheads, dashed lines), both in lateral and in medial sections. Instead, the mutant hChP epithelium is disorganized, consisting of poorly aligned cuboidal cells with several γ-tubulin-positive dots but few and sparse Arl13b-positive cilia (D,D′, red arrowheads). Medial sections (F,F′) contain a monostratified and squamous hChP epithelium, and only sparse monociliated cells (red arrowheads). (G-J) Transmission electron microscopy images of the ChP epithelial cells at E12.5; wild-type epithelial cells present several microvilli on their apical surface (G, white arrowheads) and several cilia (I, white arrowheads); however, the mutant epithelium displays sparse microvilli (H, red arrowheads) and a single cilium, when present (J, red arrowhead). (K,L) Hematoxylin and Eosin staining of E12.5 embryos shows cuboidal epithelial cells of wild-type ChP (K, black arrowheads), and squamous epithelial cells of mutant ChP (L, black arrowheads). (M) Graphs represent the number of ciliated cells in wild-type and mutant sections at E12.5. n=3/genotype. **P<0.005 (two-tailed unpaired t-test). (N) Graphs show the percentage of cells with one cilium or at least two cilia. n=3/genotype. ****P<0.0001 (two-way ANOVA with Bonferroni post-hoc analysis). Results are mean±s.d. hChP, hindbrain ChP; IV, fouth ventricle; Vz, ventricular zone. Scale bars: 20 μm in A-F; 5 μm in A′-F′; 1 μm in G-J; 10 μm in K,L.
To achieve a better resolution, we analyzed the ultrastructure of the E12.5 hChP epithelium by transmission electron microscopy. Our results reveal that whereas wild-type epithelial cells contain numerous microvilli (Fig. 6G, white arrowheads) and are multiciliated (Fig. 6I, white arrowheads), mutant cells project few, if any, microvilli (Fig. 6H, red arrowheads) and, occasionally, a single cilium (Fig. 6J, red arrowheads). At E12.5, the mutant hChP epithelium shows a sharp and significant decrease in the number of ciliated cells (Fig. 6M, n=3/genotype, **P<0.005). By counting cells with one cilium or with at least two cilia, we show that the mutant hChP displays close to 100% monociliated cells and hardly any multiciliated cells (Fig. 6N, n=3/genotype, ****P<0.0001).
Next, we analyzed the hChP at late gestation (E18.5) and postnatal stages (P18) by immunostaining frontal sections using Arl13b and γ-tubulin antibodies. At E18.5, wild-type embryos exhibit an overwhelming majority of multiciliated cuboidal cells in the hChP (Fig. S3A,A′, white arrowheads), although sparse monociliated cells are also detected in the lateral segment of the hChP (Fig. S3A,A′, white arrow). However, mutant embryos only present a minute lateral segment of the hChP, close to the Luschka foramina (Fig. S3B,B′), with cuboidal monociliated cells (Fig. S3B,B′, white arrows) and some multiciliated cells (Fig. S3B,B′, white arrowheads). In this lateral territory, the mutant epithelium has fewer ciliated cells (Fig. S3I, n=3/genotype, **P<0.005) and 20% fewer multiciliated cells than the wild type (Fig. S3L, n=3/genotype, **P<0.005). The central segment of the hChP, normally observed in the fourth ventricle of wild-type embryos (Fig. S3C,C′), was totally absent in the mutant (Fig. S3D,D′). The same defects were observed at P18 (Fig. S3E-H′).
Genes crucially involved in hindbrain patterning are dysregulated in the Zfp423 mutant
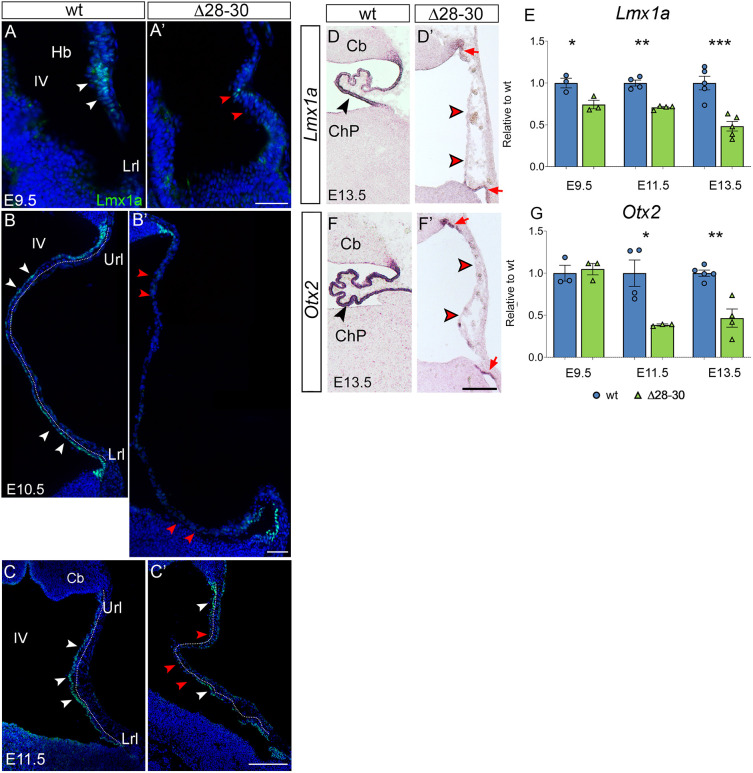
In an attempt to uncover the molecular mechanisms leading to hChP malformation in Zfp423 mutants, we analyzed the expression of established early hChP markers, with a special focus on the Lmx1a protein and the corresponding mRNA, which belong to a collection of genes differentially expressed at the onset of hChP development in the Δ28-30 mutant, as revealed by our RNA-seq analysis (Fig. S5). Lmx1a is a transcription factor that plays a key role in hChP patterning, marking the border between the neurogenic URL and the roof plate, and preventing roof-plate derivatives from adopting the fate of URL-derived cerebellar neurons. In Dreher (Lmx1a) mutants, the hChP is absent (Chizhikov et al., 2010). We conducted Lmx1a immunostaining of E9.5, E10.5 and E11.5 sagittal sections: in the wild type, at all stages, Lmx1a ‘decorates’ epithelial precursors of the dorsal hindbrain (E9.5) and hChP primordium (E10.5, E11.5) lining the ventricular lumen (Fig. 7A,B,C). At the same stages and territories, in the mutant, Lmx1a protein expression is profoundly downregulated (Fig. 7A′,B′,C′; red arrowheads indicate Lmx1a-negative cells). We confirmed these observations by RT-qPCR at E9.5 and E11.5, showing significant downregulation of Lmx1a expression at both stages (Fig. 7E).
Fig. 7.
Established markers of the developing hChP are sharply downregulated in the mutant. (A-C′) Immunostaining of parasagittal cerebellar sections to visualize Lmx1a localization at different stages of development. (A-C) In the wild type at E9.5, the protein is expressed in the progenitors of the dorsal hindbrain (white arrowheads in A), while at E10.5 and E11.5 it is present in the nascent epithelium of the hChP (white arrowheads in B,C, dotted line). (A′-C′) In the mutant, Lmx1a is almost absent at all stages analyzed, and the signal, when present, is discontinuous (white and red arrowheads in A-C and A′-C′, respectively). (D,D′,F,F′) In situ hybridization of cerebellar parasagittal sections at E13.5 with Lmx1a and Otx2 riboprobes. In the wild-type hChP, Lmx1a and Otx2 are strongly expressed (black arrowheads in D,F), while in the mutant hChP, Lmx1a (D′) and Otx2 (F′) are faintly expressed in the upper and lower rhombic lip, and are absent in the hChP, which is misfolded (red arrowheads). (E,G) RT-qPCR reveals a sharp downregulation of Lmx1a and of Otx2 in the mutant hindbrain at E9.5, E11.5 and E13.5 (E9.5, n=3/genotype; E11.5, n=4/genotype; E13.5, n=5/genotype). Data are mean±s.e.m. ***P<0.0005, **P<0.005, *P<0.05 (Welch's unequal variances t-test). Hb, hindbrain; IV, fourth ventricle; Lrl, lower rhombic lip; Url, upper rhombic lip; Cb, cerebellum; ChP, choroid plexus. Scale bars: 50 μm in A-B′; 200 μm in C,C′,D,D′,F,F′.
Moreover, by in situ hybridization, we demonstrate that Lmx1a expression in the mutant is nearly abolished in E13.5 medial sections (Fig. 7D′, red arrowheads). We also examined further stages of development, by immunostaining of E12.5 and E13.5 sagittal sections. At all stages examined, Lmx1a ‘decorates’ the epithelium of the wild-type hChP (Fig. S4A-D, white arrowheads), and is sharply downregulated in medial sections and discontinuously expressed in lateral sections of the mutant hChP epithelium (Fig. S4A′-D′, red arrowheads indicate Lmx1a-negative epithelium). We confirmed these observations by RT-qPCR at E13.5, showing a significant decrease of Lmx1a expression levels in the mutant (Fig. 7E).
Otx2 is another fundamental factor in the formation of this territory, controlling both its early development and its maintenance (Johansson et al., 2013). Our in situ hybridization results confirm that Otx2 levels are sharply reduced at E13.5 in the mutant (Fig. 7F,F′). At E9.5, its expression is not affected by Zfp423 mutation, whereas the gene is sharply downregulated at later stages (Fig. 7G). Taken together, our findings suggest that Zfp423 mutants display significant and early alterations in the expression of two transcription factors known to play crucial roles in hChP ontogenesis.
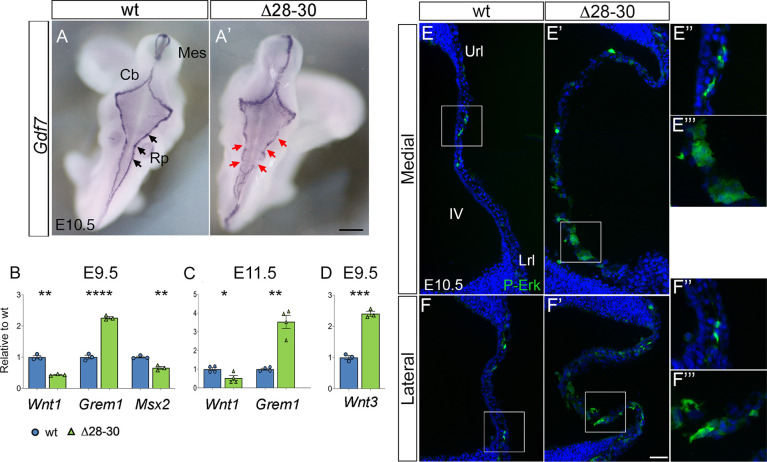
Next, we examined the expression of genes encoding extracellular morphogens released by the URL, LRL and roof plate. In fact, the hChP epithelium is lineally derived from the hindbrain roof plate, as mentioned above. By whole-mount in situ hybridization, we examined the expression of Gdf7, a highly specific roof-plate marker (Lee et al., 1998), in E10.5 embryos (Fig. 8A). Although Gdf7 is expressed in the mutant roof plate, and its overall levels are unchanged, the signal is discontinuous flanking the fourth ventricle (red arrowheads in A′). By RNA-sequencing (RNA-seq), we conducted a genome-wide transcriptome analysis (see Materials and Methods) on a 700 µm fragment (Fig. S5A) of the E9.5 hindbrain spanning rhombomeres 1-6 harvested from Zfp423 deletion mutants (Δ28-30, Casoni et al., 2017) and their wild-type littermates (Fig. S5). The results of our analysis show changes in the expression of regulatory genes crucially involved in the patterning of the dorsal hindbrain and roof plate. In particular, they indicate a significant decrease in the levels of Wnt1 and a strong upregulation of the extracellular BMP signaling antagonist Grem1. These results were confirmed by RT-qPCR at E9.5 and E11.5 (Fig. 8B,C) conducted on embryonic hindbrain mRNA. However, at the stages tested, we could not demonstrate a downregulation of the canonical Wnt signaling pathway target Axin2 or a significant decrease in Smad1,5,8 phosphorylation, a marker of BMP signaling activation (Fig. S7). Conversely, we observed a strong upregulation of Wnt3 (Fig. 8D) and Wnt8a (RNA-seq), the roles of which in hChP development have not been characterized to date. Wnt3 is expressed in the alar myelencephalon and metencephalon, particularly in the URL and LRL (Salinas and Nusse, 1992), and in the roof plate (Parr et al., 1993) at E9.5. Because Wnt3 has been found to activate a noncanonical Wnt/MAP kinase signaling pathway leading to increased ERK1/2 activation (Anne et al., 2013), we immunostained E10.5 hChP sections with a p-ERK1/2 antibody. Our results reveal a clear increase in ERK1/2 phosphorylation in the mutant hChP primordium (Fig. 8E′-F′, insets in E′″, F″′), close to the LRL, which is the site primarily involved in early stages of hChP development (Hunter and Dymecki, 2007).
Fig. 8.
The mutant hChP shows changes in markers of roof-plate patterning. (A,A′) Dorsal view of whole-mount in situ hybridization reveals a conserved but discontinuous Gdf7 expression pattern in the mutant (A′, red arrows). (B) RT-qPCR quantitation of roof-plate patterning markers at E9.5 reveals deregulated expression of three early patterning molecules involved in Wnt and BMP signaling. (C) Deregulation of Wnt1 and Grem1 persists at E11.5. (D) Wnt3 is strongly upregulated at E9.5. Data are mean±s.e.m. (B,D, n=3/genotype; C, n=4/genotype). ****P<0.0001, ***P<0.0005, **P<0.005, *P<0.05 (Welch's unequal variances t-test). (E-F‴) Parasagittal cerebellar sections at E10.5 stained for p-ERK. Both medial (E-E‴) and lateral (F-F‴) sections show an increase of p-ERK expression in the mutant hChP primordium (box in E′,F′, magnified in E‴,F‴) compared with the wild type (box in E,F; magnified in E‴,F‴). Cb, cerebellum; Mes, mesencephalon; Rp, roof plate; Url, upper rhombic lip; Lrl, lower rhombic lip; IV, fourth ventricle. Scale bars: 500 μm in A,A′; 50 μm in E-F′; 120 μm E″-F‴.
Zfp423 regulates genes crucially involved in multiciliogenesis
The ChP epithelium contains multiciliated cells, similar to those observed in the ependyma lining the ventricular cavities of the postnatal CNS, and in other tissues, including the tracheal and bronchial epithelium as well as the epithelium of male efferent ducts and fallopian tubes. In the embryonic hindbrain, proliferating monociliated radial glial progenitors lining the ventricular lumen become quiescent and uncouple centriole amplification from cell division to generate numerous centrioles and support cilia nucleation. This amplification requires the formation of intermediate structures, deuterosomes, that support massive centriole formation. Once docked at the apical plasma membrane, a process that requires Foxj1, the new centrioles become basal bodies (Spassky and Meunier, 2017).
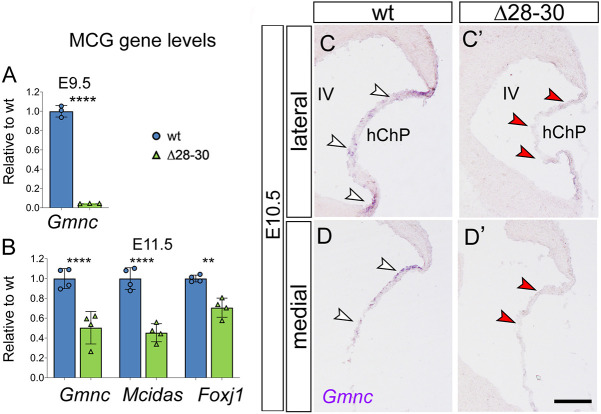
Among other differentially expressed genes identified in our transcriptome analysis, Gmnc (also known as GemC1) is the master regulator of the transition from monociliated neurogenic stem cells to multiciliated, mitotically quiescent multiciliated cells (Zhou et al., 2015). A dramatic decrease of Gmnc expression was scored in the Δ28-30 mutant hindbrain at E9.5 by RNAseq. This result was validated by RT-qPCR: Gmnc is >95% reduced in E9.5 mutants (****P<0.0001; Fig. 9A) and 50% reduced at E11.5 (****P<0.0001; Fig. 9B) and E13.5 (**P<0.005; data not shown). The results of in situ hybridization conducted at E10.5 showed that, whereas the wild-type hChP epithelium is strongly positive for Gmnc in both lateral and medial sections (Fig. 9C,D, white arrowheads), in mutants Gmnc signal is weaker and patchy in the lateral hChP epithelium (Fig. 9C′), and virtually absent in medial sections (Fig. 9D′, red arrowheads).
Fig. 9.
The mutant hChP shows changes in epithelial cell specification. (A,B) Profound downregulation of three key regulators of multiciliogenesis (see text) at different stages of development: Gmnc, Mcidas and Foxj1. (C-D′) In situ hybridization of hChP E10.5 parasagittal sections shows discontinuous expression of Gmnc in lateral sections of the mutant hChP compared with the wild type (C,C′, arrowheads), and complete lack of the transcript in medial mutant sections (D,D′, arrowheads). Data are mean±s.e.m. (A, n=3/genotype; B, n=4/genotype). ****P<0.0001, ***P<0.0005, **P<0.005. (Welch's unequal variances t-test). MCG, multiciliogenesis markers; hChP, hindbrain choroid plexus; IV, fourth ventricle. Scale bar: 200 μm in C-D′.
Gmnc functions upstream of Mcidas (also known as Linkeas or multicilin) (Spassky and Meunier, 2017; Zhou et al., 2015). RT-qPCR conducted in the E11.5 hindbrain (Fig. 9B) showed that Mcidas is significantly downregulated in the mutant (****P<0.0001). Likewise, Foxj1, a marker of multiciliated epithelia, encoding a regulator of multiciliated gene transcription and of basal body docking to the apical cytoskeleton (Gomperts et al., 2004), is also significantly downregulated at E11.5 (Fig. 9B, **P<0.005). Taken together, these results point to an overall deregulation of multiciliated cell fate determination in Zfp423 deletion mutants.
DISCUSSION
In this article, we examine the effects of Zfp423 mutation on hChP development. We find that the hChP is almost completely absent in homozygous mutants, except for a minute symmetrical vestige of this structure restricted to the lateralmost region of the fourth ventricle, right next to the foramina of Luschka. During early embryonic development, the mutant hChP is misfolded, contains very few cuboidal epithelial cells and is functionally compromised, failing to express the thyroid hormone and retinoic acid transporter transthyretin, and the water transporter aquaporin 1. Moreover, the mutant hChP loses the expression of two established markers of the embryonic hChP that are essential for its development: Lmx1a and Otx2 (Johansson et al., 2013; Millonig et al., 2000).
Fate-mapping studies have established that the hChP emerges from two postmitotic morphogenetic fields, medial and lateral, both double-positive for Wnt1 and Gdf7 (Hunter and Dymecki, 2007). Along the antero-posterior axis, the hChP is thought to develop mainly from a transition zone adjacent to the LRL, expanding caudally from a rostral bud of newly formed multiciliated cells. The caudal part of the hChP derives from rhombomeres 2-8, whereas the more rostral one derives from rhombomere 1 (Hunter and Dymecki, 2007). However, the early expression domains of some key genes in the patterning of this territory, including Wnt1, Gdf7, Lmx1a and Otx2, span the entire longitudinal expanse of the fourth ventricle. Similarly, the expression domains of genes involved in the differentiation of multiciliated epithelial cells (e.g. Gmnc and Foxj1) also extend throughout the length of the fourth ventricle. Likewise, ZFP423 protein expression spans URL and LRL starting at E9.5.
Zfp423 controls the earliest stages of hChP development
The results of studies conducted on murine and avian models clearly indicate that ChP patterning and cell fate specification starts early in development. In the mouse brain, hChP fate is determined starting between E8.5 and E9.5 (Thomas and Dziadek, 1993), 2-3 days before the hChP primordium becomes anatomically recognizable. Likewise, chicken and quail grafting studies have shown that ChP fate is determined up to 3 days before its morphological development (Wilting and Christ, 1989). Because of its precocious expression in the prospective hindbrain, starting at the open neurula stage, ZFP423 controls very early and relatively unexplored stages of hChP development.
Zfp423 and the crosstalk between rhombic lip and roof plate
Sophisticated experiments conducted in chick embryos (Broom et al., 2012) have led to the conclusion that the ectoderm-derived roof plate boundary organizer, located in the RL, signals to the roof plate itself to specify the expression of early ChP markers. In the hindbrain primordium (E7.5-E9.5, Fig. 3A-C), Zfp423 expression peaks in both structures, likely contributing to these early inductive events, Lmx1a, a key gene for the function of the roof-plate boundary organizer, is strongly downregulated in Zfp423 mutants. In the developing hindbrain, Lmx1a is a crucial regulator of cell-fate decisions and is required to segregate the roof-plate lineage from neuronal rhombic lip derivatives (Chizhikov et al., 2010).
Several other genes whose expression is significantly perturbed in the mutant are known to be crucially involved in hindbrain patterning, and include components of the BMP (Watanabe et al., 2012) and canonical Wnt signaling pathways. The gene encoding the extracellular BMP antagonist Grem1 is robustly upregulated in the mutant, but this does not result in a substantial decrease of Smad1, Smad5 and Smad8 phosphorylation at the stages tested in this work. Likewise, while significantly reduced Wnt1 expression is observed in the mutant, levels of the canonical Wnt signaling target gene Axin2 are unchanged in the mutant. In both cases, one possibility is that functionally significant changes may take place at earlier developmental stages, requiring further analyses.
Zfp423 and the crosstalk between epithelium and underlying mesenchyme
ChPs develop in response to the interaction between the monociliated radial glia, which give rise to the multiciliated epithelium, and mesodermic derivatives, including the prospective meninges, pericytes and fenestrated blood vessels. The robust (E9.5) and exclusive expression of Zfp423 in cycling epithelial cells of the developing hChP (Fig. 3D) argues strongly in favor of a cell-autonomous function of this gene in the formation of the multiciliated hChP epithelium.
The newly formed postmitotic hChP epithelium, which expresses SHH, plays an instructive role on both adjacent epithelial progenitors and underlying vascular outgrowth (Huang et al., 2009b; Nielsen and Dymecki, 2010). Interestingly, our transcriptome analysis results have revealed, in the mutant, an early upregulation of Wnt3 (Fig. 8), a gene precociously expressed in proliferating progenitors of the alar hindbrain and roof plate. The roles played by this ligand in hChP development have not been addressed to date. Although Wnt3 does not appear to activate canonical Wnt-β catenin signaling, it has been shown to activate MAPK signaling in cerebellar granule cell progenitors, antagonizing SHH-induced clonal expansion (Anne et al., 2013). Our results indicate that ERK1/2 phosphorylation is clearly activated in the mutant hChP primordium and suggest that Zfp423 may normally control cell proliferation in the hChP by suppressing Wnt3-mediated activation of the MAP kinase pathway. These results suggest an upstream role for Zfp423 within the regulatory cascades that govern roof plate and hChP patterning.
Zfp423 and the specification of multiciliated hChP cells
Zfp423 is required to activate multiciliogenesis in the hChP. In addition to the deregulation of signaling pathways controlling hindbrain patterning, Zfp423 mutants also feature a highly significant downregulation of genes promoting multiciliogenesis. Our RNA-seq analysis has revealed that, in the mutant, Gmnc expression is totally absent at E9.5, corresponding to the onset of hChP morphogenesis. Gmnc mediates the conversion of proliferating, monociliated neuroepithelium into postmitotic, multiciliated ChP epithelial cells (Arbi et al., 2016; Lalioti et al., 2019), uncoupling basal body proliferation from cell division. Gmnc controls the expression of other factors involved in the same process, namely Mcidas and Foxj1 (Lalioti et al., 2019), which are also downregulated in the mutant. Most differentially expressed genes are upregulated in the mutant, including Zfp423 itself, confirming the notion that ZFP423 acts as a transcriptional repressor (Cho et al., 2013). The fact that Gmnc transcription is drastically repressed in the mutant hindbrain suggests that it may be an indirect target of ZFP423.
The observed defect in multiciliogenesis likely represents the outcome of a broader effect of Zfp423 mutation on hChP patterning and epithelial cell type specification, also perturbing tight junction and microvillus formation. It has not been established whether or at what stages multiciliated cell differentiation is a necessary step in ChP ontogenesis. While the weak motility of these cilia has a limited effect on CSF flow, there is mounting evidence that loss of cilia leads to altered function of the ChP epithelium, as evidenced by elevated intracellular cAMP levels and increased chloride concentration in the CSF (Banizs et al., 2005). Results obtained in ciliary mutants (Tg737orpk) suggest that cilia contribute to ion transport across the ChP epithelium and control CSF production, which is dysregulated in nonobstructive forms of hydrocephalus. The loss or deformation of cilia on the surface of ChP epithelial cells may alter the function of proteins involved in ion transport and CSF production, similar to what occurs in the renal epithelia of cystic kidney diseases (Liu et al., 2005). We have preliminary evidence of altered membrane transporter gene expression in Zfp423 mutants (not shown), a finding worthy of further studies.
Finally, multiciliogenesis is partially preserved by E18.5 in two small patches located at the lateral corners of the fourth ventricle of the mutant. Although this finding may suggest delayed differentiation rather than a complete disruption in cell fate specification, it has been demonstrated that medial and lateral territories of the fourth ventricle choroid plexus are specified by different combinations of extracellular signals produced by the roof plate (Hunter and Dymecki, 2007). Thus, it is conceivable that ZFP423 may play a less crucial role in a small lateral stretch of the hChP.
Conclusions
In summary, Zfp423 is a master gene and one of the earliest known determinants of hChP development. Its mutation severely impairs hChP morphogenesis and the expression of key regulatory signals required for its development, maintenance and function. The defective production and/or secretion of key molecules by the hChP may be one causal factor of the overall phenotype observed in Zfp423 mutant mice and could contribute to the pathogenesis of cerebellar vermis hypoplasia and Joubert-like cerebellar malformations in individuals carrying ZNF423 mutations (Chaki et al., 2012).
MATERIALS AND METHODS
Animal care
All experiments described were performed in agreement with the stipulations of the San Raffaele Scientific Institute Animal Care and Use Committee (IACUC).
Mouse genetics
The Zfp423 Δ28-30 mouse lines were generated by gene targeting as previously described (Casoni et al., 2017). All experiments were performed using homozygous mutant animals starting from backcross generation N10, which carry 99.8% C57BL/6N genetic background. All studies were conducted in homozygous mutant embryos, using co-isogenic wild-type control littermates. Genotyping was carried by PCR as described previously (Corradi et al., 2003) using allele-specific primers. Primer sequences are listed in the supplementary Materials and Methods.
Tissue preparation
Embryos were collected from time-mated dams and the emergence of the copulation plug was taken as E0.5. For the preparation of embryonic samples, pregnant dams were anesthetized with Avertin (Sigma-Aldrich). Embryos were fixed for 6-8 h according to their developmental stage by immersion with 4% PFA in 1×PBS, cryoprotected overnight in 30% sucrose in 1×PBS, embedded in OCT (Bioptica) and stored at −80°C. For the preparation of postnatal brains, mice were anesthetized with Avertin (Sigma-Aldrich), transcardially perfused with 0.9% NaCl, followed by 4% PFA in 1×PBS, and prepared for freezing as above. The brains and embryos were sectioned sagittally or frontally on a cryotome (Leica CM3050S) and collected on Superfrost plus slides (VWR). Embryonic section thickness was 14 μm for immunostainings and in situ hybridization.
In no instances were embryos excluded from the analysis. The allocation of the samples in the two analysis groups were assigned trough PCR analysis (Corradi et al., 2003). No random or blinded allocation was performed.
Immunofluorescence
Sections were washed in 1×PBS, blocked and permeabilized in 10% goat serum and 0.3% Triton X-100 in 1×PBS, and incubated with primary antibodies overnight at 4°C, rinsed, then incubated with species-appropriate secondary antibodies at room temperature for 2 h (1:1000: Alexa Fluor-488, Alexa Fluor-568 or Alexa Fluor-647; Molecular Probes). Sections were counterstained with DAPI (1:5000, Sigma-Aldrich) and mounted with fluorescent mounting medium (Dako).
Antibodies
Mouse monoclonal antibodies included: ZO-1 (1:100, BD Biosciences, 610966; Martin et al., 2018), Arl13b (1:5, NeuroMab, 73-287; Hsiao et al., 2018) and PHH3 (1:1000, Abcam, ab14955; Casoni et al., 2017). Rabbit antibodies included: ZFP423/OAZ XL (1:2000, Santa Cruz Biotechnology, sc-48785; Casoni et al., 2017), Lmx1a (1:3000, Millipore AB10533, Huang et al., 2018), collagen type IV (1:500, NovusBio, NB120-6586; Velez et al., 2017), desmin (1:200, Cell Signaling Technology, 4024S; Zehendner et al., 2015), γ-tubulin (1:800, Sigma-Aldrich, T5192; Li et al., 2016), Ezrin (1:50, Upstate, 07-130; Gómez-Escudero et al., 2017), p-ERK1/2 (1:800, Cell Signaling Technology, 9101; Keyes et al., 2020), caspase 3 (1:400, BD PharMingen, 559565; Pignatti et al., 2020), γH2AX (1:200, Cell Signaling Technology, 9718; Casoni et al., 2017). A Pecam 1 rat antibody was also used (1:200, BD Biosciences, 550274; Jambusaria et al., 2020). Signals from anti-OAZ, Lmx1a and p-ERK1/2 antibodies were amplified using the Tyramide Signal Amplification Kit (Perkin Elmer; NEL701A001KT), according to the manufacturer's instructions. Antigen retrieval was performed in a citrate buffer before incubation with anti-OAZ and Lmx1a antibodies.
In situ hybridization
Digoxigenin-labeled riboprobes were transcribed from plasmids containing Ttr, Lmx1a, Otx2, Gmnc and Gdf7 cDNAs. In situ hybridization was performed as described previously (Croci et al., 2011).
RT-qPCR
E11.5 and E13.5 embryos were sacrificed and a region comprising the cerebellum and the hChP was dissected, as indicated in the schematic representation in Fig. S5A. Total RNA was extracted with RNeasy MiniKit (Qiagen), according to the manufacturer's instructions. Total RNA (1-1.5 μg) was retrotranscribed using first-strand cDNA MMLV-Retrotranscriptase (Invitrogen) and random primers. Each cDNA was diluted 1:10, and 3 μl were used for each RT-qPCR reaction. mRNA quantitation was performed with SYBR Green I Master Mix (Roche) on a LightCycler480 instrument (Roche). Each gene was analyzed in biological replicates as indicated in figure legends. The expression of the gene of interest was measured in three technical replicates, except at E9.5, owing to limiting RNA amounts.
Data analysis was performed with the 2-ΔΔC(T) method. All RNA levels were normalized based upon β-actin, H3f3a and Gapdh transcript levels.
LacZ staining
Whole embryos were removed, fixed in 4% paraformaldehyde/0.2% glutaraldehyde in phosphate buffer for 5 min and then washed in wash buffer (0.01% sodium deoxycolate, 0.02% Nonidet P40 and 2 mM magnesium chloride in phosphate buffer). The staining was performed for 2-6 h at 37°C in staining solution (50 mg X-gal, 0.106 g potassium ferrocyanide and 0.082 g potassium ferricyanide in 50 ml of wash buffer).
EdU administration and labeling
Pregnant dams were injected intraperitoneally with 5-ethynyl-2'-deoxyuridine (EdU, 50 μg/g body weight). Embryos were collected and treated as described above. EdU incorporation was revealed using the Click-iT EdU Cell Proliferation Assay kit (Life Technologies), according to the manufacturer's instructions.
Microscopy and image processing
Confocal microscopy pictures were taken on a Leica SP8 microscope. The objective used to obtain the images were the following: (1) HC PL APO CS2 20× (NA 0.75) dry; (2) HC PL APO CS 2 40× (NA 1.3) oil; and (3) HC PL APO CS 2 63× (NA 1.4) oil. Epifluorescence pictures were taken on a Zeiss Axioplan2 microscope. Images were further analyzed using FIJI/ImageJ software. Each staining was replicated on at least three different animals for each genotype. Figures of the paper were prepared using Adobe Photoshop and only contrast and brightness of the pictures were adjusted. In Fig. 7B, 8E and S1, tiling was carried out manually using Adobe Photoshop to combine different pictures, and the images were filled with black when necessary. Pictures were taken with the Zeiss Axioimager microscope. Electron microscopy analysis was performed as described previously (Croci et al., 2011).
Statistical analysis
Except where noted, statistical analysis was conducted using two-tailed unequal variance t-test (Welch t-test) for comparison of sets of quantitative values (wild type versus each mutant), or two-way ANOVA for multiple comparisons. Data were reported as the mean±s.d. or s.e.m., as indicated. All experiments were conducted on ≥3 biological replicates with the exception of Lmx1a (n=2), Otx2 (n=2) and Ttr (n=2) in situ hybridizations.
RNA-sequencing
E9.5 hindbrains spanning rhombomeres 1-6 were dissected from wild-type and mutant embryos (n=3, for details see the schematic representation in Fig. S5A) and the total mRNA was extracted (miRNeasy Micro Kit, Qiagen) to prepare sequencing libraries and, subsequently, to validate the differentially expressed gene by RTqPCR. Sequencing libraries were prepared following the SMART-Seq v4 Ultra Low Input RNA (TaKaRa) protocol. Briefly, 1 ng of total RNA was used for cDNA synthesis, followed by PCR amplification. FastQC software was used to assess the quality of the FASTQ files. Reads were aligned to the mouse genome, version mm10, using STAR_2.5.3 (Dobin et al., 2013) and the quality of the alignment assessed with Bamtools (Barnett et al., 2011). The Rsubread package (Liao et al., 2019) was used to assign read counts to the genes of the GENCODE model, version M13 (https://www.gencodegenes.org), with all isoforms of each gene considered together.
Genes with at least 1 cpm (count per million) in at least three samples were considered to be expressed and assessed for differential expression. Differentially expressed genes were identified using limma (Ritchie et al., 2015). Genes with nominal P<0.01 and absolute value of log2 fold change >1 were considered to be differentially expressed (Consortium, 2014). Geneset functional enrichment was performed with DAVID (PMID 22543366). Downstream statistics and Plot drawing were performed within the R environment. Heatmaps were generated with GENE-E (The Broad Institute of MIT and Harvard).
Supplementary Material
Acknowledgements
Image analysis was carried out at ALEMBIC, an advanced microscopy laboratory established by the San Raffaele Scientific Institute and Vita-Salute San Raffaele University. Library preparation and RNA-sequencing was conducted at the CTGB (Center for Translational Genomics and Bioinformatics, San Raffaele Scientific Institute). We thank F. Marroni and A. Pisciottani for critical discussion of the results and manuscript, and Angelo Quattrini for support with transmission electron microscopy. Special thanks to Søren Warming for making the Δ28-30 line available to us.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: F.C., L.C., G.G.C.; Methodology: F.C., L.C., F.V., P.P., G.G.C.; Formal analysis: F.C., M.R., L.M., O.C., G.G.C.; Investigation: F.C., L.C., F.V., P.P., G.G.C.; Data curation: M.R., L.M.; Writing - original draft: F.C., L.C., G.G.C.; Writing - review & editing: F.C., L.C., O.C., G.G.C.; Supervision: F.C., L.C.; Funding acquisition: O.C., G.G.C.
Funding
This work was mostly funded by the Fondazione Telethon (GGP13146 to G.G.C.); an additional contribution was provided by the Ministero della Salute Ricerca Finalizzata 2011 (PE-2011-02347716 to O.C.). S.W. was supported by the National Institutes of Health Intramural Research Program, Center for Cancer Research, National Cancer Institute. Deposited in PMC for release after 12 months.
Data availability
RNA-seq data reported in this publication have been deposited in GEO under accession number GSE160300.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.190173.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.190173.reviewer-comments.pdf
References
- Alcaraz W. A., Gold D. A., Raponi E., Gent P. M., Concepcion D. and Hamilton B. A. (2006). Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation. Proc. Natl. Acad. Sci. USA 103, 19424-19429. 10.1073/pnas.0609184103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne S. L., Govek E.-E., Ayrault O., Kim J. H., Zhu X., Murphy D. A., Van Aelst L., Roussel M. F. and Hatten M. E. (2013). WNT3 inhibits cerebellar granule neuron progenitor proliferation and medulloblastoma formation via MAPK activation. PLoS ONE 8, e81769 10.1371/journal.pone.0081769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbi M., Pefani D. E., Kyrousi C., Lalioti M. E., Kalogeropoulou A., Papanastasiou A. D., Taraviras S. and Lygerou Z. (2016). GemC1 controls multiciliogenesis in the airway epithelium. EMBO Rep. 17, 400-413. 10.15252/embr.201540882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A., Genové G., Mäe M., Nisancioglu M. H., Wallgard E., Niaudet C., He L., Norlin J., Lindblom P., Strittmatter K. et al. (2010). Pericytes regulate the blood-brain barrier. Nature 468, 557-561. 10.1038/nature09522 [DOI] [PubMed] [Google Scholar]
- Awatramani R., Soriano P., Rodriguez C., Mai J. J. and Dymecki S. M. (2003). Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat. Genet. 35, 70-75. 10.1038/ng1228 [DOI] [PubMed] [Google Scholar]
- Banizs B., Pike M. M., Millican C. L., Ferguson W. B., Komlosi P., Sheetz J., Bell P. D., Schwiebert E. M. and Yoder B. K. (2005). Dysfunctional cilia lead to altered ependyma and choroid plexus function, and result in the formation of hydrocephalus. Development 132, 5329-5339. 10.1242/dev.02153 [DOI] [PubMed] [Google Scholar]
- Barnett D. W., Garrison E. K., Quinlan A. R., Stromberg M. P. and Marth G. T. (2011). BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27, 1691-1692. 10.1093/bioinformatics/btr174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J. and Wright M. (2003). Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 15, 96-104. 10.1016/S0955-0674(02)00017-0 [DOI] [PubMed] [Google Scholar]
- Brocklehurst G. (1979). The significance of the evolution of the cerebrospinal fluid system. Ann. R Coll. Surg. Engl. 61, 349-356. [PMC free article] [PubMed] [Google Scholar]
- Broom E. R., Gilthorpe J. D., Butts T., Campo-Paysaa F. and Wingate R. J. T. (2012). The roof plate boundary is a bi-directional organiser of dorsal neural tube and choroid plexus development. Development 139, 4261-4270. 10.1242/dev.082255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casoni F., Croci L., Bosone C., D'Ambrosio R., Badaloni A., Gaudesi D., Barili V., Sarna J. R., Tessarollo L., Cremona O. et al. (2017). Zfp423/ZNF423 regulates cell cycle progression, the mode of cell division and the DNA-damage response in Purkinje neuron progenitors. Development 144, 3686-3697. 10.1242/dev.155077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaki M., Airik R., Ghosh A. K., Giles R. H., Chen R., Slaats G. G., Wang H., Hurd T. W., Zhou W., Cluckey A. et al. (2012). Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150, 533-548. 10.1016/j.cell.2012.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. E., Zhang J. and Reed R. R. (2007). The transcription factor Zfp423/OAZ is required for cerebellar development and CNS midline patterning. Dev. Biol. 307, 43-52. 10.1016/j.ydbio.2007.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov V. V., Lindgren A. G., Mishima Y., Roberts R. W., Aldinger K. A., Miesegaes G. R., Currle D. S., Monuki E. S. and Millen K. J. (2010). Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc. Natl. Acad. Sci. USA 107, 10725-10730. 10.1073/pnas.0910786107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.-W., Hong C.-J., Hou A., Gent P. M., Zhang K., Won K.-J. and Hamilton B. A. (2013). Zfp423 binds autoregulatory sites in p19 cell culture model. PLoS ONE 8, e66514 10.1371/journal.pone.0066514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium S. M.-I. (2014). A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat. Biotechnol. 32, 903-914. 10.1038/nbt.2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi A., Croci L., Broccoli V., Zecchini S., Previtali S., Wurst W., Amadio S., Maggi R., Quattrini A. and Consalez G. G. (2003). Hypogonadotropic hypogonadism and peripheral neuropathy in Ebf2-null mice. Development 130, 401-410. 10.1242/dev.00215 [DOI] [PubMed] [Google Scholar]
- Croci L., Barili V., Chia D., Massimino L., van Vugt R., Masserdotti G., Longhi R., Rotwein P. and Consalez G. G. (2011). Local insulin-like growth factor I expression is essential for Purkinje neuron survival at birth. Cell Death Differ. 18, 48-59. 10.1038/cdd.2010.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currle D. S., Cheng X., Hsu C. M. and Monuki E. S. (2005). Direct and indirect roles of CNS dorsal midline cells in choroid plexus epithelia formation. Development 132, 3549-3559. 10.1242/dev.01915 [DOI] [PubMed] [Google Scholar]
- Damkier H. H., Brown P. D. and Praetorius J. (2013). Cerebrospinal fluid secretion by the choroid plexus. Physiol. Rev. 93, 1847-1892. 10.1152/physrev.00004.2013 [DOI] [PubMed] [Google Scholar]
- Daneman R., Zhou L., Kebede A. A. and Barres B. A. (2010). Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562-566. 10.1038/nature09513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, O., Lara, R. Z., Zhang, O. R., Concepcion, D. and Hamilton, B. A (2020). ZNF423 patient variants, truncations, and in-frame deletions in mice define an allele-dependent range of midline brain abnormalities. PLoS Genet 16, e1009017 10.1371/journal.pgen.1009017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno L., Corradi A., Cobos I., Consalez G. G. and Martinez S. (2004). Ezrin gene, coding for a membrane-cytoskeleton linker protein, is regionally expressed in the developing mouse neuroepithelium. Gene Expr. Patterns 4, 749-754. 10.1016/j.modgep.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Gómez-Escudero J., Moreno V., Martín-Alonso M., Hernández-Riquer M. V., Feinberg T., Colmenar A., Calvo E., Camafeita E., Martínez F., Oudhoff M. J. et al. (2017). E-cadherin cleavage by MT2-MMP regulates apical junctional signaling and epithelial homeostasis in the intestine. J. Cell Sci. 130, 4013-4027. 10.1242/jcs.203687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts B. N., Gong-Cooper X. and Hackett B. P. (2004). Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. J. Cell Sci. 117, 1329-1337. 10.1242/jcs.00978 [DOI] [PubMed] [Google Scholar]
- Hata A., Seoane J., Lagna G., Montalvo E., Hemmati-Brivanlou A. and Massagué J. (2000). OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP- Smad and Olf signaling pathways. Cell 100, 229-240. 10.1016/S0092-8674(00)81561-5 [DOI] [PubMed] [Google Scholar]
- Hong C.-J. and Hamilton B. A. (2016). Zfp423 regulates sonic hedgehog signaling via primary cilium function. PLoS Genet. 12, e1006357 10.1371/journal.pgen.1006357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C. J., Chang C. H., Ibrahim R. B., Lin I. H., Wang C. H., Wang W. J. and Tsai J. W. (2018). Gli2 modulates cell cycle re-entry through autophagy-mediated regulation of the length of primary cilia. J. Cell Sci. 131, jcs221218 10.1242/jcs.221218 [DOI] [PubMed] [Google Scholar]
- Huang Y., Hill J., Yatteau A., Wong L., Jiang T., Petrovic J., Gan L., Dong L. and Wu D. K. (2018). Reciprocal negative regulation between Lmx1a and Lmo4 is required for inner ear formation. J. Neurosci. 38, 5429-5440. 10.1523/JNEUROSCI.2484-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Laoukili J., Epping M. T., Koster J., Hölzel M., Westerman B. A., Nijkamp W., Hata A., Asgharzadeh S., Seeger R. C. et al. (2009a). ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell 15, 328-340. 10.1016/j.ccr.2009.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ketova T., Fleming J. T., Wang H., Dey S. K., Litingtung Y. and Chiang C. (2009b). Sonic hedgehog signaling regulates a novel epithelial progenitor domain of the hindbrain choroid plexus. Development 136, 2535-2543. 10.1242/dev.033795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. L. and Dymecki S. M. (2007). Molecularly and temporally separable lineages form the hindbrain roof plate and contribute differentially to the choroid plexus. Development 134, 3449-3460. 10.1242/dev.003095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I., Shimogori T., Ohtsuka T. and Kageyama R. (2008). Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development 135, 2531-2541. 10.1242/dev.021535 [DOI] [PubMed] [Google Scholar]
- Jambusaria A., Hong Z., Zhang L., Srivastava S., Jana A., Toth P. T., Dai Y., Malik A. B. and Rehman J. (2020). Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. eLife 9, e51413 10.7554/eLife.51413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P. A., Irmler M., Acampora D., Beckers J., Simeone A. and Gotz M. (2013). The transcription factor Otx2 regulates choroid plexus development and function. Development 140, 1055-1066. 10.1242/dev.090860 [DOI] [PubMed] [Google Scholar]
- Keyes J., Ganesan A., Molinar-Inglis O., Hamidzadeh A., Zhang J., Ling M., Trejo J., Levchenko A. and Zhang J. (2020). Signaling diversity enabled by Rap1-regulated plasma membrane ERK with distinct temporal dynamics. eLife 9, e57410 10.7554/eLife.57410.sa2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M.-C., Stewart S. and Hata A. (2003). Poly(ADP-ribose) polymerase 1 interacts with OAZ and regulates BMP-target genes. Biochem. Biophys. Res. Commun. 311, 702-707. 10.1016/j.bbrc.2003.10.053 [DOI] [PubMed] [Google Scholar]
- Lalioti M. E., Arbi M., Loukas I., Kaplani K., Kalogeropoulou A., Lokka G., Kyrousi C., Mizi A., Georgomanolis T., Josipovic N. et al. (2019). GemC1 governs multiciliogenesis through direct interaction with and transcriptional regulation of p73. J. Cell Sci. 132, jcs228684 10.1242/jcs.228684 [DOI] [PubMed] [Google Scholar]
- Lee K. J., Mendelsohn M. and Jessell T. M. (1998). Neuronal patterning by BMPs: a requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 12, 3394-3407. 10.1101/gad.12.21.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen M. K. and Walsh C. A. (2011). Neurogenesis at the brain-cerebrospinal fluid interface. Annu. Rev. Cell Dev. Biol. 27, 653-679. 10.1146/annurev-cellbio-092910-154026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen M. K., Bjornsson C. S., Dymecki S. M., Gilbertson R. J., Holtzman D. M. and Monuki E. S. (2013). The choroid plexus and cerebrospinal fluid: emerging roles in development, disease, and therapy. J. Neurosci. 33, 17553-17559. 10.1523/JNEUROSCI.3258-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Xue C., Yang Q., Low B. C. and Liou Y.-C. (2016). NuSAP governs chromosome oscillation by facilitating the Kid-generated polar ejection force. Nat. Commun. 7, 10597 10.1038/ncomms10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. and Shi W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Murcia N. S., Duan Y., Weinbaum S., Yoder B. K., Schwiebert E. and Satlin L. M. (2005). Mechanoregulation of intracellular Ca2+ concentration is attenuated in collecting duct of monocilium-impaired orpk mice. Am. J. Physiol. Renal. Physiol. 289, F978-F988. 10.1152/ajprenal.00260.2004 [DOI] [PubMed] [Google Scholar]
- Lun M. P., Johnson M. B., Broadbelt K. G., Watanabe M., Kang Y.-J., Chau K. F., Springel M. W., Malesz A., Sousa A. M., Pletikos M. et al. (2015). Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production. J. Neurosci. 35, 4903-4916. 10.1523/JNEUROSCI.3081-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Veloso A., Wu J., Katrukha E. A. and Akhmanova A. (2018). Control of endothelial cell polarity and sprouting angiogenesis by non-centrosomal microtubules. eLife 7, e33864 10.7554/eLife.33864.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masserdotti G., Badaloni A., Green Y. S., Croci L., Barili V., Bergamini G., Vetter M. L. and Consalez G. G. (2010). ZFP423 coordinates Notch and bone morphogenetic protein signaling, selectively up-regulating Hes5 gene expression. J. Biol. Chem. 285, 30814-30824. 10.1074/jbc.M110.142869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimino L., Flores-Garcia L., Di Stefano B., Colasante G., Icoresi-Mazzeo C., Zaghi M., Hamilton B. A. and Sessa A. (2018). TBR2 antagonizes retinoic acid dependent neuronal differentiation by repressing Zfp423 during corticogenesis. Dev. Biol. 434, 231-248. 10.1016/j.ydbio.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millonig J. H., Millen K. J. and Hatten M. E. (2000). The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature 403, 764-769. 10.1038/35001573 [DOI] [PubMed] [Google Scholar]
- Nielsen C. M. and Dymecki S. M. (2010). Sonic hedgehog is required for vascular outgrowth in the hindbrain choroid plexus. Dev. Biol. 340, 430-437. 10.1016/j.ydbio.2010.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C., Gato A., Aparicio M. and Bueno D. (2006). Proteome analysis of chick embryonic cerebrospinal fluid. Proteomics 6, 312-320. 10.1002/pmic.200500085 [DOI] [PubMed] [Google Scholar]
- Parr B. A., Shea M. J., Vassileva G. and McMahon A. P. (1993). Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development 119, 247-261. [DOI] [PubMed] [Google Scholar]
- Pignatti E., Leng S., Yuchi Y., Borges K. S., Guagliardo N. A., Shah M. S., Ruiz-Babot G., Kariyawasam D., Taketo M. M., Miao J. et al. (2020). Beta-catenin causes adrenal hyperplasia by blocking zonal transdifferentiation. Cell Reports 31, 107524 10.1016/j.celrep.2020.107524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power D. M., Elias N. P., Richardson S. J., Mendes J., Soares C. M. and Santos C. R. A. (2000). Evolution of the thyroid hormone-binding protein, transthyretin. Gen. Comp. Endocrinol. 119, 241-255. 10.1006/gcen.2000.7520 [DOI] [PubMed] [Google Scholar]
- Raz A. and Goodman D. S. (1969). The interaction of thyroxine with human plasma prealbumin and with the prealbumin-retinol-binding protein complex. J. Biol. Chem. 244, 3230-3237. [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W. and Smyth G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas P. C. and Nusse R. (1992). Regional expression of the Wnt-3 gene in the developing mouse forebrain in relationship to diencephalic neuromeres. Mech. Dev. 39, 151-160. 10.1016/0925-4773(92)90042-I [DOI] [PubMed] [Google Scholar]
- Shao M., Ishibashi J., Kusminski C. M., Wang Q. A., Hepler C., Vishvanath L., MacPherson K. A., Spurgin S. B., Sun K., Holland W. L. et al. (2016). Zfp423 maintains white adipocyte identity through suppression of the beige cell thermogenic gene program. Cell Metab. 23, 1167-1184. 10.1016/j.cmet.2016.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N. and Meunier A. (2017). The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 18, 423-436. 10.1038/nrm.2017.21 [DOI] [PubMed] [Google Scholar]
- Sturrock R. R. (1979). A morphological study of the development of the mouse choroid plexus. J. Anat. 129, 777-793. [PMC free article] [PubMed] [Google Scholar]
- Tennyson V. M. and Pappas G. D. (1964). Fine structure of the developing telencephalic and myelencephalic choroid plexus in the rabbit. J. Comp. Neurol. 123, 379-411. 10.1002/cne.901230307 [DOI] [PubMed] [Google Scholar]
- Thomas T. and Dziadek M. (1993). Capacity to form choroid plexus-like cells in vitro is restricted to specific regions of the mouse neural ectoderm. Development 117, 253-262. [DOI] [PubMed] [Google Scholar]
- Tsai R. Y. L. and Reed R. R. (1998). Identification of DNA recognition sequences and protein interaction domains of the multiple Zinc finger protein Roaz. Mol. Cell. Biol. 18, 6447-6456. 10.1128/MCB.18.11.6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez D. O., Tsui B., Goshia T., Chute C. L., Han A., Carter H. and Fraley S. I. (2017). 3D collagen architecture induces a conserved migratory and transcriptional response linked to vasculogenic mimicry. Nat. Commun. 8, 1651 10.1038/s41467-017-01556-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R., Bretscher A. and Garbett D. (2014). Dynamics of ezrin and EBP50 in regulating microvilli on the apical aspect of epithelial cells. Biochem. Soc. Trans. 42, 189-194. 10.1042/BST20130263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Rachel R. A., Jenkins N. A. and Copeland N. G. (2006). Zfp423 is required for normal cerebellar development. Mol. Cell. Biol. 26, 6913-6922. 10.1128/MCB.02255-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Kang Y.-J., Davies L. M., Meghpara S., Lau K., Chung C.-Y., Kathiriya J., Hadjantonakis A. K. and Monuki E. S. (2012). BMP4 sufficiency to induce choroid plexus epithelial fate from embryonic stem cell-derived neuroepithelial progenitors. J. Neurosci. 32, 15934-15945. 10.1523/JNEUROSCI.3227-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting J. and Christ B. (1989). An experimental and ultrastructural study on the development of the avian choroid plexus. Cell Tissue Res. 255, 487-494. 10.1007/BF00218783 [DOI] [PubMed] [Google Scholar]
- Wurst W., Rossant J., Prideaux V., Kownacka M., Joyner A., Hill D. P., Guillemot F., Gasca S., Cado D., Auerbach A. et al. (1995). A large-scale gene-trap screen for insertional mutations in developmentally regulated genes in mice. Genetics 139, 889-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappaterra M. D., Lisgo S. N., Lindsay S., Gygi S. P., Walsh C. A. and Ballif B. A. (2007). A comparative proteomic analysis of human and rat embryonic cerebrospinal fluid. J. Proteome Res. 6, 3537-3548. 10.1021/pr070247w [DOI] [PubMed] [Google Scholar]
- Zehendner C. M., Sebastiani A., Hugonnet A., Bischoff F., Luhmann H. J. and Thal S. C. (2015). Traumatic brain injury results in rapid pericyte loss followed by reactive pericytosis in the cerebral cortex. Sci. Rep. 5, 13497 10.1038/srep13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Narasimhan V., Shboul M., Chong Y. L., Reversade B. and Roy S. (2015). Gmnc is a master regulator of the multiciliated cell differentiation program. Curr. Biol. 25, 3267-3273. 10.1016/j.cub.2015.10.062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.