Abstract
Lipidomics is a rapidly growing field, fueled by developments in analytical instrumentation and bioinformatics. To date, most researchers and industries have employed their own lipidomics workflows without a consensus on best practices. Without a community-wide consensus on best practices for the prevention of lipid degradation and transformations through sample collection and analysis, it is difficult to assess the quality of lipidomics data and hence trust results. Clinical studies often rely on samples being stored for weeks or months until they are analyzed, but inappropriate sampling techniques, storage temperatures, and analytical protocols can result in the degradation of complex lipids and the generation of oxidized or hydrolyzed metabolite artifacts. While best practices for lipid stability are sample dependent, it is generally recommended that strategies during sample preparation capable of quenching enzymatic activity and preventing oxidation should be considered. In addition, after sample preparation, lipid extracts should be stored in organic solvents with antioxidants at −20 °Cor lower in an airtight container without exposure to light or oxygen. This will reduce or eliminate sublimation, and chemically and physically induced molecular transformations such as oxidation, enzymatic transformation, and photon/heat-induced degradation. This review explores the available literature on lipid stability, with a particular focus on human health and/or clinical lipidomic applications. Specifically, this includes a description of known mechanisms of lipid degradation, strategies, and considerations for lipid storage, as well as current efforts for standardization and quality insurance of protocols.
Keywords: Lipid standardization, Lipid storage and handling, Lipidomics, Metabolite stability, Sample preservation
Overview of Lipid Stability
Lipids are ubiquitous and structurally complex molecules with diverse biological functions. The comprehensive analysis of lipids (lipidomics) has been recently useful in a plethora of plant, animal, and human studies. More notably, lipids in humans and animals have been shown over the past decade to be valuable for unmasking disease etiology (e.g., serving as biomarkers for eating patterns (Canakci, 2007) as well as several diseases, including cardiovascular disease (De Caterina, 2011; Fung et al., 2001 Halliwell, 2000; Hinterwirth et al., 2014 Massaro et al., 2008; Ridker et al., 2005; Stegemann et al., 2014; Watson, 2006), chronic kidney disease (Kwan et al., 2007; Oberg et al., 2004), specific cancers (Aiello et al., 2005; Bozza and Viola, 2010 Fernandis and Wenk, 2009; Furberg et al., 2005 Han et al., 2005; Lonning et al., 2005; Sutphen et al., 2004; Xu et al., 2007; Zhou et al., 2012), and as general markers for oxidative damage (Dalle-Donne et al., 2006)). Despite the diversity in lipidomics applications, comprehensive and standardized studies that address how lipid stability is influenced by (1) sample collection conditions, (2) pre-analytical steps, and (3) matrix-specific sample preparation procedures are lacking in the literature (Abuja et al., 2015; Zivkovic et al., 2009). Lipid stability in this context is defined as the resistance of a lipid species to change or degrade during sample collection, preparation, handling, storage, and/or analysis (pre-analytical and analytical phase of the measurement process) through enzymatic or chemical processes (i.e., in the presence of oxygen, water, light, and/or extreme temperatures). During these processes, lipids can undergo hydrolysis, oxidation, or interspecies conversion. Although lipid markers such as sphingadienine 1-phosphate (Kamlage et al., 2014; Liu et al., 2018), spingosine-1-phosphate (Kamlage et al., 2014), and lysophosphatidylcholine 18:2 (Anton et al., 2015) have been proposed as markers to assess sample quality and pre-analytical variation, these markers have not been assessed in larger cohort studies, in a panel with other lipid markers for lipid stability, or in other matrices apart from human serum and plasma. Therefore, lipid stability studies should be incorporated into the method development and validation process and matrix-dependent quality control measurements should be performed during lipidomics studies, as the degradation or interconversion of lipid species could potentially lead to misleading and/or irreproducible results.
Forms of Lipid Degradation
Lipid instability can be caused by a variety of factors such as chemical changes (e.g., oxidation) biochemical/microbial enzymatic actions (Jones et al., 2007; Wang et al., 2015), or nonoxidative heating (e.g., thermal decomposition) (Lorenz et al., 2011; Nawar, 1969). Chemical changes can trigger the release of hydrolyzed or oxidized lipid species. Hydrolysis is the enzymatic or nonenzymatic breakdown of lipids due to the presence of water.
Lipid Instability by Oxidation
Oxidation is considered a major source of lipid degradation during sample collection, preparation, and storage. Oxidized lipid species are generated via several mechanisms (e.g., autooxidation with free radicals, photooxidation, and lipoxygenase activity). The rate of oxidation is influenced by both the lipid substrate structure and extraction environment. Lipid structural components affecting the rate and types of oxidation products include the degree and location of the unsaturations on the fatty acyl chains (Yun and Surh, 2012) and lipid class (Shen and Wijesundera, 2009). During sample collection, the sample origin, the presence of water, (Yun and Surh, 2012), the temperature (Hess and O’Hare, 1950; Liu et al., 2019), light, oxidants, and antioxidants can all affect oxidation products and the rate of oxidation. In in vivo studies, the rate of oxidation is influenced by the method of anesthesia (Mohamed et al., 2020; Zhang et al., 2013) or euthanasia (Hennebelle et al., 2019; Trépanier et al., 2017). It is important to note that this difference could be tissue specific. For example, brain tissue may be affected differently than adipose tissue as suggested by Overmyer et al. (2015) in the analysis of metabolites in a mouse model.
As previously mentioned, oxidation rates are influenced by the quantity and location of double bonds in lipid species. Literature reports that double bonds located on fatty acyl chains in the sn-2 position of the glycerol backbone of triacylglycerols are less susceptible to oxidation (Shen and Wijesundera, 2009). It is postulated that the location of glycerophospholipids in membranes, the increased proportion of polyunsaturated fatty acids (PUFA) for membrane fluidity, and the relative proximity to oxidative enzymes make glycerophospholipids more susceptible to oxidation compared to triacylglycerols.(Ademowo et al., 2017) In addition, oxygen and other nonpolar oxidants can concentrate in the nonpolar region of the membrane where highly oxidizable polyunsaturated fatty acids reside. PUFA-containing cholesteryl esters have also been observed to be susceptible to oxidation when exposed to ambient air (Bowden et al., 2011), and several PUFA-derived bioactive lipids such as prostaglandin D2, peptidoleukotrienes, and 5 (6)epoxyeicosatrienoic acid are highly unstable and can undergo spontaneous nonenzymatic conversion into other metabolites (Carmella et al., 2019; Dorow et al., 2016; Maddipati and Zhou, 2011; Maskrey and O’Donnell, 2008). Conversely, many lipids that do not contain PUFA such as steroid hormones and those with saturated fatty acyl moieties appear to be less labile (Holl et al., 2008; Jane Ellis et al., 2003). Hydrogens on methylene groups adjacent to a double bond (allylic hydrogens) or two double bonds (bis-allylic hydrogens) have much lower C–H bond energies than those in unsaturated lipids, and hence can readily be abstracted by free radicals. Bis-allylic hydrogens have the weakest C–H bond and are hundreds of times more reactive with free radicals than allylic hydrogens (Min and Ahn, 2005). Therefore, an increase in the degrees of unsaturation results in a higher susceptibility to oxidation, making lipid species of marine origin more unstable due to the high number of PUFA.
Oxidized lipids can abstract hydrogens from adjacent bis-allylic hydrogens, inducing a chain oxidation reaction as lipids are often found in aggregates such as micelles and bilayers. This mechanism is termed lipid peroxidation or autoxidation (Fig. 1) and has been discussed in detail in a previous review (Metherel and Stark, 2016). Autooxidation is the oxidation of lipid species via a three-step free radical mechanism (initiation, propagation, and termination) due to the presence of oxygen and/or metals. The initial products of autooxidation (peroxides) can be further transformed into secondary oxidation products such as long-chained oxidized species (e.g., ketone, hydroxy, hydroperoxyl, and epoxy containing species) and short-chained oxidized species (e.g., species ending in a carboxylic acid or aldehyde). Photooxidation type I involves the abstraction of a hydrogen or electron from a triple state sensitizer, which yields a free radical (Shahidi and Zhong, 2010). Photooxidation type II involves the excitation of oxygen to a more reactive, excited singlet state via an energy transfer from a triplet sensitizer. Lastly, lipoxygenase involves the enzymatic conversion of polyunsaturated fatty acids to conjugated dienes, which then react with oxygen to form peroxyl radicals and hydroperoxides.
Fig 1.
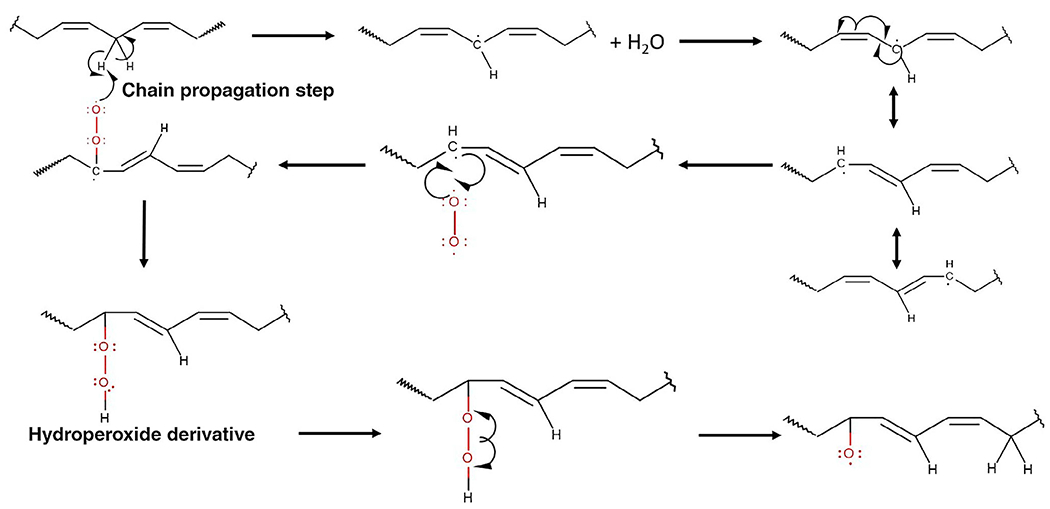
One possible mechanism of lipid peroxidation and chain propagation. Further reactions can occur, which create short-chain products (aldehydes and carboxylic acids) and other long-chain products (ketones and epoxies)
Lipid Instability by Enzymatic Activity
Based on the matrix, lipids can become subject to enzymatic degradation prior to analysis by enzymes such as lecithin cholesterol acyltransferase and phospholipases. In addition, dehydration and hydrolysis reactions under enzymatic conditions can be catalyzed by lipase activity. Common enzymatic reactions involving glycerophospholipids and glycerolipids during sample handling have been discussed elsewhere. Phospholipase A1 (PLA1) and PLA2 are responsible for the hydrolysis of glycerophospholipids in the sn-1 and sn-2 position of the glycerol backbone, respectively (Fig. 2). Failure to inhibit the activity of PLA1 and PLA2 during sample preparation results in elevated levels of lysoglycerophospholipids and free fatty acids. In addition, phospholipase D is responsible for the cleavage of glycerophospholipids into phosphatidic acids, which are converted to diacylglycerols via phosphatidic acid phosphohydrolase (Fig. 2). Alcohols such as methanol and ethanol can be used during extraction, and these alcohols in the presence of phospholipase D can act as acceptors in transphosphatidylation, leading to the ethylation of methylated lipid species (Roughan et al., 1978) (Fig. 2). The generation of transphosphatidylation products such as phosphatidylethanol (PEt) and phosphatidylmethanol (PMe) can occur at low concentrations (i.e., the generation of PEt species during alcohol consumption (Hill-Kapturczak et al., 2018)) as well as in the case of extraction where the solvent extraction mainly consists of methanol (Koelmel et al., 2018). Therefore, even trace levels of solvent contamination may generate unwanted lipid byproducts. Literature has shown lower temperatures to play a substantial role in the inactivation of enzymes, whereas samples undergoing sample preparation at ambient temperatures are subject to enzymatic activity (Hjm Jansen, 2014; Lu et al., 2017; Yang et al., 2013). Heat treatment, further discussed below, can also reduce enzymatic degradation.
Fig 2.
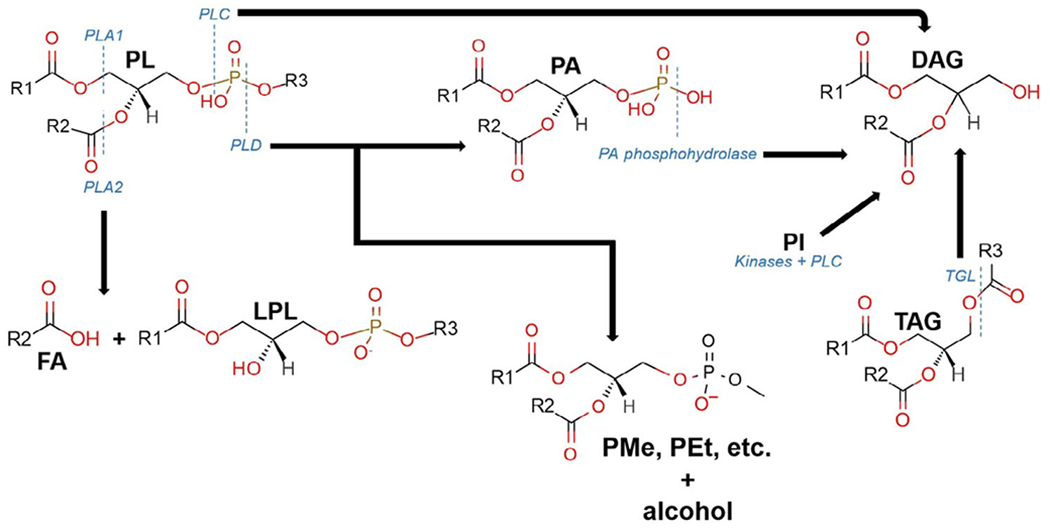
Major enzymatic degradation pathway that commonly occurs during sample processing for glycerophospholipids. The enzymes, phospholipase A1, A2, C, and D (PLA1, PLA2, PLC, and PLD, respectively) are responsible for cleaving phospholipids (PL) at ester linkages (PLA1 and PLA2), and different positions on the phosphate-containing headgroup (PLC and PLD). PLA1 and PLA2 cleave off fatty acyl chains resulting in both fatty acids (FA) and lysophospholipids (LPL). PLC and PLD generate diacylglycerols (DAG) from PL, with a phosphatidic acid intermediate in the case of PLD. DAG can also be generated via kinases and PLC from phosphatidyl inositol as well as by triacylglycerol lipases (TGL) for triacylglycerols (TAG). During extraction procedures in the presence of different alcohols (e.g., methanol or ethanol), PLD can lead to the addition of carbons onto the phosphate group as well as the removal of the other attached functional groups, diacylglycerols (DAG), generating phosphatidylmethanol (PMe), phosphatidylethanol (PEt), or other nonendogenous species
The use of structurally similar internal standards that are spiked prior to lipid extraction can help to compensate for the loss of certain lipid classes during sample preparation as well as variability due to the lipid extraction. Limitations of this approach include the lack of reliable internal standards covering all lipids of interest, differences in degradation within a lipid class (e.g., based on unsaturation location), and the high cost of standards.(Koelmel et al., 2019) In addition, reliable internal standards for certain lipid groups such as oxidized lipid species are lacking commercially. Therefore, lipid degradation should be limited to the greatest extent possible. One such way to reduce lipid degradation is through quenching enzymatic activity, which potentially halts the metabolism of lipids in an effort to maintain the original concentration of cellular lipid species. Recommendations in literature support the rapid use of cold organic solvents, such as methanol, to quench enzymatic activity early during sample preparation and to prevent the enzymatic degradation of lipid species (Kirkwood et al., 2013; Lu et al., 2017). Quenching can also be accomplished with a rapid change in temperature to either low (below −40 °C) or high temperature (above 80 °C) conditions, as well as by implementing extreme pH conditions. It should be noted that the process implemented for quenching is sample-dependent and careful consideration must be placed on the process to avoid the possible chemical degradation of certain lipid species (Gil et al., 2019) or the production of artifacts.
Various reports have highlighted the importance of utilizing plasma instead of serum in lipidomic studies through the collection of blood in the presence of EDTA, heparin, or other anticoagulants. Plasma profiles are believed to provide the most reliable representation of profiles in vivo, as significant differences in lipidomic profiles between plasma and serum have been shown (Aoki et al., 2002; Aoki et al., 2008; Aristizabal-Henao et al., 2019; Ishikawa et al., 2014). There are multiple explanations for this observation. Plasma is typically processed faster than serum, which is obtained from coagulated blood, as no clotting time is needed. Parameters that affect the clotting process in serum include the clotting time, temperature, and the type of blood collection tube (e.g., with clot activators containing glass or siliceous materials, and thrombin or with gel separation) (Arzu et al., 2019; Bowen and Remaley, 2014; Burla et al., 2018; Ng and Yeo, 2013). The effect of these parameters on clinical testing has been explained in detail elsewhere (Bowen and Remaley, 2014). Certain changes in lipid metabolism may not be detected in serum because the blood clotting process elevates the levels of particular lipids such as lysophosphatidylcholines and diacylglycerols (Ishikawa et al., 2014). It has been proposed that during the clotting process, lipids and lip-idaltering enzymes are released, which can result in the generation or degradation of various lipid species such as lysoglycerophospholipids, sphingosine-1-phosphates (S1P), prostaglandins, and oxylipins (Burla et al., 2018; Ishikawa et al., 2014). It is generally recommended for lipidomic studies to obtain plasma from venous blood as opposed to capillary blood collected from a finger prick, which is often used in point-of-care testing and screening, to avoid hemolysis as well as contamination from cosmetics, skin tissue fluid, and antiseptics (Burla et al., 2018). More specifically, cosmetics and skin tissues contain lipid species that could skew the lipid profile of interest, could cause interferences, and could potentially result in ion suppression. Plasma preparation from whole blood collected in tubes containing an anticoagulant is preferred, as anticlotting agents such as EDTA can capture heavy metals (Ferrero, 2016; Kensova et al., 2014), which are pro-oxidants. However, it has been reported that the type of anticoagulant (e.g., EDTA, lithium heparin, and citrate) used can impact lipid extraction, create interferences, or result in the degradation of certain lipid classes (Burla et al., 2018). There are currently no recommendations on which anticoagulant should be used for lipidomics studies, as there is limited research available on the effects of specific anticoagulants on the lipidome. A primary finding has suggested that the calcium-chelating effects of EDTA and citrate, but not heparin, may inhibit the calcium-dependent ex vivo formation or degradation of certain lipid classes (Gonzalez-Covarrubias et al., 2013). Therefore, the same anticoagulant should be utilized for all samples within a study and among studies that will be compared since each anticoagulant can impact the lipidome differently.
Lipid Instability by Nonoxidative Heating
While enzymatic activity can be controlled or decreased, for example, by quenching using organic solvents following sample collection and/or handling the sample at low temperatures, nonenzymatic activity regulation such as chemical degradation due to pH and temperature should be given an equal amount of attention (Gil et al., 2015; Haid et al., 2018; Sohaib et al., 2015). Nonenzymatic, nonoxidative chemical degradation ultimately affects the quality of the lipidomic datasets and the ability to infer biological responses to physiological changes because it could lead to the complete disappearance of certain lipid species or their conversion into another chemical species. For example, sample preparation procedures traditionally used for lipid analysis often involve acid/base hydrolysis under high temperatures for an extended period, which can lead to the dimerization and polymerization of unsaturated fatty acids (Nawar, 1969, 1984). In addition, very high temperatures can result in the nonoxidative decomposition of saturated fatty acids (Nawar, 1984). The effects of nonoxidative thermal degradation have been explained in detail elsewhere (Nawar, 1969).
Unfortunately, nonenzymatic chemical degradation and other types of degradation, which occur prior to extraction of the sample, cannot be corrected for via the use of spiked internal standards and thus, degradation and conversion must be prevented or accounted for prior to extraction (Gil et al., 2015). For cell culture studies, an isotopically labeled growth medium incorporated within cells (i.e., fully isotopically labeled reference material similar to that offered by the isotopic ratio outlier analysis (IROA) quantitation kit (Qiu et al., 2016; Stupp et al., 2013)), is useful in accounting for effects of degradation as it would account for the same environmental conditions across time as the endogenous lipid species. In addition to the IROA approach, lipidome isotope labeling of yeast (LILY) has been proposed as an in vivo 13C labeling technique to produce isotopically labeled eukaryotic lipid standards in yeast (Rampler et al., 2018). While these techniques have significant advantages, limitations include that this technique can be expensive, the incorporation of labels into lipids is often incomplete, and not all untargeted lipidomics studies need stable isotopes to identify biomarkers, which makes their use less common in metabolomics.
Strategies for Improving Lipid Stability
As previously mentioned, lipid stability is influenced by the sample type, sample collection workflow, sample preparation procedure, and sample storage conditions. Table 1 contains suggested recommendations that can be employed to address these lipid stability concerns. The National Institute of Standards and Technology (NIST) conducted a lipidomics survey that included 125 respondents from laboratories across 5 continents and 32 countries (Bowden et al., 2018). The survey questions targeted information on employed lipidomic methodologies, quantitation practices, and protocols related to quality controls. There was only one question that probed responses on lipid stability, “What strategies (if any) does your laboratory employ for enhancing/monitoring lipid stability (select those that apply)?” Respondents suggested the following approaches to address lipid stability: the use of internal/recovery standards (n = 96), sample preparation performed on ice (n = 76), flash freezing (n = 62), antioxidant addition (n = 54), derivatization (n = 27), the use of inhibitors (n = 14), and the use of heat treatment (n = 6). Based on the responses, it appeared that most laboratories incorporated one or more of the techniques mentioned above during a single sample preparation workflow to avoid lipid degradation. However, this same survey highlighted that only 25 out of 122 respondents had standard operating procedures (SOP) for monitoring lipid stability. It is important to include and follow optimized steps to ensure lipid stability in a laboratory SOP, while limiting the implementation of unnecessary and/or impractical preventative strategies if the lipid(s) that are being interrogated do not require it.
Table 1.
Suggested recommendations for ensuring lipid stability
| Sample collection | Pre-collection considerations | While fresh samples allow for the best representation of an unaltered lipidome, frozen samples are acceptable as long as procedures are employed to ensure lipid stability. Plasma preparation from whole blood collected in tubes containing an anticoagulant is preferred, as anti-clotting agents such as EDTA can capture heavy metals, which are pro-oxidants. Because studies have shown the type of anticoagulant used to impact lipid extraction, create interferences, and/or result in the degradation of certain lipid classes, the same anticoagulant should be utilized for all samples within a study and among studies that will be compared. |
| Sample handling | Post-collection considerations | After collection, all samples should be kept cold until sample preparation can be performed. Samples should ideally be immediately stored frozen until sample analysis at −20 °C or lower. If samples cannot be stored frozen, the time between collection and sample analysis should be minimal. |
| Sample preparation | Internal standards | If available, structurally similar internal standards should be spiked into samples before sample preparation to assess sample variability. The internal standards selected should be absent in the sample matrix and should closely represent the lipid class of interest. Lipid markers such as sphingadienine 1-phosphate, spingosine-1-phosphate, and lysophosphatidylcholine 18:2 have been proposed for use in assessing sample quality and pre-analytical variation. However, caution should be employed as these markers have not been assessed in larger cohort studies, as a panel of independent markers, or in other matrices apart from human serum and plasma. |
| Flash-freezeing | If applicable, samples can be flash frozen in liquid nitrogen to quench enzymatic activity. Sample preparation should continue to be performed in a cold environment. | |
| Additivies | The use of additives such as phenylmethanesulfonyl fluroride (PMSF) have been shown to reduce the incidences of oxidization and enzymatic degradation of lipid species. | |
| Antioxidants | The use of antioxidants is encouraged during sample preparation to prevent the degradation of lipid species by (per)oxidation. | |
| Heat-treatment | For sample matrices where additives and antioxidant addition is challenging such as with tissue samples, heat treatment, which can be directly applied to the sample or solvent, has been shown to improve the stability of phospholipids, triacylglycerols, and sphingolipids during sample preparation through the inhibition of lipases. However, it should be noted that the non-enzymatic physical and chemical degradation of lipids are not accounted for with this technique. |
|
| Storage conditions | Short-term | The short-term storage of lipid extracts at room temperature and at 4 °C should be avoided as enzymatic activity has been shown to still be present. Lipids should be stored in an environment free of water, oxygen, and/or light to avoid the chemical transformation of certain lipid species. |
| Long-term | If applicable, samples can be flash frozen in liquid nitrogen and stored at −20°C or lower. Lipid extracts should be stored absent of oxygen (under nitrogen), light, metal ions, and peroxides in an organic solvent to avoid sublimation. Lipid extracts should be stored at −20 °C or lower. |
|
| Freeze–thaw cycles | Laboratories should assess the effect of multiple freeze thaw cycles on the lipid class(es) of interest in a similar matrix as this is sample-dependent. If the lipid class of interest is affected by multiple freeze thaw cycles, biofluids should be aliquoted and tissues should be sliced appropriately to allow for multiple analyses. |
|
| Standardization efforts | Targeted | Depending on the analyte, laboratories can participate in the following QA/QC programs to ensure accuracy in lipid measurements: • [CDC] Lipid Standardization Program (LSP) for total cholesterol, triacylglycerols, high-density lipoprotein cholesterol, apolipoprotein A-I, and apolipoprotein B • [NIST and NIH ODS] Health Assessment Measurements Quality Assurance Program (HAMQAP) for free fatty acids |
| Untargeted | While no formal lipid measurement standardization/harmonization programs are currently in place for untargeted lipidomics studies, the following efforts exist: • LipidQC is a method validation tool established by NIST for robust untargeted lipidomics platforms, which can be used to compare experimental results from NIST SRM 1950 to consensus mean concentrations derived from the 2017 NIST Lipidomics Interlaboratory Comparison Exercise. • The results from various lipidomics international ring trials and interlaboratory studies have been reported in literature and can be used as a point of reference. |
Flash Freezing
Flash freezing in liquid nitrogen at −196 °C, one of the most commonly employed techniques for sample preservation, drastically reduces physical, chemical, and enzymatic degradation/transformation. Ideally, sample handling at a low temperature (−40 °C) would be maintained throughout all steps of sample storage, sample preparation, and data acquisition to prevent degradation. However, lipid solubility could be impacted even when working with organic solvents such as chloroform, causing some lipids to precipitate out of solution. Additionally, temperature regulation at low temperature that ensures quenching of enzymatic activity throughout the entire workflow is impractical for most laboratories employing common strategies or available technologies. Companies that provided cryogenic sample preparation and sample introduction are less prominent, owing to the lack of demand from the community, the high price of implementation, and the lack of awareness in the community about the impact of sample preparation on results. Therefore, additives or pre-analytical techniques that enhance lipid stability are useful as an additional step to cryogenic sample storage and handling.
Additives
Additives can be applied to reduce both physical degradation (e.g., oxidation) and enzymatic degradation. As an example of reducing enzymatic degradation, Wang et al. proposed the use of 5 mM phenylmethanesulfonyl fluoride (PMSF) as a sample pretreatment, prior to sample extraction, to increase the stability of glycerophospholipids, glycerolipids, and sphingolipids (Wang et al., 2015). The authors demonstrated higher levels of phosphatidylcholine and phosphatidylethanolamine compared to the respective lysoglycerophospholipids, thus suggesting the inhibition of phospholipase (PLA) activity. In addition, elevated levels of triacylglycerols compared to the downstream products of diacylglycerols and free fatty acids were reported, suggesting reduced hydrolysis. Reduced degradation of lipids in the presence of PMSF is likely due to the deactivation of serine hydrolase activity in proteins and enzymes via covalent binding to these active sites. Serine hydrolase is incorporated into numerous lipases, including phospholipases, owing to its nucleophilic serine residue, which can hydrolyze lipids and other molecules. Therefore, the deactivation of serine leads to a reduction in lipid enzyme degradation via hydrolysis.
Antioxidants
In addition to additives for the reduction of enzymatic activity, various antioxidants have also been reported as additive options used to decrease the incidence of oxidation during sample preparation and storage, depending on the application. Additionally, a recent primary study showed dietary supplementation with antioxidants in birds to improve the stability of lipids ex vivo (Sohaib et al., 2015). Antioxidants reduce or prevent oxidation via various mechanisms as quenchers of oxidation products and/or reactive singlet oxygen, scavengers of free radicals, metal–ion chelators, and enzyme inhibitors (Blanco and Blanco, 2017; Lü et al., 2010; Nimse and Pal, 2015). Antioxidants are generally introduced during sample preparation or added for long-term storage due to their low cost and high effectiveness. Examples of antioxidants reported in literature include butylated hydroxyanisole (BHA), butylated hydroxytoluene (BHT), ascorbic acid, carotenoids, transferrin, deferoxamine, methyl silicone, phosphoric acid, propyl gallate (PG), tocopherols, quercetin, and tertiary butylhydroquinone (TBHQ).
Heat Treatment
While additives are useful chemical methods for increasing lipid stability in plasma and urine, it is challenging to implement additives in tissue samples prior to pulverization, homogenization, and extraction. Heat treatment, a technique employed since the 1940s, has been shown to promote the stability of certain lipid species including phospholipids, neutral lipids (e.g., triacylglycerols), and sphingolipids during sample preparation via inhibition of lipases, including phospholipase A, C, and D activity (Koelmel et al., 2018; Rose et al., 2008). Heat treatment has the advantage of being applied at the time of collection, such as in environmental field studies, where access to liquid nitrogen or securing options for flash freezing is difficult. Heat treatment can be directly applied to tissues, whole animals, or serum/plasma in a vacuum-sealed chamber without solvent. In addition, the solvent can be rapidly heated during extraction (Koelmel et al., 2018). Generally, if the technology is available, the prior is desirable in that it reduces enzymatic activity earlier on during sample preparation and hence any associated enzymatic transformation of lipids. It is important to note that nonenzymatic physical and chemical transformation/degradation of lipids are not accounted for by heat treatment. While heat treatment reduces enzymatic activity, physical degradation, for example via oxidation, can still occur even though the application of heat treatment under vacuum may not increase the oxidation of lipids (Rose et al., 2008). It is often enzymatic degradation that is most significant at the small intervals between thawing and extraction, especially in tissues, whole blood, or cell cultures where the release of calcium upon cell pulverization activates enzymes to begin to degrade and transform lipids. Therefore, heat treatment is an effective approach for drastically increasing lipid stability during extraction and short-term sample handling.
Considerations for Lipid Storage
The abovementioned NIST lipidomics survey questioned 122 laboratories on temperatures used to store lipid extracts, resulting in 174 responses. Most respondents indicated that lipid extracts were stored in either a −80 °C freezer (n = 96) or −20 °C freezer (n = 55). However, a few laboratories mentioned the storage of lipids at room temperature (n = 2) or in a refrigerator (n = 14).
The stability and interconversion of lipids can be heavily impacted by external factors such as storage and freeze–thaw cycles. The type of sample that is to be stored will certainly influence the storage procedure employed to ensure lipid stability (Burtis et al., 2013). It is still unknown how storage and freeze–thaw cycles affect the entire lipidome as there appears to be sample-dependent effects (Stevens et al., 2019; Zivkovic et al., 2009). While the general idea for sample preservation is to freeze samples quickly and store at a low temperature, this principle does not consider the exact storage temperature or the physical state in which the sample should be stored (e.g., in solution, dried under nitrogen, with preservatives/additives, etc.). While literature has shown the rate of autoxidation to decrease at lower temperatures and hence cryopreservation or freezing can be used to store lipids (Hess and O’Hare, 1950; Liu et al., 2019; Velasco and Dobarganes, 2002), an exception can occur when autoxidation is induced during freezing. For example, in the case of red blood cell-containing samples, lysis can occur due to water expansion, and the resulting release of iron can drive autoxidation (Metherel et al., 2013; Metherel and Stark, 2015).
For long-term storage after sample preparation, lipid extracts should generally be stored in an airtight container at −20 °C or lower in organic solvent to avoid sublimation, an area free of light and oxygen, and in the presence of antioxidants for liquids. Lipids stored as a lyophilized material are more prone to hydrolysis and/or oxidation due to their hygroscopic behavior. There is also a lack of clear guidance between the recommendations for lipid extract storage at −20 °C or temperatures lower than −20 °C. It is suggested that organic solutions of phospholipids should not be stored at temperatures lower than −20 °C unless they are stored in glass. It is also recommended that lipid classes, such as glycerophospholipids, not be stored in aqueous solutions for extended periods of time due to the potential for hydrolysis. Plastic and/or polymer-based containers should not be used to store organic solutions of lipid extracts as these organic solvents can leach the plastics. The number of freeze–thaw cycles should remain limited as literature has shown up to a 37% variability in HDL- and LDL-cholesterol from one freeze–thaw cycle. An environment rich in water, oxygen, and/or light may cause the chemical transformation of certain lipid species despite storage at a low temperature (−20 °C or lower) (Hjm Jansen, 2014).
Literature has shown that the temperature, duration of storage, and presence of enzymatic activity all affect lipid stability differently depending on the lipid class and sample type (Hjm Jansen, 2014; Roszkowska et al., 2018; Zivkovic et al., 2009). Jansen et al. demonstrated that while HDL- and LDL-cholesterol, triacylglycerols, and apolipoprotein-A1 and B were fairly stable at −20 °C, fatty acids showed levels of degradation as high as 80% (Hjm Jansen, 2014). The authors also reported no significant differences between lipid storage at −70 and −196 °C over the course of 12 months, suggesting that a temperature lower than −20 °C may be more ideal. This observation was reflected in a fatty acid stability study (Metherel et al., 2013), where the concentrations of eicosapentaenoic acid and docosahexaenoic acid (two omega-3 PUFA) in dried-blood spots decreased more rapidly when stored at −20 °C as compared with room temperature, 4, and −80 °C. In addition, studies on the lipid profiles in human milk showed that enzymatic activity was more reduced at −70 °C or −80 °C compared to −20 °C (Fusch et al., 2015; Lev et al., 2014). Laboratories should consider an in-house freeze–thaw stability and short-term/long-term storage stability evaluation for each sample-type.
Extreme care should be taken for the lipidomic analysis of stored cell cultures. While cells can be stored at a much lower temperature in liquid nitrogen to reduce lipid instability, cells would then have to be introduced to a cryopreservant solution containing DMSO, which may cause a high background in mass spectrometry-based studies.
While written for a wide range of metabolites, the Centers for Disease Control and Prevention (CDC) has provided a resource, Improving the Collection and Management of Human Samples Used for Measuring Environmental Chemicals and Nutrition Indicators, that describes the best practices for the collection and storage (e.g., whole blood, blood cells, serum, saliva, and urine) to ensure analyte integrity (Centers for Disease Control and Prevention, 2018). The considerations presented can certainly be transferable to the design of lipidomics studies.
Standardization Efforts for Accuracy in Lipid Measurements
Quality assurance (QA) and quality control (QC) measures are necessary to ensure the harmonization of lipid measurements (Burla et al., 2018). Efforts are ongoing to establish external lipid QA and standardization programs. The CDC established the Lipids Standardization Program (LSP) to ensure the analytical accuracy and precision of select lipid measurements reported in research and clinical laboratories (Warnick et al., 2008). Blinded high-quality pooled sera samples with target values determined by generally recognized reference methods for certain lipids such as total cholesterol (TC), triacylglycerols (TAG), high-density lipoprotein cholesterol (HDL-C), apolipoprotein A-I (apo A-I), and apolipoprotein B (apo B) are provided to participants of the LSP program over a 3-month period. A statistical report that provides information about measurement consistency and whether established analytical performance criteria were met is provided to the LSP participants. Using this approach, laboratories can test the accuracy of their measurements from an in-house established protocol to ensure that these lipid measurements over time are not affected by pre-analytical factors such as storage and stability. CDC provides similar programs for other analytes such as steroid hormones and vitamin D.
In addition, a clinical or research laboratory with a lab-developed test can seek a 6-month certification by the CDC for TC through a collaboration with a Cholesterol Reference Method Laboratory Network (CRMLN) member laboratory (Myers et al., 2000). Briefly, the laboratories analyze six high-quality serum samples over a pre-determined concentration range to ensure that specific analytical criteria are met (i.e., ±3.0% maximum allowable bias to the reference method and a ≤ 3.0% CV maximum allowable imprecision) and traceability is established to the National Reference System for Cholesterol (NRS/CHOL). After meeting the certification criteria, the clinical laboratory or research laboratory is issued a 6-month valid Certificate of Traceability, which states that the analytical system (e.g., instrument model, reagent, lot, calibrator lot, and matrix) successfully demonstrated traceability to the NRS/CHOL under the tested conditions. In all of these CDC programs, procedures are used that ensure the stability of the specific lipids during processing, storage, and transport.
NIST, in collaboration with the National Institutes of Health (NIH) Office of Dietary Supplements (ODS), has been actively involved in harmonizing fatty acid measurements in biological samples, particularly blood plasma and serum. NIST and NIH ODS established Fatty Acids in Human Serum and Plasma Quality Assurance Program (FAQAP) that administered interlaboratory comparison exercises for the measurement of 24 fatty acids in selected freeze-dried and frozen plasma and serum matrices. For these studies, participants utilized their typical analytical workflows to analyze samples; the main goal was to assess laboratory performance and gain a better understanding of fatty acid measurement variability across laboratories. Moreover, NIST helped participants troubleshoot their analytical methods when requested. For the first exercise (Schantz et al., 2013), a collaboration with the CDC conducted in 2012, participants measured fatty acid concentrations in NIST SRM 2378 – Fatty Acids in Human Serum, which is composed of serum from (1)donors who had not consumed fish or flaxseed oil supplements for 1 month prior to sample collection, (2) donors who consumed flaxseed oil supplements for 1 month minimum before sample collection, and (3) donors who consumed fish oil supplements for 1 month minimum before sample collection. This first effort demonstrated the urgent need for quality control materials that can be used to increase lipid measurement comparability across laboratories. Although FAQAP formally ended in 2017, NIST and NIH ODS has since formed the Health Assessment Measurements Quality Assurance Program (HAMQAP) (Barber et al., 2018), which continues to serve the fatty acid measurement community. HAMQAP administers interlaboratory comparison exercises where participants measure a host of analytes, including fatty acids, in samples issued by NIST that represent both human intake (e.g., food, dietary supplements) and output (e.g., plasma, serum, urine) so that laboratories can assess their in-house measurements. Participation can be utilized to fulfill requirements established by accreditation bodies or to show compliance with the Food and Drug Administration (FDA) Current Good Manufacturing Practices (CGMP).
While the abovementioned approaches by the CDC and NIST/NIH ODS ensure accuracy for targeted lipid applications, NIST established a method validation tool for robust untargeted lipidomics platforms using LipidQC (Ulmer et al., 2017). Users can visually compare experimental results from the NIST Standard Reference Material (SRM) 1950, “Metabolites in Frozen Human Plasma”, against benchmark consensus mean concentrations derived from the NIST Lipidomics Interlaboratory Comparison Exercise (Bowden et al., 2017a; Bowden et al., 2017b), which was published in 2017. It is important to note that the consensus values reported are based on measurements across laboratories that as a whole demonstrated much variation due to the different methodologies employed. Therefore, consensus values may not be synonymous with a high level of measurement accuracy and instead should be used as a point of reference. Furthermore, other community-wide efforts such as the Lipidomics Standards Initiative (https://lipidomics-standards-initiative.org/ ), Lipid MAPS (http://lipidmaps.org/), and the newly formed International Lipidomics Society (https://lipidomicssociety.org/), along with various reports on pre-analytical processing, especially that from the International Ring Trial (Thompson et al., 2019), have highlighted recommendations for sample storage and freeze–thaw cycling, as well as other critical considerations for the standardization of lipidomics workflows (Burla et al., 2018; Heiskanen et al., 2013; Kirwan et al., 2018; O’Donnell et al., 2019). New bioinformatics resources capable of identifying oxidized lipids and enzymatic products such as phosphatidylmethanol lipid species in lipidomics datasets are emerging (Koelmel et al., 2017; Ni et al., 2017; Tsugawa et al., 2015) and could become an important complement in quality assurance/quality control in both targeted and untargeted analyses.
Community-wide guidelines are needed to establish best practices to reduce lipid degradation during sample preparation and storage, as there is limited consensus within the lipidomics field. In addition, rigorous studies that scrutinize the advantages and disadvantages of approaches to ensure lipid stability are needed. Further, the lipidomics community should expand the concept of lipid stability over the entire analytical workflow, which includes more comprehensive investigations into the gas phase instability of some lipid species, leading to in-source fragmentation and potentially skewed lipid profiles.
To this point, tt is important to include optimized steps across the entire lipidomics workflow (including mass spectrometric parameters) in laboratory SOP to ensure lipid stability, while limiting the implementation of unnecessary and/or impractical preventative strategies if the lipid(s) that are being interrogated do not require it. Nevertheless, participation in programs offered by the CDC, such as the LSP that ensure the accuracy and precision of select lipid measurements can aid in evaluating existing sample preparation methodologies. The procedures and approaches successfully used in the CDC and NIST programs can be adopted in untargeted lipidomics applications to ensure accuracy, reliability, and comparability in lipid measurements.
Acknowledgments
Disclaimer
Certain commercial equipment, instruments, or materials are identified in this paper to adequately specify the experimental procedures. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology; nor does it imply that the materials or equipment identified are necessarily the best for the purpose. Furthermore, the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Standards and Technology.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry. Use of trade names is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention, the Public Health Service and the US Department of Health and Human Services.
Abbreviations
- apo A-I
apolipoprotein A-I
- apo B
apolipoprotein B
- BHA
butylated hydroxyanisole
- BHT
butylated hydroxytoluene
- CDC
Centers for Disease Control and Prevention
- CGMP
current good manufacturing practices
- CRMLN
Cholesterol Reference Method Laboratory Network
- FAQAP
fatty acids in human serum and plasma quality assurance program
- FDA
Food and Drug Administration
- HAMQAP
health assessment measurements quality assurance program
- HDL-C
high-density lipoprotein cholesterol
- IROA
isotopic ratio outlier analysis
- LILY
lipidome isotope labeling of yeast
- LSP
lipids standardization program
- NIH
National Institutes of Health
- NIST
National Institute of Standards and Technology
- ODS
Office of Dietary Supplements
- PEt
phosphatidylethanol
- PG
propyl gallate
- PLA
phospholipase
- PLA1
phospholipase A1
- PMe
phosphatidylmethanol
- PMSF
phenylmethanesulfonyl fluoride
- PUFA
polyunsaturated fatty acids
- QA
quality assurance
- QC
quality control
- SOP
standard operating procedures
- SRM
standard reference material
- TBHQ
tertiary butylhydroquinone
- TC
total cholesterol
- TAG
triacylglycerol
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Abuja PM, Ehrhart F, Schoen U, Schmidt T, Stracke F, Dallmann G, … Zatloukal K (2015) Alterations in human liver metabolome during prolonged cryostorage. Journal of Proteome Research, 14:2758–2768. [DOI] [PubMed] [Google Scholar]
- Ademowo OS, Dias HKI, Burton DGA, & Griffiths HR (2017) Lipid (per) oxidation in mitochondria: An emerging target in the ageing process? Biogerontology, 18:859–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello EJ, Tworoger SS, Yasui Y, Stanczyk FZ, Potter J, Ulrich CM, … McTiernan A (2005) Associations among circulating sex hormones, insulin-like growth factor, lipids, and mammographic density in postmenopausal women. Cancer Epidemiology, Biomarkers & Prevention, 14:1411–1417. [DOI] [PubMed] [Google Scholar]
- Anton G, Wilson R, Yu Z-H, Prehn C, Zukunft S, Adamski J, . Waldenberger M (2015) Pre-analytical sample quality: Metabolite ratios as an intrinsic marker for prolonged room temperature exposure of serum samples. PLoS One, 10:e0121495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki J, Inoue A, & Okudaira S (2008) Two pathways for lysophosphatidic acid production. Biochimica et Biophysica Acta, 1781:513–518. [DOI] [PubMed] [Google Scholar]
- Aoki J, Taira A, Takanezawa Y, Kishi Y, Hama K, Kishimoto T, … Arai H (2002) Serum lysophosphatidic acid is produced through diverse phospholipase pathways. The Journal of Biological Chemistry, 277:48737–48744. [DOI] [PubMed] [Google Scholar]
- Aristizabal-Henao JJ, Fernandes MF, Duncan RE, & Stark KD (2019) Development of a rapid ultra high-performance liquid chromatography/tandem mass spectrometry method for the analysis of sn-1 and sn-2 lysophosphatidic acid Regioisomers in mouse plasma. Lipids, 54:479–486. [DOI] [PubMed] [Google Scholar]
- Arzu K, Canan T, Sevilay S, Simal Koksal C, Ezgi Co§kun Y, Fatih D, & Turan T (2019) Comparison of some biochemical tests in different blood collection tubes in hemodialysis patients. Turkish Journal of Biochemistry, 45:26–36. [Google Scholar]
- Barber CA, Benner BA, Thomas JB, Burdette CQ, Camara J, Long S, Murray JA, Phillips MM, Place BJ, Rimmer CA, Wood LJ, Yu L, Chinthalapati SKR, & Tai SS-C (2018) Health assessment measurements quality assurance program: Exercise 1 final report. NISTIR 8237. pp. 1–330 [Google Scholar]
- Blanco A, & Blanco G (2017) Antioxidants In Blanco A & Blanco G (Eds.), Medical Biochemistry (PP. 205–214). England: Academic Press. [Google Scholar]
- Bowden JA, Albert CJ, Barnaby OS, & Ford DA (2011) Analysis of cholesteryl esters and diacylglycerols using lithiated adducts and electrospray ionization-tandem mass spectrometry. Analytical Biochemistry, 417:202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden JA, Heckert A, Ulmer CZ, & Jones CM (2017b) Lipid concentrations in standard reference material (SRM) 1950: Results from an Interlaboratory Comparison Exercise for Lipidomics. NISTIR; 8185: 1–451 [Google Scholar]
- Bowden JA, Heckert A, Ulmer CZ, Jones CM, Koelmel JP, Abdullah L, … Zhou S (2017a) Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-metabolites in frozen human plasma. Journal of Lipid Research, 58:2275–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden JA, Ulmer CZ, Jones CM, Koelmel JP, & Yost RA (2018) NIST lipidomics workflow questionnaire: An assessment of community-wide methodologies and perspectives. Metabolomics, 14:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen RAR, & Remaley AT (2014) Interferences from blood collection tube components on clinical chemistry assays. Biochemia Medica (Zagreb), 24:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza PT, & Viola JP (2010) Lipid droplets in inflammation and cancer. Prostaglandins, Leukotrienes, and Essential Fatty Acids, 82:243–250. [DOI] [PubMed] [Google Scholar]
- Burla B, Arita M, Arita M, Bendt AK, Cazenave-Gassiot A, Dennis EA, … Wenk MR (2018) MS-based lipidomics of human blood plasma: A community-initiated position paper to develop accepted guidelines. Journal of Lipid Research, 59: 2001–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis CA, Ashwood ER, Bruns DE, & Tietz NW (2013) Tietz textbook of clinical chemistry and molecular diagnostics. St. Louis, MO: Saunders. [Google Scholar]
- Canakci M (2007) The potential of restaurant waste lipids as biodiesel feedstocks. Bioresource Technology, 98:183–190. [DOI] [PubMed] [Google Scholar]
- Carmella SG, Heskin AK, Tang MK, Jensen J, Luo X, Le CT, … Hecht SS (2019) Longitudinal stability in cigarette smokers of urinary eicosanoid biomarkers of oxidative damage and inflammation. PLoS One, 14:e0215853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. (2018) Improving the Collection and Management of Human Samples Used for Measuring Environmental Chemicals and Nutrition Indicators - Version 1.3, Sciences USDoHaH, Editor. National Center for Environmental Health: Division of Laboratory Sciences; pp. 1–24. [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo R, Giustarini D, & Milzani A (2006) Biomarkers of oxidative damage in human disease. Clinical Chemistry, 52:601–623. [DOI] [PubMed] [Google Scholar]
- De Caterina R (2011) N-3 fatty acids in cardiovascular disease. The New England Journal of Medicine, 364:2439–2450. [DOI] [PubMed] [Google Scholar]
- Dorow J, Becker S, Kortz L, Thiery J, Hauschildt S, & Ceglarek U (2016) Preanalytical investigation of polyunsaturated fatty acids and eicosanoids in human plasma by liquid chromatography-tandem mass spectrometry. Biopreservation and Biobanking, 14:107–113. [DOI] [PubMed] [Google Scholar]
- Fernandis AZ, & Wenk MR (2009) Lipid-based biomarkers for cancer. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences, 877:2830–2835. [DOI] [PubMed] [Google Scholar]
- Ferrero ME (2016) Rationale for the successful management of EDTA chelation therapy in human burden by toxic metals. BioMed Research International, 2016:8274504–8274504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TT, Rimm EB, Spiegelman D, Rifai N, Tofler GH, Willett WC, & Hu FB (2001) Association between dietary patterns and plasma biomarkers of obesity and cardiovascular disease risk. The American Journal of Clinical Nutrition, 73:61–67. [DOI] [PubMed] [Google Scholar]
- Furberg AS, Jasienska G, Bjurstam N, Toijesen PA, Emaus A, Lipson SF, … Thune I (2005) Metabolic and hormonal profiles: HDL cholesterol as a plausible biomarker of breast cancer risk. The Norwegian EBBA Study. Cancer Epidemiology, Biomarkers & Prevention, 14:33–40. [PubMed] [Google Scholar]
- Fusch G, Rochow N, Choi A, Fusch S, Poeschl S, Ubah AO, … Fusch C (2015) Rapid measurement of macronutrients in breast milk: How reliable are infrared milk analyzers? Clinical Nutrition (Edinburgh, Scotland), 34:465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Siegel D, Permentier H, Reijngoud DJ, Dekker F, & Bischoff R (2015) Stability of energy metabolites-an often over-looked issue in metabolomics studies: A review. Electrophoresis, 36:2156–2169. [DOI] [PubMed] [Google Scholar]
- Gil A, Zhang W, Wolters JC, Permentier H, Horvatovich P, Rebecca Heiner-Fokkema M, … Bischoff R (2019) Omics | Lipdomics and its pitfalls during the pre-analytical stage In Worsfold P, Poole C, Townshend A, & Miró M (Eds.), Encyclopedia of analytical science (3rd ed.). Oxford, England: Academic Press. [Google Scholar]
- Gonzalez-Covarrubias V, Dane A, Hankemeier T, & Vreeken R (2013) The influence of citrate, EDTA, and heparin anticoagulants to human plasma LC–MS lipidomic profiling. Metabolomics, 9: 337–348. [Google Scholar]
- Haid M, Muschet C, Wahl S, Romisch-Margl W, Prehn C, Moller G, & Adamski J (2018) Long-term stability of human plasma metabolites during storage at −80 degrees C. Journal of Proteome Research, 17:203–211. [DOI] [PubMed] [Google Scholar]
- Halliwell B (2000) Lipid peroxidation, antioxidants and cardiovascular disease: How should we move forward? Cardiovascular Research, 47:410–418. [DOI] [PubMed] [Google Scholar]
- Han C, Zhang HT, Du L, Liu X, Jing J, Zhao X, … Tian B (2005) Serum levels of leptin, insulin, and lipids in relation to breast cancer in China. Endocrine, 26:19–24. [DOI] [PubMed] [Google Scholar]
- Heiskanen LA, Suoniemi M, Ta HX, Tarasov K, & Ekroos K (2013) Long-term performance and stability of molecular shotgun lipidomic analysis of human plasma samples. Analytical Chemistry, 85:8757–8763. [DOI] [PubMed] [Google Scholar]
- Hennebelle M, Metherel AH, Kitson AP, Otoki Y, Yang J, Lee KSS, … Taha AY (2019) Brain oxylipin concentrations following hypercapnia/ischemia: Effects of brain dissection and dissection time. Journal of Lipid Research, 60:671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess PS, & O’Hare GA (1950) Oxidation of linseed oil. Industrial & Engineering Chemistry, 42:1424–1431. [Google Scholar]
- Hill-Kapturczak N, Dougherty DM, Roache JD, Karns-Wright TE, & Javors MA (2018) Differences in the synthesis and elimination of phosphatidylethanol 16:0/18:1 and 16:0/18:2 after acute doses of alcohol. Alcoholism, Clinical and Experimental Research, 42:851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinterwirth H, Stegemann C, & Mayr M (2014) Lipidomics: Quest for molecular lipid biomarkers in cardiovascular disease. Circulation. Cardiovascular Genetics, 7:941–954. [DOI] [PubMed] [Google Scholar]
- Hjm Jansen E (2014) Long term stability of parameters of lipid metabolism in frozen human serum: Triglycerides, free fatty acids, Total-, HDL- and LDL-cholesterol, apolipoprotein-A1 and B. Journal of Molecular Biomarkers & Diagnosis, 5:1–5. [Google Scholar]
- Holl K, Lundin E, Kaasila M, Grankvist K, Afanasyeva Y, Hallmans G, … Lukanova A (2008) Effect of long-term storage on hormone measurements in samples from pregnant women: The experience of the Finnish maternity cohort. Acta Oncologica, 47:406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M, Maekawa K, Saito K, Senoo Y, Urata M, Murayama M, … Saito Y (2014) Plasma and serum lipidomics of healthy white adults shows characteristic profiles by subjects’ gender and age. PLoS One, 9:e91806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jane Ellis M, Livesey JH, & Evans MJ (2003) Hormone stability in human whole blood. Clinical Biochemistry, 36:109–112. [DOI] [PubMed] [Google Scholar]
- Jones ME, Folkerd EJ, Doody DA, Iqbal J, Dowsett M, Ashworth A, & Swerdlow AJ (2007) Effect of delays in processing blood samples on measured endogenous plasma sex hormone levels in women. Cancer Epidemiology, Biomarkers & Prevention, 16:1136–1139. [DOI] [PubMed] [Google Scholar]
- Kamlage B, Maldonado SG, Bethan B, Peter E, Schmitz O, Liebenberg V, & Schatz P (2014) Quality markers addressing preanalytical variations of blood and plasma processing identified by broad and targeted metabolite profiling. Clinical Chemisty, 60:399–412. [DOI] [PubMed] [Google Scholar]
- Kensova R, Hynek D, Kynicky J, Konecna M, Eckschlager T, Adam V, … Kizek R (2014) Determination of metal ions in the plasma of children with tumour diseases by differential pulse voltammetry. International Journal of Electrochemical Science, 9:4675–4691. [Google Scholar]
- Kirkwood JS, Maier C, & Stevens JF (2013) Simultaneous, untargeted metabolic profiling of polar and nonpolar metabolites by LC-Q-TOF mass spectrometry. Current Protocols in Toxicology, 4:4.39–34.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan JA, Brennan L, Broadhurst D, Fiehn O, Cascante M, Dunn WB, … Velagapudi V (2018) Preanalytical processing and biobanking procedures of biological samples for metabolomics research: A white paper, community perspective (for “precision medicine and Pharmacometabolomics task group”-the metabolomics society initiative). Clinical Chemistry, 64:1158–1182. [DOI] [PubMed] [Google Scholar]
- Koelmel JP, Cochran JA, Ulmer CZ, Levy AJ, Patterson RE, Olsen BC, … Garrett TJ (2019) Software tool for internal standard based normalization of lipids, and effect of data-processing strategies on resulting values. BMC Bioinformatics, 20:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelmel JP, Jones CM, Ulmer CZ, Garrett TJ, Yost RA, Schock TB, & Bowden JA (2018) Examining heat treatment for stabilization of the lipidome. Bioanalysis, 10:291–305. [DOI] [PubMed] [Google Scholar]
- Koelmel JP, Kroeger NM, Ulmer CZ, Bowden JA, Patterson RE, Cochran JA, … Yost RA (2017) LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics, 18:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan BC, Kronenberg F, Beddhu S, & Cheung AK (2007) Lipoprotein metabolism and lipid management in chronic kidney disease. Journal of the American Society of Nephrology, 18:1246–1261. [DOI] [PubMed] [Google Scholar]
- Lev HM, Ovental A, Mandel D, Mimouni FB, Marom R, & Lubetzky R (2014) Major losses of fat, carbohydrates and energy content of preterm human milk frozen at —80°C. Journal of Perinatology, 34:396–398. [DOI] [PubMed] [Google Scholar]
- Liu K, Liu Y, & Chen F (2019) Effect of storage temperature on lipid oxidation and changes in nutrient contents in peanuts. Food Science & Nutrition, 7:2280–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hoene M, Yin P, Fritsche L, Plomgaard P, Hansen JS, … Lehmann R (2018) Quality control of serum and plasma by quantification of (4E,14Z)-sphingadienine-C18-1-phosphate uncovers common Preanalytical errors during handling of whole. Blood, 64:810–819. [DOI] [PubMed] [Google Scholar]
- Lonning PE, Geisler J, Krag LE, Erikstein B, Bremnes Y, Hagen AI, … Massimini G (2005) Effects of exemestane administered for 2 years versus placebo on bone mineral density, bone biomarkers, and plasma lipids in patients with surgically resected early breast cancer. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology, 23:5126–5137. [DOI] [PubMed] [Google Scholar]
- Lorenz MA, Burant CF, & Kennedy RT (2011) Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Analytical Chemistry, 83:3406–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü J-M, Lin PH, Yao Q, & Chen C (2010) Chemical and molecular mechanisms of antioxidants: Experimental approaches and model systems. Journal of Cellular and Molecular Medicine, 14:840–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Su X, Klein MS, Lewis IA, Fiehn O, & Rabinowitz JD (2017) Metabolite measurement: Pitfalls to avoid and practices to follow. Annual Review of Biochemistry, 86:277–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddipati KR, & Zhou SL (2011) Stability and analysis of eicosanoids and docosanoids in tissue culture media. Prostaglandins & Other Lipid Mediators, 94:59–72. [DOI] [PubMed] [Google Scholar]
- Maskrey BH, & O’Donnell VB (2008) Analysis of eicosanoids and related lipid mediators using mass spectrometry. Biochemical Society Transactions, 36:1055–1059. [DOI] [PubMed] [Google Scholar]
- Massaro M, Scoditti E, Carluccio MA, & De Caterina R (2008) Basic mechanisms behind the effects of n-3 fatty acids on cardiovascular disease. Prostaglandins, Leukotrienes, and Essential Fatty Acids, 79:109–115. [DOI] [PubMed] [Google Scholar]
- Metherel AH, Aristizabal Henao JJ, & Stark KD (2013) EPA and DHA levels in whole blood decrease more rapidly when stored at −20°C as compared with room temperature, 4 and −75°C. Lipids, 48:1079–1091. [DOI] [PubMed] [Google Scholar]
- Metherel AH, & Stark KD (2015) Cryopreservation prevents iron-initiated highly unsaturated fatty acid loss during storage of human blood on chromatography paper at −20°C. The Journal of Nutrition, 145:654–660. [DOI] [PubMed] [Google Scholar]
- Metherel AH, & Stark KD (2016) The stability of blood fatty acids during storage and potential mechanisms of degradation: A review. Prostaglandins, Leukotrienes, and Essential Fatty Acids, 104:33–43. [DOI] [PubMed] [Google Scholar]
- Min B, & Ahn DU (2005) Mechanism of lipid peroxidation in meat and meat products—A review. Food Science and Biotechnology, 14:152–163. [Google Scholar]
- Mohamed AS, Hosney M, Bassiony H, Hassanein SS, Soliman AM, Fahmy SR, & Gaafar K (2020) Sodium pento-barbital dosages for exsanguination affect biochemical, molecular and histological measurements in rats. Scientific Reports, 10:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers GL, Kimberly MM, Waymack PP, Smith SJ, Cooper GR, & Sampson EJ (2000) A reference method laboratory network for cholesterol: A model for standardization and improvement of clinical laboratory measurements. Clinical Chemistry, 46:1762–1772. [PubMed] [Google Scholar]
- Nawar WW (1969) Thermal degradation of lipids. Journal of Agricultural and Food Chemistry, 17:18–21. [Google Scholar]
- Nawar WW (1984) Chemical changes in lipids produced by thermal processing. Journal of Chemical Education, 61:299. [Google Scholar]
- Ng W-Y, & Yeo C-P (2013) Thrombin-accelerated quick clotting serum tubes: An evaluation with 22 common biochemical analytes. Advances in Hematology, 2013:769479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Angelidou G, Hoffmann R, & Fedorova M (2017) LPPtiger software for lipidome-specific prediction and identification of oxidized phospholipids from LC-MS datasets. Scientific Reports, 7:15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimse SB, & Pal D (2015) Free radicals, natural antioxidants, and their reaction mechanisms. RSC Advances, 5:27986–28006. [Google Scholar]
- Oberg BP, McMenamin E, Lucas FL, McMonagle E, Morrow J, Ikizler TA, & Himmelfarb J (2004) Increased prevalence of oxidant stress and inflammation in patients with moderate to severe chronic kidney disease. Kidney International, 65:1009–1016. [DOI] [PubMed] [Google Scholar]
- O’Donnell VB, Ekroos K, Liebisch G, & Wakelam M (2019) Lipidomics: Current state of the art in a fast moving field. Wiley Interdisciplinary Reviews: Systems Biology and Medicine, 12:e1466. [DOI] [PubMed] [Google Scholar]
- Overmyer KA, Thonusin C, Qi NR, Burant CF, & Evans CR (2015) Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: Studies in a C57BL/6J mouse model. PLoS One, 10:e0117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Moir R, Willis I, Beecher C, Tsai YH, Garrett TJ, … Kurland IJ (2016) Isotopic ratio outlier analysis of the S. cerevisiae metabolome using accurate mass gas chromatography/time-of-flight mass spectrometry: A new method for discovery. Analytical Chemistry, 88:2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampler E, Criscuolo A, Zeller M, El Abiead Y, Schoeny H, Hermann G, … Koellensperger G (2018) A novel Lipidomics workflow for improved human plasma identification and quantification using RPLC-MSn methods and isotope dilution strategies. Analytical Chemistry, 90:6494–6501. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Rifai N, Cook NR, Bradwin G, & Buring JE (2005) Non-HDL cholesterol, apolipoproteins A-I and B100, standard lipid measures, lipid ratios, and CRP as risk factors for cardiovascular disease in women. JAMA, 294:326–333. [DOI] [PubMed] [Google Scholar]
- Rose DJ, Ogden LV, Dunn ML, & Pike OA (2008) Enhanced lipid stability in whole wheat flour by lipase inactivation and antioxidant retention. Cereal Chemistry Journal, 85:218–223. [Google Scholar]
- Roszkowska A, Yu M, Bessonneau V, Bragg L, Servos M, & Pawliszyn J (2018) Tissue storage affects lipidome profiling in comparison to in vivo microsampling approach. Scientific Reports, 8:6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Slack CR, & RJL H (1978) Generation of phospholipid artefacts during extraction of developing soybean seeds with methanolic solvents. Lipids, 13:497–503. [Google Scholar]
- Schantz MM, Powers CD, & Schleicher RL (2013) Interlaboratory analytical comparison study of total fatty acid concentrations in human serum: results for Exercise 01: QA12FASER01. NISTIR7953, pp. 1–82. [Google Scholar]
- Shahidi F, & Zhong Y (2010) Lipid oxidation and improving the oxidative stability. Chemical Society Reviews, 39:4067–4079. [DOI] [PubMed] [Google Scholar]
- Shen Z, & Wijesundera C (2009) Effects of docosahexaenoic acid positional distribution on the oxidative stability of model triacylglycerol in water emulsion. Journal of Food Lipids, 16:62–71. [Google Scholar]
- Sohaib M, Butt MS, Shabbir MA, & Shahid M (2015) Lipid stability, antioxidant potential and fatty acid composition of broilers breast meat as influenced by quercetin in combination with alpha-tocopherol enriched diets. Lipids in Health and Disease, 14:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U, … Mayr M (2014) Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation, 129:1821–1831. [DOI] [PubMed] [Google Scholar]
- Stevens VL, Hoover E, Wang Y, & Zanetti KA (2019) Pre-analytical factors that affect metabolite stability in human urine, plasma, and serum: A review. Metabolites, 9:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp GS, Clendinen CS, Ajredini R, Szewc MA, Garrett T, Menger RF, … Edison AS (2013) Isotopic ratio outlier analysis global metabolomics of Caenorhabditis elegans. Analytical Chemistry, 85:11858–11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphen R, Xu Y, Wilbanks GD, Fiorica J, Grendys EC, LaPolla JP, . Krischer JP (2004) Lysophospholipids are potential biomarkers of ovarian cancer. Cancer Epidemiology Biomarkers & Prevention, 13:1185–1191. [PubMed] [Google Scholar]
- Thompson JW, Adams KJ, Adamski J, Asad Y, Boris D, Bowden JA, … Moseley MA (2019) International ring trial of a high resolution targeted metabolomics and Lipidomics platform for serum and plasma analysis. Analytical Chemistry, 91:14407–14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trépanier M-O, Eiden M, Morin-Rivron D, Bazinet RP, & Masoodi M (2017) High-resolution lipidomics coupled with rapid fixation reveals novel ischemia-induced signaling in the rat neurolipidome. Journal of Neurochemistry, 140:766–775. [DOI] [PubMed] [Google Scholar]
- Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, … Arita M (2015) MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nature Methods, 12:523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer CZ, Ragland JM, Koelmel JP, Heckert A, Jones CM, Garrett TJ, … Bowden JA (2017) LipidQC: Method validation tool for visual comparison to SRM 1950 using NIST Interlaboratory comparison exercise lipid consensus mean estimate values. Analytical Chemistry, 89:13069–13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco J, & Dobarganes C (2002) Oxidative stability of virgin olive oil. European Journal of Lipid Science and Technology, 104:661–676. [Google Scholar]
- Wang X, Gu X, Song H, Song Q, Gao X, Lu Y, & Chen H (2015) Phenylmethanesulfonyl fluoride pretreatment stabilizes plasma lipidome in lipidomic and metabolomic analysis. Analytica Chimica Acta, 893:77–83. [DOI] [PubMed] [Google Scholar]
- Warnick GR, Kimberly MM, Waymack PP, Leary ET, & Myers GL (2008) Standardization of measurements for cholesterol, triglycerides, and major lipoproteins. Laboratory Medicine, 39:481–490. [Google Scholar]
- Watson AD (2006) Thematic review series: Systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. Journal of Lipid Research, 47:2101–2111. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhao Z, Xiao Y, Elson P, Tan H, Plummer S, … Graham C (2007) Plasma lysophosphatidylcholine levels: potential biomarkers for colorectal cancer. Journal of Clnical Oncology, 67:1687–1687. [DOI] [PubMed] [Google Scholar]
- Yang W, Chen Y, Xi C, Zhang R, Song Y, Zhan Q, … Abliz Z (2013) Liquid chromatography-tandem mass spectrometry-based plasma metabonomics delineate the effect of metabolites’ stability on reliability of potential biomarkers. Analytical Chemistry, 85:2606–2610. [DOI] [PubMed] [Google Scholar]
- Yun J-M, & Surh J (2012) Fatty acid composition as a predictor for the oxidation stability of Korean vegetable oils with or without induced oxidative stress. Preventive Nutrition and Food Science, 17:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WL, Liu MY, Zhang ZC, & Duan CY (2013) Effect of different anesthesia methods on erythrocyte immune function in mice. Asian Pacific Journal of Tropical Medicine, 6:995–998. [DOI] [PubMed] [Google Scholar]
- Zhou X, Mao J, Ai J, Deng Y, Roth MR, Pound C, … Bigler SA (2012) Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One, 7:e48889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic AM, Wiest MM, Nguyen UT, Davis R, Watkins SM, & German JB (2009) Effects of sample handling and storage on quantitative lipid analysis in human serum. Metabolomics, 5:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]


