Abstract
Background
Protracted viral shedding is common in hospitalized patients with COVID-19 pneumonia, and up to 40% display signs of pulmonary fibrosis on computed tomography (CT) after hospital discharge. We hypothesized that COVID-19 patients with acute respiratory failure (ARF) who die in intensive care units (ICU) have a lower viral clearance in the respiratory tract than ICU patients discharged alive, and that protracted viral shedding in respiratory samples is associated with patterns of fibroproliferation on lung CT. We, therefore, conducted a retrospective observational study, in 2 ICU of Lyon university hospital.
Results
129 patients were included in the study, of whom 44 (34%) died in ICU. 432 RT-PCR for SARS-CoV-2 were performed and 137 CT scans were analyzed. Viral load was significantly higher in patients deceased as compared to patients alive at ICU discharge (p < 0.001), after adjustment for the site of viral sampling and RT-PCR technique. The median time to SARS-CoV-2 negativation on RT-PCR was 19 days [CI95 %:15–21] in patients alive at ICU discharge and 26 days [CI95 %:17-infinity] in non-survivors at ICU discharge. Competitive risk regression identified patients who died in ICU and age as independent risk factors for longer time to SARS-CoV-2 negativation on RT-PCR, while antiviral treatment was independently associated with shorter time. None of the CT scores exploring fibroproliferation (i.e., bronchiectasis and reticulation scores) were significantly associated with time to SARS-CoV-2 negativation.
Conclusions
Viral load in respiratory samples is significantly lower and viral shedding significantly shorter in ICU survivors of COVID-19 associated acute respiratory failure. Protracted viral shedding is unrelated to occurrence of fibrosis on lung CT.
Keywords: Acute respiratory failure, SARS-COV-2, COVID-19, Viral load, Viral shedding, Acute respiratory distress syndrome, Polymerase chain reaction
Background
COVID-19 infection is frequently associated with acute respiratory failure (ARF) with a median delay from symptom onset of 8–10 days [1, 2]. Approximately 14 to 28% of hospitalized patients require admission in intensive care units (ICU) [3–5]. ICU mortality is high, particularly in patients under invasive mechanical ventilation, and death is mainly related to multi-organ failure [6]. A dysregulated host-response leading to a cytokine storm has been suspected as potential mechanism involved in worsening respiratory failure and occurrence of shock [1, 7], and the optimal therapeutic regimen (antiviral, immunomodulatory or both) remains to be determined. However, protracted viral shedding is common in hospitalized patients with a median of 3 weeks [8], and may be longer in severe patients [9–12]. Whether ICU mortality is mainly related to uncontrolled viral infection, dysregulated host-response or both is currently unknown.
On the other hand, up to 40% of the patients with COVID-19 pneumonia display signs of pulmonary fibrosis on computed tomography (CT) after hospital discharge, and ICU admission has been identified as a potential risk factor [13]. Pulmonary fibrosis may be related to alveolar epithelial cell death related to viral infection, excessive immune response triggering pro-fibrotic response, and/or ventilator induced-lung injury.
We hypothesized that COVID-19 patients with ARF who die in ICU have a lower viral clearance in the respiratory tract than patients discharged alive, and that protracted viral excretion is associated with patterns of fibroproliferation on lung CT.
Hence, the primary objective of the study was to compare the time-to SARS-CoV-2 negativation assessed by real-time reverse transcriptase polymerase chain reaction (RT-PCR) in the group of patients alive at ICU discharge and in the group of patients who died in ICU. The secondary objective of the study was to evaluate the association of time-to SARS-CoV-2 negativation on RT-PCR with a CT-based score of fibroproliferation.
Methods
Study design
The study is a retrospective observational study, performed between February 2020 and June 2020, in 2 intensive care units of a university hospital (Hospices Civils de Lyon, France). This study was conducted in accordance with the amended Declaration of Helsinki and was approved by our institutional ethic committee (#20–41). Consent for data utilization was sought from the patients or their representative.
Patients
Eligible participants were adults, presenting to the ICU with ARF related to COVID-19 pneumonia confirmed by a SARS-CoV-2 positive RT-PCR in respiratory swab during the 14 days preceding ICU admission or during the 7 days following ICU admission. Exclusion criteria were negative SARS-CoV-2 RT-PCR, previous inclusion in the same study during a prior ICU stay, and lack of consent to participate.
The ICU policy during the COVID-19 epidemic was to promote sequential RT-PCR sampling and chest CT every 7–10 days. Antiviral treatments deemed active against SARS-CoV-2 were provided only in case of inclusion in antiviral clinical trials. Steroids were not administered before negativation of RT-PCR as per ICU policies at the time the study was conducted.
Data collection
The following variables were collected: demographic and anthropometric data, SAPS II [14] and SOFA scores [15] at ICU admission, immunosuppression status, fulfilment of acute respiratory distress syndrome (ARDS) criteria [16], date of first symptoms of COVID-19 infection, antiviral treatment active on SARS-CoV-2 at any time during ICU stay, respiratory and extra-respiratory organ support during ICU stay, ICU length of stay, date of chest CT performed during hospital stay and CT semi-quantitative scores (see below), date of sampling and results of each RT-PCR performed during hospital stay. Patients were considered immunocompromised if they had received chemotherapy within the last 6 months, uncontrolled HIV infection (CD4 T-cells less than 200 copies or 15%), corticosteroid therapy > 10 mg per day, immunosuppressive therapy within the last 3 months (6 months for Rituximab), aplasia, asplenia or splenectomy.
RT-PCR for SARS-CoV-2 were performed on nasopharyngeal swabs in non-intubated patients, and on nasopharyngeal swabs or lower respiratory tracts samples in patients under invasive mechanical ventilation. Due to evolution in routine diagnosis and diagnostic test shortage, RT-PCR were performed by 4 different techniques during the study period, targeting the ORF1a gene and with similar pre-analysis steps: 1-the QS technique developed by Institut Pasteur (Paris, France) was performed on 183 samples; 2; the QS technique adapted on the open access channel of the automated system Panther Fusion (Hologic ®, Marlborough, MA) was used on 180 samples (QS–HOL); 3- the assay Cobas®SARS-CoV-2 was used on the Cobas® 6800 system (Roche Diagnostics ®, Bale, Switzerland) for 57 samples; 4- the Eurobioplex SARS-CoV-2 kit (Eurobio Scientific ®, Les Ulis, France) was used on 4 samples. For all techniques, RT-PCR results were expressed in terms of the cycle threshold value, which is defined as the number of cycles required for the signal to cross the threshold (i.e., exceed the background level). Cycle threshold values are inversely related to viral load, and samples were considered positive if the cycle threshold was ≤ 45. Cycle threshold values were lacking for 8 viral swabs performed in other hospitals using unknown RT-PCR technique. Time to SARS-CoV-2 negativation on RT-PCR was computed as the elapsed time from ICU admission to the time at which the first negative RT-PCR was reported, with no subsequent positive RT-PCR. Patients without SARS-CoV-2 negativation on RT-PCR during their hospital stay were right-censored at the time of their last positive RT-PCR.
All CT scans performed on patients of center#1 during their ICU stay were analyzed. CT acquisitions were performed in the supine position with an iCT 256 or Ingenuity CT (Philips Healthcare, Eindhoven, The Netherlands) using the following settings: voltage 120 to 140 kVP, slice thickness 0.9 mm to 1.5 mm, matrix size 512 × 512. Field of view, pixel size and mAs were adapted for each patient. Each CT scan were graded simultaneously by consensus by 2 pneumologists (LF, LC) using a semi-quantitative score [17]. Five CT slices were analyzed at the following location: aortic arch, 1 cm above the dome of the right hemidiaphragm, and on three additional levels equally spaced between these two levels. At each level, each lung was divided into an anterior and a posterior zone, creating four quadrants for analysis per level. The following radiologic patterns were assessed in each quadrant: reticulation, ground-glass opacification (GGO), and intense parenchymal opacification (IPO). Spatial extension of each pattern was graded using a semi-quantitative score: 0 = no involvement; 1 = < 5% involvement of lung area on the slice; 2 = 5% to 25% involvement; 3 = 26% to 49% involvement; 4 = 50% to 75% involvement; and 5 = > 75% involvement. For each radiologic pattern, the average score (range 0–20) was calculated by summing all quadrant values of all slices, divided by the number of slices. Finally, bronchiectasis was scored as present (1) or absent (0) on each slice, with the total bronchiectasis score calculated by summing values over all slices (range 0–5). Any fibroproliferation CT feature was defined for each patient as occurrence of at least one bronchiectasis score or reticulation score > 0 during ICU stay.
Statistical analysis
The primary endpoint of the study was time to SARS-CoV-2 negativation on RT-PCR. The secondary endpoints of the study were maximal bronchiectasis, reticulation, GGO and IPO CT scores during ICU stay.
Statistical analyses were performed using R version 4.0.1 [18] and the following packages: survival [19, 20], survminer [21], cmprsk [22], powerSurvEpi [23], lme4 [24] and lmerTest [25]. A p value below 0.05 was chosen for statistical significance.
Categorical variables were expressed as count (percentage) and continuous data were expressed as medians (1st quartile-3rd quartile). Data were compared between groups with the Fisher’s exact test for categorical variables and the Mann–Whitney U test for continuous variables. 95% confidence intervals (CI95 %) for proportions were computed with the Wilson score method.
Evolution of viral load (assessed by the cycle threshold value) over time was analyzed using a linear mixed model, using time as a random slope and vital status at ICU discharge as a random intercept, and adjusting for the site of sampling (nasopharynx or lower respiratory tract) and RT-PCR technique. This adjustment was performed to take into account the potential confounding effect of site of sampling and RT-PCR technique on viral load.
The impact of ICU mortality and CT scores on time to SARS-CoV-2 negativation on RT-PCR were first analyzed without considering the competing risk of death using the Kaplan–Meier estimator and the log-rank test. Then, Fine and Gray competing risk regression analyses were performed to account for the competing risk of death on time to SARS-CoV-2 negativation on RT-PCR [26], and the proportional hazard assumption was assessed by graphical analysis of the Schoenfeld’s residuals. All multivariate analyses were performed by incorporating variables with p values ≤ 0.2 in univariate analysis and stepwise backward selection.
Results
129 patients were included in the study (97 from center #1 and 32 from center #2). Patients’ characteristics are reported in Table 1. ICU mortality amounted to 34%.
Table 1.
Patient’s characteristics at inclusion and during ICU stay
| Variable | Overall population (n = 129) | Alive at ICU discharge (n = 85) | Deceased at ICU discharge (n = 44) | p value |
|---|---|---|---|---|
| Age (yr) | 69 [59–77] | 63 [58–73] | 77 [68–82] | < 0.001 |
| Sex male | 94 (73%) | 56 (66%) | 38 (86%) | < 0.01 |
| BMI (kg m−2) | 28 [25–31] | 28 [25–31] | 28 [25–32] | NS |
| Immunosuppression | 9 (7%) | 4 (5%) | 5 (11%) | NS |
| Time between 1st symptoms and ICU admission (day) | 8 [6–11] | 9 [6–12] | 7 [5–10] | NS |
| Time between 1st RT-PCR and ICU admission (day) | 1 [0–4] | 1 [0–4] | 1 [0–2] | NS |
| SAPS2 | 39 [32–52] | 36 [28–46] | 49 [39–53] | < 0.001 |
| SOFA score at ICU admission | 4 [2–7] | 4 [2–6] | 5 [3–8] | < 0.05 |
| ARDS criteria | 87 (67%) | 51 (60%) | 34 (81%) | < 0.05 |
| Respiratory support | < 0.01 | |||
| Oxygen or NIV or HFNO only | 42 (33%) | 35 (41%) | 7 (17%) | |
| IV at any time | 87 (67%) | 50 (59%) | 34 (83%) | |
| ECMO | 6 (5%) | 1 (1%) | 5 (12%) | < 0.05 |
| Vasopressor at any time | 86 (67%) | 50 (59%) | 34 (81%) | < 0.05 |
| RRT at any time | 24 (19%) | 13 (15%) | 13 (30%) | NS |
| ICU LOS | 16 [5–31] | 17 [5–31] | 15 [5–31] | NS |
| Antiviral treatment | ||||
| None | 111 (86%) | 73 (85%) | 36 (86%) | NS |
| Immunoglobulin | 1 (1%) | 0 (0%) | 1 (2%) | |
| Lopinavir-ritonavir | 5 (4%) | 4 (5%) | 1 (2%) | |
| Lopinavir-ritonavir + IFN-β | 2 (1%) | 0 (0%) | 2 (5%) | |
| Hydroxychloroquine | 5 (4%) | 3 (4%) | 2 (5%) | |
| Remdesivir | 5 (4%) | 5 (6%) | 0 (0%) | |
Values are median [1st quartile-3rd quartile] or count (percentage)
ARDS acute respiratory distress syndrome, BMI body mass index, ECMO extracorporeal membrane oxygenation, HFNO high flow nasal oxygen, ICU intensive care unit, IFN-β interferon β, IV invasive ventilation, LOS length of stay, NS not significant, RRT renal replacement therapy, RT-PCR real-time reverse transcriptase polymerase chain reaction, SAPS2 simplified acute physiology score
Virologic tests
432 RT-PCR for SARS-CoV-2 were performed, among which 121 (28%) were sampled from the lower respiratory tract, 311 (72%) from the nasopharynx, and 301 (70%) were positive. The median number of RT-PCR per patient amounted to 3 [2–4]. 40 patients (31%) had only 1 negative RT-PCR at end of follow-up, 35 (27%) patients had at least 2 consecutive negative RT-PCR at end of follow-up, and 54 (42%) still had a positive RT-PCR at end of follow-up. Timing of RT-PCR measurement for each patient is presented in Additional file 1. Cycle threshold values significantly increased over time from ICU admission (p < 0.001, Fig. 1 and Additional file 2), and were significantly lower (i.e., viral load was significantly higher) in patients deceased as compared to patients alive at ICU discharge, after adjustment for the site of sampling and RT-PCR technique. Interestingly, the effect of the site of sampling on viral load was not statistically significant (Additional file 2), with a regression coefficient amounting to 0.39 ± 0.82 (i.e., the fitted cycle threshold values were on average 0.39 ± 0.82 higher in nasopharynx samples that in lower respiratory tract samples, p = 0.63). A sensitivity analysis using only viral samples analyzed with the QS and the QS–HOL techniques gave identical results.
Fig. 1.
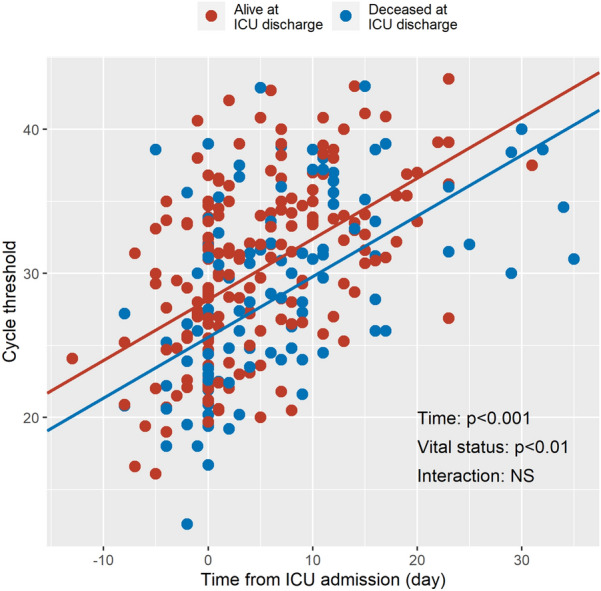
Viral load as a function of time from ICU admission in positive RT-PCR samples. Circles are individual RT-PCR measurements expressed as cycle threshold values (the lower the cycle threshold, the higher the viral load). Red and blue circles refer to patients alive and deceased at ICU discharge, respectively. Lines are regression lines for both groups, adjusted for the site of sampling (nasopharynx or lower respiratory tract) and RT-PCR technique. ICU intensive care unit, RT-PCR real-time reverse transcriptase polymerase chain reaction
Probability to change RT-PCR status
Kaplan–Meier representation of the probability of remaining positive to SARS-CoV-2 is presented in Fig. 2. Non-survivors at ICU discharge had a significantly longer viral excretion, as compared to ICU survivors (p < 0.05). The median time to SARS-CoV-2 negativation on RT-PCR was 19 days [CI95 %:15–21] in patients alive at ICU discharge, and 26 days [CI95 %:17-infinity] in non-survivors at ICU discharge.
Fig. 2.
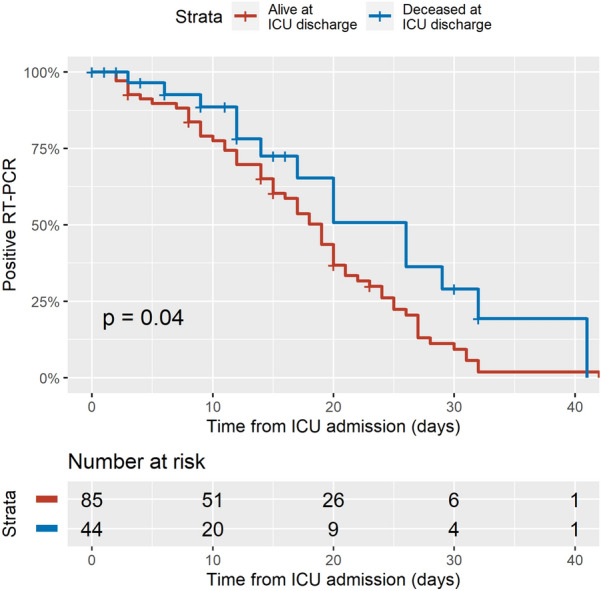
Kaplan–Meier plot of the probability of remaining RT-PCR positive for SARS-CoV-2. Blue line refers to patients deceased in ICU and red line to patients alive at ICU discharge. Right-censoring was performed at the time of the last positive RT-PCR in patients without SARS-CoV-2 negativation on RT-PCR during their hospital stay. ICU intensive care unit, RT-PCR real-time reverse transcriptase polymerase chain reaction
Univariate and multivariate competitive risk regression of the probability of SARS-CoV-2 negativation on RT-PCR are presented in Table 2 and Fig. 3. Age and vital status at ICU discharge were independently associated with lower probability of SARS-CoV-2 negativation on RT-PCR, while antiviral treatment was independently associated with higher probability. Violation of the proportional hazard assumption was not identified for all tested variables. Sensitivity analyses using either a cycle threshold value ≤ 40 to define PCR positivity (Additional file 3) or time from 1st positive RT-PCR instead of time from ICU admission (Additional file 4) provided identical results.
Table 2.
Fine and gray competing risk regression of the probability of SARS-CoV-2 RT-PCR negativation
| Variable | Univariate SHR [CI95 %] | Univariate p value | Multivariate SHR [CI95 %] | Multivariate p value |
|---|---|---|---|---|
| Vital status at ICU discharge * (ref = alive) | 0.13 [0.07–0.25] | < 0.0001 | 0.15 [0.08–0.28] | < 0.0001 |
| Age (per 1-year increment) | 0.96 [0.94–0.98] | < 0.001 | 0.98 [0.96–0.99] | < 0.01 |
| Antiviral treatment (ref = no) | 2.19 [1.20–4.01] | < 0.05 | 2.38 [1.34–4.25] | < 0.01 |
| Immunodeficiency (ref = no) | 0.24 [0.06–0.96] | < 0.05 | – | NS |
| SOFA at ICU admission (per 1-point increment) | 0.95 [0.88–1.02] | 0.16 | – | NS |
| Sex (ref = male) | 0.78 [0.49–1.22] | 0.28 | – | NS |
| Center (ref = center#1) | 2.22 [1.45–3.40] | < 0.001 | – | NS |
CI95 % 95% confidence interval, ICU intensive care unit, NS non-significant, ref reference, RT-PCR real-time reverse transcriptase polymerase chain reaction, SHR subdistribution hazard ratio
Fig. 3.
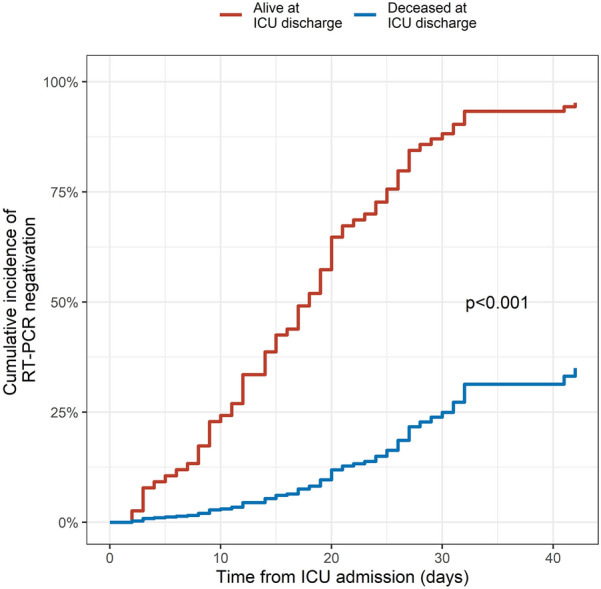
Multivariate fine and gray competitive risk regression. The model predicts the cumulative incidence of RT-PCR negativation following ICU admission. The curves represent multivariate model fit in patients deceased in ICU (blue lines) and in patients alive at ICU discharge (red lines). ICU intensive care unit, RT-PCR real-time reverse transcriptase polymerase chain reaction
CT semi-quantitative scores
137 chest CT scans were performed in 39 patients of center #1 with at least one CT scan during their ICU stay (median number of CT scans per patient: 3 [2–4]).
The first CT and last CT scans for each patient were performed 2 [1–9] days and 28 [15–39] days after ICU admission, respectively (Additional file 5). Patients included in the CT sub-study had significantly higher SAPS2 and SOFA scores, met more frequently ARDS criteria, required more often organ support, and had significantly higher ICU mortality (Additional file 6). 22 patients (56% [CI95 %:41–71%]) exhibited patterns of fibroproliferation during their ICU stay (40% in the group of patients alive at ICU discharge and 74% in patients deceased in ICU, p = 0.05, Table 3). Maximal bronchiectasis and IPO CT scores during ICU stay were associated with ICU mortality, while maximal reticulation and GGO scores were not (Table 3).
Table 3.
CT scores as a function of ICU mortality
| Variables | Overall population (n = 39) | Alive at ICU discharge (n = 20) | Deceased at ICU discharge (n = 19) | p value |
|---|---|---|---|---|
| Maximal bronchiectasis score | 1 [0–3] | 0 [0–1] | 2 [0–3] | < 0.05 |
| Maximal reticulation score | 0 [0–1] | 0 [0–0] | 0 [0–1] | 0.15 |
| Any fibroproliferation CT featurea | 22 (56%) | 8 (40%) | 14 (74%) | 0.05 |
| Maximal GGO score | 14 [11–15] | 14 [13–15] | 12 [9–15] | 0.16 |
| Maximal IPO score | 9 [8–11] | 8 [7–9] | 11 [9–12] | < 0.01 |
Values are median [1st quartile-3rd quartile] or count (percentage)
CT computed tomography, GGO ground glass opacification, ICU intensive care unit, IPO intense parenchymal opacification
aAny fibroproliferation CT feature was defined as occurrence of at least one bronchiectasis score or reticulation score > 0 during ICU stay
Fine and Gray competing risk regression analysis of the relationship between the probability of SARS-CoV-2 negativation on RT-PCR and maximal CT scores during the ICU stay is presented in Fig. 4. The IPO score was associated with a lower probability of SARS-CoV-2 negativation on RT-PCR (i.e., the higher the score, the longer the time to negate RT-PCR), while the GGO score was associated with a higher probability of SARS-CoV-2 negativation on RT-PCR. None of the CT scores exploring fibroproliferation (i.e., bronchiectasis and reticulation scores) were significantly associated with time to SARS-CoV-2 negativation on RT-PCR. Multivariate analyses were not performed owing to the small number of patients in this analysis.
Fig. 4.
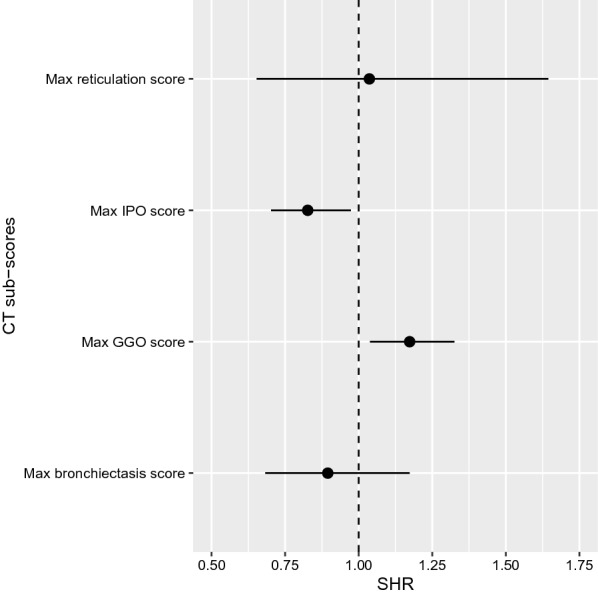
Fine and grey competitive risk regression of CT derived-scores to predict SARS-CoV-2 negativation on RT-PCR. The plot presents SHR estimates of CT-derived sub-scores to predict SARS-CoV-2 negativation on RT-PCR. Closed circles are SHR estimates and bars are their 95% confidence interval. Broken lines represent SHR equal to one. CT computed tomography, GGO ground glass opacification, IPO intense parenchymal opacification, SHR subdistribution hazard ratio, RT-PCR real-time reverse transcriptase polymerase chain reaction
Discussion
The main findings of the study are the following: 1- Time to SARS-CoV-2 negativation on RT-PCR is significantly longer and viral load significantly higher in respiratory samples of COVID-19 patients with ARF who died in ICU; 2- CT patterns of fibroproliferation are identified in more than 50% of the patients, but are not significantly associated with duration of viral shedding in respiratory samples; 3- The IPO CT score is associated with longer time to SARS-CoV-2 negativation on RT-PCR.
Regarding external validity, age, sex ratio in the present study were similar to previous ICU reports [1, 27–29]. Median time from symptoms to ICU admission was in the range of previously published studies on ICU patients [1, 27, 30]. Rate of patients requiring invasive ventilation, vasopressor administration and ICU mortality were similar to those observed in large ICU cohorts [28–30]. The relationship between duration of viral shedding and age has been previously observed in univariate analysis [8], and has been linked to immunosenescence. Demonstration of a lower duration of viral shedding in patients treated with antiviral agents is not unexpected as preliminary data suggest a positive effect on clinical outcome at least for remdesivir [31]. However, interpretation of the effect of antiviral treatments on viral shedding in the present study should be cautious as the number of patients is limited, the antiviral treatments were highly heterogeneous, and none of them has clearly shown any clinical efficacy apart remdesivir.
To our knowledge, no previously published study has related ICU mortality and duration of viral shedding in patients with ARF. Oppositely, a study on 191 hospitalized patients identified a significantly shorter viral shedding in non-survivors, but this study did not account for the competitive risk of death and mixed ICU and non ICU patients [11]. High viral load has been previously related to disease severity [12, 32], in line with the finding of the present study. It may be hypothesized that the longer duration of viral shedding in the non-survivors may be a consequence of the higher viral load at ICU admission. The longer viral shedding in non survivors may also be the consequence of ineffective immune response in this group, as our group has recently shown that up to 20% of ICU patients with COVID-19 and ARF presented with a sustained abrogation of type I interferon production [33], a major component of the innate immune system in the defense against viral infection. Taken together, these data suggest that efficient antiviral therapies are warranted to improve ICU mortality.
Several studies have analyzed chest CT with semi quantitative scores in non-severe patients with SARS-CoV-2 infection. In 21 repeatedly-studied patients from the early stage to the late phase of the disease, ground glass opacities were predominant 0–4 days after symptoms onset, crazy paving pattern was predominant 5–8 days after symptom onset, while the extent of IPO peaked at 10 days after symptoms onset [34]. Consistently with this finding, 100% of our patients exhibited IPO, as virtually all CT were performed more than 10 days after symptom onset. On 83 patients including 25 presenting with severe patients, total CT score and presence of IPO on CT were associated with disease severity [35]; a result in line with our results on more severely ill patients. In 3 studies, bronchus distortion was observed in 30% to 50% of the patients [36–38], a value similar to the 56% observed in the present study. We were unable to demonstrate an association between CT scores of fibroproliferation (reticulation and bronchiectasis scores) and protracted viral shedding, suggesting that lung fibrosis following COVID-19 may be related to host factors or external factors (such as ventilator-induced or patient self-inflicted lung injury) rather than to the intensity of viral aggression. However, the strong association between IPO score and duration of viral shedding suggests that patients with extensive IPO on CT are strong candidate for antiviral therapies. On the other hand, the association of GGO extent on CT and lower duration of viral shedding may suggest that GGO reflect recruitment of inflammatory cells in lung parenchyma, although this remains speculative.
Some limitations of the present study should be acknowledged. First, the estimated duration of viral shedding may have been overestimated by the relatively wide interval between RT-PCR samples (7–10 days), although this would be similar in both groups of patients (alive and deceased at ICU discharge). Second, we performed a majority of nasopharyngeal swabs for RT-PCR and this may have biased the results as false-negative RT-PCR may be less frequent in lower-respiratory-tract samples of ICU patients [39]. Third, 4 different RT-PCR techniques were used with expected impact on cycle threshold values, although this was accounted for by adjusting the multivariate analysis for the site of measurement and RT-PCR technique. Fourth, the cycle threshold defining RT-PCR positivity may be questionable, as the virus may be not cultivable anymore at such high values [40], and as positivity at this level might reflect the immune system deficiency and the impossibility to completely clear the virus. Nevertheless, the primary judgment criterion of the present study was unchanged using a lower cycle threshold (i.e., 40) in sensitivity analyses. Fifth, only 27% of the patients had at least 2 consecutive negative RT-PCR, and duration of viral shedding may have been underestimated by false negative measurements [8]. Finally, the CT score was computed in only 5 CT slices as previously performed in non-resolving ARDS [17], and a selection bias could not be ruled out, although the methodology was identical in all analyzed CT.
Nevertheless, the multicenter feature of the study and the relatively large sample size are important factors limiting the risk of bias. Although only a subset of patients could be analyzed by computed tomography, the number of CT scan analyzed ensured that the temporal time course of COVID-19 lung lesion has been correctly addressed. The demonstration of fibroproliferation on CT in the majority of the patients of the study suggest that anti-fibrotic therapies may be of interest to decrease duration of mechanical ventilation and prevent lung sequelae after ICU discharge.
Conclusions
Viral load in respiratory samples is significantly lower and viral shedding significantly shorter in ICU survivors of COVID-19 associated respiratory failure, and is unrelated to occurrence of fibrosis on lung CT. Lung fibrosis is frequent in the most severe ICU patients, suggesting that anti-fibrotic therapies may be of interest to improve prognostic. The extension of intense parenchymal opacities on CT is associated with duration of viral shedding in respiratory samples suggesting that ICU patients presenting this pattern are strong candidate for potent antiviral therapies against SARS-CoV-2.
Supplementary information
Additional file 1. Timing of rt-pcr sampling.
Additional file 2. Multivariate linear regression of variables associated with cycle threshold values (the inverse of viral load) in positive RT-PCR samples.
Additional file 3. Sensitivity analysis using a cycle threshold value ≤ 40 to define pcr positivity. multivariate fine and gray competitive risk regression of the probability of SARS-CoV-2 RT-PCR negativation.
Additional file 4. Sensitivity analysis using time from 1st positive RT-PCR instead of time from ICU admission. Multivariate fine and gray competitive risk regression of the probability of SARS-CoV-2 RT-PCR negativation.
Additional file 5. Timing of chest computed tomography.
Additional file 6. Patient characteristics at ICU admission as a function of their inclusion in the chest ct sub-study.
Acknowledgements
The authors wish to thank: Loredana BABOI for her help in data acquisition, Hodane YONIS, Guillaume DAVID, Judith PROVOOST, Clotilde BETTINGER, Pauline BERNON, Pierre LE CAM and Paul CHABERT for their implication in inclusion of patients.
Abbreviations
- ARDS
Acute respiratory distress syndrome
- ARF
Acute respiratory failure
- CI95 %
95% confidence interval
- CT
Computed tomography
- GGO
Ground glass opacity
- ICU
Intensive care unit
- IPO
Intense parenchymal opacity
- RT-PCR
Real-time reverse transcriptase polymerase chain reaction
Authors’ contributions
JCR had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. LB, FD, LC and FW contributed substantially to the study design, data acquisition, data analysis and interpretation, and the writing of the manuscript.’ EF, MM, STA, AB, and BL contributed substantially to the study design, data analysis and interpretation, and the writing of the manuscript. All authors read and approved the final manuscript.
Funding
None
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was conducted in accordance with the amended Declaration of Helsinki and was approved by our institutional ethic committee (#20–41). Consent for data utilization was sought from the patients or their representative.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13613-020-00783-4.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020. [DOI] [PMC free article] [PubMed]
- 4.Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argenziano MG, Bruce SL, Slater CL, Tiao JR, Baldwin MR, Barr RG, et al. Characterization and clinical course of 1000 patients with coronavirus disease 2019 in New York: retrospective case series. BMJ. 2020;369:m1996. doi: 10.1136/bmj.m1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Y, Tu L, Zhu P, Mu M, Wang R, Yang P, et al. Clinical Features of 85 Fatal Cases of COVID-19 from Wuhan. A Retrospective Observational Study. Am J Respir Crit Care Med. 2020;201:1372–1379. doi: 10.1164/rccm.202003-0543OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao AT, Tong YX, Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed]
- 9.Fang Z, Zhang Y, Hang C, Ai J, Li S, Zhang W. Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Q-L, Li G-M, Ji F, Ma S-H, Zhang G-F, Xu J-H, et al. Clinical course and treatment efficacy of COVID-19 near Hubei Province, China: a multicentre, retrospective study. Transbound Emerg Dis. 2020. [DOI] [PMC free article] [PubMed]
- 11.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Yan L-M, Wan L, Xiang T-X, Le A, Liu J-M, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–657. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei J, Lei P, Yang H, Fan B, Qiu Y, Zeng B, et al. Analysis of thin-section CT in patients with coronavirus disease (COVID-19) after hospital discharge. J Xray Sci Technol. 2020;28(3):383–389. doi: 10.3233/XST-200685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Gall JR, Lemeshow S, Saulnier F. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270:2957–2963. doi: 10.1001/jama.1993.03510240069035. [DOI] [PubMed] [Google Scholar]
- 15.Vincent JL, Moreno R, Takala J, Willatts S, De Mendonca A, Bruining H, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22:707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 16.ARDS Definition Task Force, Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–33. [DOI] [PubMed]
- 17.Burnham EL, Hyzy RC, Paine R, Kelly AM, Quint LE, Lynch D, et al. Detection of fibroproliferation by chest high-resolution CT scan in resolving ARDS. Chest. 2014;146:1196–1204. doi: 10.1378/chest.13-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team. R: A language and environment for statistical computing [Internet]. Vienna, Austria: R Foundation for statistical Computing; 2020. Available from: http://www.R-project.org.
- 19.Therneau TM, Grambsch PM. Modeling survival data: extending the cox model. New York: Springer; 2000. [Google Scholar]
- 20.Therneau TM. A Package for Survival Analysis in R. [Internet]. 2020. http://CRAN.R-project.org/package=survival.
- 21.Kassambara A, Kosinski M, Biecek P. survminer: Drawing Survival Curves using “ggplot2”. 2019. https://CRAN.R-project.org/package=survminer.
- 22.Gray B. cmprsk: Subdistribution Analysis of Competing Risks. 2019. https://CRAN.R-project.org/package=cmprs.
- 23.Qiu W, Chavarro J, Lazarus R, Rosner B, Ma J. powerSurvEpi: Power and Sample Size Calculation for Survival Analysis of Epidemiological Studies. 2018. https://CRAN.R-project.org/package=powerSurvEpi.
- 24.Bates D, Maechler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 25.Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest Package: tests in Linear Mixed Effects Models. J Stat Softw. 2017;82:1–26. doi: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- 26.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. doi: 10.1080/01621459.1999.10474144. [DOI] [Google Scholar]
- 27.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auld SC, Caridi-Scheible M, Blum JM, Robichaux C, Kraft C, Jacob JT, et al. ICU and Ventilator Mortality Among Critically Ill Adults With Coronavirus Disease 2019. Crit Care Med. 2020;48(9):e799–e804. doi: 10.1097/CCM.0000000000004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitra AR, Fergusson NA, Lloyd-Smith E, Wormsbecker A, Foster D, Karpov A, et al. Baseline characteristics and outcomes of patients with COVID-19 admitted to intensive care units in Vancouver, Canada: a case series. CMAJ. 2020;192(26):E694–E701. doi: 10.1503/cmaj.200794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, et al. Remdesivir for the Treatment of Covid-19 - Preliminary Report. N Engl J Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J, et al. Impact of SARS-CoV-2 Viral Load on Risk of Intubation and Mortality Among Hospitalized Patients with Coronavirus Disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trouillet-Assant S, Viel S, Gaymard A, Pons S, Richard J-C, Perret M, et al. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol. 2020;146(1):206–208. doi: 10.1016/j.jaci.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, et al. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K, Wu J, Wu F, Guo D, Chen L, Fang Z, et al. The Clinical and Chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol. 2020;55:327–331. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou S, Wang Y, Zhu T, Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am J Roentgenol. 2020;214:1287–1294. doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- 37.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest ct findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. AJR Am J Roentgenol. 2020;214:1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 38.Ding X, Xu J, Zhou J, Long Q. Chest CT findings of COVID-19 pneumonia by duration of symptoms. Eur J Radiol. 2020;127:109009. doi: 10.1016/j.ejrad.2020.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z, et al. SARS-CoV-2 viral load in clinical samples from critically ill patients. Am J Respir Crit Care Med. 2020;201:1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Timing of rt-pcr sampling.
Additional file 2. Multivariate linear regression of variables associated with cycle threshold values (the inverse of viral load) in positive RT-PCR samples.
Additional file 3. Sensitivity analysis using a cycle threshold value ≤ 40 to define pcr positivity. multivariate fine and gray competitive risk regression of the probability of SARS-CoV-2 RT-PCR negativation.
Additional file 4. Sensitivity analysis using time from 1st positive RT-PCR instead of time from ICU admission. Multivariate fine and gray competitive risk regression of the probability of SARS-CoV-2 RT-PCR negativation.
Additional file 5. Timing of chest computed tomography.
Additional file 6. Patient characteristics at ICU admission as a function of their inclusion in the chest ct sub-study.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


