Abstract
Osteoarthritis (OA) is a degenerative joint disease, and inflammation within an arthritic joint plays a critical role in disease progression. Proinflammatory cytokines, specifically IL-1 and TNF-α, induce aberrant expression of catabolic and degradative enzymes and inflammatory cytokines in OA and result in a challenging environment for cartilage repair and regeneration. MicroRNAs (miRNAS) are small, non-coding RNAs and are important regulatory molecules that act by binding to target mRNAs to reduce protein synthesis and have been implicated in many diseases, including OA. The goal of this study was to understand the mechanisms of miRNA regulation of the transcriptome of tissue engineered cartilage in response to IL-1β and TNF-α using an in vitro murine induced pluripotent stem cell (miPSC) model system. We performed miRNA and mRNA sequencing to determine the temporal and dynamic responses of genes to specific inflammatory cytokines as well as miRNAs that are differentially expressed in response to both cytokines or exclusively to IL-1β or TNF-α. Through integration of mRNA and miRNA sequencing data, we created networks of miRNA-mRNA interactions which may be controlling the response to inflammatory cytokines. Within the networks, hub miRNAs, miR-29b-3p, miR-17–5p, and miR-20a-5p, were identified. As validation of these findings, we found that delivery of miR-17–5p and miR-20a-5p mimics significantly decreased degradative enzyme activity levels while also decreasing expression of inflammation-related genes in cytokine treated cells. This study utilized an integrative approach to determine the miRNA interactome controlling the response to inflammatory cytokines and novel mediators of inflammation-driven degradation in tissue-engineered cartilage.
Keywords: microRNA, RNA-sequencing, inflammation, tissue engineering, induced pluripotent stem cell
Introduction
Osteoarthritis (OA) is a debilitating joint disease that affects over 32 million adults in the United States alone (1). OA is characterized not only by the degeneration of articular cartilage, but also by subchondral bone thickening, osteophyte formation, and modification of synovial fluid composition, which leads to joint pain, inflammation, and stiffness (2). These processes are mediated in part by pro-inflammatory cytokines, such as interleukin-1 (IL-1) and tumor necrosis factor-α (TNF-α). These cytokines are elevated in the synovial fluid, synovial membrane, subchondral bone, and cartilage of patients with OA, and act by both suppressing synthesis of cartilage matrix components and inducing expression of catabolic and degradative enzymes and inflammatory cytokines (3–5). The presence of inflammation within an OA joint provides a challenging environment for articular cartilage repair, as pro-inflammatory cytokines will cause degeneration of native and tissue engineered cartilage, as well as prevent chondrogenesis of stem cells (6, 7).
Thus, a number of approaches have targeted the IL-1 or TNF-α pathways as a therapy for OA (8–11) or as a means of inhibiting inflammatory effects on tissue-engineered cartilage (12–14). Although these studies show promise for targeted anti-inflammatory therapies, clinical studies have not shown consistent and clear effects on OA (15). Furthermore, although IL-1 and TNF-α use many shared signaling pathways and mediators, it has been shown that they have distinct roles in OA. IL-1 plays a role in sustained cell infiltration and cartilage destruction, while TNF-α is important for early joint swelling and driving the inflammatory cascade (3, 16). Because there are no effective disease modifying treatments available that address both the symptoms and structural changes of OA (17, 18), a further understanding of the molecular mechanisms regulating cartilage response to inflammation is needed for the development of alternative therapeutic approaches.
In this regard, multiple studies suggest that interaction between microRNAs (miRNAs) and mRNAs may play a critical role in the regulation of chondrocyte biology [reviewed by Swingler et al. (19) and by Asahara (20)]. miRNAs are small, endogenous, non-coding RNAs that play an important role in the regulation of many biological processes, such as proliferation, differentiation, and diseases including OA. They act by binding to target mRNAs which results in suppressed protein synthesis by translational inhibition, or, primarily, mRNA degradation (21–23). miRNAs have unique expression profiles in different tissues, cell types, developmental stages, and diseases states (24), and are attractive therapeutic targets because of their ability to regulate multiple genes and target pathways in complex diseases. miRNAs have been shown to participate in chondrogenesis, chondrocyte homeostasis, and osteoarthritis (25–28). A number of experiments have shown differentially expressed miRNAs from OA affected tissues compared to normal cartilage (29–31), and some have explored the effects of IL-1β treatment on miRNA changes. However, these studies have not determined the specific roles of miRNAs in regulating chondrocyte response to IL-1 or TNF-α while controlling for factors such as stage of disease, body mass index, or location of tissue harvest (26). The deregulation of miRNAs is an important factor in the pathogenesis of OA and understanding their specific roles and regulatory networks in response to inflammatory cytokines is important to guide miRNA-based therapies.
In a recent study, an integrative analysis of miRNA and mRNA sequencing data comparing preserved (e.g., undamaged) and lesioned human osteoarthritic cartilage revealed the miRNA interactome that potentially regulates the chondrocyte transcriptome in healthy or pathologic cartilage (32). Integration of RNA-sequencing data allows for an unbiased and comprehensive study of the miRNA-mRNA interactions, which could reveal previously unknown pathways or mediators in complex disease processes. Application of this technique to chondrogenically-differentiated stem cells and tissue-engineered cartilage would provide a better understanding of how tissues designed to replace damaged cartilage are going to respond within a diseased joint. Furthermore, induced pluripotent stem cells (iPSCs) provide a source of large numbers of genetically-defined cells that are capable of chondrogenic differentiation for cartilage repair or in vitro OA disease modeling (33–35).
The goal of this study was to determine the mechanisms of miRNA regulation of tissue engineered cartilage in response to inflammatory cytokines using an in vitro murine induced pluripotent stem cell (miPSC) model system previously developed in our lab (33, 34). This three-dimensional culture system also allows for the study of precise experimental factors while maintaining miPSC-derived chondrocytes in a rounded shape within the cartilaginous extracellular matrix, which is required to maintain their phenotype and as well as the cell-matrix interactions that can significantly influence the response to exogenous stimuli (33, 34, 36). Furthermore, this model system allows for the control of many parameters that have confounded other miRNA-based studies (37), leading to inconsistencies among different studies on key miRNAs and mechanisms of action. We performed an integrative analysis of miRNA and mRNA sequencing data to functionally characterize the miRNA regulatory networks driving IL-1β and TNF-α mediated responses of tissue-engineered cartilage (Figure 1A). We identify key miRNA mediators driving novel mechanisms in the inflammatory response of articular chondrocytes and explore their ability to attenuate inflammation in a miPSC model system of OA. This study provides a thorough description of the miRNA-mRNA network following cytokine exposure, which can serve as the basis for designing new therapies to prevent inflammation-driven degradation. Furthermore, these findings can be applied to tissue engineering strategies for the treatment of OA, providing a better understanding of the response of this tissue to the inflammatory environment of an OA joint, such that therapies can be designed to improve replacement of damaged tissues and prevent disease progression.
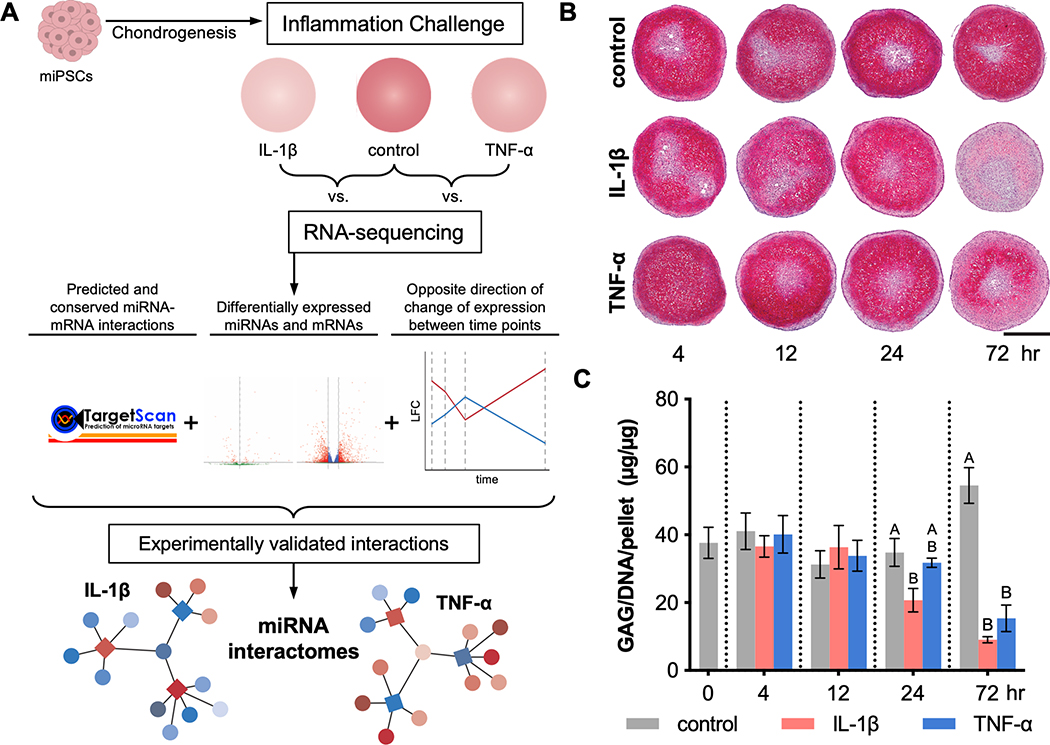
Figure 1.
Overview of experimental and analytical approach to evaluate tissue level responses of miPSC-derived cartilage to inflammation. (A) miPSCs were chondrogenically differentiated and then exposed to inflammatory cytokines, IL-1β or TNF-α, for 4, 12, 24, and 72 hours and then analyzed by RNA-sequencing. Predicted and conserved miRNA-mRNA interactions in which the miRNA and mRNA were differentially expressed and had opposite direction of change of expression level from one time point to the next were selected. MiRNA-mRNA pairs were further filtered for those that have been experimentally validated, and miRNA interactomes were constructed. At each time point, (B) pellet sections were stained for safranin-O/hematoxylin/fast green (scale bar = 500μm) and (C) sGAG content was measured, normalized to DNA content per pellet (mean±SEM; n=3). At each time point, differences among groups with different letters are statistically significant (p<0.05).
Materials and Methods
Cell culture and differentiation
Murine iPSCs (miPSCs), derived from mouse primary fibroblasts from adult C57BL/6 mice using a non-integrating system, were purchased from the Gates Center for Regenerative medicine (University of Colorado, Denver). iPSCs were cultured in KnockOut™ Dulbecco’s Modified Eagles Medium (DMEM) (Gibco), 20% lot-selected FBS (Atlanta Biologicals), 100 nM MEM nonessential amino acids (NEAA, Gibco), 55 µM 2-mercaptoethanol (2-me, Gibco), 2 mM Glutamax (Gibco), 1% penicillin/streptomycin (P/S, Gibco) and maintained on mitomycin C-treated mouse embryonic fibroblasts (Millipore).
miPSCs were differentiated towards a mesenchymal state using a high-density micromass culture (33). Differentiation medium contained DMEM-high glucose (DMEM-HG, Gibco), 1% culture medium supplement containing recombinant human insulin, human transferrin, and sodium selenite (ITS+, BD), 100 nM MEM NEAA, 55 µM 2-me, 24 ng/mL gentamicin, 50 μg/mL L-ascorbic acid, and 40 μg/mL L-proline. On days 3–5, this medium was supplemented with 100 nM dexamethasone and 50 ng/mL bone morphogenetic protein 4 (BMP-4, R&D Systems). After 15 days of culture, the micromasses were dissociated with pronase (Millipore Sigma) and collagenase type II (Worthington Biochemical) and the pre-differentiated iPSCs (PDiPSCs) were plated on gelatin-coated dishes in expansion medium containing DMEM-HG, 10% lot-selected FBS, 1% ITS+, MEM NEAA, 55 µM 2-me, 1% P/S, 50 µg/mL L-ascorbic acid, 40 µg/mL L-proline, and 4 ng/mL of basic fibroblast growth factor (bFGF, R&D Systems). Passage 2 PDiPSCs were pelleted by centrifugation of 250–300k cells and cultured for 21 days in chondrogenic medium consisting of DMEM-HG, 1% ITS+, 100nM MEM NEAA, 55 µM 2-me, 1% P/S, 50 µg/mL L-ascorbic acid, 40 µg/mL L-proline, 100 nM dexamethasone, and 10 ng/mL TGF-β3 (R&D Systems).
Inflammatory challenge
After 21 days of chondrogenic culture, pellets underwent inflammatory challenge for 72 hours, which involved culture in chondrogenic media with 1 ng/mL IL-1β, 20 ng/mL TNF-α, or control media without cytokines. TGF-β and dexamethasone were removed during this period. To capture the short- and long-term responses of cells to inflammation, pellets were collected after 4, 12, 24, and 72 hours, rinsed in PBS, and stored at −80ºC for RNA sequencing or at −20ºC for biochemical analysis.
RNA-sequencing
To extract RNA from tissue engineered cartilage, frozen pellets were homogenized with a miniature bead beater. Total cellular RNA was isolated using the mirVana miRNA Isolation Kit (Invitrogen) for small RNA sequencing or Total RNA Purification Plus Kit (Norgen) for mRNA sequencing. Three pellets were pooled per sample and each sample was sequenced on an Illumina HiSeq3000 (n=3). RNA-sequencing reads were trimmed (Trimmomatic (38), Cutadapt (39)), aligned (STAR aligner (40) to map small RNA-sequencing reads to a custom murine miRNA database based on miRBase v22 (41) and RNA-sequencing reads to mm10 from Gencode vM15), counted (featureCounts (42)), and only protein coding genes were included in downstream mRNA sequencing analysis.
Differential expression analysis and miRNA-mRNA network construction
The R package, DESeq2 (43), was used to determine differentially expressed miRNAs and genes through pairwise comparisons at each time point. Networks of miRNA-mRNA interactions were constructed through the following selection criteria: 1. conserved and predicted miRNA-mRNA target pairs (TargetScan Mouse release 7.2 (44)); 2. differential expression (DE) at any time point (miRNA: padj < 0.05; mRNA: padj < 0.05, |log2(FoldChange) (LFC)| > 1; padj is the false discovery rate computed by the Benjamini-Hochberg multiple testing correction); 3. opposite direction of change of expression level between time points for miRNA and mRNA based on the assumption that miRNA targeting of an mRNA will lead to mRNA degradation (22, 23, 32); 4. miRNA-mRNA interaction is experimentally validated (miRTarBase v7.0 (45) and TarBase v7 (46) databases). K-means clustering was applied to the normalized counts (RPKM, reads per kilobase per million mapped reads) of DE mRNAs at any time point in RStudio. The number of clusters, 5, was determined by the sum of squared error method. Enrichr (47, 48) was used for KEGG pathway analysis. Interactive networks were visualized on a web application with the R package, Shiny (49) (https://guilaklab.shinyapps.io/validatednetwork/).
In vitro experiments
Day 21 pellets were digested with collagenase type II and chondrocyte-like cells were plated in monolayer in media containing DMEM-HG, 10% FBS, 1% ITS+, 100nM MEM NEAA, 55 µM 2-me, 1% P/S. After 4 – 5 days, cells were transfected with mirVana miRNA mimics and antagomirs (Invitrogen): controls [mimic (M), mimic Negative Control #1; inhibitor (I), inhibitor Negative Control #1]; miR-29b-5p [M, MC10103; I, MH10103]; miR-17–5p [M, MC12412; I, MH12412]; miR-20a-5p [M, MC10057; I, MH10057]. Lipofectamine RNAiMAX Transfection Reagent (Invitrogen) was used according to manufacturer’s protocol and FBS and P/S were removed from media prior to transfection. 24 hours after transfection, cells were subject to an inflammatory challenge of 0.5 ng/mL IL-1β, 20 ng/mL TNF-α, or control media without cytokines. Cell culture media was collected and stored at −20°C and cells were lysed with buffer RL and stored at −80°C for analysis at 24 and 48 hours.
Histological and biochemical analysis of pellet cultures
After 72 hours of inflammatory challenge, pellets were washed with PBS and fixed in 10% neutral buffered formalin for 24 hours, paraffin embedded, and sectioned at 8 μm thickness. Slides were stained for Safranin-O/hematoxylin/fast green (50).
Pellets were digested overnight in 125 μg/mL papain at 65°C for biochemical analysis. DNA content was measured with PicoGreen assay (Thermo Fisher) and total sulfated glycosaminoglycan (sGAG) content was measured using a 1,9-dimethylmethylene blue assay at 525 nm wavelength (51).
Gene expression validation with quantitative real time PCR
For mRNA gene expression analysis, isolated, total cellular RNA was reverse transcribed using Superscript VILO cDNA master mix (Invitrogen). Quantitative, reverse transcription polymerase chain reaction (qRT-PCR) was performed using Fast SYBR master mix (Applied Biosystems) following the manufacturer’s protocol. Primer pairs were synthesized by Integrated DNA Technologies, Inc.: Ccl2 [forward (F), 5’-GGCTCAGCCAGATGCAGTTAA-3’; reverse (R), 5’-CCTACTCATTGGGATCATCTTGCT-3’[; Mmp13 [F, 5’-GGGCTCTGAATGGTTATGACATTC-3’; R, 5’-AGCGCTCAGTCTCTTCACCTCTT-3’]; r18s [F, 5’-CGGCTACCACATCCAAGGAA-3’; R, 5’-GGGCCTCGAAAGAGTCCTGT-3’]. Data are reported as fold changes and were calculated using the ΔΔCT method and are shown relative to the non-transfected control media group at each time point and ribosomal 18s is used as the reference gene.
MMP activity assay
MMP activity in culture media was measured by detecting the quenching of a fluorogenic substrate Dab-Gly-Pro-Leu-Gly-Met-Arg-Gly-Lys-Flu (Sigma-Aldrich)(52). Latent MMP’s were activated using p-aminophenylmercuric acetate, and total MMP activity was measured as the difference between a broad-spectrum MMP inhibitor, GM6001 (EMD Biosciences Inc.) and a scrambled negative control peptide (EMD Biosciences Inc.). Fluorescence was measured at 485 nm excitation and 535 nm emission and is reported as relative fluorescence units (RFU).
Statistical analysis
Statistical analysis was performed with the JMP Pro software package. A two-way ANOVA with Tukey’s HSD post-hoc test was used to analyze all biochemistry, qRT-PCR, and MMP activity assay data (α = 0.05).
Results
Histological and biochemical response of tissue engineered cartilage to inflammation
After 21 days of culture in chondrogenic media, the resulting cartilaginous pellets showed a spherical shape, and pellets in control media without cytokines exhibited robust safranin-O staining (Figure 1B). However, when treated with inflammatory cytokines, either 1 ng/ml of IL-1β or 20 ng/ml of TNF-α, pellets exhibited reduced safranin-O staining by 72 hours, indicating a decrease in the primary cartilage extracellular matrix component, aggrecan (Figure 1B). Biochemical analysis by DMMB assay, normalized to dsDNA content, showed a corresponding decrease in sulfated glycosaminoglycans (sGAGs) at 24 and 72 hours in groups receiving IL-1β or TNF-α, as compared to the pellets in control media (Figure 1C).
Differentially expressed miRNAs and genes in response to inflammatory cytokines
Day 21 pellets were subject to IL-1β and TNF-α inflammatory challenge and collected for RNA-seq at 4, 12, 24, and 72 hours. The IL-1β or TNF-α stimulated pellets were compared pairwise to control at all time points in order to determine differentially expressed (DE) miRNAs and genes (mRNAs) at each time point. Over time, there were increasing numbers of DE miRNAs and genes. From 4 to 72 hours following treatment, the number of significantly DE miRNAs increased from 4 (4 up-regulated, 0 down-regulated) to 86 (37 up-regulated, 49 down-regulated) in IL-1β treated pellets and 4 (4 up-regulated, 0 down-regulated) to 168 (100 up-regulated, 68 down-regulated) in TNF-α treated pellets (Figure 2A). Similarly, the number of DE mRNAs from 4 to 72 hours increased from 772 (481 up-regulated, 291 down-regulated) to 1781 (849 up-regulated, 932 down-regulated) in IL-1β treated pellets and 612 (516 up-regulated, 96 down-regulated) to 1147 (635 up-regulated, 512 down-regulated) in TNF-α treated pellets.
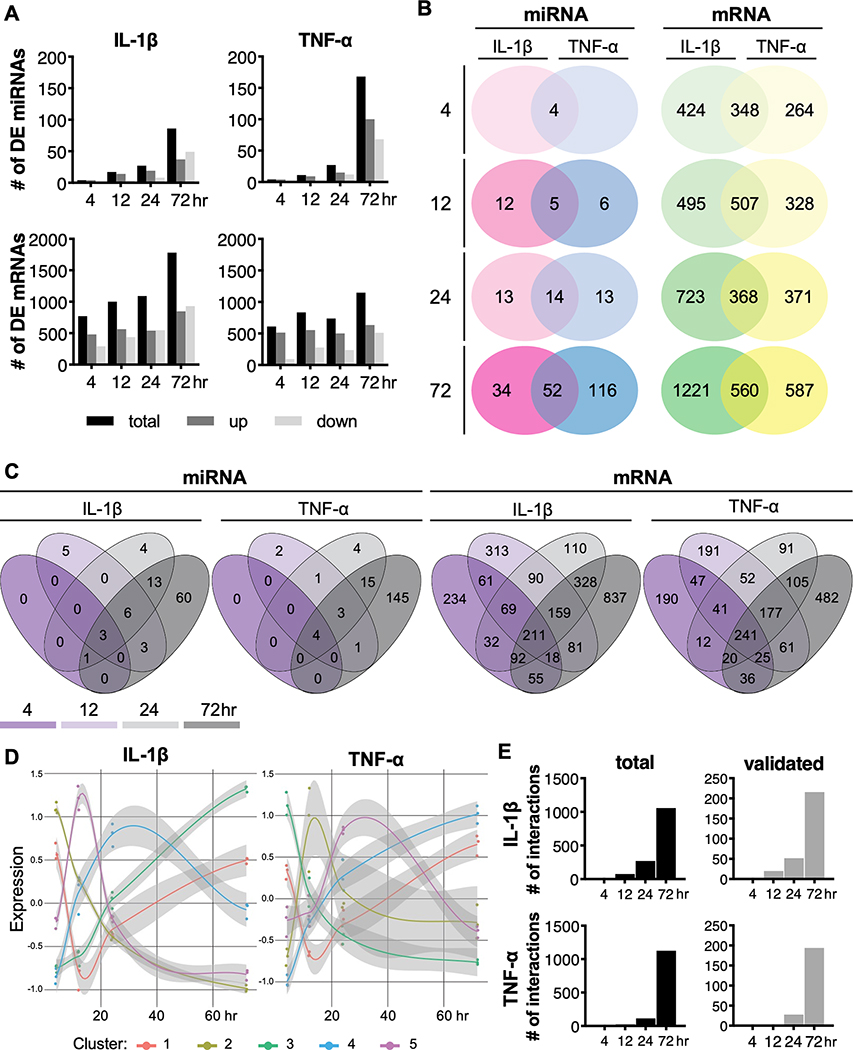
Figure 2.
Differential expression (DE) analysis of miRNAs and mRNAs. (A) The number of up- and down-regulated miRNAs and mRNAs in response to IL-1β or TNF-α over time. Distribution of shared or unique miRNAs or mRNAs (B) with each inflammatory cytokine over time or (C) at each time point. (D) Expression patterns of clusters of differentially expressed miRNAs. Points on graph represent the scaled reads per kilobase per million mapped reads (RPKM) values for the centers of each sample in the cluster (n=3). (E) The number of total and validated miRNA-mRNA interactions for IL-1β and TNF-α following RNA- and small RNA-sequencing data integration.
While IL-1β and TNF-α groups shared the same DE miRNAs at 4 hours, over time there was an increasing number of both shared miRNAs as well as miRNAs that were uniquely up- or down-regulated with each cytokine. However, with respect to the mRNA transcriptome, we identified many DE genes that were specific to each cytokine at early time points. At later times, there were increasing numbers of shared and unique DE genes in response to either IL-1β or TNF-α (Figure 2B). There were 348 shared, 424 IL-1β specific, and 264 TNF-α specific DE genes at 4 hours, which increased to 560 shared, 1221 IL-1β specific, and 587 TNF-α specific DE genes at 72 hours.
Analysis of the differentially expressed miRNAs over time showed that most of the miRNAs that are expressed at early time points (4 and 12 hours) are also expressed at later time points (24 and 72 hours) for both IL-1β and TNF-α treated groups (Figure 2C). On the other hand, there were more differentially expressed genes unique to each time point in addition to the genes that were differentially expressed at multiple time points (Figure 2C). Cluster analysis revealed 5 unique expression profiles of genes over time for both IL-1β and TNF-α pellets (Figure 2D), and KEGG pathway analysis shows that each of these clusters are enriched for different pathways (Table S1 and S2). These results show that there are unique miRNA and mRNA profiles with some overlapping genes and miRNAs, and others that are dependent on IL-1β or TNF-α.
Differential expression analysis reveals miRNAs that are differentially expressed compared to controls at each time point (Figure S1). At 4 hours, miR-222–5p (IL-1β, TNF-α: LFC (padj), 1.27 (1.6×10−18), 1.57 (8.3×10−30)) and −3p (IL-1β, TNF-α: LFC (padj), 0.84 (1.5×10−12), 0.72 (6.2×10−9)), miR-92a-1–5p (IL-1β, TNF-α: LFC (padj), 1.43 (1.2×10−7), 1.02 (2.5×10−3)), and miR-155–5p (IL-1β, TNF-α: LFC (padj), 0.52 (2.3×10−3), 0.67 (2.2×10−6)) were up-regulated in response to both IL-1β and TNF-α. At 72 hours, among the most highly upregulated miRNAs in both IL-1β and TNF-α treated pellets were miR-146a-5p (IL-1β, TNF-α: LFC (padj), 3.05 (2.0×10−15), 4.10 (3.0×10−28)), miR-7a-5p (IL-1β, TNF-α: LFC (padj), 3.79 (7.6×10−3), 4.86 (1.1×10−4)), and miR-155–5p (IL-1β, TNF-α: LFC (padj), 1.57 (1.4×10−40), 1.43 (1.6×10−33)); those most highly down regulated with both cytokines include: miR-341–5p (IL-1β, TNF-α: LFC (padj), −2.53 (4.0×10−27), −1.85 (1.7×10−16)), miR-410–5p (IL-1β, TNF-α: LFC (padj), −1.81 (1.1×10−2), −1.82 (1.0×10−2)), and miR-665–3p (IL-1β, TNF-α: LFC (padj), −1.61 (8.0×10−11), −1.11 (1.5×10−5)). Several miRNAs were found to be regulated exclusively by IL-1β, such as miR-138–1-3p and miR-154–3p (Figure S3A), or by TNF-α, such as miR-7678–3p, miR-6972–3p, miR-147–3p, miR-210–5p, and miR-421–5p (Figure S3B). Similarly, differentially expressed genes were visualized with volcano plots (Figure S2), and KEGG pathway analysis was conducted on significant DE mRNAs at each time point to determine enriched pathways over time for both IL-1β (Table S3) and TNF-α (Table S4) groups. The differentially expressed mRNAs in response to both cytokines were significantly enriched for genes involved in TNF signaling pathway, cytokine-cytokine receptor interaction, and IL-17 signaling pathways and characterized by genes such as Ptgs2, Nfkb1 and 2, MMP’s, and chemokine CC and CXC subfamily genes. Additional pathways of interest that were involved at various time points were rheumatoid arthritis, protein digestion and absorption, and PI3K-Akt signaling pathways, characterized by genes such as Tgfb3, MMP’s, Ccl2, and Col2a1 and other collagens.
miRNA interactomes in response to inflammatory cytokines
To determine the functional roles that miRNAs have on predicted target genes, RNA and small RNA-sequencing data sets were integrated such that networks could be constructed to visualize relationships between miRNA-mRNA predicted target pairs that are conserved (TargetScan database), contain a miRNA and mRNA that are each differentially expressed, and whose levels of expression are changing in opposite directions from one time point to the next (Figure S4; Figures 3 and 4). The total number of interactions increased over time in both the IL-1β and TNF-α groups from 15 to 1059 and 1 to 1128, respectively. There were 550 genes and 49 miRNAs in the IL-1β network and 445 genes and 78 miRNAs in the TNF-α network at 72 hours. Upon further filtering for only interactions that have been previously experimentally validated from the miRTarBase or TarBase databases, the number of interactions within networks reduced to 216 (153 genes and 28 miRNAs) in the IL-1β and 194 (128 genes and 24 miRNAs) in the TNF-α network at 72 hours (Figures 2E).
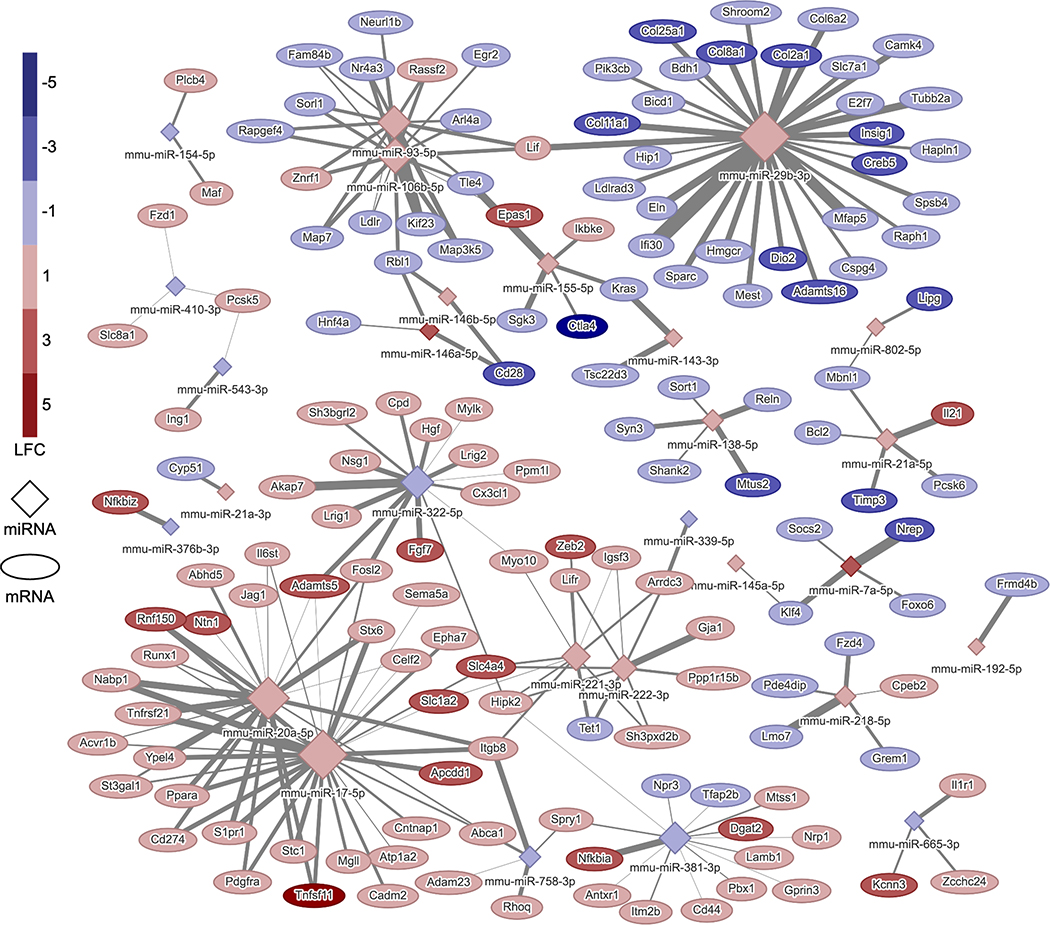
Figure 3.
IL-1β miRNA-mRNA interactome at 72 hours: network of conserved and predicted miRNA-mRNA target pairs that are differentially expressed, are changing expression levels in opposite directions from 24 to 72 hours, and have been experimentally validated in response to 1 ng/mL IL-1β. The color of the node indicates the LFC of the gene or miRNA, and the size of the node is proportional to the number of connections. The thickness of the line corresponds to the context++ score from the TargetScan database. LFC, log2(FoldChange).
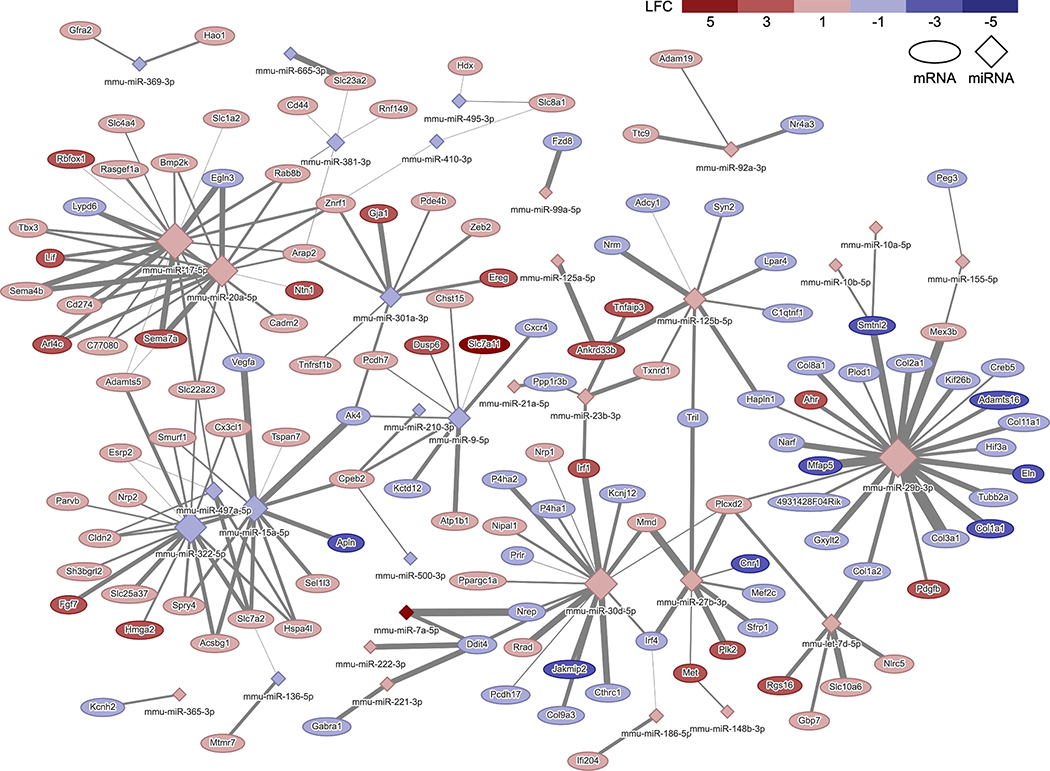
Figure 4.
TNF-α miRNA-mRNA interactome at 72 hours: network of conserved and predicted miRNA-mRNA target pairs that are differentially expressed, are changing expression levels in opposite directions from 24 to 72 hours, and have been experimentally validated in response to 20 ng/mL TNF-α. The color of the node indicates the LFC of the gene or miRNA, and the size of the node is proportional to the number of connections. The thickness of the line corresponds to the context++ score from the TargetScan database. LFC, log2(FoldChange).
To determine which miRNAs within the networks regulate the inflammatory responses, KEGG pathway analysis was conducted on the genes within the networks including all interactions (Tables 1 and 2). Genes in inflammation and OA related pathways (PI3K-AKt signaling pathway, ECM-receptor interaction, cytokine-cytokine receptor interaction, MAPK signaling, protein digestion and absorption, TNF signaling pathway JAK-STAT signaling pathway, and IL-17 signaling pathway; Tables 1–3) were overlaid onto the network and quantified the number of connections from each miRNA. Hub miRNAs, miR-17–5p and miR-20a-5p and miR-29b-3p, were identified in both IL-1β and TNF-α networks and are potentially controlling the miRNA response to these cytokines. miR-17–5p was connected to 52 genes (29 validated interactions) in the IL-1β network and 36 genes (21 validated interactions) in the TNF-α network at 72 hours. miR-20a-5p was connected to 52 genes (23 validated interactions) in the IL-1β network and 36 genes (17 validated interactions) in the TNF-α network at 72 hours. miR-29b-3p was connected to 79 genes (30 validated interactions) in the IL-1β network and 54 genes (23 validated interactions) in the TNF-α network at 72 hours. Inflammation and cartilage related interactions with these hub miRNAs are listed in Table 3.
Table 1.
Enriched pathways of genes within the IL-1β network at 72 hours.
| Pathway | P-value | Genes |
|---|---|---|
| PI3K-Akt signaling pathway | 1.3×10−9 | Lama3; Lamc2; Il6ra; Pik3cb; Lpar4; Thbs2; Fgf1; Egfr; Gys1; Fgf7; Reln; Gng2; Ibsp; Erbb4; Itgb8; Pdgfra; Hgf; Lamb1; Igf1; Ngf; Pgf; Ereg; Col2a1; Kitl; Col6a2; Fgf18; Col9a1; Bcl2; Sgk3; Kras; Fgfr3; Creb5; Fgf10 |
| Rap1 signaling pathway | 3.5×10−7 | Pdgfra; Prkcb; Hgf; Pik3cb; Lpar4; Igf1; Adcy1; Ngf; Fgf1; Egfr; Pgf; Fgf7; Plcb4; Cnr1; Kitl; Fgf18; Kras; Rapgef5; Fgfr3; Rapgef4; Fgf10 |
| Focal adhesion | 2.7×10−6 | Pdgfra; Prkcb; Hgf; Lama3; Lamc2; Lamb1; Pik3cb; Igf1; Thbs2; Egfr; Pgf; Mylk; Reln; Col2a1; Ibsp; Col6a2; Col9a1; Bcl2; Itgb8 |
| ECM-receptor interaction | 2.7×10−6 | Reln; Sv2c; Col2a1; Ibsp; Col6a2; Lama3; Col9a1; Itgb8; Lamc2; Lamb1; Thbs2; Cd44 |
| Pathways in cancer | 5.8×10−6 | Epas1; Il23r; Lama3; Lamc2; Il6ra; Pik3cb; Lpar4; Adcy1; Fgf1; Egfr; Fgf7; Gng2; Ednrb; Il13ra1; Fzd1; Pdgfra; Jag1; Prkcb; Fzd4; Hgf; Lamb1; Igf1; Pgf; Runx1; Nfkbia; Bmp2; Plcb4; Kitl; Fgf18; Bcl2; Kras; Il6st; Fgfr3; Fgf10 |
| Protein digestion and absorption | 6.5×10−6 | Col17a1; Col15a1; Col27a1; Col2a1; Mme; Col11a1; Eln; Fxyd2; Col6a2; Col9a1; Atp1a2; Slc8a1 |
| Cytokine-cytokine receptor interaction | 2.1×10−5 | Il21; Gdf10; Mstn; Il1r1; Il23r; Il34; Lif; Lifr; Il6ra; Tnfrsf11b; Ngf; Acvr1b; Cxcl2; Cx3cl1; Cxcl10; Bmp3; Bmp2; Il18rap; Tnfsf11; Il6st; Tnfrsf21; Il13ra1 |
| cGMP-PKG signaling pathway | 2.3×10−5 | Oprd1; Mef2c; Kcnj8; Atp1a2; Adcy1; Adra2c; Slc8a1; Adra2a; Mylk; Nppc; Plcb4; Ednrb; Fxyd2; Adora1; Pde3a; Creb5 |
| Insulin secretion | 2.3×10−5 | Rims2; Adcyap1; Plcb4; Prkcb; Fxyd2; Atp1a2; Cck; Kcnn3; Adcy1; Rapgef4; Creb5 |
| MAPK signaling pathway | 2.3×10−5 | Pdgfra; Mef2c; Prkcb; Il1r1; Hgf; Igf1; Ngf; Dusp8; Fgf1; Egfr; Pgf; Ereg; Cacna1g; Cacna1i; Fgf7; Erbb4; Kitl; Fgf18; Kras; Fgfr3; Map3k5; Fgf10 |
KEGG pathway analysis of the top enriched pathways, ranked by p-value, and their associated genes.
Table 2.
Enriched pathways of genes within the TNF-α network at 72 hours.
| Pathway | P-value | Genes |
|---|---|---|
| Protein digestion and absorption | 1.1×10−12 | Col18a1; Col15a1; Col27a1; Col11a1; Eln; Col22a1; Col11a2; Atp1b1; Slc8a1; Col1a1; Col3a1; Col1a2; Col2a1; Col5a1; Slc7a8; Col4a4; Col9a1; Col9a3 |
| ECM-receptor interaction | 3.9×10−10 | Itga2; Lamc2; Thbs2; Col1a1; Reln; Col1a2; Col2a1; Ibsp; Col4a4; Itga11; Col9a1; Col9a3; Itgb7; Cd44; Itga9 |
| PI3K-Akt signaling pathway | 2.2×10−9 | Csf1r; Csf1; Pdgfb; Lamc2; Lpar4; Thbs2; Fgf7; Reln; Ibsp; Itgb7; Itga2; Vegfc; Ngf; Prlr; Ereg; Vegfa; Col1a1; Col1a2; Col2a1; Ppp2r2c; Col4a4; Itga11; Ddit4; Col9a1; Col9a3; Fgfr3; Met; Creb5; Itga9 |
| Focal adhesion | 4.0×10−9 | Cav3; Itga2; Pdgfb; Vegfc; Lamc2; Parvb; Thbs2; Vegfa; Mapk10; Col1a1; Reln; Col1a2; Col2a1; Ibsp; Col4a4; Itga11; Col9a1; Col9a3; Itgb7; Met; Itga9 |
| TNF signaling pathway | 1.6×10−7 | Cebpb; Edn1; Csf1; Ccl20; Lif; Tnfaip3; Traf1; Tnfrsf1b; Cxcl2; Cx3cl1; Mapk10; Cxcl10; Irf1; Creb5 |
| Human papillomavirus infection | 5.8×10−7 | Notch3; H2-q7; Lamc2; Thbs2; Prkcz; Reln; Ibsp; Itgb7; Ikbke; Fzd2; Fzd4; Itga2; Fzd8; Vegfa; Col1a1; Col1a2; Col2a1; Ppp2r2c; Col4a4; Irf1; Itga11; Col9a1; Col9a3; Creb5; Itga9 |
| Transcriptional misregulation in cancer | 7.6×10−7 | Csf1r; Arnt2; Cebpb; Mef2c; Gadd45a; Igfbp3; Hmga2; Etv1; Traf1; Etv5; Dusp6; Nr4a3; Tspan7; Bmp2k; Itgb7; Erg; Met |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 4.2×10−6 | Cacng8; Gja1; Cacnb3; Des; Itga2; Itga11; Dmd; Itgb7; Slc8a1; Itga9 |
| Hypertrophic cardiomyopathy (HCM) | 2.1×10−5 | Cacng8; Edn1; Cacnb3; Des; Itga2; Itga11; Dmd; Itgb7; Slc8a1; Itga9 |
| Pathways in cancer | 2.8×10−5 | Camk2b; Csf1r; Notch3; Pdgfb; Cxcr4; Lamc2; Lpar4; Adcy1; Fgf7; Egln1; Arnt2; Egln3; Edn1; Fzd2; Gadd45a; Fzd4; Txnrd1; Itga2; Fzd8; Vegfc; Traf1; Vegfa; Mapk10; Bmp2; Traf4; Col4a4; Fgfr3; Met |
KEGG pathway analysis of the top enriched pathways, ranked by p-value, and their associated genes.
Table 3.
Inflammation and cartilage related genes* connected to hub miRNAs in the IL-1β and TNF-α networks at 72 hours.
| IL-1β | TNF-α | |
|---|---|---|
| miR-29b-3p | Adamts16; Col11a1; Col15a1; Col25a1; Col27a1; Col2a1; Col6a2; Col8a1; Col9a1; Creb5; Dio2; Eln; Ibsp; Lif; Mstn; Mybl2; Pik3cb | Adam19; Adamts16; Adamts20; Col11a1; Col15a1; Col19a1; Col1a1; Col1a2; Col22a1; Col27a1; Col2a1; Col3a1; Col5a1; Col8a1; Col9a1; Creb5; Eln; Hif3a; Ibsp; Itga11; Mstn; Pdgfb |
| miR-17–5p | Acvr1b; Adamts5; Atp1a2; Bmp2; Il6st; Itgb8; Jag1; Pdgfra; Ppara; Runx1; Tnfrsf21; Tnfsf11 | Adamts5; Bmp2; Bmp2k; Col4a4; Ereg; Itgb7; Lif; Vegfa |
| miR-20a-5p | Acvr1b; Adamts5; Atp1a2; Bmp2; Il6st; Itgb8; Jag1; Pdgfra; Ppara; Runx1; Tnfrsf21; Tnfsf11 | Adamts5; Bmp2; Bmp2k; Col4a4; Ereg; Itgb7; Lif; Vegfa |
All interactions are listed, and interactions that have been experimentally validated are bolded.
In vitro validation of miRNA-mRNA interactome targets
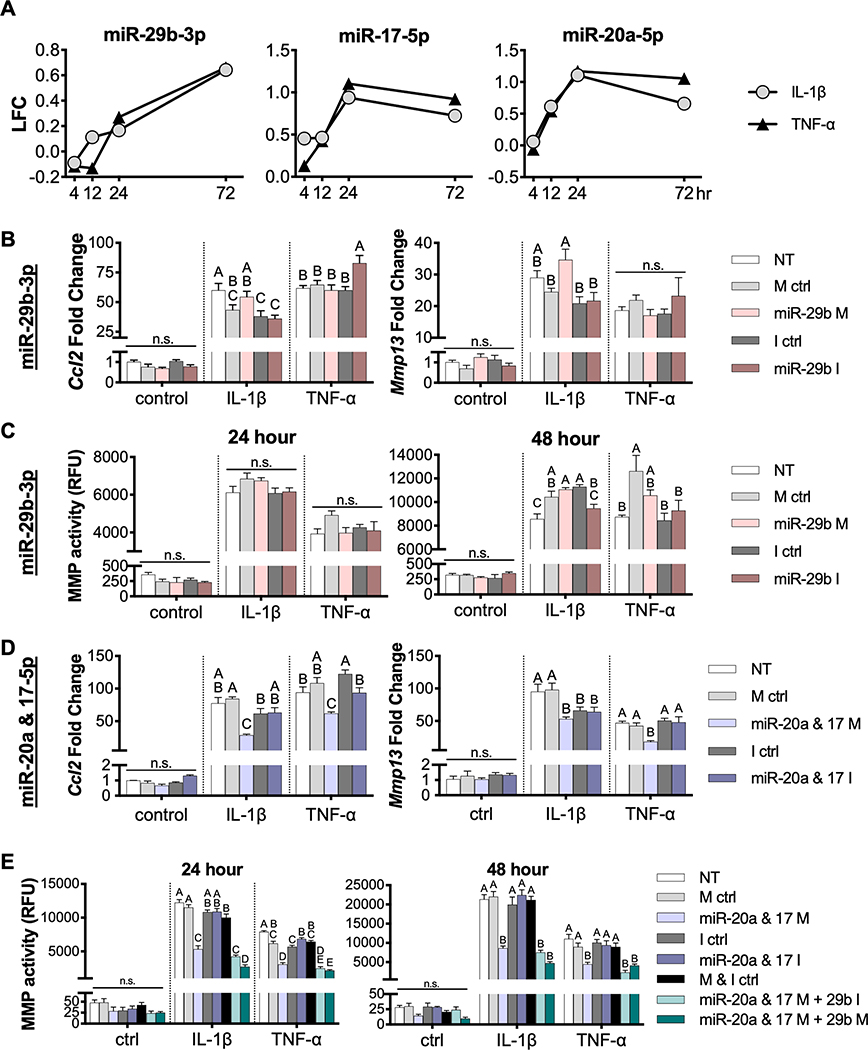
To probe specific miRNA-mRNA interactions and to determine the therapeutic potential of hub miRNAs within the networks, miRNA mimics or inhibitors were delivered to monolayer chondrocyte-like cells prior to challenge with inflammatory cytokines. The miR-29b-3p hub had the most connections to inflammation and cartilage/OA related genes in both the IL-1β and TNF-α networks, and RNA-sequencing analysis shows that miR-29b-3p expression increased over time (Figure 5A). miR-29b-3p mimics and inhibitors alone had no significant effect on the overall inflammatory status of the cells in response to either cytokine. Expression levels of Ccl2 and Mmp13, markers of inflammation and matrix degradation in OA, were not significantly decreased with miR-29b-3p mimic or inhibitor delivery 24 hours after inflammatory cytokines were given to cells, as compared to their respective controls (Figure 5B). Furthermore, miR-29b-3p mimics and had no effect on the MMP activity levels at 24 and 48 hours in TNF-α groups. Likewise, there were no significant differences in MMP activity levels after 24 hours in IL-1β exposed cells; however, at 48 hours, miR-29b inhibitor resulted in significantly lower MMP activity levels as compared to the inhibitor control, but this effect was not significant when compared to the non-transfected control (Figure 5C).
Figure 5.
In vitro evaluation of hub miRNA mimic and inhibitor delivery. (A) RNA-sequencing analysis showed expression of hub miRNAs miR-29b-3p, miR-17–5p, and miR-20a-5p in response to inflammatory cytokines, 0.5 ng/mL IL-1β or 20 ng/mL TNF-α. LFC, log2(FoldChange). miR-29b-3p mimics (M) and inhibitors (I) were delivered and cells were challenged with 0.5 ng/mL IL-1β or 20 ng/mL TNF-α to evaluate (B) Ccl2 and Mmp13 gene expression after 24 hours as measured by qRT-PCR and (C) MMP activity in cell culture media after 24 and 48 hours (n=4; mean±SEM). miR-17–5p and miR-20a-5p mimics (M) and inhibitors (I) were delivered in combination and cells were challenged with 0.5 ng/mL IL-1β and 20 ng/mL TNF-α to evaluate (D) Ccl2 and Mmp13 gene expression after 24 hours as measured by qRT-PCR and (E) MMP activity levels in cell culture media, determined with and without the further addition of miR-29b-3p mimics and inhibitors. (n=4; mean±SEM). Fold changes were determined relative to NT (non-transfected) control group. For each cytokine, groups with different letters are statistically significant (p<0.05): n.s., not significant.
In addition to the miRNA-29b-3p hub, network analysis revealed that miR-20a-5p and miR-17–5p, which are miRNAs expressed from the miR-17~92 cluster, may also be controlling the inflammatory response of tissue engineered cartilage to inflammation. RNA-sequencing results show that both miR-20a-5p and miR-17–5p increase from 4 to 24 hours, but slightly decrease from 24 to 72 hours in both IL-1β and TNF-α stimulated pellets (Figure 5A). With the addition of miRNA-20a & 17–5p mimics, there is a significant decrease in Mmp13 and Ccl2 expression at 24 hours after IL-1β and TNF-α delivery (Figure 5D). Additionally, there is a significant decrease in MMP activity in culture media at both 24 and 48 hours when miRNA-20a & 17–5p mimics are delivered to IL-1β and TNF-α stimulated pellets. The addition of miR-29b-3p mimics further decreases MMP activity levels in cell culture supernatant at 24 hours in both IL-1β and TNF-α groups as compared to miRNA-20a & 17–5p mimics alone, while the addition of miR-29b-3p inhibitor did not show significantly lower levels of MMP activity with either cytokine or at any time point.
Discussion
In this study, we quantified the transcriptomic changes that occur over time in tissue engineered cartilage in response to inflammatory cytokines IL-1β and TNF-α. Integration of mRNA and miRNA RNA-seq data sets in response to these cytokines revealed regulatory networks, in which the mechanisms of action for previously studied and novel hub miRNAs can be explored as well as their ability to modulate inflammation. Our results show the presence of distinct and common miRNA signatures in response to these cytokines, resulting in complex changes in the miRNA-mRNA interactome over time. These findings provide further data in support of the use of iPSCs-derived cartilage as a system for in vitro disease modeling and have important implications for the development of new approaches to target the chondrocyte inflammatory response.
Through RNA-seq analysis, we determined the temporal and dynamic response of genes to specific inflammatory cytokines. There have been a number of previous studies exploring the differentially expressed genes in OA tissues as compared to controls (53–55), and a few that explore the time course of OA (56–58). However, these studies are on the scale of days-weeks, where previous studies have shown specific, dynamic patterns of gene expression in response to inflammatory stimuli within hours (59). Here, we found that different genes were up- and down-regulated at various time points, with genes expressed at each time point exclusively and expressed across multiple time points (Figures 2A and 2C). Over time, differentially expressed genes in response to either IL-1β and TNF-α were found to be involved in similar inflammation and matrix degradation signaling pathways (Table 2 and 3). Furthermore, cluster analysis revealed the patterns of temporal expression of genes in response to inflammatory cytokines (Figure 2D). These findings provide a comprehensive exploration of the transcriptomic changes of tissue-engineered cartilage, as well as time course of these responses, both of which are important for guiding therapies to inhibit both chronic inflammation and flares that occur within a diseased joint.
Similarly, we characterized the temporal response of miRNAs to IL-1β and TNF-α and found that most of the differentially expressed miRNAs were up- and down-regulated at the 24 and 72 hour time points. There were a number of shared miRNAs, such as miR-155–3p and miR-146a-5p, which have previously been implicated in inflammatory responses of various cells (60–63), and have significantly increased expression in response to both cytokines in these findings. Interestingly, miR-155–5p and miR-222–3p were miRNAs that were significantly up-regulated at 4 hours. In bovine articular cartilage, miR-222–3p has been shown to have higher expression in weight-bearing regions of articular cartilage condyles and therefore, is thought to play a role in mechanotransduction (64). Superficial zone cartilage shows a higher expression of miR-222 compared to middle zone chondrocytes, and during dedifferentiation and subsequent redifferentiation, expression of miR-222 increased and then decreased, respectively (65). Furthermore, in developing human cartilage, miR-222 was found to be more highly expressed in hypertrophic compared to precursor chondrocytes (28). These findings implicate a role for miR-222–3p in chondrogenesis. Our results suggest that this miRNA is also rapidly responsive to inflammatory cytokines.
We also identified several miRNAs that showed specific responses to IL-1β or TNF-α. For example, miR-210–5p expression was significantly decreased in response to TNF-α but not IL-1β. Previous studies of animal models of OA have shown that delivery of miR-210 can reduce inflammation in the joint space by inhibiting NF-κB (66). Another example is miR-140, which is highly expressed in articular cartilage and a critical regulator of skeletal development and cartilage homeostasis (67, 68). Some studies, however, show that miR-140 is increased, while others show decreased expression in OA cartilage (69). In our study, miR-140–3p was the third most abundant miRNA across all samples and was significantly decreased only in response to TNF-α at 72 hours. This finding could explain the confounding results for miR-140 in OA studies as this miRNA appears to be both stimulus- and time-dependent. miRNAs that have not been previously implicated in healthy or diseased cartilage were also discovered through this analysis, such as miR-341–5p and miR-665–3p which were significantly decreased with both inflammatory cytokines, miR-154–3p which was significantly downregulated with IL-1β treatment only, and miR-421–5p, miR-7678–3p, miR-6972–3p, and miR-147–3p which were TNF-α specific.
To obtain a more complete understanding of the specific molecular mediators and networks regulating OA, studies are now using integrative -omics approaches (genomics, transcriptomics, proteomics, metabolomics, lipidomics, and epigenomics) (70). However, few studies have investigated the miRNA interactome by correlating miRNA expression levels to protein or mRNA changes in human OA tissues (29, 32). Here, we sought to provide an in-depth analysis of the specific contributions of IL-1β and TNF-α, as well as determine the response of tissue engineered cartilage to the inflammatory environment of an OA joint. Thus, we created networks of miRNA-mRNA interactions which may be controlling the response to inflammatory cytokines IL-1β and TNF-α (Figures 3 and 4) through integration of mRNA and miRNA sequencing data. Using this approach, we were able to identify key miRNAs of interest, miR-29b-3p, miR-17–5p, and miR-20a-5p. miR-29b has been reported to be increased in both joint tissues in a murine model of OA and human OA cartilage tissues and negatively regulates Smad, NF-κB, and canonical Wnt signaling pathways (71). In this study, miR-29b-3p was significantly increased in response to both IL-1β and TNF-α and has the most connections to inflammation related genes in both validated networks (Figures 3 and 4). However, miR-29b-3p mimics or inhibitors alone were not able to attenuate inflammation in cells (Figures 5B and 5C).
The other hub miRNAs identified in this study were miR-20a-5p and miR-17–5p, which are members of the miR-17~92 cluster. The miR-17~92 family has been extensively studied as an oncogene, however its role in other processes such as proliferation, development, and differentiation have also been explored (72). While the role of this miRNA cluster in OA has not been reported, miR-17 and miR-20a showed significantly increased expression in hypertrophic chondrocytes as compared to precursor and differentiated chondrocytes in developing human cartilage (28). Additionally, in humans with mutations in MIR17HG (the gene encoding the miR-17~92 family), display features of Feingold syndrome, including skeletal abnormalities (73) and mice with hemizygous deletion of the miR-17~92 cluster replicate the features seen in humans. Furthermore, this cluster has recently been implicated in the response of fibroblast-like synoviocytes, the resident cells of the synovial membrane, to inflammation in rheumatoid arthritis (74, 75). Here, we found a novel role for the miR-17~92 cluster, specifically miR-17–5p and miR-20a-5p, in the ability to modulate the inflammatory response of tissue engineered cartilage. We show that delivery of miR-20a-5p and miR-17–5p mimics significantly decreases the MMP activity level, while also decreasing Mmp13 and Ccl2 expression levels in IL-1β and TNF-α treated cells (Figure 5D). Furthermore, the addition of miR-29b-3p further decreased MMP activity levels, showing that targeting multiple hubs within the networks can increase the efficacy of miRNA targeted therapies (Figure 5E). Connected genes which may be direct targets and controlling this response are Adamts5, TGF-β signaling pathway members (Bmp2, Acvr1b), and inflammation pathway related genes (Tnfsf11, Il6st, Acvr1b, Tnfrsf21, Pdgfra, Lif, Vegfa).
In comparison to a previously reported OA miRNA-mRNA interactome (32), hub miRNAs from this study were also significantly DE in lesioned compared to preserved OA cartilage. Additionally, genes connected to miR-20a-5p and miR-17–5p, such as Adamts5, Bmp2, Vegfa, and Lif, and genes connected to miR-29b-3p, interestingly, many collagen genes (Col2a1, Col6a2, Col8a1, Col9a1, Col11a1, Col15a1, Col25a1, and Col27a1), were also DE in lesioned compared to preserved OA cartilage. However, this study did not find strong inverse correlations for any of these interactions, which could be due to the different experimental systems.
In summary, this study utilized an integrative approach to determine the miRNA-mRNA interactome controlling the response of tissue-engineered cartilage to inflammatory cytokines. The miRNA hubs identified through this analysis, miR-17–5p and miR-20a-5p, were further validated by showing that delivery of mimics for these miRNAs significantly decreased degradative enzyme production, as well as the expression of inflammation related genes. Furthermore, we found that targeting multiple miRNAs further decreased inflammation in cells, highlighting the importance of comprehensively assessing the entire miRNA interactome. Importantly, several of the hub miRNAs and connected genes identified in this study are consistent with those previously shown to be DE in human OA tissues, providing further support for the use of this iPSC-based system as an in vitro model of human disease. This integrative analysis allowed us to discover novel mediators and mechanisms of inflammation in tissue-engineered cartilage and provides information to guide future studies seeking novel miRNA targeted therapies for arthritic diseases.
Supplementary Material
Acknowledgments
This study was supported in part by the Nancy Taylor Foundation for Chronic Diseases, the Arthritis Foundation, NIH (AG46927, AG15768, AR76820, P30 AR74992, P30 AR073752), the Philip and Sima Needleman Fellowship for Regenerative Medicine from the Washington University Center of Regenerative Medicine, and the NSF Graduate Research Fellowship Program (DGE-1745038). Funding for this project and technical support was also provided by the Bioinformatics Research Core at the Washington University Center of Regenerative Medicine. We sincerely thank Sara Oswald for editorial assistance and Dr. Audrey McAlinden for advice and discussion on this study.
GSE Accession Number
The RNA-sequencing data in this manuscript can be retrieved from GEO (GSE147833, reviewer access code qpufocuctxkztkb).
Nonstandard Abbreviations List
- OA
osteoarthritis
- RNA
ribonucleic acid
- miRNA
miR: microRNA
- mRNA
messenger RNA
- iPSC
induced pluripotent stem cell
- miPSC
murine induced pluripotent stem cell
- PDiPSCs
Pre-differentiated iPSCs
- IL-1
interleukin-1
- TNF-α
tumor necrosis factor-alpha
- sGAG
sulfated glycosaminoglycan
- MMP
matrix metalloproteinase
- RNA-seq
RNA-sequencing
- DE
Differentially expressed
- NF-κB
Nuclear Factor kappa-light-chain-enhancer of activated B cells
References
- 1.Cisternas MG, et al. , Alternative methods for defining osteoarthritis and the impact on estimating prevalence in a US population‐based survey. Arthritis Care Res. (Hoboken). 68, 574–580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldring MB, Goldring SR, Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann. N. Y. Acad. Sci. 1192, 230–237 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier J-P, Fahmi H, Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 7, 33 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Goldring MB, Otero M, Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 23, 471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastiaansen-Jenniskens Y, Saris D, Creemers LB, “Pro-and anti-inflammatory cytokine profiles in osteoarthritis” in Cartilage, (Springer, 2017), pp. 81–97. [Google Scholar]
- 6.Ousema PH, et al. , The inhibition by interleukin 1 of MSC chondrogenesis and the development of biomechanical properties in biomimetic 3D woven PCL scaffolds. Biomaterials 33, 8967–8974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wehling N, et al. , Interleukin-1beta and tumor necrosis factor alpha inhibit chondrogenesis by human mesenchymal stem cells through NF-kappaB-dependent pathways. Arthritis Rheum. 60, 801–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraus VB, et al. , Effects of intraarticular IL1-Ra for acute anterior cruciate ligament knee injury: a randomized controlled pilot trial (NCT00332254). Osteoarthr. Cartil. 20, 271–278 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Furman BD, et al. , Targeting pro-inflammatory cytokines following joint injury: acute intra-articular inhibition of interleukin-1 following knee injury prevents post-traumatic arthritis. Arthritis Res. Ther. 16, R134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fioravanti A, Fabbroni M, Cerase A, Galeazzi M, Treatment of erosive osteoarthritis of the hands by intra-articular infliximab injections: a pilot study. Rheumatol. Int. 29, 961–965 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Grunke M, Schulze-Koops H, Successful treatment of inflammatory knee osteoarthritis with tumour necrosis factor blockade. Ann. Rheum. Dis. 65, 555–556 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunger JM, Zutshi A, Willard VP, Gersbach CA, Guilak F, Genome engineering of stem cells for autonomously regulated, closed-loop delivery of biologic drugs. Stem cell reports 8, 1202–1213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a Glass K, et al. , Tissue-engineered cartilage with inducible and tunable immunomodulatory properties. Biomaterials, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moutos FT, et al. , Anatomically shaped tissue-engineered cartilage with tunable and inducible anticytokine delivery for biological joint resurfacing. Proc. Natl. Acad. Sci. 113, E4513–E4522 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thysen S, Luyten FP, Lories RJU, Targets, models and challenges in osteoarthritis research. Dis. Model. Mech. 8, 17–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joosten LA, TNF alpha and IL-1 beta are separate targets in chronic arthritis. Clin. Exp. Rheumatol. 17, S105–14 (1999). [PubMed] [Google Scholar]
- 17.Matthews GL, Hunter DJ, Emerging drugs for osteoarthritis. Expert Opin. Emerg. Drugs 16, 479–491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldring MB, Berenbaum F, Emerging targets in osteoarthritis therapy. Curr. Opin. Pharmacol. 22, 51–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swingler T, et al. , The function of microRNAs in cartilage and osteoarthritis. Clin. Exp. Rheumatol. 37, 40–47 (2019). [PubMed] [Google Scholar]
- 20.Asahara H, Current status and strategy of microRNA research for cartilage development and osteoarthritis pathogenesis. J. bone Metab. 23, 121–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP, MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Guo H, Ingolia NT, Weissman JS, Bartel DP, Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichhorn SW, et al. , mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 56, 104–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown BD, et al. , Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 25, 1457–1467 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Vicente R, Noël D, Pers Y-M, Apparailly F, Jorgensen C, Deregulation and therapeutic potential of microRNAs in arthritic diseases. Nat. Rev. Rheumatol. 12, 211 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Shen J, Abu-Amer Y, O’Keefe RJ, McAlinden A, Inflammation and epigenetic regulation in osteoarthritis. Connect. Tissue Res. 58, 49–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi T, et al. , Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc. Natl. Acad. Sci. 105, 1949–1954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAlinden A, Varghese N, Wirthlin L, Chang L-W, Differentially expressed microRNAs in chondrocytes from distinct regions of developing human cartilage. PLoS One 8, e75012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A, Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One 3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crowe N, et al. , Detecting new microRNAs in human osteoarthritic chondrocytes identifies miR-3085 as a human, chondrocyte-selective, microRNA. Osteoarthr. Cartil. 24, 534–543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haseeb A, Makki MS, Khan NM, Ahmad I, Haqqi TM, Deep sequencing and analyses of miRNAs, isomiRs and miRNA induced silencing complex (miRISC)-associated miRNome in primary human chondrocytes. Sci. Rep. 7, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coutinho De Almeida R, et al. , RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann. Rheum. Dis. 78, 270–277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diekman BO, et al. , Cartilage tissue engineering using differentiated and purified induced pluripotent stem cells. Proc. Natl. Acad. Sci. U. S. A. 109, 19172–19177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willard VP, et al. , Use of cartilage derived from murine induced pluripotent stem cells for osteoarthritis drug screening. Arthritis Rheumatol. 66, 3062–3072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adkar SS, et al. , Genome engineering for personalized arthritis therapeutics. Trends Mol. Med. 23, 917–931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caron MMJ, et al. , Redifferentiation of dedifferentiated human articular chondrocytes: comparison of 2D and 3D cultures. Osteoarthr. Cartil. 20, 1170–1178 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Cong L, Zhu Y, Tu G, A bioinformatic analysis of microRNAs role in osteoarthritis. Osteoarthr. Cartil. 25, 1362–1371 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Bolger A, Giorgi F, Trimmomatic: a flexible read trimming tool for illumina NGS data. URL http//www.usadellab.org/cms/index.php (2014).
- 39.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 40.Dobin A, et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozomara A, Birgaoanu M, Griffiths-Jones S, miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general-purpose read summarization program. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal V, Bell GW, Nam J-W, Bartel DP, Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou C-H, et al. , miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 44, D239–D247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vlachos IS, et al. , DIANA-TarBase v7. 0: indexing more than half a million experimentally supported miRNA: mRNA interactions. Nucleic Acids Res. 43, D153–D159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen EY, et al. , Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.V Kuleshov M, et al. , Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang W, Cheng J, Allaire JJ, Xie Y, Mcpherson J, shiny: web application framework for R. R Package Version 1.4. 0. 2019.
- 50.Estes BT, Diekman BO, Gimble JM, Guilak F, Isolation of adipose-derived stem cells and their induction to a chondrogenic phenotype. Nat. Protoc. 5, 1294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farndale RW, Buttle DJ, Barrett AJ, Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim. Biophys. Acta (BBA)-General Subj. 883, 173–177 (1986). [DOI] [PubMed] [Google Scholar]
- 52.Wilusz RE, Weinberg JB, Guilak F, McNulty AL, Inhibition of integrative repair of the meniscus following acute exposure to interleukin‐1 in vitro. J. Orthop. Res. 26, 504–512 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fisch KM, et al. , Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthr. Cartil. 26, 1531–1538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinberg J, et al. , Integrative epigenomics, transcriptomics and proteomics of patient chondrocytes reveal genes and pathways involved in osteoarthritis. Sci. Rep. 7, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soul J, et al. , Stratification of knee osteoarthritis: two major patient subgroups identified by genome-wide expression analysis of articular cartilage. Ann. Rheum. Dis. 77, 423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loeser RF, et al. , Disease progression and phasic changes in gene expression in a mouse model of osteoarthritis. PLoS One 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Löfgren M, Svala E, Lindahl A, Skiöldebrand E, Ekman S, Time-dependent changes in gene expression induced in vitro by interleukin-1β in equine articular cartilage. Res. Vet. Sci. 118, 466–476 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Svala E, et al. , An inflammatory equine model demonstrates dynamic changes of immune response and cartilage matrix molecule degradation in vitro. Connect. Tissue Res. 56, 315–325 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Hao S, Baltimore D, The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 10, 281 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamasaki K, et al. , Expression of MicroRNA‐146a in osteoarthritis cartilage. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 60, 1035–1041 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taganov KD, Boldin MP, Chang K-J, Baltimore D, NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. 103, 12481–12486 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soyocak A, et al. , miRNA-146a, miRNA-155 and JNK expression levels in peripheral blood mononuclear cells according to grade of knee osteoarthritis. Gene 627, 207–211 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Kriegsmann M, et al. , Expression of miR-146a, miR-155, and miR-223 in formalin-fixed paraffin-embedded synovial tissues of patients with rheumatoid arthritis and osteoarthritis. Virchows Arch. 469, 93–100 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Dunn W, DuRaine G, Reddi AH, Profiling microRNA expression in bovine articular cartilage and implications for mechanotransduction. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 60, 2333–2339 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Hong E, Reddi AH, Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs-221/−222,−140, and-143/145 expression. Tissue Eng. Part A 19, 1015–1022 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Zhang D, Cao X, Li J, Zhao G, MiR-210 inhibits NF-κB signaling pathway by targeting DR6 in osteoarthritis. Sci. Rep. 5, 12775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyaki S, et al. , MicroRNA‐140 is expressed in differentiated human articular chondrocytes and modulates interleukin‐1 responses. Arthritis Rheum. 60, 2723–2730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakamura Y, Inloes JB, Katagiri T, Kobayashi T, Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol. Cell. Biol. 31, 3019–3028 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McAlinden A, Im G, MicroRNAs in orthopaedic research: Disease associations, potential therapeutic applications, and perspectives. J. Orthop. Res. 36, 33–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ratneswaran A, Rockel JS, Kapoor M, Understanding osteoarthritis pathogenesis: a multiomics system-based approach. Curr. Opin. Rheumatol. 32, 80–91 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Le LTT, et al. , The microRNA-29 family in cartilage homeostasis and osteoarthritis. J. Mol. Med. 94, 583–596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bai X, Hua S, Zhang J, Xu S, The MicroRNA Family Both in Normal Development and in Different Diseases: The miR-17–92 Cluster. Biomed Res. Int. 2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Pontual L, et al. , Germline deletion of the miR-17∼ 92 cluster causes skeletal and growth defects in humans. Nat. Genet. 43, 1026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Philippe L, Alsaleh G, Bahram S, Pfeffer S, Georgel P, The miR-17∼ 92 cluster: a key player in the control of inflammation during rheumatoid arthritis. Front. Immunol. 4, 70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Philippe L, et al. , MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann. Rheum. Dis. 72, 1071–1079 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.