Abstract
Background & Aims:
Chronic atrophic gastritis can lead to gastric metaplasia and increase risk of gastric adenocarcinoma. Metaplasia is a precancerous lesion associated with an increased risk for carcinogenesis, but the mechanism(s) by which inflammation induces metaplasia are poorly understood. We investigated transcriptional programs in mucous neck cells and chief cells as they progress to metaplasia mice with chronic gastritis.
Methods:
We analyzed previously generated single cell RNA-sequencing (scRNA-seq) data of gastric corpus epithelium to define transcriptomes of individual epithelial cells from healthy BALB/c mice (controls) and TxA23 mice, which have chronically inflamed stomachs with metaplasia. Chronic gastritis was induced in B6 mice by Helicobacter pylori infection. Gastric tissues from mice and human patients were analyzed by immunofluorescence to verify findings at the protein level. Pseudotime trajectory analysis of scRNA-seq data was used to predict differentiation of normal gastric epithelium to metaplastic epithelium in chronically inflamed stomachs.
Results:
Analyses of gastric epithelial transcriptomes revealed that gastrokine 3 (Gkn3) mRNA is a specific marker of mouse gastric corpus metaplasia (spasmolytic polypeptide expressing metaplasia, SPEM). Gkn3 mRNA was undetectable in healthy gastric corpus; its expression in chronically inflamed stomachs (from TxA23 mice and mice with Helicobacter pylori infection) identified more metaplastic cells throughout the corpus than previously recognized. Staining of healthy and diseased human gastric tissue samples paralleled these results. Although mucous neck cells and chief cells from healthy stomachs each had distinct transcriptomes, in chronically inflamed stomachs, these cells had distinct transcription patterns that converged upon a pre-metaplastic pattern, which lacked the metaplasia-associated transcripts. Finally, pseudotime trajectory analysis confirmed the convergence of mucous neck cells and chief cells into a pre-metaplastic phenotype that ultimately progressed to metaplasia.
Conclusions:
In analyses of tissues from chronically inflamed stomachs of mice and humans, we expanded the definition of gastric metaplasia to include Gkn3 mRNA and GKN3-positive cells in the corpus, allowing a more accurate assessment of SPEM. Under conditions of chronic inflammation, chief cells and mucous neck cells are plastic and converge into a pre-metaplastic cell type that progresses to metaplasia.
Keywords: gastric cancer, atrophy, SPEM, immune response
INTRODUCTION
Gastric cancer is a significant cause of global morbidity and mortality.1 Chronic inflammation, generally due to Helicobacter pylori (H. pylori) infection, is the most common disease process precipitating gastric carcinogenesis.2 While autoimmune gastritis (AIG) has been traditionally associated with neuroendocrine tumors rather than gastric adenocarcinoma, recent studies demonstrate that AIG patients develop atrophy and metaplasia, and have a significantly increased risk of adenocarcinoma similar to patients with chronic H. pylori infection.3–8 Gastric cancer development, whether during chronic autoimmunity or H. pylori infection, is strongly associated with several pre-neoplastic events thought to be critical for eventual malignant transformation: parietal cell loss (atrophy), mucous neck cell hyperplasia, spasmolytic polypeptide-expressing metaplasia (SPEM), and intestinal metaplasia.9, 10 The cellular and molecular biology of SPEM has been given particular attention, as it is considered a potential cellular origin of intestinal metaplasia, dysplasia, and eventually adenocarcinoma.11–13 There is a significant possibility that the programs induced by inflammation in gastric epithelium to promote metaplastic change is conserved in other organ systems and cancer types.14
SPEM can arise from mature chief cells that undergo cellular reprogramming (recently termed paligenosis) that involves cell structure degradation, progenitor-associated gene activation, and re-entry into the cell cycle.14 This process likely serves as an acute reparative lesion in injury models such as DMP-777, L-635, acid-induced ulcers, and high dose tamoxifen (HDT).14–18 However, it is possible that mucous neck cells also demonstrate plasticity and contribute to SPEM lesions during chronic inflammation, a critical question to resolve since most human SPEM arises in the context of chronic gastritis.19 Mucous neck cell plasticity has not been adequately studied in the context of SPEM, and our previous work using single cell RNA-sequencing (scRNA-seq) demonstrated that SPEM could not be distinguished from hyperplastic/hypertrophic mucous neck cells by the expression of mature murine chief cell transcripts such as Gif.20 This indicates that SPEM cells are not necessarily limited to the base of corpus glands where Gif expression is highest, but are also likely to be present throughout the neck region of the gland normally considered mucous neck cell hyperplasia/hypertrophy. This suggests that epithelial plasticity during tissue damage may not be limited to chief cells, but that multiple epithelial lineages along the gland axis could contribute to SPEM, particularly mucous neck cells.
These data raise two fundamental questions concerning the nature of SPEM: 1) How can SPEM be differentiated from other cells in the gastric corpus during chronic inflammation?; 2) What are the most likely cellular origin(s) of SPEM based on the effect of chronic inflammation on the transcriptomes of mucous neck and chief cells? Here, we analyzed the transcriptional programs of gastric epithelial cells from healthy and chronically inflamed gastric corpus to address these critical gaps in knowledge.
We identified mucous neck and chief cells from the healthy gastric corpus as well as mucous neck cells, chief cells, and SPEM cells from chronically inflamed gastric corpus. Transcriptional and immunofluorescent analyses determined that gastrokine-3 (Gkn3) transcripts, absent from the healthy corpus, were expressed specifically by SPEM cells in the gastric corpus resulting from both autoimmune gastritis and H. pylori infection. Immunofluorescent staining of human gastric tissue samples paralleled our murine results and determined that GKN3 is expressed in the glandular cells of healthy human antrum, is not detected in the healthy corpus, but expression is induced in the corpus during chronic atrophic gastritis. Additionally, during inflammation, both mucous neck cells and chief cells transition to a mucinous phenotype that lacks SPEM-associated or intestinal transcripts, indicating that these cells are not yet SPEM but may constitute a “pre-SPEM” cell type. Finally, pseudotime trajectory analysis was used to calculate, in an unsupervised manner, the lineage relationships between healthy and inflamed mucous neck and chief cells and bona fide SPEM. This machine-learning algorithm predicted that both mucous neck and chief cells contribute to SPEM that develops in a setting of chronic inflammation. These studies identified the mucous neck and chief cell-specific responses to inflammation and determined that both cell types are likely contributors to SPEM, marked specifically by Gkn3 transcripts. This broadens the understanding of the origins of inflammation induced SPEM and reveals additional epithelial plasticity within the metaplastic corpus glands.
RESULTS
Identifying mucous neck cells, chief cells, and SPEM cells in single cell transcriptional data sets
In order to measure the impact of chronic inflammation on the transcriptional programs of mucous neck cells and chief cells, we analyzed single cell datasets of epithelial cells from healthy and chronically inflamed gastric corpora in mice.20 Briefly, these libraries contain transcriptional data from more than 18,000 individual epithelial cells from the corpus region of healthy BALB/c and stomachs which had chronic inflammation for three months from TxA23 mice, a mouse model of autoimmune gastritis demonstrated to develop parietal cell atrophy, SPEM, and gastric intraepithelial neoplasia.21–24 Mucous neck cells and chief cells were identified using uniform manifold approximation and projection (UMAP), a recently developed dimensionality reduction algorithm that provides higher reproducibility and more meaningful cluster organization than t-distributed stochastic neighbor embedding (tSNE) for single cell transcriptional datasets.25 UMAP clustering of epithelial cell libraries from healthy and chronically inflamed stomachs identified 30 clusters of cells, successfully separating mucous neck cells, foveolar cells, chief cells, parietal cells, ECL cells, and neuroendocrine cells (G cells, D cells, etc.) (Figure 1A, Supplemental Figure 1). Clusters containing mucous neck cells and chief cells were identified based on expression of the murine lineage-defining genes Mucin 6 (Muc6) for mucous neck cells and gastric intrinsic factor (Gif) for chief cells. Muc6 was detected at much higher levels in the mucous neck cell cluster and nearly absent from healthy chief cells while Gif was strongly associated with the chief cell cluster and only sporadically detected at much lower levels in the mucous neck cell cluster (Figure 1B). The cluster containing SPEM cells was identified by having the highest expression level for Tff2, also known as trefoil factor 2/spasmolytic polypeptide, from which the term SPEM was coined,26 as well as other known SPEM transcripts such as: Cd44, Cftr, and Wfdc2.20 Analysis of cluster-defining transcripts revealed that that the SPEM cluster was also most strongly associated with Gkn3 transcript expression which was absent or expressed at very low levels in any other cluster of cells (Figure 1C). While Muc6 and Gif transcripts are both strongly expressed in the SPEM cluster, these transcripts are also highly expressed in other clusters that lacked SPEM-associated transcripts, making them poor criteria for the definition of SPEM in single cell data sets (Supplemental Figure 1C). A small subset of foveolar cells in cluster 11 demonstrated a low level of Gkn3 transcript expression, but only the SPEM cell cluster was transcriptionally defined by the expression of Gkn3. These data demonstrate the separation of chief cells, mucous neck cells, and SPEM cells using single cell transcriptional data.
Figure 1. Identification of mucous neck cells, chief cells, and SPEM cells in single cell transcriptional data sets.
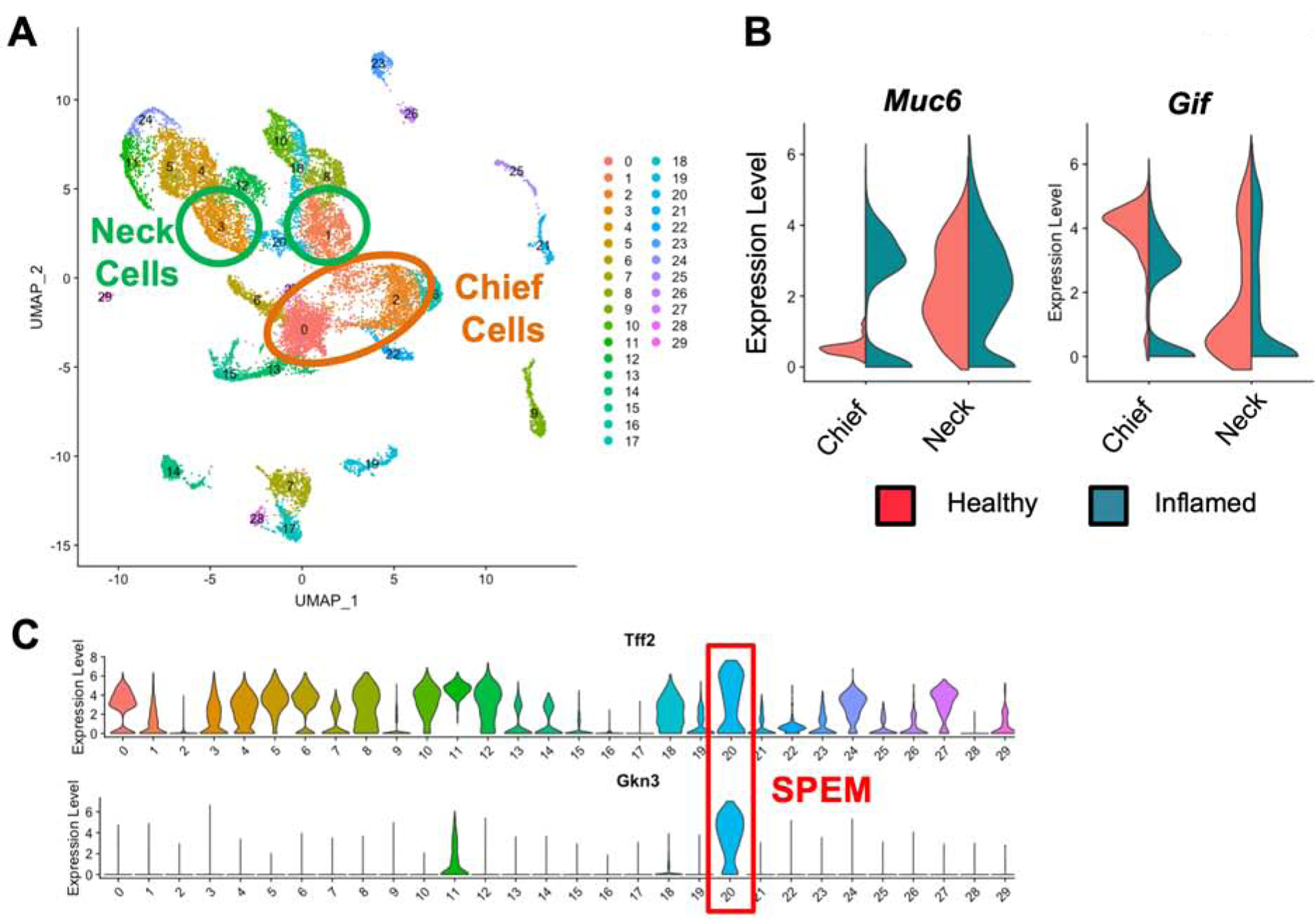
(A) Uniform manifold approximation and projection (UMAP) based unbiased clustering of gastric epithelial cell suspensions isolated from healthy BALB/c and chronically inflamed TxA23 mice. Cells are colored by cluster identity. Clusters corresponding to mucous neck cells are circled in green and clusters corresponding to chief cells are circled in orange. (B) Violin plots of mucous neck cell and chief lineage specific gene expression within the mucous neck and chief cell clusters identified in Figure 1A. Violin plots are divided in half by the library the cell was identified in, left side showing gene expression in healthy BALB/c cells (red), right side showing gene expression in chronically inflamed cells (green). (C) Violin plots of Tff2 and Gkn3 gene expression levels in all identified clusters of the combined healthy and inflamed scRNA-seq libraries. The cluster identified as SPEM is outlined in red.
GKN3 is expressed specifically by SPEM in the murine gastric corpus
Single cell transcriptional analyses determined that the Gkn3 transcript identified the SPEM cell cluster more specifically than any other transcript. Gastrokine-3 (Gkn3) is normally expressed in the murine gastric antrum but little is known about the function(s) of this protein. It has been previously associated with gastric corpus atrophy and metaplasia,27, 28 but the precise cellular source(s) for Gkn3 has not been described. SPEM is thought to coincide with “antralization” of the gastric corpus, and it has been demonstrated that SPEM cells can aberrantly express antral genes such as Pdx1.29 We hypothesized that aberrant expression of the antral gene Gkn3 was specific to SPEM in the gastric corpus. To test this, we used a multicolor RNA in situ hybridization technique, RNAscope (Figure 2A). Healthy antrum, healthy corpus, and chronically inflamed corpus from TxA23 mice were stained with probes for Gkn3 (cyan), Tff2 (yellow), and Muc6 (red) transcripts. As expected, Gkn3 was detected in the antrum but not detected in the healthy gastric corpus. In chronically inflamed atrophic mucosa, Gkn3 emerged in areas of Muc6+Tff2+ SPEM at the base of gastric glands. GKN3 protein expression in SPEM was measured by immunofluorescent staining with antibodies to Gif (red), GSII (green), anti-TFF2 (magenta), and anti-GKN3 (yellow) in gastric corpus tissue from healthy BALB/c mice (top), TxA23 mice with chronic autoimmune gastritis (middle), and B6 mice chronically infected with H. pylori (bottom). Healthy mice have normal staining patterns of GIF in chief cells, GSII and TFF2 in mucous neck cells, with no GKN3 detected. Mice with GIF+GSII+Tff2+ SPEM in areas of corpus atrophy, caused either by autoimmune gastritis or chronic H. pylori infection, expressed GKN3 (Figure 2B). These studies confirm the single cell RNA sequencing data and show that Gkn3/GKN3 is specific for SPEM cells in the gastric corpus of mice with chronic atrophic gastritis caused by either autoimmunity or chronic infection with H. pylori. Critically, this transcript is expressed at high levels and enables the identification cells running the SPEM transcriptional program and differentiates SPEM from non-SPEM.
Figure 2. GKN3 is expressed by SPEM cells in the murine gastric corpus.
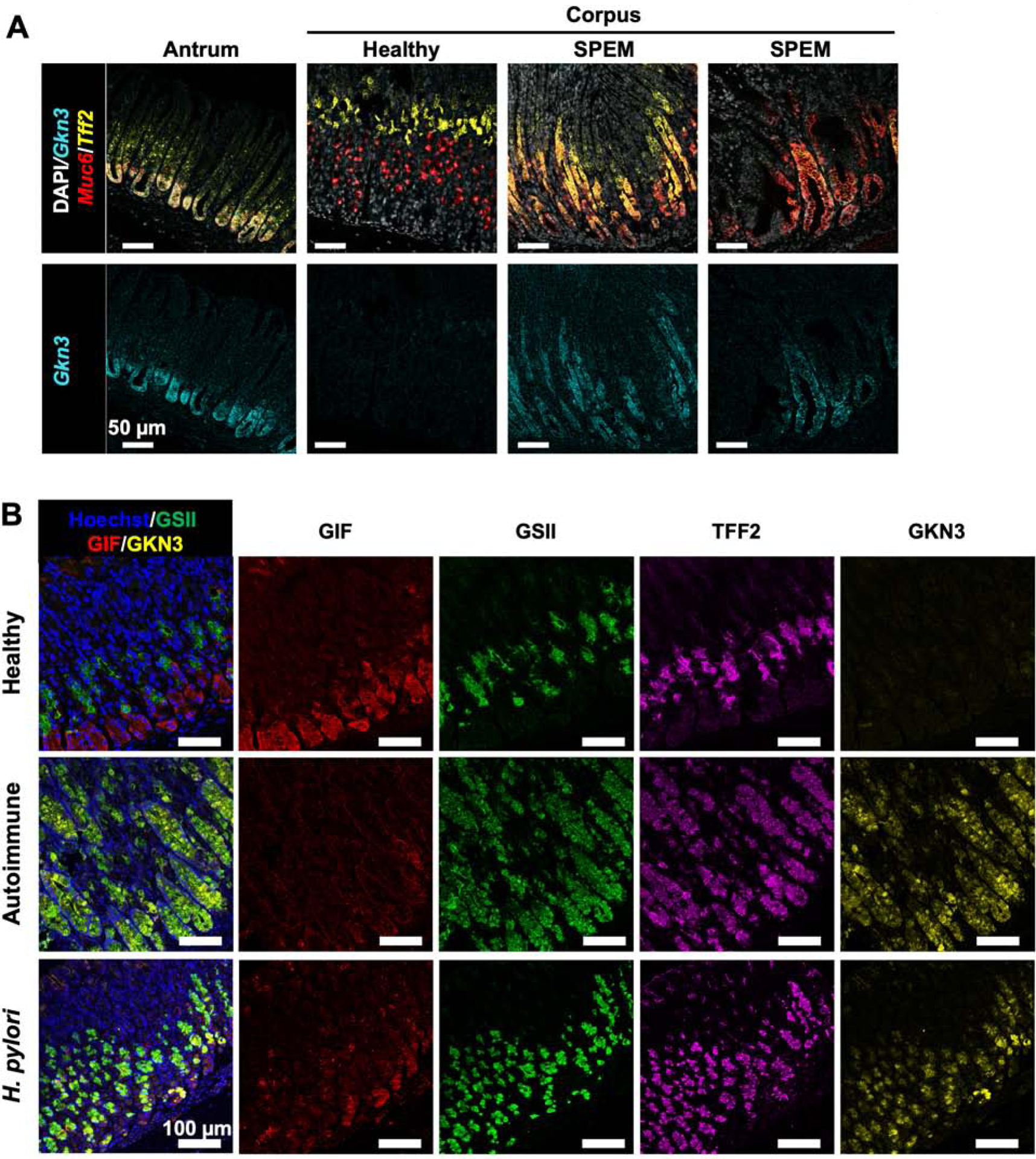
(A) Representative RNAscope images of healthy antrum, healthy corpus, and atrophic corpus from TxA23 mice with chronic gastritis. Nuclei identified with DAPI (white). The identified transcripts were Gkn3 (cyan), Tff2 (yellow), and Muc6 (red). Fluorescent images are representative of 3–5 mice per group from 1–2 separate experiments. (B) Representative fluorescent images gastric corpus from healthy mice, mice with chronic autoimmune gastritis (“Autoimmune”) and mice with chronic Helicobacter pylori infection (“H. pylori”). Tissue is stained with anti-GIF (red), GSII (green), anti-TFF2 (magenta), and anti-GKN3 (yellow).
GKN3 is expressed in normal human antrum and induced in the human corpus during gastritis
The determination that GKN3 is a specific marker of SPEM cells in the murine corpus is a critical finding, but human expression of GKN3 has not been determined. Based on predictions from sequencing analyses of the GKN3 locus it has been hypothesized that GKN3 may be a pseudogene in humans; however, there are a limited number of non-tolerated mutations in the locus and there is a possibility of alternatively spliced transcripts, both of which are inconsistent with pseudogenicity.27, 28 Additionally, protein expression has not been rigorously assessed. We used immunofluorescent staining with Hoechst (blue), GSII (green), and anti-GKN3 rabbit sera (red) to analyze GKN3 expression in healthy antrum, healthy corpus, and biopsies of chronic atrophic gastritis. In these analyses we stained 3 healthy human antra, 5 healthy human corpora, and 16 biopsies of corpus gastritis from different patients. Staining of healthy antrum and corpus determined that GKN3 expression was detected in antral glands but largely absent from healthy corpus, recapitulating the staining pattern of this protein in mice (Figure 3A). Critically, staining of biopsies from the corpus of patients with chronic atrophic gastritis revealed that GKN3 expression was detected in the 4 patients with metaplasia development. Furthermore, this staining is primarily increased at gland bases in inflamed corpus tissue, a pattern identical to that of metaplasia development. Menheniott et al. predicted that perhaps only the African population might be expected to produce GKN3 based on the retention of an ancestral variant of the GKN3 in this group.27 However, the group of 4 positive patients we identified contained 3 Caucasian patients and 1 patient of African descent, suggesting GKN3 expression outside of the African population. These results demonstrate that the same anti-sera that identifies GKN3 in murine gastric tissue also stains human gastric tissue in a conserved pattern. This suggests that GKN3 may be expressed in healthy human antrum but induced in the corpus when chronic atrophic gastritis develops, similar to observations in mice.
Figure 3. GKN3 is expressed in normal human antrum and in the inflamed human corpus.
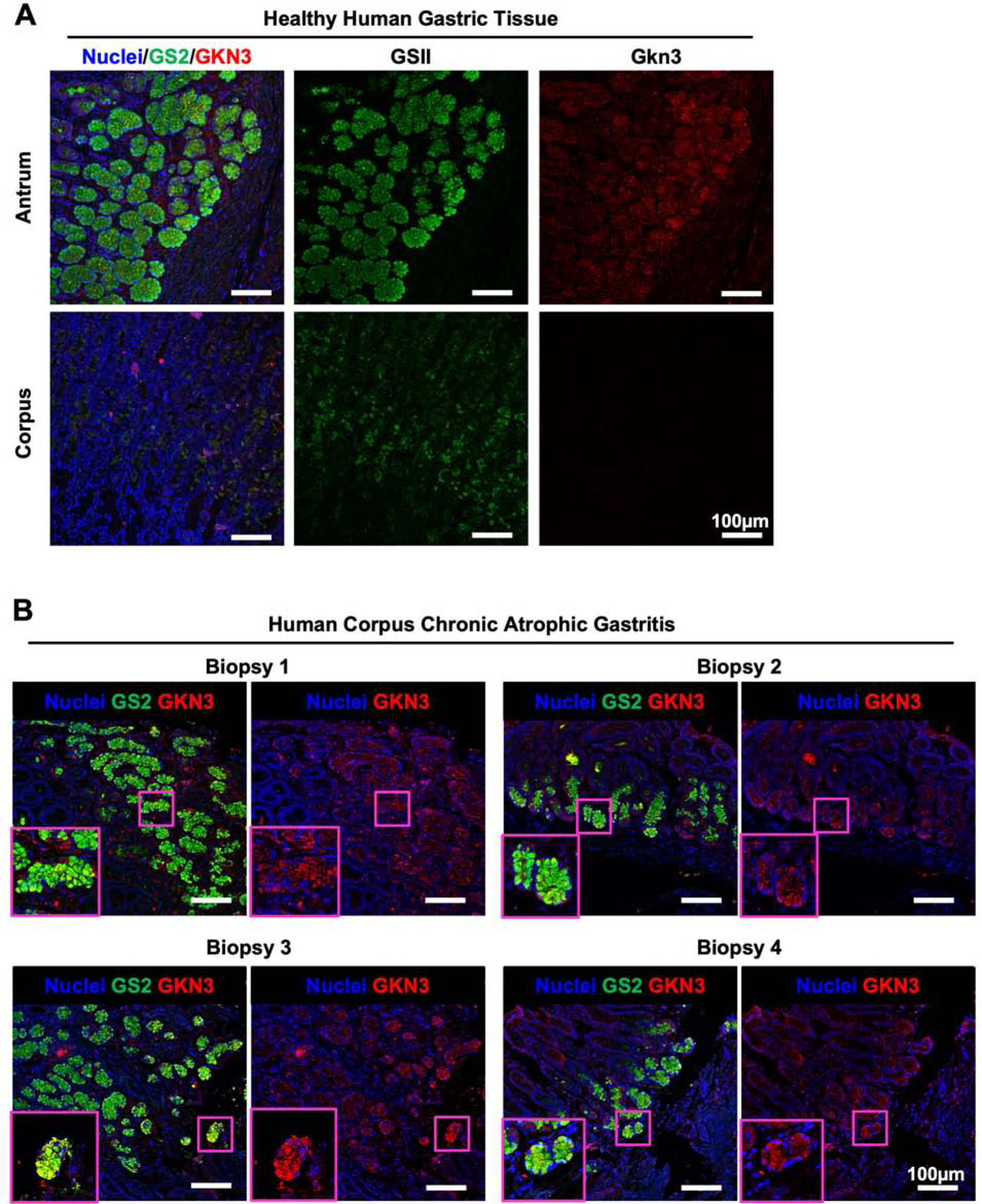
(A) Representative fluorescent images of normal human antrum and corpus. Tissue is stained with GSII (green) and anti-GKN3 (red). (B) Representative fluorescent images of corpus biopsies from human patients diagnosed with corpus gastritis. Tissue is stained with GSII (green) and anti-GKN3 (red). Magenta boxes indicate high magnification inserts demonstrating GSII+GKN3+ cells in human gastritis biopsies.
Mucous neck cells and chief cells enact a mucinous transcriptional program in response to inflammation
To determine how mucous neck and chief cells respond to chronic inflammation, we separated the transcriptomes of mucous neck and chief cells (previously identified in Figure 1) for independent analyses. The unsupervised UMAP algorithm grouped mucous neck and chief cells separately from one another, and within the mucous neck cell and chief cell clusters were cells from both healthy and inflamed stomachs. We hypothesized that comparing the transcriptomes of healthy and inflamed epithelial cells in these clusters would identify the early epithelial transcriptional responses to inflammation. We compared the transcriptomes of mucous neck and chief cells from chronically inflamed corpus to the normal mucous neck and cell transcriptomes that were placed in the same clusters. The average per-cell expression level of all detected transcripts of 2109 mucous neck cells from inflamed corpus was compared to 744 mucous neck cells from healthy stomachs in the same cluster, revealing that mucous neck cells upregulated several inflammatory response genes (e.g. H2-Eb1 (MHC class II), Cd74, H2-K1 (MHC class I), Pigr, B2m, S100a11, etc. Supplemental Table 1). Additionally, mucus production (Muc6) was also significantly increased in mucous neck cells from inflamed stomachs. Of note, there was no significant increase in the expression of SPEM associated transcripts (e.g. Gkn3, Cftr, Cd44, Wfdc2) in this subset of inflamed mucous neck cells, although there was significant upregulation of the long non-coding RNA Xist, which has been previously associated with SPEM during H. pylori infection (Figure 4A–B, Supplemental Table 1).30 The early chief cell response to inflammation was similarly assessed by comparing the per-cell average expression level of all detected transcripts between 1900 chief cells from inflamed corpus and 1357 chief cells from normal corpus. In contrast to mucous neck cells, chief cells exposed to inflammation demonstrated significant transcriptional reprogramming by adopting a transcriptional signature very similar to that of mucous neck cells during inflammation. This involved a significant upregulation of both inflammatory genes (e.g. H2-Eb1, Cd74, H2-K1, Pigr, B2m, S100a11, etc., Supplemental Table 2) and genes associated with mucus production such as Muc6, Agr2, and Tff2. While there was a small decrease in the proportion of cells expressing some of these inflammatory genes such as B2m, average expression levels of these genes was significantly higher in the cell population from the inflamed condition. Notably, there was no induction of SPEM-associated transcripts (e.g. Gkn3, Cftr, Cd44, Wfdc2) in these cells (Figure 4C–D, Supplemental Table 2). These results indicate that, during chronic inflammation, both mucous neck cells and chief cells enact transcriptional programs indicative of an inflammatory response but distinct from SPEM.
Figure 4. Mucous neck cells and chief cells adopt a mucinous transcriptional program in response to inflammation.
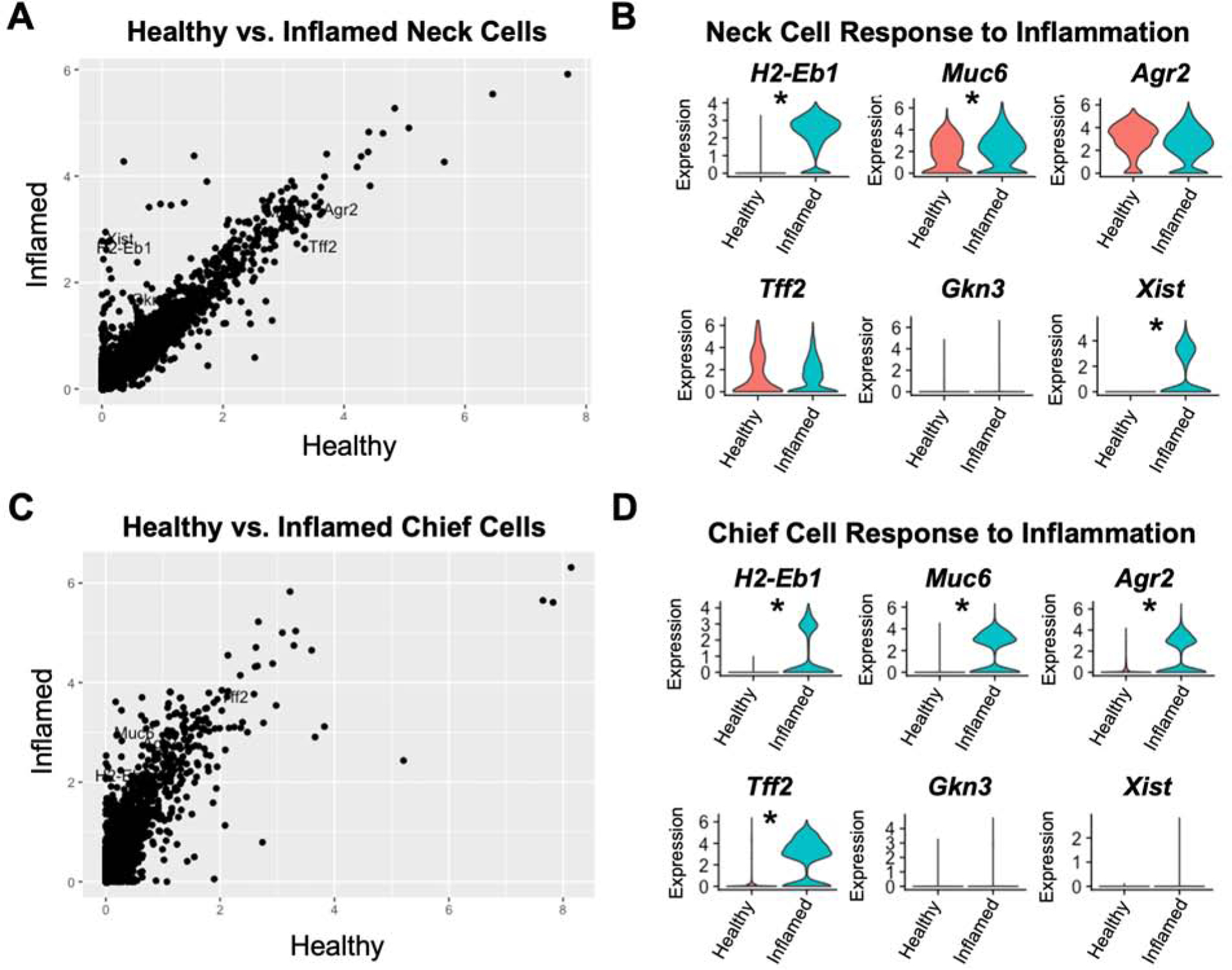
(A) Dot plot of the relative expression of all detected transcripts calculated from the per-cell average relative expression from scRNA-seq data sets of Mucous neck cells from healthy stomachs vs mucous neck cells from inflamed stomachs. (B) Violin plots of select gene expression levels in healthy (red) and inflamed (green) mucous neck cells. (C) Dot plot of the relative expression of all detected transcripts calculated from the per-cell average relative expression from scRNA-seq data sets of chief cells from healthy stomachs vs chief cells from inflamed stomachs. (D) Violin plots of select gene expression levels in healthy (red) and inflamed (green) chief cells. * indicates p value <0.001 by Wilcoxon rank sum test with Bonferroni post-test correction.
Mucous neck and chief cells adopt a “pre-SPEM” phenotype type during chronic inflammation
Having established that mucous neck cells and chief cells undergo distinct transcriptional changes in response to inflammation, we compared the transcriptional profiles of mucous neck cells and chief cells from inflamed corpus to SPEM cells. To do this we generated a data set that included the previously analyzed mucous neck and chief cells from the chronically inflamed corpus as well as 271 SPEM cells identified by having the highest expression level of SPEM-associated transcripts including: Tff2, Gkn3, Xist, Wfdc2, and Cftr (Supplemental Table 3 and 4). No SPEM cells were co-classified as either mucous neck cells or chief cells. The average per-cell expression level of all detected transcripts of mucous neck cells isolated from inflamed stomach was compared to SPEM cells to determine the significant transcriptional differences. This analysis revealed that, compared to mucous neck cells, SPEM cells from the chronically inflamed corpus expressed significantly higher levels of Muc6 and Tff2, but also upregulated SPEM-associated transcripts such as Gkn3, Cftr, and Wfdc2 (Figure 5B). Surprisingly, while Xist is associated with SPEM, Xist expression in SPEM cells was similar to mucous neck cells from chronically inflamed stomachs. (Figure 5A–B, Supplemental Table 3). This indicates that Xist is not necessarily a SPEM-associated transcript as previously thought but could potentially represent a transcript activated by inflammation in mucous neck cells that is retained in SPEM cells. Next, the gene expression profiles of chief cells from inflamed corpus were compared to SPEM. Both chief cells exposed to inflammation and SPEM cells expressed Muc6 and Tff2, but SPEM cells had significantly higher expression of metaplastic transcripts (e.g. Cftr, Wfdc2 and Gkn3, Supplemental Table 4). Unlike mucous neck cells, however, chief cells did not express Xist (Figure 5C–D, Supplemental Table 4). This identifies Xist as a transcript potentially specific to the inflammatory transcriptional program of mucous neck cells. Similar to mucous neck cells, this comparison determined that the inflammatory transcriptional program in chief cells, while distinct from healthy cells, did not yet express SPEM-specific transcripts. Combined with previous results showing transcriptional differences induced by inflammation in mucous neck and chief cells, these data indicate that both mucous neck cells and chief cells adopt a “pre-SPEM” phenotype in the inflamed gastric corpus defined by transcripts that differ significantly from both healthy mucous neck and chief cells and SPEM.
Figure 5. Mucous neck cells and chief cells transition to a “pre-SPEM” cell type in response to inflammation.
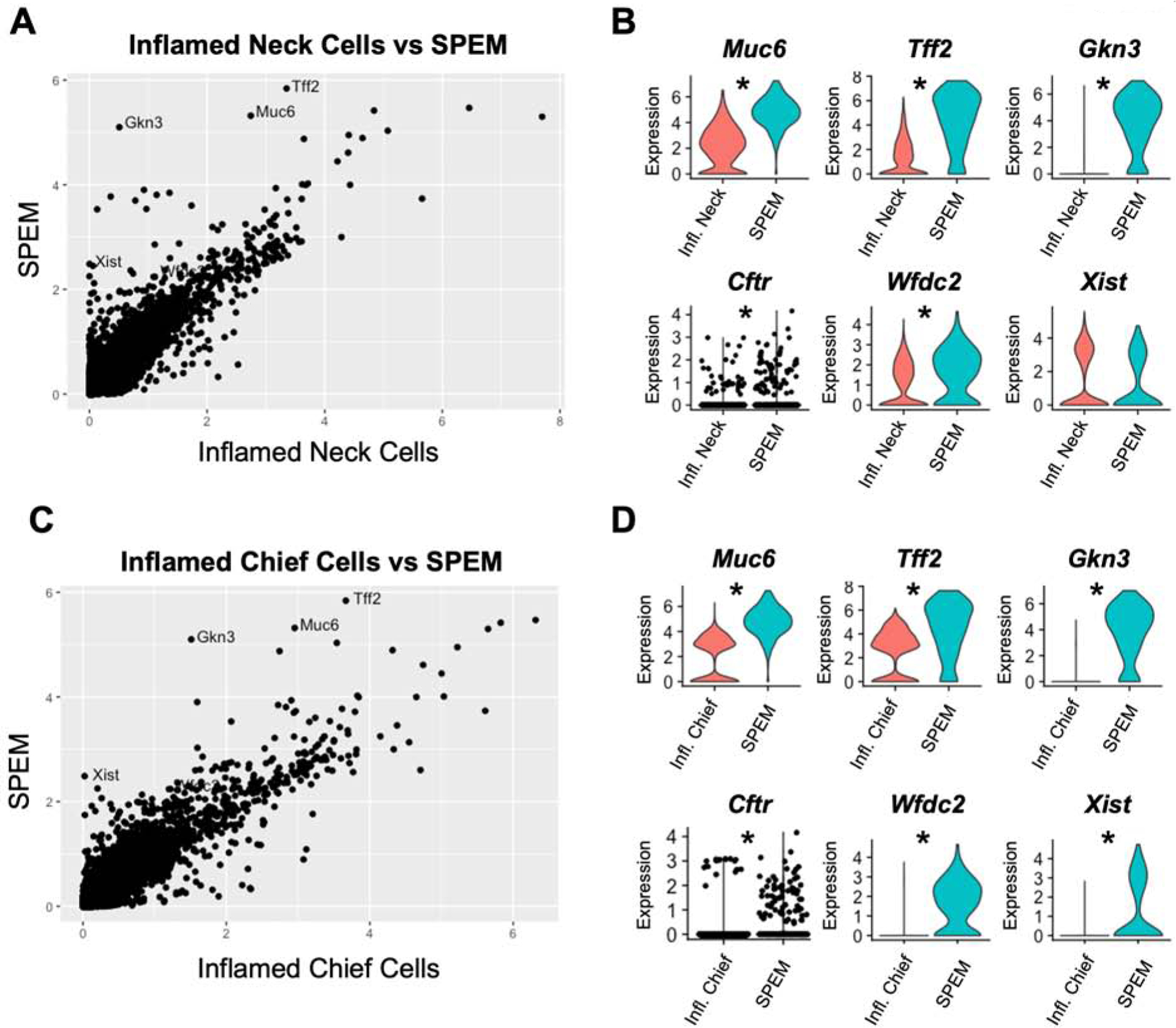
(A) Dot plot of the relative expression of all detected transcripts calculated from the per-cell average relative expression from scRNA-seq data sets of mucous neck cells from inflamed stomachs vs SPEM cells. (B) Violin plots of select gene expression levels in inflamed mucous neck cells (red) and SPEM cells (green). Violin plot for Cftr has individual cell points visualized as black dots overlaid on the violin distribution. (C) Dot plot of the relative expression of all detected transcripts calculated from the per-cell average relative expression from scRNA-seq data sets of chief cells from inflamed stomachs vs SPEM cells. (D) Violin plots of select gene expression levels in inflamed chief cells (red) and SPEM cells (green). Violin plot for Cftr has individual cell points visualized as black dots overlaid on the violin distribution. * indicates p value <0.001 by Wilcoxon rank sum test with Bonferroni post-test correction.
Pseudotime analysis traces the origin of inflammation induced SPEM to both mucous neck cells and chief cells
Based on the transcriptional data indicating that mucous neck cells and chief cells respond to inflammation by activating a metaplastic transcriptional program, we hypothesized that both mucous neck cells and chief cells contribute to inflammation induced SPEM. To test this, we used a machine learning approach, pseudotime trajectory analysis, in combination with the newly identified murine SPEM-specific transcript, Gkn3. Pseudotime analysis makes use of the fact that cells do not differentiate synchronously, meaning that cells collected at the same timepoint are likely to be at various stages of differentiation, in this case, developing into SPEM. It employs machine learning to calculate the likely differentiation trajectory of a group of cells and calculates each cell’s distance from the start of the trajectory using a unit called pseudotime.31 This computational approach was recently used to establish the lineage relationships of gastric corpus isthmal stem cells32 but has not yet been used to infer the lineage relationship between mucous neck cells, chief cells, and SPEM. We subjected mucous neck cells and chief cells from healthy and inflamed stomachs and SPEM to pseudotime trajectory analysis to elucidate the likely differentiation of SPEM during chronic inflammation. Critically, this unsupervised algorithm identified both mucous neck cells and chief cells from healthy stomach as cellular origins of Muc6+Tff2+Gkn3+Gif+/− SPEM (Figure 6A–B). Additionally, both mucous neck and chief cells are related to SPEM via a shared “pre-SPEM” trajectory of differentiation comprised of mucous neck and chief cells from inflamed gastric corpus. Finally, the algorithm also inferred that the trajectory terminated in two different SPEM subsets. However, branch pathway analysis of the terminal SPEM branches determined that the divergence of the trajectory was driven by differences in inflammatory response genes (e.g. H2-Eb1, Cd74, H2-K1, Pigr, B2m, S100a11, etc.) rather than metaplasia associated transcripts (Figure 6A, C). These results are consistent with a model in which both chief cells and mucous neck cells contribute to SPEM during chronic inflammation.
Figure 6. Pseudotime trajectory analysis traces the origins of inflammation-induced SPEM to both mucous neck cells and chief cells.
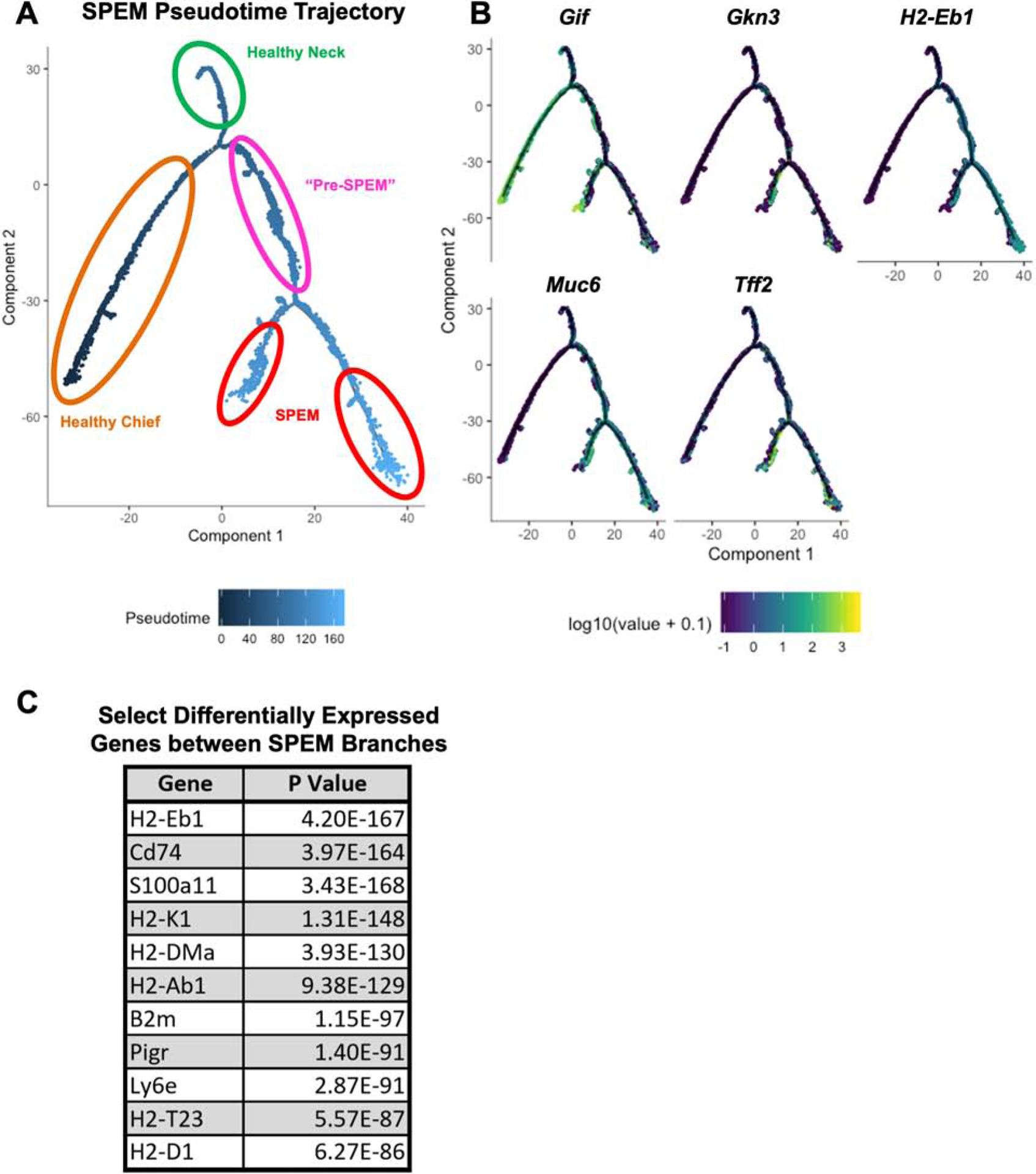
(A) Calculated pseudotime trajectory based on differentially expressed genes in healthy and inflamed mucous neck cells, healthy and inflamed chief cells, and SPEM cells. Cells are colored according to computed pseudotime coordinate, with darker cells denoting earlier time points in the trajectory and lighter cells denoting later time points. Branches of the timeline containing healthy chief cells (orange), healthy mucous neck cells (green), “Pre-SPEM” comprised of inflamed mucous neck and chief cells (pink), and SPEM cells (red) are circled. (B) Pseudotime trajectory as shown in Figure 4A, but cells are now colored according to gene expression for Gif, Gkn3, Muc6, Tff2, and H2-Eb1. Darker cells indicate lower expression, lighter cells indicate higher expression. (C) Select significant differentially expressed genes between the terminal SPEM branches shown in 4A.
METHODS
Murine and Human Tissue
Healthy BALB/c background mice were originally purchased from the Jackson Laboratory and bred in our animal facility. TxA23 mice express a transgenic T cell receptor specific for a peptide from H+/K+ ATPase alpha chain on a BALB/c background and have been previously described.23, 24, 33, 34 All mice were maintained in our animal facility and cared for in accordance with institutional guidelines. Studies were performed on a mixed group of male and female mice with co-housed littermate controls. Formalin fixed paraffin embedded tissue from Helicobacter pylori infected B6 mice were as previously described.35 All human gastric specimens were obtained with approval from the institutional review boards of the Washington University and Saint Louis University Schools of Medicine (St. Louis, MO).
Gastric Epithelial Single Cell Libraries
Analyses were performed on 10x Genomics single cell gene expression libraries from healthy BALB/c and inflamed TxA23 mice. Cell isolation and library preparation for these samples was described previously.20 Briefly, the libraries are comprised of over 18,000 corpus gastric epithelial cells from 5 unmanipulated 8–12 week old BALB/c mice and 5 chronically inflamed 12 week old TxA23 mice per group, sequenced in two separate reactions for each cohort.
Data Processing & Statistical Analysis
Raw data were processed through the CellRanger 3.0 pipeline (10x Genomics) and secondary clustering and differential expression analysis were conducted in Seurat/R.36 Pseudotime trajectory inference was performed in Monocle.31 Detailed discussion of the analysis pipeline can be found in the supplementary methods.
Immunofluorescence
Stomachs were prepared, stained, and imaged using methods modified from Ramsey et. al.37 Briefly, paraffin-embedded specimens were cut into 5 μm sections, deparaffinized and rehydrated. After antigen retrieval with 10 mM sodium citrate (pH 6.0), sections were washed with phosphate-buffered saline (PBS) and blocked in 1% bovine serum albumin and 0.3% Triton X-100 in PBS followed by overnight incubation with primary antibodies. The primary antibodies used for immunostaining were previously described rabbit anti-GKN3 serum (1:400),28 rabbit anti-TFF2 (1:100, ab49536, Abcam), and goat anti-GIF (1:10,000, gift from David Alpers, Washington University in St. Louis). After washing, sections were incubated with secondary antibodies and GS-II lectin (1:500, L21415, ThermoFisher). Sections were washed, stained with Hoechst (62249, ThermoFisher) 1:20,000 in PBS and mounted in ProLong Gold Antifade mountant (P36934, ThermoFisher).
Multicolor RNA- In Situ Hybridization (RNAscope)
Tissue fixation and RNAscope assay (Advanced Cell Diagnostics, Hayward, CA) was performed according to manufacturer recommendations. Briefly, gastric tissue was harvested from BALB/c and TxA23 mice and fixed for 24 hours in 4% paraformaldehyde at 4 degrees Celsius. Stomach tissue was then moved to 10% sucrose in PBS until tissue sank to the bottom, approximately 18 hours. Tissue was then moved to a 30% sucrose solution in PBS until sinking to the bottom of the tube, about 18 hours. Sections were pretreated using protease and incubated with probes targeting Tff2 (Cy3.5), Gkn3 (FITC), and Muc6 (Cy5.5). Slides were washed thoroughly using wash buffer (Advanced Cell Diagnostics) after each hybridization step at room temperature. Staining was performed on antrum and corpus tissue from 3 mice per group.
DISCUSSION
Gastric adenocarcinoma almost always develops in the context of SPEM, which is strongly associated with the loss of acid-secreting parietal cells during chronic inflammation.12, 13, 38 Therefore, understanding the cellular responses to inflammation that give rise to SPEM is critical for understanding the pathogenesis of adenocarcinoma. Chief cells respond to acute tissue damage by losing zymogenic functions and reprogramming into mucinous TFF2-expressing SPEM cells.15–17, 39 Chief cell reprogramming into SPEM reflects a multi-organ general cell response termed “paligenosis” and has also been observed in pancreatic acinar-to-ductal metaplasia.14 While these studies have been critical in understanding the cellular mechanisms by which chief cells alter their cellular phenotype in the setting of injury, it is unclear whether chief cells are the only population capable of plasticity during chronic inflammatory responses that last months or years compared to acute drug induced models that act over days or weeks.19 We previously found that SPEM cannot be separated from mucous neck cell hyperplasia by the expression of mature chief cell transcripts such as Gif and that the metaplastic program operating in acute SPEM cells is highly conserved in chronic inflammatory SPEM.20 The goal of these studies was to determine the early lineage-specific transcriptional responses to inflammation and their relationship to the SPEM transcriptional program.
We identified healthy and inflamed mucous neck and chief cells using unsupervised clustering of over 18,000 gastric corpus epithelial cells from healthy and chronically inflamed stomachs from our previously generated scRNA-seq data set.20 Importantly, unsupervised clustering placed both healthy and inflamed cells belonging to the same lineage together within clusters. This indicated that, despite being isolated from a chronically inflamed environment, the overall transcriptional programs of mucous neck and chief cells from inflamed stomachs were still similar enough for an unbiased algorithm to place them in the same cluster as mucous neck and chief cells from healthy stomachs. This clustering result determined that these cells are mature mucous neck and chief cells undergoing the early responses to inflammation rather than a separate lineage of metaplastic cells. Additionally, these are unlikely to be cells transitioning normally out of the low neck into the high base as these would have a hybrid phenotype between mucous neck and chief rather than increased mucus production with no induction of chief cell transcripts.
Importantly, we identified Gkn3 as a SPEM-specific transcript in the murine gastric corpus. Gkn3 encodes for a secreted protein normally expressed with Tff2 in the gastric antrum was reported to be upregulated in response of atrophy and metaplasia.27 Our dataset of 18,000 epithelial cells from healthy and inflamed corpus region of stomachs detected Gkn3 only in the SPEM cluster. To verify that this transcript could be used to faithfully identify SPEM, we stained for the presence of Gkn3 transcript and GKN3 protein in healthy and metaplastic gastric corpus. These analyses detected Gkn3/GKN3 only in the TFF2+GSII+GIF+/− SPEM glands in chronically inflamed corpus. While previous immunofluorescent studies identified GKN3 protein in the healthy gastric corpus,27, 28 this could be accounted for by differences in the polyclonal antibodies used from the two different studies. Of note, earlier studies showed relatively rare cells at the transition of the neck and base regions of the gastric unit, so there may be low-level GKN3 expression in normal corpus in cells that are poised for pre-metaplastic conversion. GKN3-specific reagents are commercially restricted, and the use of immunized animal sera could have batch-to-batch variation. Regardless, the mRNA-based data shown here indicate that Gkn3 transcripts are specific for SPEM in scRNA-seq data sets in a way that would be very useful for identifying SPEM lesions in future studies. This is particularly important because chief cells can lose Gif expression in the transition to SPEM, meaning that identifying SPEM as Muc6+Tff2+Gif+ cells only identifies a subset of SPEM that has not yet lost Gif expression. Tff2 is a normal feature of healthy gastric corpus, but SPEM can be identified pathologically when TFF2+ cells are found at the base of the gland which is normally reserved for GIF+TFF2− chief cells. In scRNA-seq data sets, the in situ location is unknown. Therefore, the identification of a SPEM-specific transcript expressed at high levels, like Gkn3, is critical for the successful separation of SPEM cells from hyperplastic/hypertrophic mucous neck cells for analysis.
Critically, combined with our previous work demonstrating that Gif expression does not uniquely identify SPEM cells,20 these results indicate that Gkn3+ SPEM cells are found both in the neck region of the gland and at the base, necessitating an expansion of which cells are considered metaplastic during chronic inflammation. The fact that immunofluorescent staining of human gastric tissue with the same polyclonal anti-sera mirrored these results suggest that GKN3 may not be a pseudogene in humans.27, 28 As discussed in Geahlen et al., several aspects of the GKN3 locus do not fit with pseudogenicity. For example: although predictive sequence analysis of potential genetic loci for GKN3 suggest a large portion of the population have stop codon in the predicted 3rd codon of GKN3, the predicted stop codon was not found in all human sequences. Furthermore, analyses of the human GKN3 sequence revealed a lack of non-tolerated mutations across the human population, which is inconsistent with the notion that GKN3 has evolved into a pseudogene.28 Interesting, GKN3 has been identified in proteomic databases of human tissue (https://www.proteomicsdb.org/proteomicsdb/#protein/proteinDetails/52447/projects).40, 41 Further molecular studies are necessary in order to definitively establish whether GKN3 is expressed in humans, including the identification of alternatively spliced transcripts that could be responsible for the staining pattern we observed in human tissue. Thus, there are several interesting features of this gene locus to resolve with future work in mice and humans; with respect to both its role in normal physiology of the antrum and in the corpus epithelial response to inflammation.
Transcriptional comparisons of mucous neck cells from healthy and inflamed stomachs determined that, in addition to significant upregulation of inflammatory response genes (e.g. H2-Eb1 (MHC class II), Cd74, H2-K1 (MHC class I), Pigr, B2m, S100a11, etc.), both cell types adopted a mucinous phenotype that lacked metaplastic gene expression. These transcriptional changes in chief cells are consistent with paligenosis, a cellular process by which chief cells redifferentiate from the mature zymogenic cell into mucin-secreting proliferative SPEM cells. These results suggest that even during chronic inflammatory responses chief cell reprogramming contributes to SPEM. Additionally, the role of epithelial cell antigen presentation to both CD4 and CD8 T cells via inflammation induced expression of major histocompatibility complexes I and II in progression of gastritis to metaplasia is an important question of critical interest. These findings suggest that gastric epithelial cells may be capable of presenting antigens from self or non-self (e.g. H. pylori) proteins, and activating CD4+ and CD8+ T cells in the tissue.20
Characterizing the mucous neck cell and chief cell responses to inflammation indicated that both of these cell types contribute to the bona fide SPEM lesions that develop during chronic inflammation. We used pseudotime trajectory analysis to computationally infer the differentiation relationships from healthy and inflamed mucous neck cells and chief cells to SPEM. Critically, this unsupervised, unbiased approach generated a trajectory in which healthy chief and mucous neck cells are on a shared differentiation pathway that progresses to SPEM. The trajectory further supports the hypothesis that chief cells dedifferentiate/transdifferentiate into SPEM, as both mucous neck cells and chief cells share the “pre-SPEM” trajectory that leading to the SPEM terminus rather than approaching SPEM from separate paths. Understanding that these two different cell types can contribute to SPEM through two unique cellular processes is critical for determining which signals and molecular mechanisms regulate metaplastic development induced by inflammation - it is possible that the signals which induce chief cell paligenosis are unique from those that drive mucous neck cells into the SPEM phenotype. Finally, the pseudotime analysis identified two terminal SPEM branches based on the level of a given cell’s inflammatory response. This likely identifies cells isolated from somewhat less inflamed regions of the gastric corpus during the initial single cell preparation. Since metaplasia-associated transcripts are not differentially expressed between the two branches of SPEM, it is unlikely that these represent functionally unique cell types from a metaplastic standpoint.
These studies provide critical transcriptional, pathological, and computational evidence in support of the hypothesis that both mucous neck cells and chief cells are capable of plasticity during inflammation and differentiate into SPEM via a “pre-SPEM” transcriptional phenotype. The possibility of multiple origins of SPEM during inflammation is a critical finding with significant implications for the understanding of carcinogenesis, necessitating increased study of what factors and cellular processes are important for mucous neck cell transition to SPEM and how these differ from chief cells. A better understanding of the cell-type specific responses to inflammation that drive plasticity, metaplastic change, and carcinogenesis is critical across all organ systems in which inflammation drives neoplastic transformation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Washington University in St. Louis Genome Technology Access Center/McDonnell Genome Institute (GTAC/MGI) for assistance with 10X Genomics Chromium Single Cell RNA isolation and Illumina sequencing. Additionally, we thank Dr. Grant Kolar, Barbara Nagel, and Caroline Murphy of the Saint Louis University Research Microscopy and Histology Core for assistance with fluorescent microscopy assays and Dr. Joel C. Eissenberg for critical reading of the manuscript.
Grant Support:
KAB was supported by an NIH/NIDDK NRSA Predoctoral fellowship (F30 DK118873). RJD and JCM were supported by the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK110406). JCM was supported by the NIH National Cancer Institute (R01 CA246208). RJD was previously supported by the American Cancer Society (RSG-12-171-01-LIB), the American Gastroenterological Association Funderburg Award, and the Arthritis National Research Foundation.
Abbreviations:
- SPEM
spasmolytic polypeptide-expressing metaplasia
- PC
parietal cell
- MUC
Mucin
- Gif
gastric intrinsic factor
- Tff2
trefoil factor 2/spasmolytic polypeptide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors declare no potential conflicts of interest.
Transcript Profiling: https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP227356
REFERENCES
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- 2.Fox JG, Wang TC. Inflammation, atrophy, and gastric cancer. J Clin Invest 2007;117:60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landgren AM, Landgren O, Gridley G, et al. Autoimmune disease and subsequent risk of developing alimentary tract cancers among 4.5 million US male veterans. Cancer 2011;117:1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeong S, Choi E, Petersen CP, et al. Distinct metaplastic and inflammatory phenotypes in autoimmune and adenocarcinoma-associated chronic atrophic gastritis. United European Gastroenterol J 2017;5:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elsborg L, Mosbech J. [Pernicious anemia as a risk factor for development of stomach cancer]. Ugeskr Laeger 1979;141:773–6. [PubMed] [Google Scholar]
- 6.Vannella L, Lahner E, Osborn J, et al. Systematic review: gastric cancer incidence in pernicious anaemia. Aliment Pharmacol Ther 2013;37:375–82. [DOI] [PubMed] [Google Scholar]
- 7.Toh BH. Diagnosis and classification of autoimmune gastritis. Autoimmun Rev 2014;13:459–62. [DOI] [PubMed] [Google Scholar]
- 8.Hemminki K, Liu X, Ji J, et al. Effect of autoimmune diseases on mortality and survival in subsequent digestive tract cancers. Ann Oncol 2012;23:2179–84. [DOI] [PubMed] [Google Scholar]
- 9.Correa P A human model of gastric carcinogenesis. Cancer Res 1988;48:3554–60. [PubMed] [Google Scholar]
- 10.Correa P, Piazuelo MB. The gastric precancerous cascade. J Dig Dis 2012;13:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi H, Goldenring JR, Kaminishi M, et al. Identification of spasmolytic polypeptide expressing metaplasia (SPEM) in remnant gastric cancer and surveillance postgastrectomy biopsies. Dig Dis Sci 2002;47:573–8. [DOI] [PubMed] [Google Scholar]
- 12.Lennerz JK, Kim SH, Oates EL, et al. The transcription factor MIST1 is a novel human gastric chief cell marker whose expression is lost in metaplasia, dysplasia, and carcinoma. Am J Pathol 2010;177:1514–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halldorsdottir AM, Sigurdardottrir M, Jonasson JG, et al. Spasmolytic polypeptide-expressing metaplasia (SPEM) associated with gastric cancer in Iceland. Dig Dis Sci 2003;48:431–41. [DOI] [PubMed] [Google Scholar]
- 14.Willet SG, Lewis MA, Miao ZF, et al. Regenerative proliferation of differentiated cells by mTORC1-dependent paligenosis. EMBO J 2018;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam KT, Lee HJ, Sousa JF, et al. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology 2010;139:2028–2037 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldenring JR, Ray GS, Coffey RJ, et al. Reversible drug-induced oxyntic atrophy in rats. Gastroenterology 2000;118:1080–93. [DOI] [PubMed] [Google Scholar]
- 17.Huh WJ, Khurana SS, Geahlen JH, et al. Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology 2012;142:21–24 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertaux-Skeirik N, Wunderlich M, Teal E, et al. CD44 variant isoform 9 emerges in response to injury and contributes to the regeneration of the gastric epithelium. J Pathol 2017;242:463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayakawa Y, Fox JG, Wang TC. Isthmus Stem Cells Are the Origins of Metaplasia in the Gastric Corpus. Cell Mol Gastroenterol Hepatol 2017;4:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bockerstett KA, Lewis SA, Wolf KJ, et al. Single-cell transcriptional analyses of spasmolytic polypeptide-expressing metaplasia arising from acute drug injury and chronic inflammation in the stomach. Gut 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osaki LH, Bockerstett KA, Wong CF, et al. Interferon-gamma directly induces gastric epithelial cell death and is required for progression to metaplasia. J Pathol 2019;247:513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bockerstett KA, Osaki LH, Petersen CP, et al. Interleukin-17A Promotes Parietal Cell Atrophy by Inducing Apoptosis. Cell Mol Gastroenterol Hepatol 2018;5:678–690 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TL, Khurana SS, Bellone CJ, et al. Autoimmune gastritis mediated by CD4+ T cells promotes the development of gastric cancer. Cancer Res 2013;73:2117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHugh RS, Shevach EM, Margulies DH, et al. A T cell receptor transgenic model of severe, spontaneous organ-specific autoimmunity. Eur J Immunol 2001;31:2094–103. [DOI] [PubMed] [Google Scholar]
- 25.Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt PH, Lee JR, Joshi V, et al. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest 1999;79:639–46. [PMC free article] [PubMed] [Google Scholar]
- 27.Menheniott TR, Peterson AJ, O’Connor L, et al. A novel gastrokine, Gkn3, marks gastric atrophy and shows evidence of adaptive gene loss in humans. Gastroenterology 2010;138:1823–35. [DOI] [PubMed] [Google Scholar]
- 28.Geahlen JH, Lapid C, Thorell K, et al. Evolution of the human gastrokine locus and confounding factors regarding the pseudogenicity of GKN3. Physiol Genomics 2013;45:667–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leys CM, Nomura S, Rudzinski E, et al. Expression of Pdx-1 in human gastric metaplasia and gastric adenocarcinoma. Hum Pathol 2006;37:1162–8. [DOI] [PubMed] [Google Scholar]
- 30.Nomura S, Baxter T, Yamaguchi H, et al. Spasmolytic polypeptide expressing metaplasia to preneoplasia in H. felis-infected mice. Gastroenterology 2004;127:582–94. [DOI] [PubMed] [Google Scholar]
- 31.Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S, Fink J, Jorg DJ, et al. Defining the Identity and Dynamics of Adult Gastric Isthmus Stem Cells. Cell Stem Cell 2019;25:342–356 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen TL, Dipaolo RJ. A new mouse model of inflammation and gastric cancer. Oncoimmunology 2013;2:e25911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen TL, Makhlouf NT, Anthony BA, et al. In vitro induced regulatory T cells are unique from endogenous regulatory T cells and effective at suppressing late stages of ongoing autoimmunity. PLoS One 2014;9:e104698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saenz JB, Vargas N, Mills JC. Tropism for Spasmolytic Polypeptide-Expressing Metaplasia Allows Helicobacter pylori to Expand Its Intragastric Niche. Gastroenterology 2019;156:160–174 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramsey VG, Doherty JM, Chen CC, et al. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development 2007;134:211–22. [DOI] [PubMed] [Google Scholar]
- 38.Mills JC, Goldenring JR. Metaplasia in the Stomach Arises From Gastric Chief Cells. Cell Mol Gastroenterol Hepatol 2017;4:85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radyk MD, Burclaff J, Willet SG, et al. Metaplastic Cells in the Stomach Arise, Independently of Stem Cells, via Dedifferentiation or Transdifferentiation of Chief Cells. Gastroenterology 2018;154:839–843 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilhelm M, Schlegl J, Hahne H, et al. Mass-spectrometry-based draft of the human proteome. Nature 2014;509:582–7. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt T, Samaras P, Frejno M, et al. ProteomicsDB. Nucleic Acids Res 2018;46:D1271–D1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


