Abstract
TFR2 cells limit antibody responses, but the underlying mechanisms remain largely unknown. Here, we identify Fgl23 as a soluble TFR cell effector molecule through single-cell gene expression profiling. Highly expressed by TFR cells, Fgl2 directly binds to B cells, especially light-zone germinal center B cells, as well as to TFH4 cells and directly regulates B cells and TFH in a context-dependent and type 2 antibody isotype-specific manner. In TFH cells, Fgl2 induces the expression of Prdm15 and a panel of checkpoint molecules including PD16, TIM37, LAG38 and TIGIT9, resulting in TFH cell dysfunction. Mice deficient in Fgl2 had dysregulated antibody responses at steady state and upon immunization. In addition, loss of Fgl2 results in expansion of autoreactive B cells upon immunization. Consistent with this observation, aged Fgl2−/− mice spontaneously developed autoimmunity associated with elevated autoantibodies. Thus, Fgl2 is a TFR cell effector molecule that regulates humoral immunity and limits systemic autoimmunity.
Introduction
B cell-mediated antibody production is a major component of adaptive immunity. Multiple processes that contribute to optimal antibody responses depend on a histologically specialized site called the GC10 located in B cell zones of secondary lymphoid organs. Such processes include B cell affinity maturation, class switch recombination, plasma cell differentiation and memory B cell generation (1, 2). GC formation, maintenance and antibody class switching depend on help from specialized CD4+ TFH cells (3), which were first described as CD4+ T cells expressing a high level of the chemokine receptor CXCR5 that drives TFH migration to B cell follicles in response to the chemokine CXCL13 (4–6). TFH cells express the transcription factor Bcl611, a master regulator that mediates a unique TFH transcriptional program, while the transcription factor Blimp1 (encoded by the gene Prdm1) antagonizes Bcl6 and inhibits TFH differentiation and help (7–9). In addition to the expression of Bcl6, TFH cells express transcription factor cMaf12, which induces IL21 production (10) and Ascl213, which is critical for the expression of CXCR5 and trafficking of TFH cells into the GCs (11). TFH cells also express high levels of the co-stimulatory molecule ICOS and the co-inhibitory molecule PD1. These cells provide help to B cells through co-stimulatory molecules and cytokines, including CD40L, IL21 and IL4 (6, 10, 12–18).
Recently, a CXCR5+Foxp3+ CD4 T cell population was shown to mediate inhibitory effects on germinal center reactions and antibody responses, and thus the cells were defined as T follicular regulatory (TFR) T cells (19–21). Moreover, the cells have been shown to limit autoimmunity responses in animal models of influenza infection and Sjogren’s disease (22, 23). Unlike TFH cells, however, Bcl6 and Blimp1 are co-expressed in TFR cells. Blimp1 has been previously shown to influence Treg14 function by inducing an effector regulatory phenotype (24). It is expressed by Treg at mucosal sites and by a small subset of splenic Treg cells that produce IL10 in a Blimp115 (25). Similar to these Treg cells, TFR cells share high expression of IL10, GITR16 and ICOS. Thus, TFR cells were suggested to be the follicular counterparts of the Blimp1+ IL10+ effector Treg cells found at mucosal surfaces (25, 26) and in addition, because of their presence in GCs, to regulate T cell dependent antibody production by B cells. Recent studies suggested that TFR cells are able to durably alter multiple metabolic pathways in B cells through epigenetic changes, resulting in their diminished antibody production (27). Expression of the co-inhibitory receptor CTLA417 on TFR cells was demonstrated to be important for TFR cell effector functions as the loss of CTLA4 on TFR cells led to their reduced capacity to suppress antibody responses (28, 29). However, the effector mechanisms utilized by TFR cells to suppress B cells, DCs or other helper T cells, has not been fully investigated.
We performed population and single-cell RNA-seq assays of CD4+ T cells and computationally analyzed their transcriptome to identify soluble molecules that are differentially expressed in TFR/effector Treg cells in comparison to TFH cells or conventional Treg cells. We identified Fgl2 as a TFR cell effector molecule and a direct regulator of antibody responses. The molecule binds to B cells, particularly light-zone GC B cells, and TFH cells, and thereby directly modulates class-switch recombination in B cells and cytokine production from TFH cells. The inhibitory functions of Fgl2 were partly due to its effects on TFH cells as it induces expression of Prdm1 and a panel of “co-inhibitory” checkpoint molecules. Fgl2-deficient mice have dysregulated homeostatic and immunization-induced antigen-specific antibody responses. Fgl2-deficient TFR cells failed to suppress IgG1 production in vitro and in vivo. Moreover, aged Fgl2-deficient mice showed signs of autoimmunity with elevated autoantibody level. Together these findings demonstrate that Fgl2, a TFR effector molecule, regulates TFH cell and B cell functions and loss of Fgl2 results in dysregulated autoantibody responses and development of systemic autoimmunity.
Materials and Methods
Mice.
Wild-type CH7 black 6 (C57BL/6J), Fcgr2b−/−, Fcgr3−/− and Prdm1fl/fl mice were purchased from Jackson Laboratories. FoxP3IRES-GFP and Fgl2−/− mice on the C57BL/6 background have been published previously (30, 31). Fgl2−/− with FoxP3IRES-GFP reporter mice were bred in the lab. CD28−/− mice on the C57BL/6 background were from Arlene Sharpe lab. All mice were between 6 and 8 weeks of age at the time of experiments unless specific ages were mentioned in particular experiments. All mice used in each experiment were age-matched and gender-matched. The experiments were conducted in accordance with animal protocols approved by the Harvard Medical Area Standing Committee on Animals or BWH IACUC.
Immunizations.
Mice were subcutaneously immunized with 100 ug NP-OVA18 (Biosearch Technologies) emulsified in Complete Freund’s Adjuvant (BD) in the mouse flanks as previously described (32). For immunization to induce autoreactive B cells, additional heat killed dried Mycobacterium tuberculosis was added to CFA to the final concentration of 4 mg/mL. Mice were sacrificed later, and inguinal lymph nodes were harvested.
Antibodies.
The following antibodies were used for surface staining: anti-CD4 (RM4-5), anti-CD19 (6D5), anti-CXCR5 biotin (2G8), anti-PD1 (RMP1-30), anti-Fas (15A7), GL7 (GL7), anti-IgM (RMM1), anti-IgD (11-26c.2a), anti-B220 (RA3-6B2), anti-CD38 (90), anti-CD138 (281-2), anti-CXCR4 (L276F12), anti-CD86 (GL1), anti-polyhistidine tag (His tag) (AD1.1.10) and anti-Tim3 (5D12). Secondary staining for biotinylated primary antibody was done using streptavidin (BioLegend). For intracellular staining samples were fixed with Fixation/Perm solution kit (BD) for intracellular Ig or FoxP3 Fix/Perm buffer set (eBioscience) for staining transcription factors according to manufacturer’s instruction. Samples were then intracellularly stained with anti-IgG1 (RMG1-1), anti-IgG2b (RMG2b1), anti-IgE (RME1), anti-IgA (goat anti-mouse polyclonal, Southern Biotech), anti-FoxP3 (FJK-16S) and anti-Bcl6 (K112-91). For Fc block experiments, 10 ng/mL purified anti-mouse CD16/32 antibody (BioLegend) was used. For anti-IL10 blocking experiments, 500 ng/mL of purified rat anti-mouse IL10 (BD) was used.
Sorting.
Single cell suspensions were diluted in PBS supplemented with 3% FBS with 2mM EDTA. CD4+ cells were enriched by magnetic positive selection (Miltenyi Biotec). CD4+ enriched cells were then stained and sorted as follows: TFH (CD4+ CD19−ICOS+CXCR5+FoxP3−), TFR (CD4+ CD19−ICOS+CXCR5+FoxP3+). B cells were isolated from flow-through from CD4+ selection and sorted as CD19+CD4−. Single sorting was used in all of the experiments, except for gene expression profiling ones, in which double sorting was used.
Droplet-based single-cell RNA-Seq.
Inguinal lymph nodes from wild-type mice immunized with NP-OVA/CFA (s.c.) for 7 days were isolated and CD4+ cells by magnetic positive selection (Miltenyi Biotec). Cells were then sorted based on CD19−CD4+CXCR5+PD1+ with permissive thresholds for CXCR5 and PD1 gating. Cells were then subjected to droplet-based single-cell RNA-seq using the Chromium Single Cell Gene Expression Solution platform (10x Genomics).
Population RNA-Seq.
Samples were sorted as described above. RNAseq library preparations were performed. Briefly, RNA was isolated using RNeasy Mini Kit (Qiagen). The cDNA libraries were prepared based on modified SMARTseq2 protocol as previously described with 8 amplification cycles(33, 34). The library quality was confirmed using BioAnalyzer high sensitivity DNA chip (Agilent Technologies). RNA Sequencing reactions were sequenced on an Illumina HiSeq 2000 or Illumina NextSeq sequencer (Illumina) according to manufacturer’s instructions, sequencing 50bp reads.
In vitro culture assays.
In vitro culture assays were performed as previously described(35). Briefly, 5x104 total CD19+CD4− B cells or GC B cells (CD19+CD4−Fas+GL-7+) 3x104 TFH cells and/or 1.5x104 TFR cells were plated in 96-well plates along with 25 ng LPS, 5 ug anti-CD40 (BioLegend) or 2ug/ml anti-CD3 and 5ug/ml anti-IgM (Jackson Immunoresearch). Polarizing cytokines for T cells and B cells class-switch recombination are described in specific experiments. Cultures were harvested after 3-6 days as specified in specific experiments. Recombinant tetrameric mouse Fgl2-His (R&D Systems) was added into the culture as described. For analysis, B cells were gated as CD19+CD4− cells while specific isotype staining was done by intracellular staining described earlier, TFH cells were gated as CD4+CD19−FoxP3− cells, and TFR cells were gated as CD4+ CD19−FoxP3+ cells.
ELISA.
ELISA measurements of IgG from culture supernatants and sera were performed as described previously(32, 36). For autoantibody ELISA, mouse anti-nuclear antigens (ANA/ENA) Ig’s (total (A+G+M)), anti-dsDNA Ig’s (Total A+G+M) and anti-dsDNA IgG1-specific ELISA kits were used according to the manufacturer’s instruction.
Bead-based immunoassays.
Soluble mouse Ig isotype measurements were performed using mouse immunoglobulin isotyping kit (BD) while soluble cytokine measurements were performed using LEGENDplex kit (BioLegend) according to manufacturers’ instructions. The mouse immunoglobulin isotyping kit provides no standard curve assessment so the results should be considered more as qualitative. The LEGENDplex kit, however, provides standard curve assessment.
Autoantigen microarray.
Lupus autoantigen microarrays were constructed and developed as described(37–39). Briefly, 33 were spotted onto Epoxy slides (Arrayit Corporation, Sunnyvale, CA, USA). The microarrays were blocked for 1h at 37°C with 1% bovine serum albumin and incubated for 2 hr at 37°C with a 1:100 dilution of the test serum in blocking buffer. The arrays were then washed and incubated for 45 minutes at 37°C with a 1:500 dilution of a goat anti-mouse IgG detection antibody conjugated to Cy3 and anti-mouse IgM detection antibody conjugated to Cy5 (Jackson ImmunoResearch, West Grove, PA). The arrays were scanned with a ScanArray 4000X scanner (GSI Luminomics, Billerica, Massachusetts, USA).
ELISPOT.
Ethanol-activated MultiScreenHTS IP filer plates (EMDMillipore) were coated with either 100 μL of filtered (with 0.45 μM filters) Salmon Sperm DNA (5 μg/mL g/mL; ThermoFisher) or NP-OVA (10 μg/mL, Biosearchtech) overnight at 4°C. The plates were then washed with PBS and blocked for 2 hours with blocking buffer (5% FCS, 3% BSA in PBS) before they were re-washed and let dry. 50 μL of splenocytes in clone media (started with 5 x 105 cells per well with 1/3 serial dilutions) were plated and 50 μL of ACD40 (10 μg/mL) was added per well (final concentration of 5 μg/mL). The cells were incubated at 37°C for 24 hours. After that, cells and unbound cytokines were washed by incubating with PBS Tween-20 buffer for 10 minutes and then thoroughly re-washed. 50 μL of biotin-conjugated goat anti-mouse Ig (1:350 dilution in clone media, SouthernBiotech) was then added and incubated overnight at 37°C for primary staining. After rewashing, secondary staining was done for 1 hour using 50 μL of streptavidin-ALP (1:1000 dilution in 1% BSA in PBS, Mabtech). The plates were then washed and developed using 50 μL BCIP/NBT-plus substrate (Mabtech). When spots are clearly visible under a dissecting microscope, stop the development by discarding the substrate and rinse plates with tap water thoroughly. Spots were counted manually with a dissecting microscope.
Nanostring.
nCounter platform (NanoString Technologies) was used according to the manufacturer’s instruction. A codeset of T cells associated genes and 4 additional house-keeping genes were custom-made.
Histology and Immunohistochemistry.
Spleens, lymph nodes, lungs, kidneys, livers, guts and patches of skin were harvested and fixed in 10% formalin, paraffin-embedded, and sectioned. Representative sections were stained with hematoxylin & eosin. For immunohistochemistry, avidin-biotin immunohistochemical staining was performed on the sections with rabbit anti-mouse CD3 (Abcam, Cambridge, UK) and rat anti-mouse CD45R (B220; BD Biosciences) using reagents from Vector Laboratories (Burlingame, CA).
Statistical analysis for non-RNA-Seq data.
Statistical analysis was performed using Prism 7 (GraphPad). Differences between two groups were compared using either Kruskal-Wallis tests followed by post hoc Dunn’s tests for multiple comparisons or two-tailed unpaired Welch’s T tests (n.s; * p <0.05, ** p<0.01 and *** p<0.001). All figures show the means ± SD.
Data analysis for scRNA-Seq data.
10x sequencing outputs were processed with Cell Ranger (10x Genomics), and loaded into a Seurat object (40), which was used to scale the data, regress out unwanted axes of variation (number of UMIs and ratio of mitochondrial UMIs per library), and cluster the cells with the SLM algorithm. Default parameter values were used unless specified otherwise. Differential expression between cell clusters was performed with Seurat’s implementation of a Wilcoxon-based procedure followed by Bonferroni adjustment for multiple comparisons.
Computational transcriptomic signatures.
Computational signatures (Fig. 1 a) were computed as described in (PMC4671824, PMC6763499). Briefly, a signature is list of genes positively or negatively associated with a cellular state of interest with some weight (here, always +1 or −1). Let v be a column vector where the coordinate of a given gene is its weight or 0 if the gene is not part of the signature. Given a gene expression CPM matrix G (cells X genes), the product vector Gv gives the scores for each cell with respect to the signature.
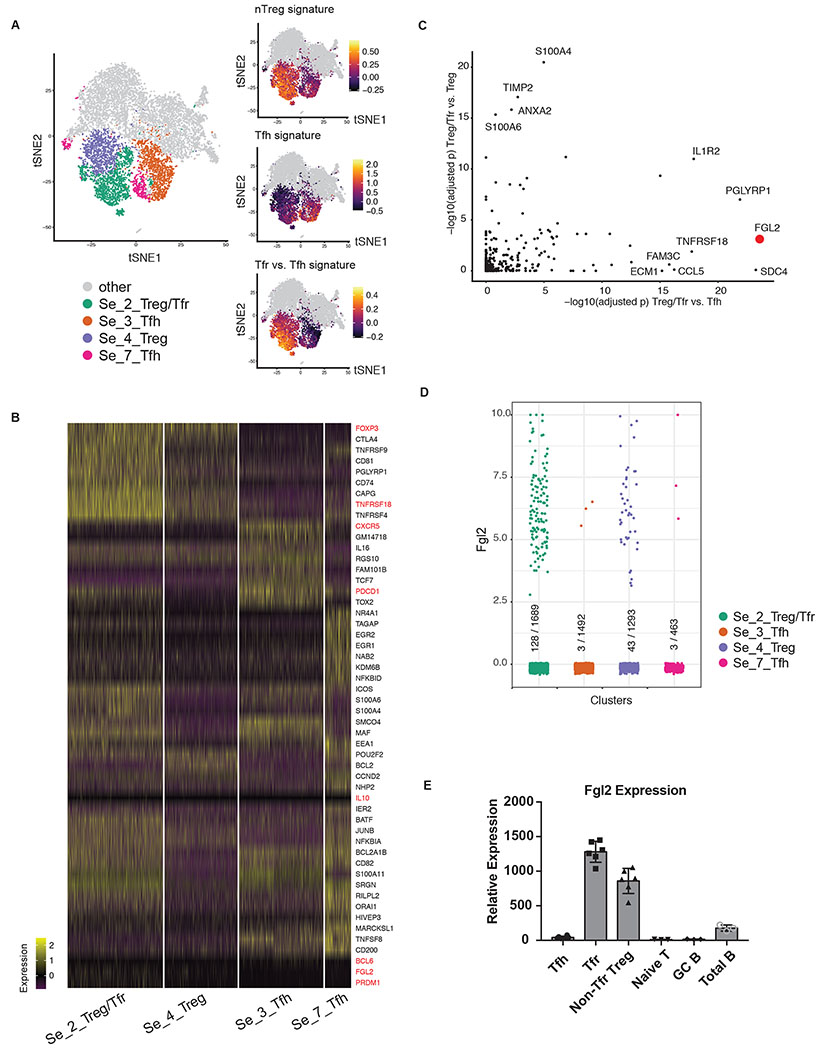
Figure 1.
Fgl2 is a distinguishing marker of TFR cells. (a) tSNE visualization of 10x single-cell transcriptomes. Four relevant clusters of interest (left) are shown whereas the grey dots correspond to cells belonging to other clusters. The other three panels present values of computational signatures allowing one to assign identities to the clusters (Methods). (b) Genes that are differentially expressed between the clusters are shown. All of the genes, except for the ones in red, are the top differentially expressed genes in the corresponding clusters. The genes in red, on the other hand, are assigned manually as they are known TFH/Treg marker genes, as well as Fgl2. (c) Markers of TFR cells should be enriched in Treg/TFR cluster compared with both TFH and non-TFR Treg clusters. Each dot corresponds to a gene associated with an extracellular secreted product (GO:0005576). Its x- and y-values are BH-adjusted p-values for hypergeometrically testing whether detections of the gene are enriched in the given comparisons. (d) Fgl2 expression in the 4 clusters. (e) High expression of Fgl2 by TFR cells was confirmed by Taqman qPCR. The single-cell RNA-seq experiment was performed once with samples run on 3 different lanes. The qPCR results are representative of 3 independent experiments with the plot showing the means ± SD.
Here, we addressed 10x sparsity with respect to key signature transcripts by partitioning the cells into small, mutually exclusive groups (“micropools” of ~5 cells each) and averaging the gene expression profiles of cells in the pool following the method outlined in (PMC6763499). We then used that average profile to compute a signature value and assigned that value as the signature score for all cells in the pool.
We defined three transcriptomic signatures. The first was an unsupervised signature based on TFR vs. TFH cells (Fig. 1 c; Table S1) by calling differentially expressed (DE) genes in the population RNA libraries described in the results section. Libraries were aligned with Tophat2(41), reads per transcript were counted with featureCounts(42) and DE genes were then called with DESeq2(43) using the thresholds FDR <= 10% and |B| >= 10% where B is the moderated B-statistic. Signature weights were set to +1 for upregulated and −1 for downregulated genes in a TFR vs. TFH comparison. The second signature (Table S1) consisted of a list of manually curated TFH-related genes (all set to have the same weight +1). The third signature (Table S1) was adapted from PMC4671824 and was based on a bulk RNA-Seq comparison of nTreg cells to other T helper subsets (GSE14308), with +1,−1 weights for genes upregulated or downregulated in nTreg, respectively.
Results
Fgl2 is a distinguishing marker for TFR cells
To assess potential effector molecules downstream of TFR cells, we performed a high-throughput 10x single-cell RNA-Seq assay on CD19−CD4+CXCR5+PD1+ T cells from draining lymph nodes of wild-type mice immunized with NP-OVA/CFA for 7 days. We used permissive thresholds for CXCR5 and PD1 gating (fig. S1A) in order to include cells like non-TFR Treg cells, which would be useful for comparative analyses. The permissive gating also allows us to gain a complete statistical representation of TFR cells states (44). More restrictive gating would preferentially exclude TFR states where CXCR5 or PD1 are present but are expressed at low levels. The inclusion of such transitional state is crucial for identifying novel regulators that drive development of different cell states. The use of droplet-based scRNA-Seq, which typically sequences thousands of cells in a single run assured that TFR cells would stochastically be represented in the data despite the permissive CXCR5 and PD1 gating thresholds. We applied a standard 10x quality control and data processing pipeline (Methods) to quantify the transcriptome of 12,628 cells, which we subsequently clustered with the unsupervised SLM algorithm (45).
For each of the cells, we computed quantitative signatures of T cell identity (34) and visualized them with t-SNE plots19. We identified 4 pertinent clusters of interest, one corresponding to TFR cells with enrichment in both Treg and TFH signatures (Fig. 1 a; Se_2_Treg/TFR in green), one corresponding to non-TFR Treg cells with no TFH signature enrichment (Fig. 1 a; Se_4_Treg in purple) and two corresponding to TFH cells (Fig. 1 a; Se_3_TFH and Se_7_TFH in orange and pink). We confirmed the identity of these clusters using three computational transcriptomic signatures (Fig 1 a; Methods; Table S1). The first signature was derived from differential expression in population RNA-seq of sorted TFR and TFH cells (CD19−CD4+CXCR5+PD1+FoxP3+ and CD19−CD4+CXCR5+PD1+FoxP3− respectively) from draining lymph nodes of Focp3IRES-eGFP reporter mice immunized with NP-OVA (Fig. S1 b–c). The second signature was a hand-curated list of TFH-associated genes, and the third signature was a computational signature distinguishing nTreg cells from other Th subsets. Next, we verified the identification of these clusters through genes differentially enriched in them. As expected, Se_2_Treg/TFR and Se_4_Treg clusters express Foxp3 while TFH-associated genes, including Cxcr5, Programmed cell death 1(Pdcd1), Icos, Maf and, to a less extent, Bcl6, are only enriched in the Se_2_Treg/TFR cluster. It is likely that the Se_2_Treg/TFR cluster not only includes TFR cells but also non-TFR effector Treg cells with enriched expression of Prdm1 (encoding BLIMP1) and Tnfrsf18 (encoding GITR). The two TFH clusters, on the other hand, are enriched in the TFH marker genes with minimal Foxp3 enrichment (Fig. 1 b and S1e). We hypothesized that the transcriptomic program of TFR cells should reflect a combination of TFH and Treg elements. Indeed, the differential expression between Se2 (Treg/TFR) and Se4 (Treg) was well aligned with the differential expression between TFH and non-TFH cells (Se3 and Se7 vs. the “other” cluster shown in Fig. 1 a) (Fig. S1f). Two notable exceptions were Cxcr5 and Crip1. Cxcr5 distinguished TFH from non-TFH but not Se2 from Se4. Crip1 was significantly downregulated in TFH compared to non-TFH but upregulated in Se2 compared to Se4. Interestingly, HIF1a which is associated with an effector over a tolerant Th phenotype (46, 47), was significantly upregulated in Se2 (Treg and TFR) over Se4 (Treg).
To screen for potential novel TFR effector molecules, we sought genes that are a) preferentially detected in the Se_2_Treg/TFR cluster compared with the two TFH clusters b) preferentially detected in the Se_2_Treg/TFR cluster compared with the Se_4_Treg cluster and c) associated with an extracellular secreted product (GO:0005576). Through these criteria, we identified Fgl2 among the top genes (Fig. 1, c and d). Additionally, we investigated the population RNA-seq data on TFR cells and TFH cells (Fig. S1 c) and confirmed that Fgl2 was among the top genes encoding secreted proteins differentially expressed by TFR cells when compared to TFH cells (Fig. S1 d). qPCR20 results confirmed that TFR cells expressed a high level of Fgl2 in comparison with TFH cells while non-TFR Treg cells also expressed high levels of Fgl2 as described previously (48). On the other hand, naïve T cells, total CD19+ CD4− B cells and germinal center B cells expressed low levels of Fgl2 if any (Fig. 1 e).
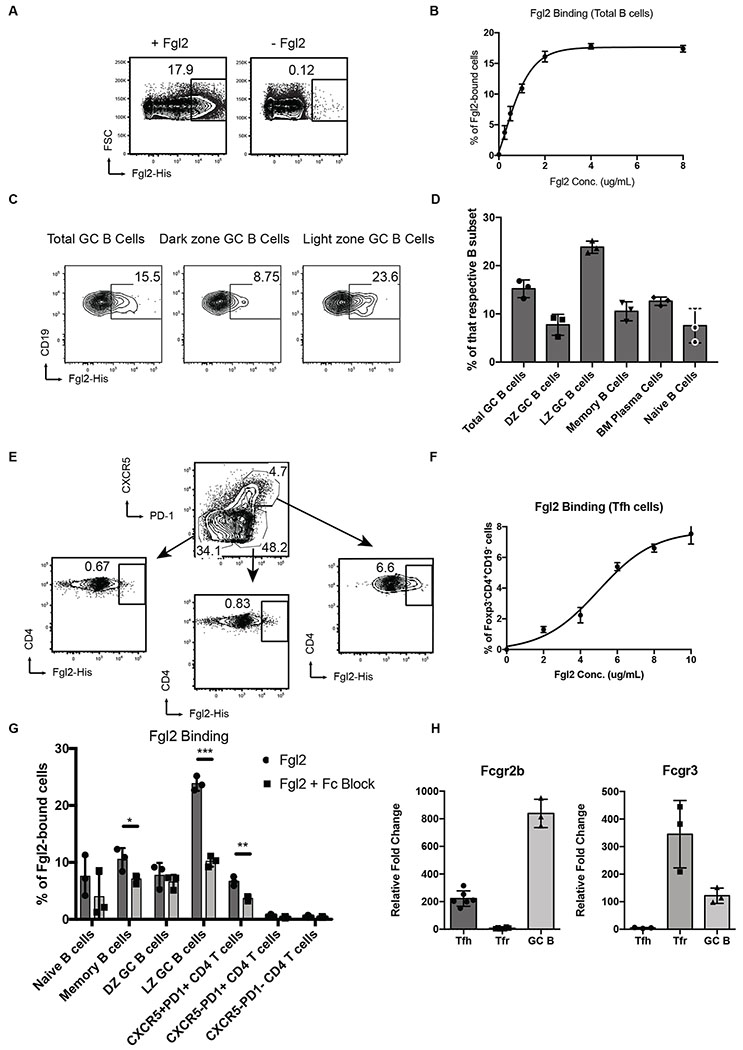
Fgl2 directly binds B cells and TFH cells
To test the hypothesis that Fgl2 is an effector molecule of TFR cells, we first investigated whether Fgl2 directly binds to B cells and TFH cells, as Fgl2 was previously demonstrated to bind to DCs, peritoneal macrophages and total splenic B cells (49, 50). Using recombinant Fgl2 protein with a His-tag, we showed that Fgl2 also binds in a dose-dependent fashion to total CD19+ B cells in draining lymph nodes of NP-OVA/CFA immunized mice for 7 days (Fig. 2, a and b) while His-tag alone does not (Fig. S2A). Further analysis of B cell subsets revealed preferential binding of Fgl2 to LZ21 germinal center B cells, defined as CD19+CD38−Fas+GL-7+CXCR4−CD86+ cells (51) (Fig. 2, c and d and Fig. S2, b and c). The preferential binding to germinal center B cells, particularly LZ GC B cells, implies its relevance to TFR functions as suppressors of GCs. Fgl2 also preferentially binds to TFH cells, defined as CD19−CD4+Foxp3−CXCR5+PD1+ cells, in a dose-dependent fashion (Fig. 2, e and f).
Figure 2.
Fgl2 directly binds and regulates B cells and TFH cells in vitro. (a-d) Draining lymph node cells or bone marrow cells from NP-OVA/CFA immunized mice for 14 days were stained and treated with recombinant Fgl2 with His-tag followed by secondary anti-His antibodies with fluoroflore (APC or PE). (a-b) Fgl2 binds to total B cells (CD19+CD4−) from NP-OVA/CFA immunized mice for 7 days with the titration curve showing a dose-dependent binding. (c-d) Fgl2 binds to different B cell subsets gated as followed: total GC B cells (CD19+CD38−Fas+GL-7+), DZ GC B cells (CD19+CD38−Fas+GL-7+CXCR4+CD86−), LZ GC B cells cells (CD19+CD38−Fas+GL-7+CXCR4−CD86+). Gating for the other B cell subsets and representative FACS plot can be found in Figures S2 a–c (e-f) Fgl2 preferentially binds to TFH cells (CD19+CD4−Foxp3−CXCR5+PD1+) with the titration curve showing a dose-dependent binding. (g) Fgl2 binding was partially abolished in both LZ GC B cells and TFH cells in the presence of Fc block. (h) Expression of Fcgr2b and Fcgr3 were assessed by Taqman qPCR in sorted TFH cells, TFR cells and GC B cells from mice immunized with NP-OVA/CFA 7 days earlier. The summary results were pooled from 2-3 independent experiments. Differences between two groups were compared using two-tailed unpaired Welch’s T tests (n.s; * p <0.05, ** p<0.01 and *** p<0.001). All plots show the means ± SD.
Fgl2 has been reported to bind two receptors: Fcgr2b22 and Fcgr323 (49). Since Fgl2 can bind to both B cells and TFH cells, we studied if the binding was dependent on the two receptors and how the receptors are expressed. Treatment with Fc block, which antagonized both receptors, partially suppressed Fgl2 binding both on LZ GC B cells and TFH cells (Fig. 2 g and Fig. S2, b to d). Moreover, we found that, at RNA level, Fcgr2b is highly expressed on GC B cells and, at a lower level, on TFH cells while Fcgr3 is expressed on TFR cells and, to a lower extent, on GC B cells (Fig. 2 h). We were unable to properly detect the two receptors at protein level as available antibodies for flow cytometry were not able to discriminate Fcgr2b and Fcgr3.
Fgl2 directly regulates B cells
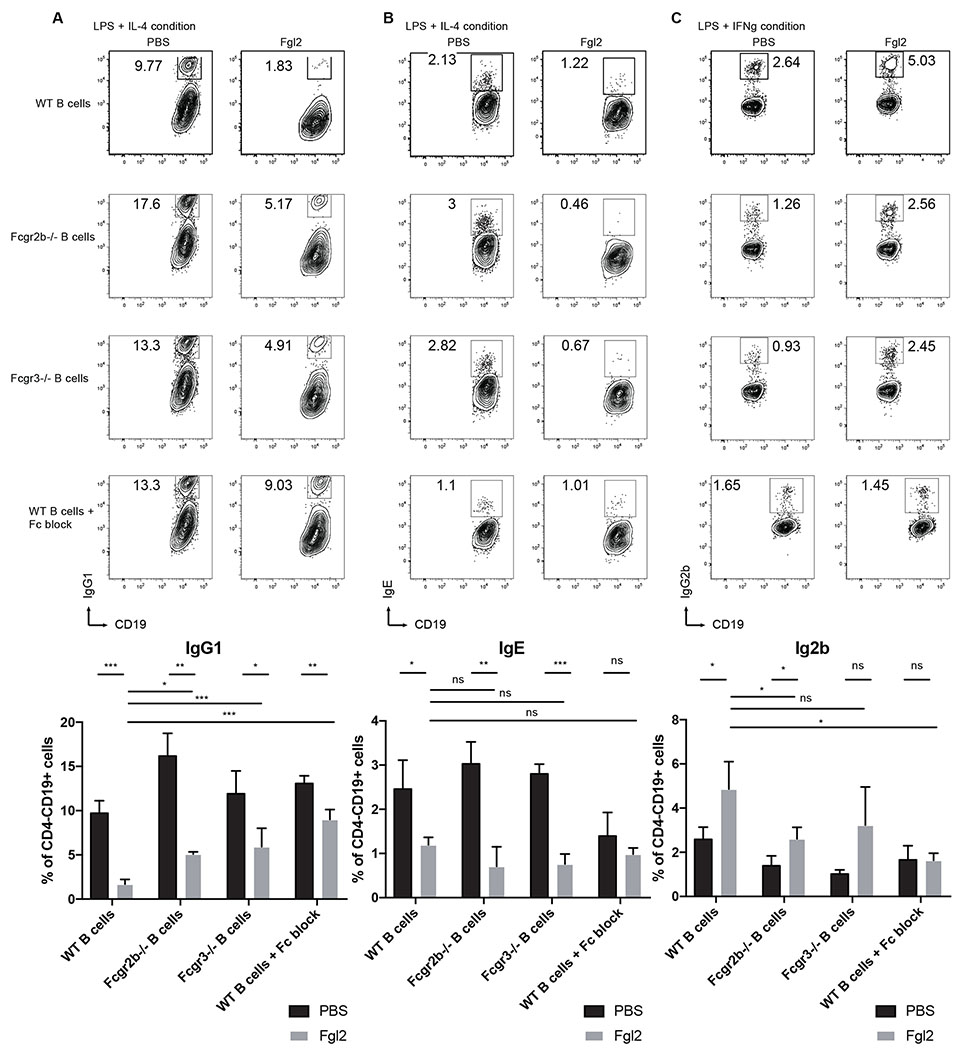
Next, we analyzed the effects of Fgl2 on B cells and TFH cells. We cultured sorted total splenic CD19+CD4− B cells from non-immunized mice in different conditions in the presence of Fgl2 and found that Fgl2 limits B cells survival and proliferation under anti-IgM24 condition but not in LPS and anti-CD40 conditions (Fig. S3, a to c).
To investigate if Fgl2 influences B CSR25, we cultured sorted total CD19+CD4− splenic B cells from non-immunized mice in cytokine-polarizing conditions in the presence of LPS with or without recombinant Fgl2 for 4 days. The presence of recombinant Fgl2 inhibited IgG1 and IgE CSR26 under IL4-polarizing conditions, but modestly enhanced IgG2b under IFN-g-polarizing conditions as assayed by intracellular staining and cytometric bead array. Similar results were also observed when total CD19+CD4− splenic B cells from mice immunized with NP-OVA/CFA for 7 days were used (Fig. 3, a to c and Fig. S4, a and b), as well as when sorted GC B cells (CD19+CD4−Fas+GL-7+) from mice immunized with NP-OVA/CFA for 14 days were cultured in the presence of anti-CD40 antibodies (Fig. S4 c). These findings suggested that B cell CSR regulation by Fgl2 is context-dependent and isotype-selective. The impact of Fgl2 on B cell CSR was not dependent solely on either Fcgr2b or Fcgr3 as Fgl2 still retained its effects on single-knockout B cells. However, while we could not generate double-knockout mice due to the close proximity of loci encoding the two receptors, the presence of Fc block abrogated the effects of Fgl2 on B cell CSR but never reached the wild-type level, suggesting that there may be additional receptors through which Fgl2 must act or that the FC-block does not completely block the receptors (Fig. 3, a to c and Fig. S4 c).
Figure 3.
Fgl2 regulates B cell CSR in vitro (a-c) Effect of Fgl2 on B cell CSR: In vitro class switching assay. Sorted total B cells (CD19+CD4−) nonimmunized wild-type, Fcgr2b−/− or Fcgr3−/− mice for 7 days were cultured in LPS + IL4 (IgG1 and IgE) or LPS + IFNG (IgG2b) for 4 days in the presence or absence of Fc block. Switched isotypes were detected by flow cytometry. Differences between two groups were compared Kruskal-Wallis tests followed by post hoc Dunn’s tests for multiple comparisons (n.s; * p <0.05, ** p<0.01 and *** p<0.001). All plots show the means ± SD with n = 3 in each group.
Fgl2 directly regulates TFH cells
To test the effects of Fgl2 on TFH cells, we sorted TFH cells and cultured them with plate-bound anti-CD3 and anti-CD28. The addition of Fgl2 suppressed production of most secreted cytokines tested, including IFNg27, IL2, IL4, IL10, IL17A, IL21 and IL13 in vitro (Fig. S5 a). However, these data should be interpreted with caution as expression of TFH-associated genes, including Bcl6, Maf, Cxcr5 and Icos, was lost upon in vitro activation by anti-CD3/anti-CD28 in the absence of B cells (Fig S5 b). The addition of total CD19+CD4− B cells from the same immunized mice we harvested TFH cells in the TFH/B cell co-culture in the presence of soluble anti-CD3 and IgM (32, 35) resulted in only secreted IL13 and IL5, but not IFNG, being suppressed in the presence of soluble Fgl2 (Fig. 4 a). Interestingly, upon Fgl2 treatment, Il4 mRNA was decreased whereas Il21 was significantly upregulated in TFH cells (Fig. 4b). B cell survival in the presence of TFH cells was not affected (Fig. S3 d) unless B cells and TFH cells were from immunization with different antigens (Fig. S3 e). This selective suppression of type-2 cytokines was associated with a selective decrease in IgG1 production (Fig. 4 a). However, whether the decrease in IgG1 production was due to the altered cytokine profile of the TFH cells is unclear, as Fgl2 was demonstrated earlier to directly suppress IgG1 CSR on B cells (Fig. 3 a).
Figure 4.
Effect of Fgl2 on B cells in the context of TFH presence. (a) Effects of Fgl2 on TFH/B cell co-culture: sorted TFH cells were co-cultured with sorted total B cells from the same immunized mice in the presence of soluble ACD3 and AIgM with or without Fgl2 for 3 days. Cytokines were detected by bead-based LEGENDplex kit and antibody isotypes were detected by bead-based mouse immunoglobulin isotyping kit. (b) Sorted TFH cells were co-cultured with sorted total B cells from immunized mice in the presence of soluble ACD3 and AIgM with or without Fgl2 for 3 days. T cells were then re-sorted and subjected to NanoString gene expression profiling. Barchart exhibiting Il4 and Il21 mRNA normalized expression. (c) Wild-type TFH cells or Fcgr2b−/− TFH cells were co-cultured with wild-type B cells from immunized mice in cells in the presence of soluble ACD3 and AIgM with or without Fgl2 for 3 days. IgG1 in B cells was detected by flow cytometry. (d) Heatmap showing the filtered genes based on differential expression over 2 folds while p < 0.05 usingNanoString gene expression profiling. (e) Prdm1 expression was evaluated by Taqman qPCR. (f) TFH cells were sorted from wild-type or Fgl2−/− mice and were subjected to RNA-seq. Some of the top differentially-expressed genes were shown in the volcano plot. Prdm1 was among the top genes differentially expressed and its expression was confirmed by Taqman qPCR (g). (h) Same experiment settings as in (c). However, T cells were then resorted and subjected to Taqman qPCR to measure Prdm1 expression. (i) Control TFH cells (Prdm1fl/fl or Prdm1fl/+ with CD4cre−) or Prdm1+/− (Prdm1fl/+ with CD4cre+) TFH cells were co-cultured with wild-type B cells from immunized mice in cells in the presence of soluble ACD3 and AIgM with or without Fgl2 for 3 days. IgG1 in B cells was detected by flow cytometry. (j) Same experiment settings as in (a). Resorted T cells were subjected to Taqman qPCR to measure Prdm1, Havcr2, Lag3 and Tigit expression. (k) Same experiment settings as in (a). Protein expression of PD1, TIM3, LAG3 and TIGIT were detected by flow cytometry. The summary results were pooled from 2-4 independent experiments, except for the NanoString gene expression and RNA-seq profiling, which were performed once with n = 3 in each experiment group. Differences between two groups in all but gene expression profiling data were compared using two-tailed unpaired Welch’s T tests except for (g) in which Kruskal-Wallis test followed by post hoc Dunn’s test for multiple comparisons was used (n.s; * p <0.05 ; ** p<0.01 and *** p<0.001). All plots show the means ± SD.
Fgl2 regulates antibody responses through TFH cells in vitro
To investigate whether the role of Fgl2 in regulating antibody responses is dependent on TFH cells, we used a well characterized co-culture system previously described (35). As TFH cells only express Fcgr2b but not Fcgr3, we tested whether the Fgl2 would regulate IgG1 suppression through TFH cells deficient in Fcgr2b. Addition of Fgl2 to wild type B cells and TFH cultures resulted in significant inhibition of IgG1 production. While co-culturing wild-type B cells with Fcgr2b−/− TFH cells showed no significant difference in IgG1 level in the absence of Fgl2, the suppression by exogenous Fgl2 was partially rescued when Fcgr2b−/− TFH cells were present in the culture (Fig. 4 c and Fig. S6, a and b), suggesting that the Fcgr2b receptor expression on TFH cells can regulate IgG1 CSR in the presence of exogenous Fgl2.
To further investigate potential downstream molecules of TFH cells that modulate IgG1 CSR in response to Fgl2, we re-sorted TFH cells after the co-culture with B cells and Fgl2 and analyzed gene expression profiling using the Nanostring platform. We failed to see inhibition of TFH-related genes, including Bcl6, Pdcd1, and IL21; these genes were in fact up-regulated. There was induction of genes associated with TFH functional suppression, including Ctla4 and Prdm1 (Fig. 4, d and e). The effect of Fgl2 on Prdm1 expression in TFH cells was also observed in vivo as Prdm1 was among the top genes differentially expressed genes when expression analysis was undertaken between wild-type and Fgl2−/− derived TFH cells and the reduced Prdm1 expression in TFH cells from Fgl2−/− mice was confirmed by qPCR (Fig. 4, f, g and Fig. S5, c and d). Prdm1 induction by Fgl2 in TFH cells was dependent on Fcgr2b on the cells as Fcgr2b-deficient TFH cells failed to upregulate Prdm1 in response to Fgl2 treatment as measured by qPCR from T cells resorted from TFH/B cell co-culture experiments (Fig. 4 h).
To test if Prdm1 is downstream of Fgl2 in TFH cells, we co-cultured wild-type B cells with Prdm1-deficient TFH cells. Notably, TFH cells deficient in both Prdm1 copies (Prdm1−/−) failed to induce IgG1 in B cells (Fig. S6, c to e) even though the gene was shown to be antagonistic of Bcl6 (7). Thus, we hypothesized that such functions might require some level of Prdm1 expression, and the regulation is rather dependent on some Prdm1 level but not in the context of its total absence. In fact, mice deficient only in one copy of Prdm1 were previously shown to possess phenotype distinct from those deficient in both (52). Using TFH cells from mice deficient in one copy of Prdm1 (Prdm1+/−), we found that those TFH cells were as capable of inducing IgG1 in B cells as wild-type control TFH cells while they conferred modest but significant resistance to Fgl2-mediated suppression of IgG1 B cells upon Fgl2 treatment (Fig. 4 i and Fig. S6, f and g), suggesting that the effects of Fgl2 on TFH cells have some dependency on Prdm1. Notably, the rescues of inhibition mediated by Fgl2 in Prdm1+/− TFH cells is modest, suggesting that other molecules are likely to be involved in addition to Prdm1. In fact we have previously shown that multiple transcription factors cooperate to mediating a defined phenotype/function and we predict that modest effects observed by loss of Prdm1 in abrogating Fgl2 derived inhibition may be due to 1) use of Prdm1 heterozygous mice and 2) other transcription factors might be playing a compensatory or additive role in mediating inhibitory function of Fgl2 (53); 3) Fgl2 may utilize other mechanisms in addition to induction of Prdm1 in mediating its inhibitory function.
As Prdm1 was previously shown to be involved in regulating expression of IL-10 and co-inhibitory gene module (53), we also checked if Fgl2 had effects on checkpoint molecules on TFH cells. We found that Fgl2 induced upregulation of multiple co-inhibitory, check-point molecules, including PD1, TIM3, LAG3 and TIGIT on TFH cell co-cultured with B cells measured at both mRNA and protein levels (Fig. 4, j and k and Fig. S6 h). Furthermore, while we found a decreased expression of Il10 in Fgl2−/− TFH cells (Fig. S6i), addition of Fgl2 to the cultures had no effect on IL-10 expression (Fig. S6j). To study the role of IL10 on B cells, we co-cultured TFH and B cells in presence or not of Fgl2 and found that IL-10 blockade did not prevent humoral response inhibition upon Fgl2 treatment (Fig. S6k). This indicates an IL10-independent role of Fgl2 on B cell responses in vitro. Since we have previously observed that Prdm1 is critical for the induction of module of “checkpoint” molecules, expression of the module further supports that Prdm1 and molecules downstream of Prdm1 are also induced by Fgl2. Taken together, the results suggest that Fgl2 can partially suppress IgG1 in B cells through modulation of Prdm1 level in TFH cells, potentially antagonizing Bcl6 (7) in an Fcgr2b-dependent fashion while the molecule also induces expression co-inhibitory molecules on TFH cells.
Fgl2 regulates antibody responses in vivo
In order to determine if Fgl2 modulates antibody responses in vivo, we analyzed antibody production in Fgl2-deficient mice. While young, 8-week-old, Fgl2−/− showed no significant differences in total serum antibody isotypes (Fig. S7 a), 20-week-old Fgl2−/− mice had significantly elevated total serum IgG1, IgG2a, IgA and IgE spontaneously (Fig. 5 a). We also analyzed PP28 where TFH cells and GCs are present at steady state and we found that Fgl2−/− mice had increased PP IgA+ B cells, GC B cells, TFH cells and TFR cells, but a decreased total number of Treg cells (which include TFR cells) (Fig. 5, b and c). Elevated free fecal IgA was also observed even though the frequency of IgA-coated bacteria was reduced (Fig. 5 b and Fig. S7 b). These findings suggested that antibody responses at steady state were dysregulated with age in the absence of Fgl2 in vivo.
Figure 5.
Fgl2 regulates antibody responses in vivo. (a) Serum antibody isotypes in 20-week-old wild-type and Fgl2−/− mice were detected by bead-based mouse immunoglobulin isotyping kit and ELISA (for IgE). The mouse immunoglobulin isotyping kit provides no standard curve assessment so the results should be considered more as qualitative. (b-c) Peyer’s Patches of 20-month-old wild-type and Fgl2−/− mice were immunophenotyped by flow cytometry for IgA+ B cells, GC B cells, TFH cells, TFR cells and total Treg cells. Free fecal IgA was measured by ELISA. (d) Wild-type and Fgl2−/− mice were immunized with NP-OVA (s.c.) in CFA for 21 days and NP-specific isotypes were detected by ELISA. The results were pooled from 3 independent experiments. Differences between two groups were compared using two-tailed unpaired Welch’s T tests (n.s; * p <0.05, ** p<0.01 and *** p<0.001). All plots show the means ± SD.
Next we wanted to see if Fgl2 could affect antigen-specific responses. We immunized wild-type and Fgl2−/− mice with NP-OVA emulsified in CFA. At day 21, we detected significantly enhanced NP-specific IgG1 but not NP-specific IgG2b by ELISA in Fgl2−/− mice (Fig. 5 d). Interestingly, examination at an earlier time point (day 10) when GC B cells and TFH cells were still present showed no significant difference in the frequency of total GC B cells and, surprisingly, significant decrease in the frequency of NP-specific GC B cells (Fig. S7 c).
Fgl2 from TFR cells regulates antibody responses in vitro and in vivo
Because Fgl2 affected the numbers of both TFH cells and TFR cells in vivo (Fig. S7 d and e) and Fgl2 may come from other cellular sources including conventional Treg cells, we utilized co-culture and transfer experiments to study the contribution of Fgl2 from TFR cells in the contexts where the number of TFH cells and TFR cells are equivalent. To address whether Fgl2 from TFR cells modulates antibody responses in vitro, sorted total CD19+ B cells from the immunized mice were co-cultured with TFH cells and wild-type or Fgl2−/− TFR cells in the presence of soluble anti-CD3 and anti-IgM antibodies for 3 days. We used Fgl2−/− B cells in this system to make sure that the main source of Fgl2 will come from TFR cells as B cells could produce some level of Fgl2 (Fig. 1 e). As expected, the addition of wild-type TFR cells suppressed IgG1 CSR as shown by intracellular IgG1 staining and Ig bead array on secreted IgG1 while using Fgl2−/− TFR cells partially rescued it. The addition of exogenous Fgl2 also further suppressed IgG1 CSR in all conditions (Fig. 6, a to c). Non-TFR Treg cells, which express Fgl2 (Fig. 1 e), did not significantly suppress IgG1 CSR and Fgl2 depletion in non-TFR Treg cells resulted in no significant difference in the suppression (Fig. 6, d to f), suggesting the presence of TFR-specific effects that render Fgl2 functional at physiological concentration. Such findings demonstrated that Fgl2 from TFR cells suppress IgG1 responses in vitro.
Figure 6.
Fgl2 from TFR cells regulates antibody responses in vitro and in vivo. (a-c) In vitro co-culture assay; 50,000 Fgl2−/− sorted total CD19+ B cells from immunized mice were co-cultured with 30,000 TFH cells and 3,000 wild-type or Fgl2−/− TFR cells in the presence of soluble anti-CD3 and anti-IgM antibodies. Recombinant Fgl2 was added in certain conditions. (d-f) In vitro co-culture assay with the same settings as (a-c) while conditions with 3,000 wild-type or Fgl2−/− non-TFR Treg cells were included. (g-i) Sorted 50,000 WT or Fgl2−/− CD19−CD4+CXCR5+PD1+ T cells (TFH + TFR cells) were transferred into CD28−/− mice. The mice were then immunized with NP-OVA/CFA for 14 days and NP-specific IgG1 was detected by ELISA (h). After 30 days, recall responses were induced and cellular composition of inguinal lymph nodes were analyzed by flow cytometry (i). The results were pooled from 2-5 independent experiments. Differences between two groups were compared using Kruskal-Wallis tests followed by post hoc Dunn’s tests for multiple comparisons (n.s; * p <0.05, ** p<0.01 and *** p<0.001). All plots show the means ± SD.
Next we sought to investigate if Fgl2 from TFR cells regulates antibody responses in vivo. The experimental scheme is shown in Fig. 6 g. In short, we sorted TFH cells and TFR cells from mice immunized 7 days earlier with NP-OVA/CFA. Wild-type TFH cells were co-transferred with either wild-type TFR cells or Fgl2−/− TFR cells into CD28−/− recipient mice, which lacked endogenous TFH/TFR cells as previously described (19, 32). The recipient mice, along with TFH-only transfer and no-transfer controls, were then immunized with NP-OVA/CFA. At day 21, serum NP-specific IgG1 levels were significantly enhanced in mice receiving Fgl2−/− TFR cells (Fig. 6 h). GC B cells, TFH cells and TFR cells were analyzed on day 7 after recall immunization. The frequency of NP-specific GC B cells in mice receiving Fgl2−/− TFR cells was slightly but significantly elevated while the frequency of TFH cells and TFR cells were not significantly different (Fig. 6 i). Collectively, the data suggested that Fgl2 from TFR cells regulates IgG1 and GC responses in vivo without affecting gross TFH/TFR cell frequencies.
Fgl2 is required for autoantibody controls
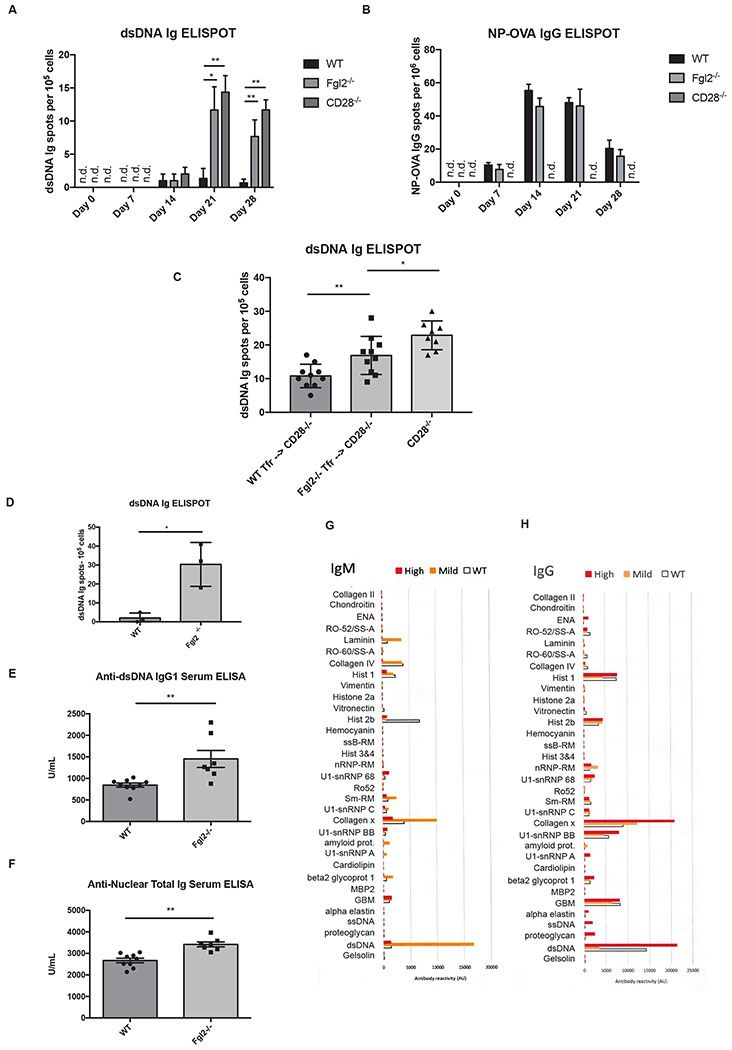
We then asked whether NP-OVA/CFA immunization alone would be sufficient to induce autoreactive B cells in young Fgl2−/− mice as influenza infection was previously shown to induce anti-dsDNA antibodies in mice specifically deficient in TFR cells (22). ELISPOT29 results showed that Fgl2−/− mice and CD28−/− control mice, which completely lacked both TFH and TFR cells, had a significant increase in inguinal lymph node anti-dsDNA (total Ig) ASCs30 with the peak at day 21 before the level started to go down (Fig. 7 a) while only wild-type and Fgl2−/− mice, but not CD28−/− control mice, have elevated inguinal lymph node anti-NP-OVA (total IgG) ASCs starting from day 7 (Fig. 7 b). Moreover, no significant differences in systemic levels of autoantibodies were observed as shown by the level of serum anti-dsDNA and ANA31. The data suggested that while Fgl2−/− mice still have functional total IgG ASCs against a foreign antigen despite some alterations in IgG isotypes demonstrated earlier, they failed to control local and transient expansion of autoreactive B cells during inflammation. This enhancement of autoreactive B cell responses strongly supports Fgl2 as a TFR effector molecule that controls autoimmunity.
Figure 7.
Fgl2 is important for limiting autoantibodies. (a-b) Wild-type and Fgl2−/− mice were immunized by NP-OVA/CFA (with additional heat killed dried Mycobacterium tuberculosis). Inguinal lymph nodes were harvested at the time points indicated and the cells were subjected to ELISPOT assay to detect dsDNA Ig and NP-OVA IgG ASCs. (c) Sorted 10,000 WT or Fgl2−/− CD19−CD4+CXCR5+PD1+Foxp3+ TFR cells were transferred into CD28−/− mice. The mice were then immunized with NP-OVA/CFA (with additional heat killed dried Mycobacterium tuberculosis) for 21 days and dsDNA Ig ASCs were detected by ELISPOT (d) Splenocytes from 12-month-old wild-type and Fgl2−/− mice were subjected to dsDNA Ig ELISPOT. (e-f) Anti-dsDNA IgG1 and ANA in sera from 12-month-old wild-type and Fgl2−/− mice were measured by ELISA. (g-h) An array of lupus IgM and IgG autoantibodies were measured in sera from 12-month-old wild-type (n = 6) and Fgl2−/− mice (n = 9) using autoantigen microarray. Fgl2−/− mice with mild diseases (n = 5) had either normal histology or mild dermatitis while the one with high, severe diseases (n = 4) had inflammation in multiple organs, including severe dermatitis, reactive changes in the spleen, ileitis, enlarged Peyer’s Patches and increased lymphoid clusters in lung. The results were pooled from 2-3 independent experiments. The autoantigen microarray results for (g-h) are from one independent experiment. Differences between two groups shown were compared using Kruskal-Wallis tests followed by post hoc Dunn’s tests for multiple comparisons for (a) to (c) and two-tailed unpaired Welch’s T tests for (d) to (f) (n.s; * p <0.05, ** p<0.01 and *** p<0.001). All plots show the mean ± SD.
To test if Fgl2 from TFR cells was relevant to the phenotype we saw in Fgl2−/− mice, we asked if transfer of TFR cells into CD28−/− mice, which lacked TFR cells and failed to control expansion of anti-dsDNA ASCs, would reverse the phenotype and whether the reversion was Fgl2-dependent. In fact, transfer of 10,000 wild-type TFR cells in to CD28−/− mice significantly suppressed expansion of anti-dsDNA ASCs upon NP-OVA/CFA immunization as measured by ELISPOT on day 21. However, the transfer of Fgl2−/− TFR cells in the same scheme resulted in significantly inferior suppression of such expansion (Fig. 7 c), suggesting that Fgl2 from TFR cells can at least partially suppress autoreactive B cells in vivo.
Aged Fgl2−/− mice (7-12 months) deficient in Fgl2 have previously been shown to have impaired Treg function and develop glomerulonephritis (31). However, autoantibody levels have not been investigated. To assess whether there were active autoreactive B cells in aged 12-month-old Fgl2−/− mice with no external perturbation, we performed ELISPOT experiments to detect anti-dsDNA ASCs using total splenocytes from aged Fgl2−/− mice and age-matched wild-type controls and found that aged Fgl2−/− mice had elevated splenic anti-dsDNA (total Ig) ASCs (Fig. 7 d). To determine if the elevation of autoantibodies was systemic, we measured serum antibodies and found elevated levels of anti-dsDNA IgG1 and ANA in aged Fgl2−/− mice as compared to age-matched wild-type controls (Fig. 7, e and f). A more comprehensive detection was performed using Lupus-associated autoantigen microarrays. We observed elevation of multiple autoantibodies against Lupus-associated autoantigens, including ssDNA, alpha elastin, β2 glycoprotein, dsDNA, U1 snRNP32 and collagen x, in aged Fgl2−/− mice, and the distribution of IgM/IgG isotypes also reflected disease severity based on histological data (Fig. 7, g and h). Although this increase in autoantibodies was not seen with all autoantigens and in fact for some antigens like Histone 2b, the autoantibody level was decreased. This suggests that the loss of Fgl2 does not uniformly increase in the production of antibodies to all autoantigens.
Aged Fgl2−/− Mice Developed Inflammatory, Lupus-like Autoimmunity
Forty-two percent of the aged, 12-month-old, Fgl2−/− mice in our cohort spontaneously developed a skin-associated phenotype that included patches of hair loss, hyperkeratosis and dermatitis (Fig. 8, a and b). Histological analysis of the skin showed signs of surface ulceration, hyperkeratosis, elongation of rete ridges, dermal scarring, epidermolysis, follicular plugging (Fig. 8 c, arrow) and basal cell discohesion (Fig. 8 c, arrow). Infiltration of T and B cells into the skin was also observed (Fig. 8 c). 18% of the aged mice also had spontaneous germinal center cell phenotype in spleens (Fig. 8, a and d). The results suggested that the loss of Fgl2 led not only to systemic elevation of Lupus-associated autoantigens but also clinical manifestations of inflammatory diseases.
Figure 8.
Aged Fgl2−/− mice developed inflammatory, lupus-like features. (a-b) A portion of aged, 12-month-old, Fgl2−/− mice spontaneously developed skin disease with varied severity. (c) Histological analysis shows signs of surface ulceration, hyperkeratosis, elongation of rete ridges, dermal scarring, epidermolysis, follicular plugging (arrow) and basal cell discohesion (arrow). Infiltration of T cells and B cells was also observed. (d) Cellular composition of mice with spontaneous reactive splenic GCs showed increased in GC B cells and TFH cells. The assessment histological phenotype and its frequencies is based on one large single cohort of mice with the numbers shown in (a) while representative results were shown in (b-d).
Discussion
We have used single-cell transcriptome analysis and population RNA-seq to identify Fgl2 as a top-ranking gene in follicular regulatory T cells. Moreover, we showed that Fgl2 is preferentially expressed in TFR cells as compared to TFH cells, naïve T cells and GC B cells and it is critical in controlling type-2 antibody responses and autoreactive B cell responses spontaneously arising during inflammation. Aged Fgl2−/− mice develop spontaneous skin inflammation and Lupus-like phenotypes with elevation of autoantibodies, highlighting the critical role of Fgl2 in regulating autoimmunity. We showed that Fgl2 directly acts on B cells and TFH cells and regulates antibody production and class switch recombination, and TFR derived Fgl2 is critical for these processes, thus demonstrating that Fgl2 is a TFR effector molecule both in vitro and in vivo.
Upon their discoveries, TFR cells were shown express Treg-associated genes along with TFH-associated ones (19–21). Our data, based on transcriptome at the resolution of single cells through a clustering scheme, showed that TFR cells are similar to effector Treg cells as they are in the same cluster in tSNE. The finding is consistent with what was observed earlier among Treg cells (54). Our analysis, however, also took well-defined TFH cells into account. The results, therefore, demonstrate that TFR cells are still fundamentally Treg cells that express some TFH-associated genes rather than simply Treg/TFH hybrid cells.
The suppression of type-2 antibody responses by Fgl2 was unexpected as Fgl2 was previously shown to suppress effector TH1 and TH17 but not TH2 cells (48). However, our data also demonstrated that the effects of Fgl2 are, in fact, context and cell type-dependent (Fig. 4 a and Fig. S5 a). The presence of B cells, which is required for maintaining the TFH phenotype (Fig. S5 b), dictates this bias, as in the absence of B cells we observed extensive cytokine suppression effects in non-TFH effector cells cultured in vitro. It is important to note that even though Fgl2 suppresses type-2 antibody responses directly on B cells, its effect can be exerted through TFH cells via the Fcgr2b receptor (Fig. 4 b), suggesting a nuanced mechanism. Thus, it is not surprising that the contexts where Fgl2 is expressed and the cells it acts on could lead to different effector outcomes.
On TFH cells, we have shown that Fgl2 induces Prdm1 in an Fcgr2b-depedent fashion and the pathway partly exerts inhibition of TFH-mediated IgG1 CSR on B cells. Expression of Prdm1 is potentially critical at multiple levels including antagonization of Bcl6 (7) and induction of a panel of check-point molecules (PD1, TIM3, LAG3 and TIGIT). It is possible that dysregulation of check-point molecule expression might account for some autoimmune phenotypes we observed in aged Fgl2-deficient mice. In fact, it was recently observed that germline HAVCR233 (encoding TIM3) mutations were associated with subcutaneous panniculitis-like T cell lymphomas with some patients developing Lupus-like disease with anti-DNA antibodies (55).
While Fgl2 produced by Treg cells has been previously shown to inhibit effector T cell responses, its functions based on TFR cells have not been studied. Here we demonstrated the relevance of Fgl2 in TFR cells to regulate antibody responses both in vitro and in vivo. In addition, mice deficient in Fgl2, unlike total Treg-deficient mice with severe multi-organ inflammation at young age (56, 57), were generally healthy at a young age while immunization led to altered isotypes against the immunized antigen. Notably, such immunization also induced local induction of autoreactive B cells producing anti-dsDNA. The phenotype was similar to that observed in TFR-deficient mice (22, 23) where alterations in humoral responses against foreign antigens were subtle while the mice only developed autoimmunity at an older age or upon induction. To what extent the spontaneous development of autoimmune phenotype, particularly skin inflammation, in aged Fgl2-deficient mice is dependent on Fgl2 from TFR cells, however, is still not known. The observation, nonetheless, collectively supports the relevance of Fgl2 as a key regulator of tissue inflammation and autoimmunity, however, what needs to be resolved is the relative roles that Fgl2 plays in inducing autoimmunity when produced by Tregs vs. TFR cells.
Recent studies have implicated TFR cells in limiting autoimmunity (22, 23). Our observations that NP-OVA immunization was sufficient to induce transient and local induction of autoreactive B cells in Fgl2-deficient mice. Furthermore, the Fgl2-deficient mice spontaneously developed dermatitis and multiple types of autoantibodies against Lupus-associated autoantigens with age suggested that Fgl2 is an effector molecule used by TFR cells to control autoreactive B cells. TFR cells might directly interact with autoreactive B cells themselves as their TCR repertoire is skewed toward self-antigen (58). Our data, in addition, shows that Fgl2 is only able to limit B cell survival and proliferation in the absence of cognate TFH help (Fig. S3 e) or another co-stimulatory pathway like TLR4 by LPS stimulation (Fig. S3 b). This observation implies that, through Fgl2, TFR cells might specifically target autoreactive B cells due to their lack of help from cognate TFH cells. Moreover, dysregulation in co-stimulatory pathways such as TLR pathways that leads to autoreactive B cell dysregulation (59, 60) might be partially due to their resistance to TFR suppression.
The presence of autoantibodies against lupus-associated autoantigens in aged Fgl2−/− mice lead to the hypothesis that Fgl2 may be a relevant molecule in lupus and/or other humoral autoimmune diseases like Sjogren’s syndrome. In fact, mice with Fcgr2b deficiency develop a spontaneous lupus-like disease with high titer autoantibodies and B cells from the mice can lead to the development of lupus-like disease (61). Furthermore, polymorphisms in both low-affinity Fc receptors known to bind to Fgl2 have been shown to be associated with susceptibility to SLE in humans (62–66). These studies not only underscore the role of these Fc receptors but may also suggest that the effects are partly due to loss of Fgl2 signaling.
Fgl2−/− mice have been reported previously to develop glomerulonephritis, which we did not observe in our cohort (31). However, we were able to see spontaneous germinal center cell phenotype in spleens in a small portion of the observed group. This suggests that additional environmental factors, including possible variation in the microbiome in our housing facility, likely modulate disease. As we hypothesize that the phenotype observed with age were due to the accumulated insults over the lifespan, the variability in the frequency and the underlying natures of such insults, like host-microbiome interaction, might result in manifestation of different clinical phenotypes in Fgl2-deficient mice.
Taken together, our work uncovered Fgl2 as a TFR cell effector molecule that directly acts on B cells and TFH cells. This finding provides a novel path for targeting antibody responses by modulating TFR effector function, which could potentially be useful for vaccine development and, through supplementation, for treating systemic autoimmune diseases.
Supplementary Material
Key points.
Fgl2 is a soluble effector molecule highly expressed by TFR cells.
Fgl2 directly binds to B cells and TFH cells regulating humoral responses.
Fgl2-deficient mice spontaneously develop an Inflammatory, Lupus-like Autoimmunity.
Acknowledgements
We thank Deneen Kozoriz (Evergrande Center) for cell sorting. We are thankful to Mary Collins for advice and editing of the manuscript. We also thank Michelle Weinstein for editing of the manuscript.
This research was funded by the following grants: NIH-NIAID National Institute of Allergy and Infectious Diseases (5P01AI073748-09, 5P01AI39671, 2P01AI56299, 5P01AI129880) and William E. Paul Distinguished Innovator award from the Lupus Research Alliance (594584) supports the work performed in the Kuchroo Lab. C.W. is supported by a Career Transition Fellowship from National Multiple Sclerosis Society (TA-1605-08590).
Footnotes
Conflict of Interest
V.K.K is a member of the scientific advisory boards for Compass Therapeutics, Tizona Therapeutics and Bicara Therapeutics, which have interests in cancer immunotherapy. V.K.K. has an ownership interest in and is a member of the scientific advisory board for Tizona Therapeutics and Bicara Therapeutics. V.K.K. is a co- founder of and has an ownership interest in Celsius Therapeutics. V.K.K.’s interests were reviewed and managed by the Brigham and Women’s Hospital and Partners Healthcare in accordance with their conflict of interest policies. V.K.K. is a named inventor on patents related to TIM3 and TIGIT.
Follicular regulator T
Fibrinogen-like protein 2
T follicular helper cells
PR domain zinc finger protein 1
Programmed cell death protein 1
T-cell immunoglobin mucin-3
Lymphocyte-activation gene 3
T cell immunoreceptor with Ig and ITM domains
Germinal center
B-cell lymphoma 6 protein
Cellular musculoaponeurotic fibrosarcoma proto-oncogene
Achaete-scute complex homolog 2
Regulatory t cell
B lymphocyte-induced mutation protein-1 dependent manner
Glucocorticoid-induced TFNR family related gene
Cytotoxic T-lymphocyte associated protein 4
Nitrophenyl hapten conjugated to chicken Ovalbumin
t-Distributed stochastic neighbor embedding
Quantitative polymerase chain reaction
Light-zone
Fc gamma receptor 2b
Fc gamma receptor 3
Immunoglobin M
Cell class-switch recombination
Class switch recombination
Interferon Gamma
Peyer’s Patches
Enzyme-linked immune absorbent spot
Antibody-secreting cells
Anti-nuclear antibodies
Small nuclear Ribo nuclear proteins
Hepatitis A virus Cellular Receptor 2
References
- 1.De Silva NS, and Klein U. 2015. Dynamics of B cells in germinal centres. Nature reviews. Immunology 15: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Victora GD, and Nussenzweig MC. 2012. Germinal centers. Annual review of immunology 30: 429–457. [DOI] [PubMed] [Google Scholar]
- 3.Crotty S 2011. Follicular helper CD4 T cells (TFH). Annual review of immunology 29: 621–663. [DOI] [PubMed] [Google Scholar]
- 4.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, and Moser B. 2000. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med 192: 1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, and Butcher EC. 2001. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med 193: 1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, and Forster R. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med 192: 1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, and Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science (New York, N.Y.) 325: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, and Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science (New York, N.Y.) 325: 1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, and Vinuesa CG. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31: 457–468. [DOI] [PubMed] [Google Scholar]
- 10.Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, and Kuchroo VK. 2009. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol 10: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, Nakatsukasa H, Neelapu SS, Chen W, Clevers H, Tian Q, Qi H, Wei L, and Dong C. 2014. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature 507: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAdam AJ, Greenwald RJ, Levin MA, Chernova T, Malenkovich N, Ling V, Freeman GJ, and Sharpe AH. 2001. ICOS is critical for CD40-mediated antibody class switching. Nature 409: 102–105. [DOI] [PubMed] [Google Scholar]
- 13.Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC 3rd, Liu C, Schwartzberg PL, and Leonard WJ. 2002. A critical role for IL-21 in regulating immunoglobulin production. Science (New York, N.Y.) 298: 1630–1634. [DOI] [PubMed] [Google Scholar]
- 14.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, and Dong C. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinhardt RL, Liang HE, and Locksley RM. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol 10: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, and Crotty S. 2010. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J Immunol 185: 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, and Crotty S. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34: 932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu H, Li X, Liu D, Li J, Zhang X, Chen X, Hou S, Peng L, Xu C, Liu W, Zhang L, and Qi H. 2013. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature 496: 523–527. [DOI] [PubMed] [Google Scholar]
- 19.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, Fagarasan S, Liston A, Smith KG, and Vinuesa CG. 2011. Foxp3+ follicular regulatory T cells control the germinal center response. Nature medicine 17: 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, and Dong C. 2011. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nature medicine 17: 983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wollenberg I, Agua-Doce A, Hernandez A, Almeida C, Oliveira VG, Faro J, and Graca L. 2011. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J Immunol 187: 4553–4560. [DOI] [PubMed] [Google Scholar]
- 22.Botta D, Fuller MJ, Marquez-Lago TT, Bachus H, Bradley JE, Weinmann AS, Zajac AJ, Randall TD, Lund FE, Leon B, and Ballesteros-Tato A. 2017. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat Immunol 18: 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu W, Liu X, Lin X, Feng H, Sun L, Li S, Chen H, Tang H, Lu L, Jin W, and Dong C. 2018. Deficiency in T follicular regulatory cells promotes autoimmunity. The Journal of Experimental Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, and Calame K. 2006. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol 7: 457–465. [DOI] [PubMed] [Google Scholar]
- 25.Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, and Kallies A. 2011. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol 12: 304–311. [DOI] [PubMed] [Google Scholar]
- 26.Cretney E, Kallies A, and Nutt SL. 2013. Differentiation and function of Foxp3(+) effector regulatory T cells. Trends in immunology 34: 74–80. [DOI] [PubMed] [Google Scholar]
- 27.Sage PT, Ron-Harel N, Juneja VR, Sen DR, Maleri S, Sungnak W, Kuchroo VK, Haining WN, Chevrier N, Haigis M, and Sharpe AH. 2016. Suppression by TFR cells leads to durable and selective inhibition of B cell effector function. Nat Immunol 17: 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sage PT, Paterson AM, Lovitch SB, and Sharpe AH. 2014. The coinhibitory receptor CTLA-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity 41: 1026–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wing JB, Ise W, Kurosaki T, and Sakaguchi S. 2014. Regulatory T cells control antigen-specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA-4. Immunity 41: 1013–1025. [DOI] [PubMed] [Google Scholar]
- 30.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, and Kuchroo VK. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441: 235–238. [DOI] [PubMed] [Google Scholar]
- 31.Shalev I, Liu H, Koscik C, Bartczak A, Javadi M, Wong KM, Maknojia A, He W, Liu MF, Diao J, Winter E, Manuel J, McCarthy D, Cattral M, Gommerman J, Clark DA, Phillips MJ, Gorczynski RR, Zhang L, Downey G, Grant D, Cybulsky MI, and Levy G. 2008. Targeted deletion of fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. J Immunol 180: 249–260. [DOI] [PubMed] [Google Scholar]
- 32.Sage PT, Francisco LM, Carman CV, and Sharpe AH. 2013. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol 14: 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, and Regev A. 2014. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Current protocols in molecular biology 107: 4.22.21-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, Kuchroo VK, Park H, and Regev A. 2015. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell 163: 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sage PT, and Sharpe AH. 2015. In vitro assay to sensitively measure T(FR) suppressive capacity and T(FH) stimulation of B cell responses. Methods Mol Biol 1291: 151–160. [DOI] [PubMed] [Google Scholar]
- 36.Sage PT, Alvarez D, Godec J, von Andrian UH, and Sharpe AH. 2014. Circulating T follicular regulatory and helper cells have memory-like properties. J Clin Invest 124: 5191–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintana FJ, Hagedorn PH, Elizur G, Merbl Y, Domany E, and Cohen IR. 2004. Functional immunomics: microarray analysis of IgG autoantibody repertoires predicts the future response of mice to induced diabetes. Proc Natl Acad Sci U S A 101 Suppl 2: 14615–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li QZ, Xie C, Wu T, Mackay M, Aranow C, Putterman C, and Mohan C. 2005. Identification of autoantibody clusters that best predict lupus disease activity using glomerular proteome arrays. J Clin Invest 115: 3428–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu HY, Quintana FJ, and Weiner HL. 2008. Nasal anti-CD3 antibody ameliorates lupus by inducing an IL-10-secreting CD4+ CD25− LAP+ regulatory T cell and is associated with down-regulation of IL-17+ CD4+ ICOS+ CXCR5+ follicular helper T cells. J Immunol 181: 6038–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao Y, Smyth GK, and Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner A, Regev A, and Yosef N. 2016. Revealing the vectors of cellular identity with single-cell genomics. Nature Biotechnology 34: 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waltman L, and van Eck NJ. 2013. A smart local moving algorithm for large-scale modularity-based community detection. [Google Scholar]
- 46.Cho SH, Raybuck AL, Blagih J, Kemboi E, Haase VH, Jones RG, and Boothby MR. 2019. Hypoxia-inducible factors in CD4(+) T cells promote metabolism, switch cytokine secretion, and T cell help in humoral immunity. Proceedings of the National Academy of Sciences of the United States of America 116: 8975–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, and Pan F. 2011. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 146: 772–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V, Sharpe AH, Quintana FJ, Mathis D, Benoist C, Hafler DA, and Kuchroo VK. 2014. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity 40: 569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H, Shalev I, Manuel J, He W, Leung E, Crookshank J, Liu MF, Diao J, Cattral M, Clark DA, Isenman DE, Gorczynski RM, Grant DR, Zhang L, Phillips MJ, Cybulsky MI, and Levy GA. 2008. The FGL2-FcgammaRIIB pathway: a novel mechanism leading to immunosuppression. Eur J Immunol 38: 3114–3126. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, Zhang L, Cybulsky M, Gorczynski R, Crookshank J, Manuel J, Grant D, and Levy G. 2006. Identification of the receptor for FGL2 and implications for susceptibility to mouse hepatitis virus (MHV-3)-induced fulminant hepatitis. Adv Exp Med Biol 581: 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, and Nussenzweig MC. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143: 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, and Wherry EJ. 2009. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity 31: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, Nyman J, Marjanovic ND, Kowalczyk MS, Wang C, Kurtulus S, Law T, Etminan Y, Nevin J, Buckley CD, Burkett PR, Buenrostro JD, Rozenblatt-Rosen O, Anderson AC, Regev A, and Kuchroo VK. 2018. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558: 454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zemmour D, Zilionis R, Kiner E, Klein AM, Mathis D, and Benoist C. 2018. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol 19: 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gayden T, Sepulveda FE, Khuong-Quang D-A, Pratt J, Valera ET, Garrigue A, Kelso S, Sicheri F, Mikael LG, Hamel N, Bajic A, Dali R, Deshmukh S, Dervovic D, Schramek D, Guerin F, Taipale M, Nikbakht H, Majewski J, Moshous D, Charlebois J, Abish S, Bole-Feysot C, Nitschke P, Bader-Meunier B, Mitchell D, Thieblemont C, Battistella M, Gravel S, Nguyen V-H, Conyers R, Diana J-S, McCormack C, Prince HM, Besnard M, Blanche S, Ekert PG, Fraitag S, Foulkes WD, Fischer A, Neven B, Michonneau D, de Saint Basile G, and Jabado N. 2018. Germline HAVCR2 mutations altering TIM-3 characterize subcutaneous panniculitis-like T cell lymphomas with hemophagocytic lymphohistiocytic syndrome. Nature Genetics. [DOI] [PubMed] [Google Scholar]
- 56.Godfrey VL, Wilkinson JE, Rinchik EM, and Russell LB. 1991. Fatal lymphoreticular disease in the scurfy (sf) mouse requires T cells that mature in a sf thymic environment: potential model for thymic education. Proc Natl Acad Sci U S A 88: 5528–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hadaschik EN, Wei X, Leiss H, Heckmann B, Niederreiter B, Steiner G, Ulrich W, Enk AH, Smolen JS, and Stummvoll GH. 2015. Regulatory T cell-deficient scurfy mice develop systemic autoimmune features resembling lupus-like disease. Arthritis Research & Therapy 17: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maceiras AR, Almeida SCP, Mariotti-Ferrandiz E, Chaara W, Jebbawi F, Six A, Hori S, Klatzmann D, Faro J, and Graca L. 2017. T follicular helper and T follicular regulatory cells have different TCR specificity. Nature communications 8: 15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B, Duramad O, and Coffman RL. 2005. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med 202: 1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, and Shlomchik MJ. 2006. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity 25: 417–428. [DOI] [PubMed] [Google Scholar]
- 61.Bolland S, and Ravetch JV. 2000. Spontaneous autoimmune disease in Fc(gamma)RIIB-deficient mice results from strain-specific epistasis. Immunity 13: 277–285. [DOI] [PubMed] [Google Scholar]
- 62.Willcocks LC, Carr EJ, Niederer HA, Rayner TF, Williams TN, Yang W, Scott JA, Urban BC, Peshu N, Vyse TJ, Lau YL, Lyons PA, and Smith KG. 2010. A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc Natl Acad Sci U S A 107: 7881–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen JY, Wang CM, Ma CC, Luo SF, Edberg JC, Kimberly RP, and Wu J. 2006. Association of a transmembrane polymorphism of Fcgamma receptor IIb (FCGR2B) with systemic lupus erythematosus in Taiwanese patients. Arthritis and rheumatism 54: 3908–3917. [DOI] [PubMed] [Google Scholar]
- 64.Chu ZT, Tsuchiya N, Kyogoku C, Ohashi J, Qian YP, Xu SB, Mao CZ, Chu JY, and Tokunaga K. 2004. Association of Fcgamma receptor IIb polymorphism with susceptibility to systemic lupus erythematosus in Chinese: a common susceptibility gene in the Asian populations. Tissue antigens 63: 21–27. [DOI] [PubMed] [Google Scholar]
- 65.Kyogoku C, Dijstelbloem HM, Tsuchiya N, Hatta Y, Kato H, Yamaguchi A, Fukazawa T, Jansen MD, Hashimoto H, van de Winkel JG, Kallenberg CG, and Tokunaga K. 2002. Fcgamma receptor gene polymorphisms in Japanese patients with systemic lupus erythematosus: contribution of FCGR2B to genetic susceptibility. Arthritis and rheumatism 46: 1242–1254. [DOI] [PubMed] [Google Scholar]
- 66.Siriboonrit U, Tsuchiya N, Sirikong M, Kyogoku C, Bejrachandra S, Suthipinittharm P, Luangtrakool K, Srinak D, Thongpradit R, Fujiwara K, Chandanayingyong D, and Tokunaga K. 2003. Association of Fcgamma receptor IIb and IIIb polymorphisms with susceptibility to systemic lupus erythematosus in Thais. Tissue antigens 61: 374–383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.