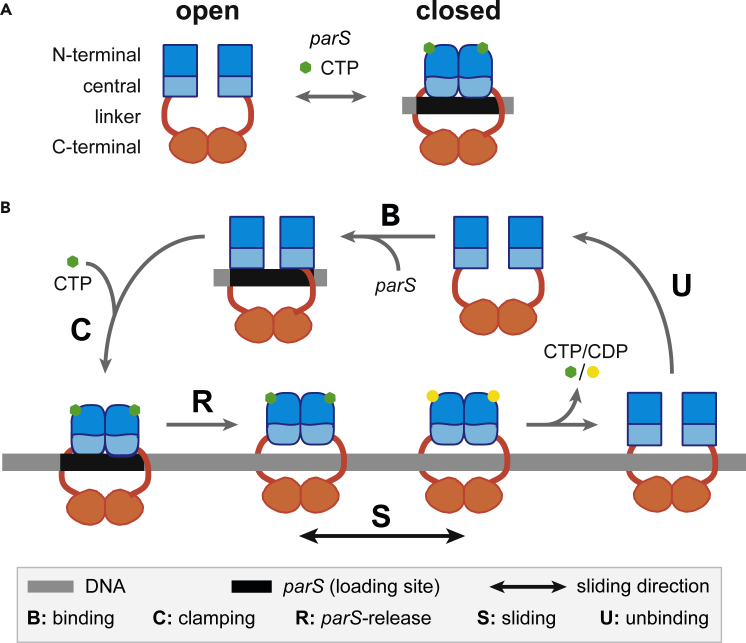
Figure 1.
Schematic Cycle for ParB Clamping and Sliding By Diffusion Along DNA
(A) Open and closed conformations mediated by CTP and parS DNA. ParB is a homodimer composed of a C-terminal dimerization domain (orange) linked to the central (light blue) and N-terminal (dark blue) domains by a flexible linker (red). The central domain contains the two DNA binding motifs for parS binding (Sanchez et al., 2013). The N-terminal part contains the ParA interaction domain, the arginine-like motif, the CTP binding motif, and the multimerization domain (Ah-Seng et al., 2009; Soh et al., 2019; Surtees and Funnell, 1999). In the presence of parS and CTP, ParB dimer forms a clamp around the DNA.
(B) Schematic representation for the “clamping and sliding” model displaying the five key steps. The open conformation of ParB dimer enables DNA binding. Upon specific binding to parS centromere (step B), ParB undergoes a conformational change promoting CTP binding which subsequently induces ParB to form a clamp around parS (step C). Clamping promotes its release from parS allowing the ParB clamp to slide away from parS by diffusion (step S) and to free the parS loading site for the next round of loading. The parameters used in the physical modeling B, R, D, and U correspond to the parS binding (B), ParB clamping (C) and release (R), free diffusion during sliding (S), and DNA unbinding (U) steps, respectively. Note that (I) R is the total release rate from parS which is, to a first approximation, equal on both sides of parS (not represented on the schematic representation for simplicity); therefore, ParB clamps are loaded on each side at a rate R/2, and (ii) the stage at which CTP hydrolysis occurs was not determined in the original clamping models (Jalal et al., 2020; Soh et al., 2019).