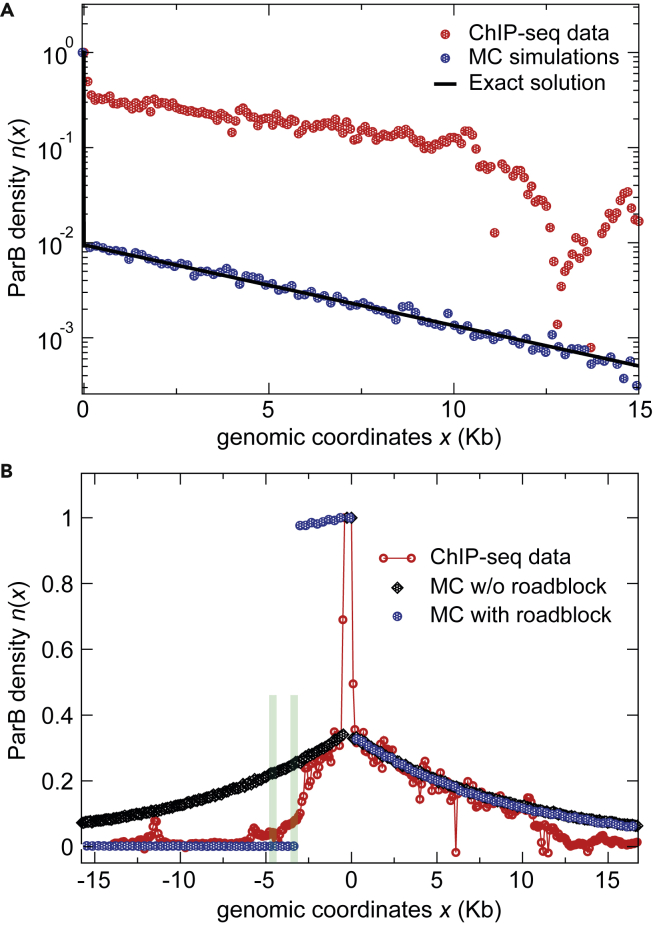
Figure 2.
Comparisons between the “Clamping and Sliding” Model and the ParBF DNA Binding Pattern
High-resolution ChIP-sequencing data from a previous study (Sanchez et al., 2015) display the average number of reads per 100-bp windows as a function of the genomic coordinates (red circles). Note that the signal at parSF (coordinate 0) is normalized to 1 by averaging the number of reads over the 550-bp centromere locus.
(A) The model from Equation (2) contains the three kinetic parameters D = 4.3105 bp2 s−1 (Guilhas et al., 2020), R = 0.1 s−1, and U = 0.017 s−1 (Figures S1B–S1D). The exact solution (black line) and the Monte Carlo (MC; blue dots) simulation data are plotted on the right side of parSF. The theoretical prediction is compared to the ChIP-seq data (Sanchez et al., 2015). The large discrepancy between the model and the ChIP-seq data is essentially due to the amplitude (value of R).
(B) Simulation in the absence (black diamonds) and in the presence (blue circles) of a roadblock compared to the ChIP-seq data with the optimized parameters (R = 1.91 s−1 and U = 0.0047 s−1). The two strong roadblocks (green bars), present at ∼3-kb and 4.5-kb on the left of parSF, are due to the binding of RepE to incC and ori2 iterons, respectively (Rodionov et al., 1999; Sanchez et al., 2015). Note that the two MC simulations overlap on the right side of parSF.
See also Figure S2.