Abstract
The skin’s physical barrier is reinforced by an arsenal of immune cells that actively patrol the tissue and respond swiftly to penetrating microbes, noxious agents, and injurious stimuli. When unchecked, these same immune cells drive diseases such as psoriasis, atopic dermatitis, and alopecia. Rapidly-advancing microscopy, animal modeling, genomic, and computational technologies have illuminated the complexity of the cutaneous immune cells and their functions in maintaining skin health and driving disease. Here we discuss the recent technology-driven breakthroughs that have transformed our understanding of skin immunity and highlight burgeoning areas that hold great promise for future discoveries.
Introduction
Owing to its exteriority, the skin has captivated human imagination since ancient Roman and Egyptian civilizations. Yet, modern day experimental dermatology and immunology did not take root till the late 19th and 20th centuries, respectively. In parallel, experimental dermatologists and immunologists studied contact hypersensitivity, graft rejection and histocompatibility, and adjuvant responses (Chase, 1985). These foundational works revealed the immune underpinnings of skin diseases, while still viewing the skin as an epithelial barrier that recruited immune allies only under duress.
Advancing technologies illuminated the myriad of immune cells that reside in and continually patrol the skin, shifting the view that the skin is simply an inert barrier (Kobayashi et al., 2019a). Inspired by the 2019 Montagna Biology of Skin Symposium, we discuss the remarkable discoveries in skin immunity that have resulted from imaging, tissue processing, and genomic techniques (Figure 1). We also highlight the importance of these tools to understanding immune dysfunction in inflammatory skin disease. Finally, we explore emerging technologies and their potential for further expanding knowledge of skin immunity.
Figure 1:
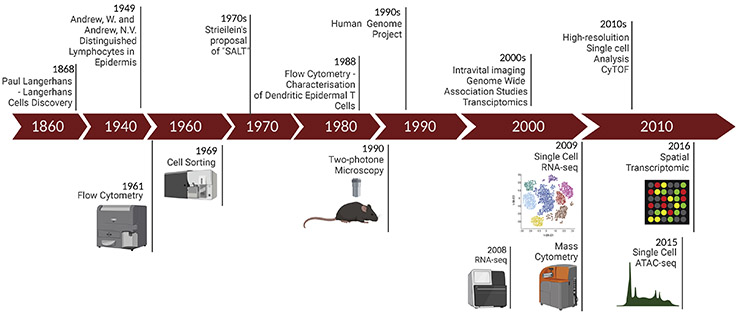
Timeline of the technological advances, their implementation, and seminal discoveries in skin immunity. The top row summarizes seminal discoveries and implemented technologies, and the bottom row presents innovations and their precise year of development.
Seeing is believing
In 1868 Paul Langerhans, enabled by rudimentary light microscopy, uncovered cells with dendrites in the epidermis (Langerhans, 1868), which he concluded were epidermal nerves. His discovery of Langerhans cells was the first known observation of immune cells in normal skin. In 1949 Andrews and Andrews, distinguished lymphocytes in normal epidermis using light microscopy (Andrew and Andrew, 1949). A few decades later, Streilein built upon these and other works to propose that the skin had a dedicated immune component, which he termed “Skin Associated Lymphoid Tissue (SALT)” (Streilein, 1978).
Since then, sophisticated imaging techniques, most notably fluorescence microscopy, have been widely used to illuminate the immune microanatomy of the skin (Kabashima et al., 2019). Fluorescence microscopy enabled the simultaneous visualization of multiple cells types and their expressed factors at higher resolution than simple light microscopy (Sanderson et al., 2014). Initially used to detect epidermal-resident dendritic epidermal T cells (DETCs) and Langerhans cells (LCs) (Havran and Allison, 1990, Kissenpfennig et al., 2005, Steiner et al., 1988), microscopic analyses have revealed the tightly controlled spatial distribution of the myriad of immune cells in the skin (Kabashima et al., 2019). LCs and intraepithelial lymphocytes (DETCs (Havran and Allison, 1990), CD8+ resident memory T cells (TRM) (Schenkel and Masopust, 2014)) and innate lymphoid cells (ILCs) (Kobayashi et al., 2019b) capable of traversing the basement membrane reside in the epidermis. The upper dermis houses several dendritic cell (DC) subsets (Tamoutounour et al., 2013), γδT cells (Gray et al., 2011), CD4 T helper (Adachi et al., 2015) and regulatory T cells (Ali et al., 2017), and ILCs (Kobayashi et al., 2019b). These cells are enriched around hair follicles (Adachi et al., 2015), highlighting this region is a key immunological hub in skin. The lower dermis houses various macrophage subsets in close apposition to vasculature, nerves and adipocytes (Silva et al., 2019). In addition to immune localization, dynamic imaging has divulged immune surveillance function in normal skin. Images of LCs extending their dendrites through the epidermis captured their homeostatic uptake of external antigens (Ouchi et al., 2011). LCs and other DCs migrate to the lymph nodes to induce T effector and regulatory cells and/or provide homeostatic signals to maintain these populations in normal skin (Naik et al., 2015, Seneschal et al., 2012).
Pioneered in 1990, multiphoton microscopy, allowed for deeper tissue penetration and opened the door to intravital imaging (Denk et al., 1990). Coupled with the generation of fluorescence reporter animals, multiphoton imaging was used to live image immune cells (Kabashima and Egawa, 2014). This enabled 3D reconstruction of immune niches, and revealed the interaction of leukocytes with the skin’s structural components (Kabashima and Egawa, 2014). Intravital imaging is also a powerful tool to visualize the induction, and propagation of inflammatory responses (Obeidy et al., 2018). A key feature of inflammatory responses in the skin is a compromised barrier, particularly in the case of infectious agents or tissue injury. Live imaging identified neutrophils as “first responders”, infiltrating within hours of epidermal breach (Obeidy et al., 2018, Peters et al., 2008) and the kinetics of DC migration to the lymph nodes under stress, illustrating that functionally specialized DC subsets migrate with specific kinetics to induce adaptive responses (Tamoutounour et al., 2013).
Quantitative imaging combined with pathways specific modulation of cell-cell interactions, cell-extracellular matrix (ECM) interactions, or motility has unearthed therapeutic targets in inflammation (Matheu et al., 2008, Overstreet et al., 2013). For instance, perivascular lymphocytes and DCs form clusters in an interleukin (IL)-1R-dependent manner to drive contact dermatitis (Natsuaki et al., 2014). Imaging studies provided insight into how innate and adaptive immune cells control skin tumors. To this end, a role for CD8+ TRMs and innate immune cells in restraining melanoma and epithelial neoplasms has been identified (Caulin et al., 2007, Park et al., 2019). Thus, imaging techniques have provided invaluable insights into the location, migration, interactions, and functions of immune cells in skin health and disease.
Cytometry and Genomic technologies widen the lens
Perhaps the most underappreciated and widely-used methodology in skin immunology, is the ability to efficiently extract viable cells from the skin while preserving expression of surface proteins for phenotypic analysis. This was first accomplished by employing a serine protease, trypsin, to digest ECM and sever cell-cell interactions to obtain DETCs and LCs cells (Havran and Allison, 1990, Steiner et al., 1988). Since then, sophisticated enzymes with minimal non-specific activity have become available, and are used to prepare cell suspensions for a number of downstream analysis platforms (Botting et al., 2017, Clark et al., 2006).
Flow cytometry has been the cornerstone of immunology for many decades and is a ubiquitously used to analyze cells from healthy and diseased skin (Adan et al., 2017). Antibodies raised to specific protein moieties (surface markers, cytokines, transcription factors and signaling components) are coupled with fluorescent indicators and have empowered researchers to examine multiple cellular parameters simultaneously and quantitatively. In 1969, Herzenberg published a new technique to obtain highly purified cell populations called fluorescence activated cell sorting (FACS) (Hulett et al., 1969). Cells purified from directly from the skin with FACS have been used for functional in vitro studies (Seneschal et al., 2012), in vivo cell transfer experiments (Schenkel and Masopust, 2014), and downstream tissue and cell-specific genomic analysis (Cheng et al., 2018).
However, fluorescent indicators have restricted analysis to the visible-light and ultraviolet (UV) spectrum and limited the number of parameters that could be measured simultaneously. In 2009, Tanner and colleagues overcame these limitations by developing mass cytometry (CyTOF) (Bandura et al., 2009). CyTOF blends flow cytometry with mass spectrometry, using metal-conjugated antibodies to dramatically increase the number of analytes from as few as 10,000 cells, enabling efficient analysis of small patient samples (Bandura et al., 2009, Yao et al., 2014). Multiparametric CyTOF analysis of normal skin and in inflammatory disease, revealed a remarkable intraindividual heterogeneity in homeostatic DC populations and highly polarizing impact of inflammatory diseases on immune subsets (Alcantara-Hernandez et al., 2017, Farrera et al., 2020).
Genome-based analysis has radically transformed our understanding of skin immunity. Spurred by the human genome project (Collins et al., 2003), the ability to sequence and compile whole human genomes uncovered genetic susceptibility loci underlying a number of complex inflammatory skin diseases (Paternoster et al., 2011, Tsoi et al., 2017). These studies provided key insights into the molecular and cellular drivers of complex multifactorial diseases. For instance, genome wide association studies (GWAS) of Alopecia areata were instrumental in identifying the key innate and adaptive immune drivers of hair follicle destruction (Petukhova et al., 2010). Similarly, the IL23 and NF-κB immune pathways were linked to psoriasis with GWAS (Nair et al., 2009).
Microarray technology and, more recently, RNA sequencing has provided a global picture of gene expression from skin tissue and purified immune cells (Li et al ., 2014, Nirschl et al., 2017). Transcriptional analysis has also been instrumentational in revealing the unique, universal, and synergistic cellular programs induced by inflammatory cytokines (Mehta et al., 2017, Swindell et al., 2018). Mechanistic studies using cell culture systems and animal models have defined the causal contributions factors identified by GWAS and transcriptional studies (Billi et al., 2020, Hawkes et al., 2017) paving the way for development of targeted therapeutics.
The power of evaluating the gene expression of a single cell was harnessed by next generation sequencing platforms to evaluate transcriptomes at cellular resolution (Tang et al., 2009). There has since been an explosion in the use of single cell RNA sequencing (scRNAseq) by skin immunologist to study cellular heterogeneity (Shook et al., 2018), identify rare cell populations (Kobayashi et al., 2019b) and map the developmental (Popescu et al., 2019) and functional trajectories of distinct cell lineages (Tan et al., 2019). Comparing immune cells in psoriasis, atopic dermatitis, vitiligo, and bullous skin disease (Cheng et al., 2018, Travis K Hughes, 2019) has revealed heterogeneity not only in immune cells but also in the functionally responsive stromal cells that they engage. While scRNAseq has been instrumental in mapping the cellular ecology of cutaneous immunity, just as the genomic techniques that came before, functional followup studies will be essential to determine causality and meaningful cellular interactions. Perhaps the most exciting application of scRNAseq is its use in rapid diagnosis, particularly in diseases that lack a clear mechanism. Nagao and colleagues recently used scRNAseq to effectively diagnose and treat a patient with Drug-induced hypersensitivity syndrome, a disease with an elusive pathophysiology (Kim et al., 2020)
Emerging technology and future promise
Many emerging technologies are melding methods to evaluate multiple modalities in the same sample. For instance, cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) (Stoeckius et al., 2017) combines antibody-based protein detection with scRNAseq allowing for simultaneous evaluation of gene transcript and its protein product within a single cell. Similarly, concurrent assessment of epigenetics state, including chromatin accessibility, DNA and histone modifications and 3D chromatin structure, and the transcriptional landscape of a single cell may provide a more nuanced understanding of regulatory genomic elements that underlie distinct cell states (Jia et al., 2018). One of the most exciting techniques on the horizon is spatial transcriptomics (Moncada et al., 2020), a method that provides gene expression coupled with spatial distribution in a tissue. Spatial transcriptomics will be particularly useful to evaluate microanatomical heterogeneity in disease, for instance the tumor-stromal interface, or the edge and bed of a non-healing wound. Widely implementing these technologies will undoubtedly require tremendous computational power and the use of machine learning. An added challenge posed by these techniques is the integration of large data sets and dissemination for downstream functional validation. Nevertheless, these advances present a tantalizing toolbox with which cutaneous biologists can compose rich portraits skin immune health and disease.
Acknowledgements
We thank our friends and colleagues who work has inspired this perspective; in particular, the speakers of the 2019 Montagna Biology of Skin Symposium. We apologize, due to space constraints we were not able to include all relevant papers published in skin immunology, which are comprehensively reviewed by Kobayashi and colleagues(Kobayashi et al., 2019a). Our figure was made using Biorender (www.biorender.com). This work was supported by NIAID (1K22AI135099-01 S.N.).
Footnotes
Conflict of Interest
The authors state no conflict of interest
Data Availability Statement
No datasets were generated or analyzed during the current study
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, et al. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat Med 2015;21(11): 1272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adan A, Alizada G, Kiraz Y, Baran Y, Nalbant A. Flow cytometry: basic principles and applications. Crit Rev Biotechnol 2017;37(2):163–76. [DOI] [PubMed] [Google Scholar]
- Alcantara-Hernandez M, Leylek R, Wagar LE, Engleman EG, Keler T, Marinkovich MP, et al. High-Dimensional Phenotypic Mapping of Human Dendritic Cells Reveals Interindividual Variation and Tissue Specialization. Immunity 2017;47(6):1037–50e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong HA, Lai K, et al. Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 2017;169(6):1119–29 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew W, Andrew NV. Lymphocytes in the normal epidermis of the rat and of man. Anat Rec 1949;104(2):217–41. [DOI] [PubMed] [Google Scholar]
- Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem 2009;81(16):6813–22. [DOI] [PubMed] [Google Scholar]
- Billi AC, Ludwig JE, Fritz Y, Rozic R, Swindell WR, Tsoi LC, et al. KLK6 expression in skin induces PAR1-mediated psoriasiform dermatitis and inflammatory joint disease. J Clin Invest 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botting RA, Bertram KM, Baharlou H, Sandgren KJ, Fletcher J, Rhodes JW, et al. Phenotypic and functional consequences of different isolation protocols on skin mononuclear phagocytes. J Leukoc Biol 2017;101(6):1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin C, Nguyen T, Lang GA, Goepfert TM, Brinkley BR, Cai WW, et al. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J Clin Invest 2007;117(7):1893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW. Immunology and experimental dermatology. Annu Rev Immunol 1985;3:1–29. [DOI] [PubMed] [Google Scholar]
- Cheng JB, Sedgewick AJ, Finnegan AI, Harirchian P, Lee J, Kwon S, et al. Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep 2018;25(4):871–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RA, Chong BF, Mirchandani N, Yamanaka K, Murphy GF, Dowgiert RK, et al. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J Invest Dermatol 2006;126(5):1059–70. [DOI] [PubMed] [Google Scholar]
- Collins FS, Green ED, Guttmacher AE, Guyer MS, Institute USNHGR. A vision for the future of genomics research. Nature 2003;422(6934):835–47. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science 1990;248(4951):73–6. [DOI] [PubMed] [Google Scholar]
- Farrera C, Melchiotti R, Petrov N, Weng Teng KW, Wong MT, Loh CY, et al. T-cell phenotyping uncovers systemic features of atopic dermatitis and psoriasis. J Allergy Clin Immunol 2020;145(3):1021–5 e15. [DOI] [PubMed] [Google Scholar]
- Gray EE, Suzuki K, Cyster JG. Cutting edge: Identification of a motile IL-17-producing gammadelta T cell population in the dermis. J Immunol 2011. ;186(11 ):6091–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havran WL, Allison JP. Origin of Thy-1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature 1990;344(6261):68–70. [DOI] [PubMed] [Google Scholar]
- Hawkes JE, Gudjonsson JE, Ward NL. The Snowballing Literature on Imiquimod-Induced Skin Inflammation in Mice: A Critical Appraisal. J Invest Dermatol 2017;137(3):546–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett HR, Bonner WA, Barrett J, Herzenberg LA. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science 1969; 166(3906):747–9. [DOI] [PubMed] [Google Scholar]
- Jia G, Preussner J, Chen X, Guenther S, Yuan X, Yekelchyk M, et al. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun 2018;9(1):4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K, Egawa G. Intravital multiphoton imaging of cutaneous immune responses. J Invest Dermatol 2014;134(11):2680–4. [DOI] [PubMed] [Google Scholar]
- Kabashima K, Honda T, Ginhoux F, Egawa G. The immunological anatomy of the skin. Nat Rev Immunol 2019;19(1):19–30. [DOI] [PubMed] [Google Scholar]
- Kim D, Kobayashi T, Voisin B, Jo JH, Sakamoto K, Jin SP, et al. Targeted therapy guided by single-cell transcriptomic analysis in drug-induced hypersensitivity syndrome: a case report. Nat Med 2020;26(2):236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissenpfennig A, Henri S, Dubois B, Laplace-Builhe C, Perrin P, Romani N, et al. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity 2005;22(5):643–54. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Naik S, Nagao K. Choreographing Immunity in the Skin Epithelial Barrier. Immunity 2019a;50(3):552–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Voisin B, Kim DY, Kennedy EA, Jo JH, Shih HY, et al. Homeostatic Control of Sebaceous Glands by Innate Lymphoid Cells Regulates Commensal Bacteria Equilibrium. Cell 2019b;176(5):982–97 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerhans P Über die Nerven der menschlichen Haut. Archiv für pathologische Anatomie und Physiologie und für klinische Medicin 1868;44:325–37. [Google Scholar]
- Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol 2014;134(7):1828–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu MP, Beeton C, Garcia A, Chi V, Rangaraju S, Safrina O, et al. Imaging of effector memory T cells during a delayed-type hypersensitivity reaction and suppression by Kv1.3 channel block. Immunity 2008;29(4):602–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta NN, Teague HL, Swindell WR, Baumer Y, Ward NL, Xing X, et al. IFN-gamma and TNF-alpha synergism may provide a link between psoriasis and inflammatory atherogenesis. Sci Rep 2017;7(1): 13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol 2020. [DOI] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 2015;520(7545):104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet 2009;41(2):199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsuaki Y, Egawa G, Nakamizo S, Ono S, Hanakawa S, Okada T, et al. Perivascular leukocyte clusters are essential for efficient activation of effector T cells in the skin. Nat Immunol 2014;15(11):1064–9. [DOI] [PubMed] [Google Scholar]
- Nirschl CJ, Suarez-Farinas M, Izar B, Prakadan S, Dannenfelser R, Tirosh I, et al. IFNgamma-Dependent Tissue-Immune Homeostasis Is Co-opted in the Tumor Microenvironment. Cell 2017;170(1):127–41 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeidy P, Tong PL, Weninger W. Research Techniques Made Simple: Two-Photon Intravital Imaging of the Skin. J Invest Dermatol 2018;138(4):720–5. [DOI] [PubMed] [Google Scholar]
- Ouchi T, Kubo A, Yokouchi M, Adachi T, Kobayashi T, Kitashima DY, et al. Langerhans cell antigen capture through tight junctions confers preemptive immunity in experimental staphylococcal scalded skin syndrome. J Exp Med 2011;208(13):2607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet MG, Gaylo A, Angermann BR, Hughson A, Hyun YM, Lambert K, et al. Inflammation-induced interstitial migration of effector CD4(+) T cells is dependent on integrin alphaV. Nat Immunol 2013;14(9):949–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SL, Buzzai A, Rautela J, Hor JL, Hochheiser K, Effern M, et al. Tissue-resident memory CD8(+) T cells promote melanoma-immune equilibrium in skin. Nature 2019;565(7739):366–71. [DOI] [PubMed] [Google Scholar]
- Paternoster L, Standl M, Chen CM, Ramasamy A, Bonnelykke K, Duijts L, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat Genet 2011. ;44(2): 187–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NC, Egen JG, Secundino N, Debrabant A, Kimblin N, Kamhawi S, et al. In vivo imaging reveals an essential role for neutrophils in leishmaniasis transmitted by sand flies. Science 2008;321(5891):970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 2010;466(7302): 113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu DM, Botting RA, Stephenson E, Green K, Webb S, Jardine L, et al. Decoding human fetal liver haematopoiesis. Nature 2019;574(7778):365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ, Smith I, Parker I, Bootman MD. Fluorescence microscopy. Cold Spring Harb Protoc 2014;2014(10):pdb top071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity 2014;41(6):886–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneschal J, Clark RA, Gehad A, Baecher-Allan CM, Kupper TS. Human epidermal Langerhans cells maintain immune homeostasis in skin by activating skin resident regulatory T cells. Immunity 2012;36(5):873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook BA, Wasko RR, Rivera-Gonzalez GC, Salazar-Gatzimas E, Lopez-Giraldez F, Dash BC, et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 2018;362(6417). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva HM, Bafica A, Rodrigues-Luiz GF, Chi J, Santos PDA, Reis BS, et al. Vasculature-associated fat macrophages readily adapt to inflammatory and metabolic challenges. J Exp Med 2019;216(4):786–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner G, Koning F, Elbe A, Tschachler E, Yokoyama WM, Shevach EM, et al. Characterization of T cell receptors on resident murine dendritic epidermal T cells. Eur J Immunol 1988;18(9):1323–8. [DOI] [PubMed] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14(9):865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streilein JW. Lymphocyte traffic, T-cell malignancies and the skin. J Invest Dermatol 1978;71(3):167–71. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Beamer MA, Sarkar MK, Loftus S, Fullmer J, Xing X, et al. RNA-Seq Analysis of IL-1B and IL-36 Responses in Epidermal Keratinocytes Identifies a Shared MyD88-Dependent Gene Signature. Front Immunol 2018;9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoutounour S, Guilliams M, Montanana Sanchis F, Liu H, Terhorst D, Malosse C, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 2013;39(5):925–38. [DOI] [PubMed] [Google Scholar]
- Tan L, Sandrock I, Odak I, Aizenbud Y, Wilharm A, Barros-Martins J, et al. Single-Cell Transcriptomics Identifies the Adaptation of Scart1(+) Vgamma6(+) T Cells to Skin Residency as Activated Effector Cells. Cell Rep 2019;27(12):3657–71 e4. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009;6(5):377–82. [DOI] [PubMed] [Google Scholar]
- Hughes Travis K MHWI, Gierahn Todd M, Tran Do, David Weiss, Andrade Priscilla R., Feiyang Ma, de Andrade Silva Bruno J., Shuai Shao, Tsoi Lam C, Jose Ordovas-Montanes, Gudjonsson Johann E, Modlin Robert L, Love J Christopher, Shalek Alex K. Highly Efficient, Massively-Parallel Single-Cell RNA-Seq Reveals Cellular States and Molecular Features of Human Skin Pathology. bioRxiv 2019. [Google Scholar]
- Tsoi LC, Stuart PE, Tian C, Gudjonsson JE, Das S, Zawistowski M, et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat Commun 2017;8:15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Liu R, Shin MS, Trentalange M, Allore H, Nassar A, et al. CyTOF supports efficient detection of immune cell subsets from small samples. J Immunol Methods 2014;415:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]


