SUMMARY
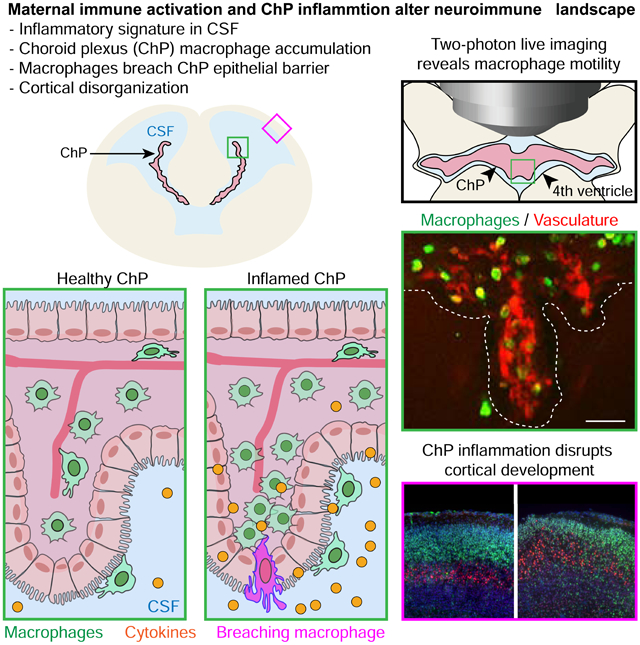
The choroid plexus (ChP) regulates brain development by secreting instructive cues and providing a protective brain barrier. Here, we show that polyI:C-mediated maternal immune activation leads to an inflammatory response in the developing embryonic mouse brain that manifests as pro-inflammatory cerebrospinal fluid (CSF) and accumulation of ChP macrophages. Elevation of CSF-CCL2 was sufficient to drive ChP immune cell recruitment, activation, and proliferation. In addition, ChP macrophages abandoned their regular tiling pattern and relocated to the ChP free margin where they breached the weakened epithelial barrier. We further found that these immune cells entered from the ChP into the brain via anatomically specialized “hotspots” at the distal tips of ChP villi. In vivo two-photon imaging demonstrated that surveillance behaviors in ChP macrophages had already emerged at this early stage of embryogenesis. Thus, the embryonic ChP forms a functional brain barrier that can mount an inflammatory response to external insults.
Keywords: Choroid plexus, cerebrospinal fluid, maternal immune activation, two-photon imaging, immune cells, neural development, CCL2, cytokines
Graphical Abstract
eTOC blurb
Cui et al. find that the choroid plexus barrier propagates maternal inflammation to the embryonic brain by secreting inflammatory signals into the cerebrospinal fluid and gating macrophage entry into the brain via “hotspots,” contributing to an altered neuroimmune landscape. Two-photon imaging uncovers diverse surveillance behaviors of embryonic choroid plexus macrophages.
INTRODUCTION
Immune system disturbances during development increase the susceptibility of offspring to multiple neurodevelopmental disorders (Ashwood et al., 2006; Estes and McAllister, 2016; Knuesel et al., 2014; Patterson, 2011). In particular, maternal infection and elevated levels of maternal serum cytokines during pregnancy are linked to a higher incidence of psychiatric disorders (Jones et al., 2016; Rudolph et al., 2018). In mice, maternal infection can be mimicked by mid-gestational exposure to a synthetic viral genome mimetic, polyinosine-polycytidylic acid (polyI:C). Cytokine signaling during maternal immune activation (MIA) disrupts placental function and fetal brain development. Such disruptions can result in postnatal behavioral deficits that model certain aspects of Autism Spectrum Disorders (ASDs) (Choi et al., 2016; Hsiao and Patterson, 2011; Malkova et al., 2012; Smith et al., 2007). Additionally, dysregulation of brain parenchymal microglial development and function has been reported in MIA offspring, implicating an altered neuroimmune landscape (Bilbo et al., 2018; Cunningham et al., 2013; Matcovitch-Natan et al., 2016). However, it is not well understood how the inflammatory signals propagate from the peripheral maternal-fetal barrier to the developing fetal brain.
The choroid plexus (ChP) forms the blood-cerebrospinal fluid (CSF) barrier, and secretes signals into the CSF that instruct brain development (Fame and Lehtinen, 2020; Ghersi-Egea et al., 2018; Lehtinen et al., 2011). In adults, the ChP also provides a gateway for immune cell entry into the CNS, and orchestrates the interplay between the central nervous system (CNS) and peripheral immune system (Engelhardt et al., 2017; Reboldi et al., 2009; Schwartz and Baruch, 2014). Aging-induced type-I interferon signaling at the ChP affects cognitive function and adult neurogenesis, highlighting the active roles of ChP in regulating brain function by orchestrating neuroimmune responses (Baruch et al., 2014). In contrast, much less is known about ChP immune cell behaviors during development. Indeed, it even remains unclear if embryonic ChP immune cells are capable of responding to external insults (Fame and Lehtinen, 2020; Kierdorf et al., 2019). Nonetheless, the embryonic ChP contains multiple immune cell populations expressing classic immune response machinery (Dani et al., 2019). A key question is whether the embryonic ChP contributes to disruptions of normal brain development associated with immune system activation (Ben-Reuven and Reiner, 2019). In particular, we asked if changes in the maternal environment caused by MIA alter embryonic ChP signaling, thereby compromising normal brain development.
Here, we show that the embryonic ChP is a site of inflammation in polyI:C-induced MIA. MIA triggered the accumulation of phagocytic macrophages and elevated chemotactic CCL2-CCR2 signaling at the embryonic ChP-CSF interface. These changes were paralleled by the emergence of a pro-inflammatory CSF signature and disruptions of ChP barrier integrity. We developed a two-photon imaging approach to characterize embryonic ChP macrophages in vivo, and observed diverse surveillance behaviors. Together, our findings demonstrate that the ChP helps establish an inflammatory state in the embryonic brain in response to maternal inflammation.
RESULTS
MIA triggers accumulation of activated macrophages at the embryonic ChP
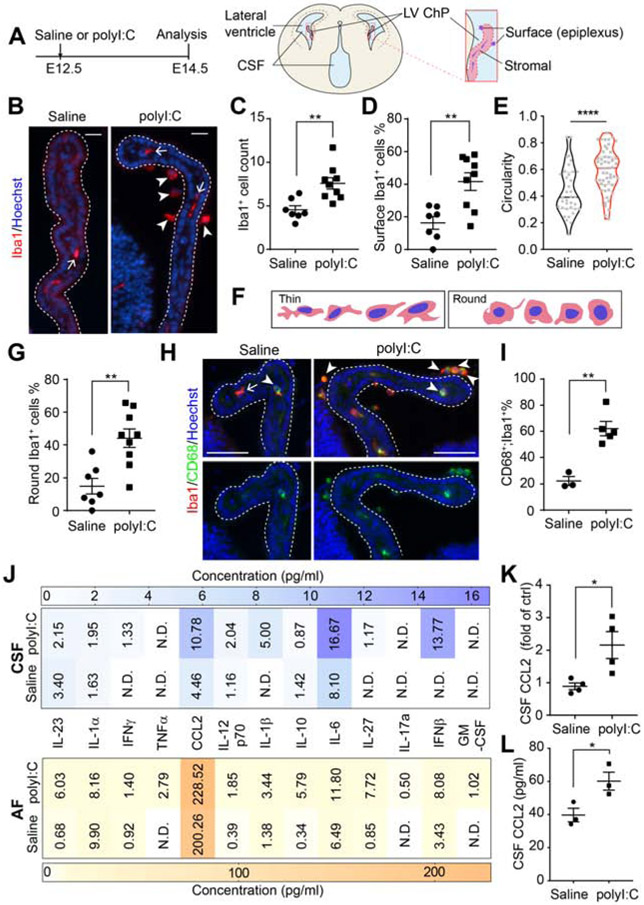
MIA induced by intraperitoneal delivery of polyI:C, (20 mg/kg) to pregnant dams at embryonic day (E) 12.5 (Figure 1A) triggered an accumulation of Iba1+ macrophages at the ChP at E14.5 (Figure 1B-C). Most Iba1+ cells localized to the apical, CSF-facing surface of the ChP (e.g. epiplexus location, Figure 1D). Morphological analyses and double-staining of Iba1+ macrophages with the phagocytic marker CD68 revealed the putative activation states of these cells. MIA caused ChP Iba1+ cells to exhibit a morphological shift to a “round” shape with increased circularity (>0.4) and solidity (Figure 1E-G, Figure S1F, G). Co-staining with the phagocytic marker, CD68, further demonstrated increased phagocytic activity of ChP macrophages during MIA (Figure 1H-I).
Figure 1. MIA triggers macrophage accumulation at the embryonic ChP and modifies CSF composition.
(A) Schematic of experimental paradigm and E14.5 brain. (B) LV ChP macrophages from saline- or polyI:C conditions. Arrow: macrophages in ChP stromal space; arrowheads: macrophages at ChP surface. Scale = 20 μm. (C) Iba1+ macrophage numbers per LV ChP. **P = 0.0030, unpaired t-test. (D) Percent of epiplexus ChP macrophages. **P = 0.0029, unpaired t-test. (E) Iba1+ macrophage circularity. ****P < 0.0001, unpaired t-test. Dashed lines: median and quartile values. Data points: individual cells. (F) Schematic depicting major morphological states of Iba1+ macrophages at E14.5. (G) Round ChP Iba1+ macrophages (% of total). **P = 0.0020, unpaired t-test. See also Figure S1F. (H) CD68+ macrophages from MIA E14.5 LV ChP Arrows: “thin” macrophage, CD68−;Iba1+; arrowheads: “round” macrophages, CD68+;Iba1+. Scale = 50 μm. (I) Macrophages (Iba1+) that are phagocytic (CD68+;Iba1+, in %) in (H) (% of total). **P = 0.0021, unpaired t-test. (J) Cytokines and chemokines in E14.5 CSF and AF. n ≥ 4 litters per group. (K) Relative changes in CCL2 in CSF samples from (J). *P = 0.025, unpaired t-test. (L) E14.5 CSF [CCL2] analyzed by sandwich ELISA. n = 3 litters per group, *P = 0.0039, unpaired t-test. All data presented as mean ± S.E.M., except in (J) where mean values are presented. See also Figure S1, Tables S1-S2.
Because phenotypes in the MIA model depend on the molecular size of the polyI:C reagent (Kowash et al., 2019), we validated our polyI:C quality in control experiments (Figure S1A). We also confirmed the presence of gut commensal segmented filamentous bacteria (critical for IL17-mediated, polyI:C-induced neurodevelopmental defects, Kim et al., 2017) in our colony (Figure S1B). As expected, MIA led to the upregulation of maternal serum cytokines 3 hours following polyI:C challenge (Figure S1C), and induced cortical disorganization phenotypes in offspring at birth (Figure S1D, (Choi et al., 2016; Shin Yim et al., 2017)). At baseline, the lateral ventricle (LV) and the fourth ventricle (4V) ChP had comparable Iba1+ cell numbers (Figure S1E).
MIA is accompanied by increased CCL2 and other inflammatory mediators in embryonic CSF
Because the early CSF distributes instructive factors and provides a biomarker signature of the developing brain (Chau et al., 2015, 2018; Fame et al., 2019), we searched for evidence of inflammation in the cytokine profile of E14.5 CSF. We observed increased levels of proinflammatory cytokines including CCL2 (C-C motif Chemokine Ligand-2), interferons (IFNβ and IFNγ), and interleukins (IL1β and IL6), indicating that MIA shifted the embryonic CSF to a pro-inflammatory state (Figure 1J, K, Table S1). IL17a, which mediates aspects of neurodevelopmental defects in offspring in response to maternal inflammation (Choi et al., 2016), was not detected in E14.5 CSF following MIA using this assay (13-plexed FACS-ELISA). We hypothesized that CSF cytokines participate in recruiting Iba1+ macrophages to the ChP In particular, the most significantly increased factor, CCL2, is known to recruit circulating myeloid cells to sites of inflammation in adult disease models (Garzia et al., 2018; Howe et al., 2017; Lu et al., 1998). We confirmed CSF-CCL2 elevation using sandwich ELISA (Figure 1L). Then we compared CCL2 levels across different body fluids at E14.5 (Figure S1H) and found a similar elevation in CCL2 in serum as in CSF (Figure S1I). As cytokine signaling at the placenta is upregulated by MIA (Hsiao and Patterson, 2011), we also tested if cytokine levels in the amniotic fluid (AF) reflected these changes. We did not observe changes in AF-CCL2 levels in response to MIA (Figure S1J, Table S2). These data demonstrate that MIA triggers a pro-inflammatory cytokine response in the developing embryo.
MIA upregulated CCL2-CCR2 signaling at the ChP-CSF interface
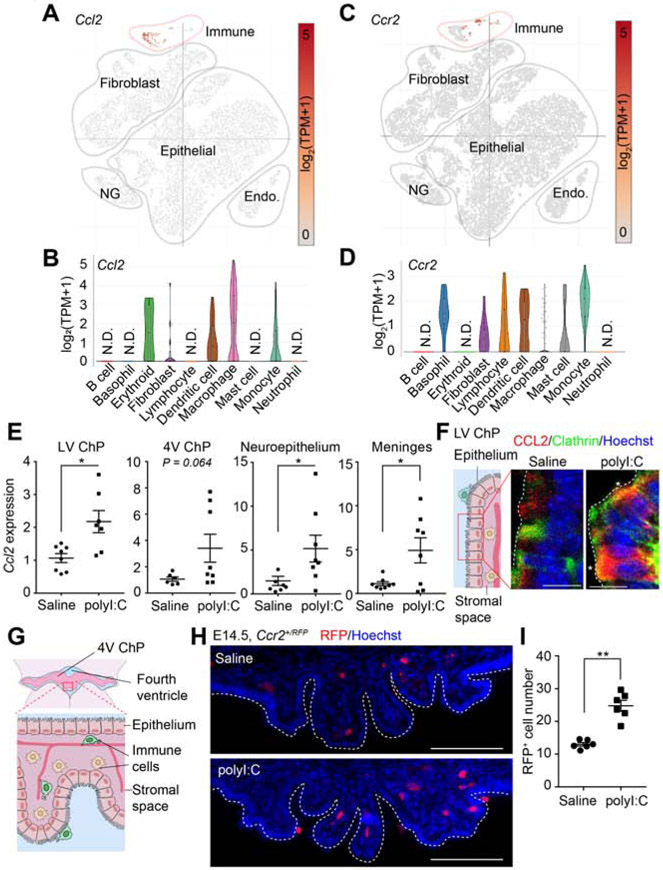
Given the accumulation of ChP macrophages and elevation of CSF-CCL2, we investigated whether ChP contributes to CSF-CCL2 upregulation. Analyses of single cell transcriptomics data (Dani et al., 2019) indicated that, at baseline, embryonic ChP macrophages express Ccl2 (Figure 2A, B, S2A). qRT-PCR and mRNA in situ hybridization of E14.5 ChP revealed an increase in Ccl2 expression in response to MIA (Figure 2E, S2B). Further, intracellular CCL2 was increased in the ChP (Figure 2F, S2C, (Lu et al., 1998)). We cannot rule out contributions by other CSF-contacting cell types (e.g., developing meningeal immune cells and neuroepithelial progenitors) to overall CSF-CCL2 levels (Figure 2E). Nevertheless, our data highlight the ChP as a brain tissue that likely contributes to elevated CSF-CCL2 levels during MIA.
Figure 2. MIA is accompanied by increased CCL2-CCR2 signaling at the ChP-CSF barrier.
(A) Ccl2 and (B) Ccr2 expression in E16.5 ChP from Dani et al., bioRxiv, 2019. Endo: endothelial cells; NG: neuroglia. (C-D) Violin plots showing Ccl2 (C) and Ccr2 (D) expression in embryonic ChP immune cell subclusters from Dani et al., bioRxiv, 2019. (E) Ccl2 expression (qRT-PCR) in CSF-contacting tissue following MIA, normalized to saline. LV ChP: *P = 0.016; Neuroepithelium, *P = 0.031; Meninges: *P = 0.033 Welch’s t-test. One outlier in polyI:C LV ChP sample and one outlier in saline neuroepithelium sample were removed using the ROUT method (Q = 1%). (F) ChP schematic and representative images of CCL2 cellular distribution in LV ChP. Asterisks: co-localization of CCL2 and Clathrin. Scale = 10 μm. See also Figure S2D for antibody validation. (G) E14.5 4V ChP schematic. (H) Ccr2+/RFP monocytes in 4V ChP of MIA E14.5 Ccr2+/RFP samples. Scale = 100 μm. (I) Ccr2+/RFP monocyte numbers (per LV ChP) for (H). ****P < 0.0001, unpaired t-test. All data presented as mean ± S.E.M. See also Figure S2.
Because CCL2 can signal through the cell surface receptor, CCR2, to recruit circulating CCR2-expressing monocytes to sites of inflammation (Lu et al., 1998), we tested if a similar mechanism could contribute to ChP macrophage accumulation following MIA. Analysis of Ccr2 expression in single cell transcriptomic data (Dani et al., 2019) uncovered a small subset of CCR2-expressing monocytes in ChP at baseline (Figure 2C, D). Subsequent immunostaining in Ccr2+/RFP mice revealed an increase in Ccr2+/RFP monocytes in ChP following MIA (Figures 2G-I, S2D). Together, these data support the model that during MIA, increases in CCL2 in the ChP and CSF recruit CCR2-expressing cells to the ChP.
The upregulation of certain chemokines in AF and CSF correlates with neurodevelopmental disorders including ASD (Abdallah et al., 2012; Vargas et al., 2005). Given the relevance of clinical implications of the rodent MIA model in ASD pathology, we performed gene expression analyses of the embryonic LV ChP, focusing on the following chemokines: CCL3, CCL4, CCL5, CXCL1, and CXCL10. We found that the embryonic ChP had very low baseline expression of Ccl5 and Cxcl1 (Ct > 35, not shown). ChP Cxcl10 expression was upregulated by MIA, whereas the expression of Ccl3 and Ccl4 was unaffected (Figure S2E). Thus, MIA elevates the expression of additional chemokines as part of the ChP inflammatory response, mirroring the inflammatory profiles of the CSF and brain in ASD patients.
CSF-CCL2 augmentation stimulates Iba1+ cell accumulation at the ChP
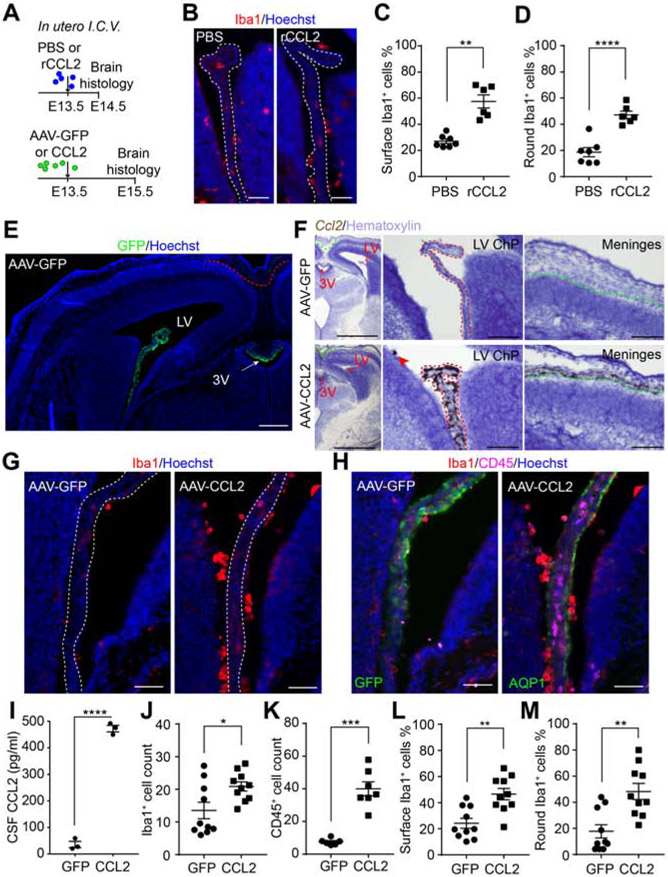
To test if transient CSF-CCL2 elevation is sufficient to stimulate macrophage accumulation at the ChP, we delivered mouse recombinant CCL2 (rCCL2) by a single intracerebroventricular injection (in utero ICV) at E13.5 and analyzed ChP immune cells 24 hours later (Figure 3A, B). Compared to vehicle controls, rCCL2 increased the number of Iba1+ cells at the ChP-CSF surface and the proportion of “round” Iba1+ cells, suggesting that the transient elevation of CCL2 in the CSF altered the activation status of these cells (Figure 3C, D).
Figure 3. CSF-CCL2 supplementation stimulates immune cell accumulation at the ChP.
(A) Schematic of in utero ICV injection paradigm. (B) ChP macrophages in PBS or rCCL2. (C-D) Cell location (C) and shape (D) of macrophages in (B). (C) Epiplexus macrophages (% of total). **P = 0.0011, Welch’s t-test. (D) Round macrophages (% of total). ****P < 0.0001, unpaired t-test. (E) Representative image of E15.5 AAV-GFP brain, 48 h post infection. Red dashed line: meninges where no GFP signal was observed using the viral transduction paradigm illustrated in (A). Scale = 200 μm. (F) Representative image of Ccl2 RNAscope in situ hybridization of untreated or AAV-CCL2 E15.5 section. Representative 4x images of brain sections for AAV-GFP or AAV-CCL2 condition (left; Scale = 1 mm) and representative 20x images showing LV ChP (middle; scale = 100 μm) and meninges (right; scale = 100 μm). Red dashed line: epithelial cells of LV ChP where Ccl2 transcript was most enriched in AAV-CCL2 samples; green dashed line: meninges where Ccl2 transcript was detected but at lower levels than in LV ChP epithelial cells from AAV-CCL2 samples. (G-H) LV ChP macrophages (G) and immune cells (H) in AAV-GFP or -CCL2 conditions. (I) CSF CCL2 concentrations; n = 3 litters/condition. ****P < 0.0001, unpaired t-test. (J-K) LV ChP Iba1+ (J) and CD45+ (K) cell numbers in each condition. ***P = 0.0002, Welch’s t-test. *P = 0.0195, unpaired t-test. (L-M) Location (L) and shape (M) of Iba1+ cell in (G). (L) Epiplexus macrophages (% of total). **P = 0.011, unpaired t-test. (M) Round macrophages (% of total). **P = 0.012, unpaired t-test. Scale = 50 μm. All data presented as mean ± S.E.M. See also Figure S3.
We next used an adeno-associated viral approach (AAV) to determine if sustained production of CCL2 by ChP epithelial cells alters the brain’s immune environment. AAV was delivered by in utero ICV at E13.5, and samples were collected at E15.5 (Figure 3A). We confirmed AAV tropism for ChP epithelial cells in AAV-GFP controls (Figure 3E, (Chen et al., 2020; Haddad et al., 2013; Kaiser et al., 2020)). RNAscope in situ hybridization analyses revealed an increase in ChP Ccl2 expression (Figure 3F). However, AAV-CCL2 additionally induced Ccl2 upregulation in the meninges (Figure 3F, right), likely reflecting a broader secondary inflammatory response caused by CCL2 overexpressed by the ChP. CSF-CCL2 was increased ten-fold in AAV-CCL2 samples compared to AAV-GFP controls (Figure 3I). AAV-CCL2 ChP samples showed increased numbers of Iba1+ macrophages and CD45+ leukocytes (Figure 3G, H, J, K) with elevated prevalence of a round morphology (Figure 3L, M). An increase of Iba1+ cells at the apical CSF surface of the ventricular zone of the developing cerebral cortex was also observed in AAV-CCL2 samples (Figure S3), suggesting a global immune activation in the brain. Taken together, these results demonstrate that enhanced CSF-CCL2 availability during early brain development, either via direct delivery of CCL2 into the CSF or sustained overexpression of CCL2 by the ChP, is sufficient to promote immune cell accumulation at the ChP and ventricle walls.
Two-photon imaging reveals embryonic ChP macrophage motility and mobility
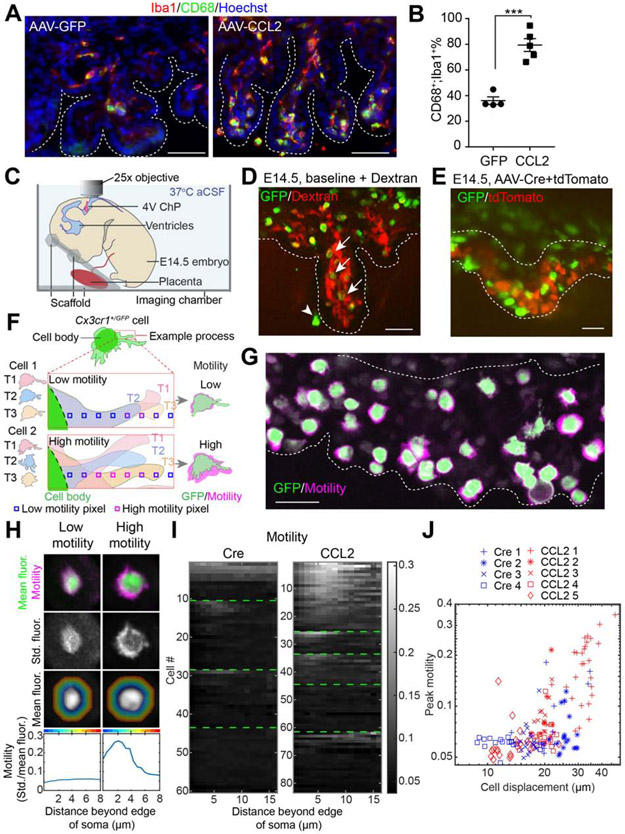
We recently captured the movements of mature ChP macrophages in adult mice at baseline and following local and systemic insults (Shipley et al., 2020). In fixed tissues, the expression of CD68 is commonly exploited as a marker of increased macrophage activity and was increased in fixed AAV-CCL2 samples (Figure 4A, B). Here, we established a live embryo imaging platform for 3D imaging of ChP macrophages (Figure 4C). We imaged the ChP in 4V via the cisterna magna, as it was more readily accessible than LV ChP, while demonstrating similar immune cell accumulation and activation (Figure 4A, Figure S4A). To help preserve tissue health, embryos remained attached to the excised placenta and retained circulation for an average of one hour while bathed in warm (37°C) artificial CSF (not shown). To improve image clarity, 4V ChP was carefully exposed to avoid causing hemorrhage. Comparative histological analyses indicated that acute 4V ChP exposure did not induce any immediate additional immune response or cell death in the ChP during the experimental time period (Figure S4B-G). We used Cx3cr1+/GFP reporter mice for live imaging of GFP fluorescence in embryonic ChP macrophages (Dani et al., 2019; Shipley et al., 2020). In addition, to help distinguish ChP from adjacent tissues, and to distinguish between ChP surface macrophages (epiplexus) and those located within the stromal space, we also imaged ChP vasculature (via liver injection of Texas Red dextrans; Figure 4D, Video S1). Alternatively, we labeled ChP epithelial cells using AAV-tdTomato (in utero ICV delivery, 1 day prior to imaging; Figure 4E).
Figure 4. Visualizing immune cells at the developing ChP.
(A) Phagocytic macrophages at E15.5 4V ChP Scale = 50 μm. (B) Phagocytic 4V ChP macrophages (CD68+;Iba1+; % of total Iba1+), mean ± S.E.M. ***P = 0.0002, unpaired t-test. (C) Schematic of imaging setup. (D) Z-projection of E14.5 4V ChP across time-averaged imaging volume of GFP+ cells and Dextran-labeled ChP vasculature from a naive Cx3cr1+/GFP sample. Arrows: GFP+ cells in the stromal region of 4V ChP. Arrowhead: GFP+ cell at 4V ChP tissue surface (epiplexus cell). Scale = 50 μm. (E) Z-projection image of E14.5 4V ChP GFP+ macrophages and tdTomato+ epithelial cells from a Cx3cr1+/GFP sample. Images taken 1 day after in utero ICV delivery of AAV-Cre (control) and AAV-tdTomato (mixture ratio of Cre : tdTomato = 5 : 1). Data are from recordings averaging 30-minutes. Scale = 50 μm. Dashed white lines: 4V ChP boundaries. (F) Schematic of cells exhibiting low (Cell 1) or high (Cell 2) motility. Low motility cells showed limited movement of processes, leading to a smaller motility “halo” than high-motility cells. (G) ChP macrophages with estimated motility “halos” (see below), from an AAV-CCL2 + AAV-tdTomato Cx3cr1+/GFP sample. (H) Procedure for estimating motility in low- and high-motility cells. Top row: mean cell body fluorescence (green) and standard deviation of bandpass filtered fluorescence across time, at each pixel (motility “halo,” magenta). Second row: standard deviation of bandpass-filtered fluorescence. Third row: Mean cell body fluorescence. Regions with the same pseudocolor hue indicate pixels with a similar distance from the outer edge of the cell body (up to 8 μm away; see also hollow squares in F). Bottom row: for all pixels at a given distance from the outer edge of the cell (cf. colors at top of panel), we calculated the average of the standard deviation (third row) normalized by mean fluorescence (second row). This showed that the example cell at right (a “High motility” cell from the CCL2 condition) had elevated motility of processes peaking at 2-3 μm from the cell border. (I) Heatmap of motility (same estimate as bottom row of H) for every imaged cell in Cre and CCL2 conditions (1 cell / row). For each sample, rows were sorted by the cell’s peak motility. Green lines separate data from different samples (Cre: n = 66 cells from 4 individuals; CCL2: n = 89 cells from 5 individuals). Cells from the first sample of each condition correspond to cells in Video S7. Gray scale indicates motility score. (J) Relationship between cell displacement and peak motility for each cell for all samples (with a different symbol for each individual) and conditions (CCL2 and Cre; see Methods). We noted a trend to greater displacement and mobility for CCL2 vs. Cre conditions. However, these differences may be due, in part, to overall variability across individuals. Indeed, we could not determine whether the CCL2 vs. control condition had a significant effect on motility (P = 0.33) or displacement (P = 0.67) when accounting differences across individuals using a normally distributed generalized linear mixed effects model (GLMM) with an identity link function, with the category type as a predictive variable and the sample number as a fixed effect. We also asked whether displacement and motility were correlated and found that the level of motility strongly predicted that of displacement (P < 0.0001) even when accounting for sample number (using a normally distributed GLMM with an identity link function, with the displacement as a predictive variable and both the sample number as a fixed effect). See also Figure S4, Videos S1-7.
We characterized the motility and mobility of ChP Cx3cr1+/GFP macrophages. Consistent with a role in immune surveillance, embryonic ChP macrophages exhibited motile processes in control conditions (Video S2). Notably, these processes were shorter and less ramified than in adult (Shipley et al., 2020). Processes from neighboring cells sometimes contacted each other and lingered (Video S3). To quantitatively characterize cell behaviors, we first developed a custom analysis pipeline to stabilize the 3D volumes and to measure the local displacement of macrophage cell bodies (Figure S4H). Having estimated the movement of each macrophage, we then aligned a small movie centered on the macrophage, followed by estimation of the motility of macrophage processes (Figure 4F). The standard deviation of fluorescence intensity across time of each pixel in a region of interest (ROI) surrounding the cell was averaged across pixels in the ROI and normalized to the mean fluorescence of each cell body to obtain an estimate of cell process “motility” (Figure 4G, Figure S4I). Across individual cells, motility at ~ 3 μm from the edge of the cell body showed the greatest differences between cells (Figure 4H).
We next tested if CSF-CCL2 augmentation affected ChP immune cell motility and mobility. We confirmed CSF-CCL2 levels were upregulated 24 h post viral transduction, which was the timepoint selected for embryo imaging (Figure S4J). Histological analyses also showed that immune cells started to accumulate at the ChP at this time (Figure S4K-N). We observed somewhat faster moving processes in cells in the CCL2 condition than in controls (Videos S2, 4, 5). Further, some cells exhibited clear displacement, with cell bodies that moved toward other cells (Video S6). Some ChP macrophages appeared to respond to CSF-CCL2 elevation, as reflected by trends towards greater displacement and peak motility of macrophage processes (Video S7, Figure 4I), albeit to a varying degree across cells and samples. Further, cell displacement and cell process motility were significantly positively correlated (Figure 4J). Overall, the high-resolution live imaging approach revealed substantial macrophage motility in the developing ChP, indicating surveillance behavior.
CSF-CCL2 promotes immune cell infiltration and proliferation at the embryonic ChP
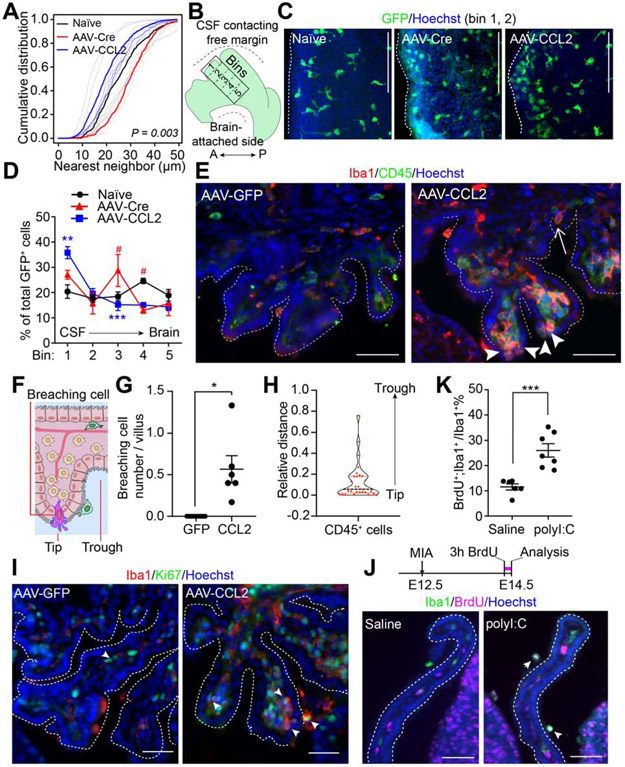
In adult mice, macrophages are evenly tiled across the ChP, and survey their “territory” in baseline conditions (Shipley et al., 2020). Here, we found that embryonic ChP macrophages also tiled the LV ChP at regular intervals at baseline (Figure 5A, S5A). The regularity of this tiling was disrupted by elevated CSF-CCL2 (Figure S5B). Nearest-neighbor analyses revealed that CSF-CCL2 shifted the macrophage distribution toward an irregular pattern involving an increased number of cells per tissue area (Figure 5A, Figure S5B-D). Further, we noticed a more dense clustering of macrophages along the CSF contacting free margin of the ChP (Figure 5B-D), suggesting immune cell recruitment at this site in the ChP.
Figure 5. ChP barrier properties are altered by CSF composition.
(A-E) Macrophage distribution analysis of LV ChP explants of naïve (untreated), AAV-Cre (AAV control), and AAV-CCL2 Cx3cr1+/GFP conditions. (A) Cumulative distribution of nearest-neighbor distances across GFP+ immune cells indicates that differences between groups are statistically significant (studentized permutation test, P = 0.003). See also Figure S5C. (B) Schematic depicting LV ChP explant and binned areas for cell counting. A, Anterior; P, Posterior. (C) Cx3cr1+/GFP LV ChP explants from each condition. (D) GFP+ cell numbers from bin #1 (CSF-contacting region) to bin #5 (brain-proximal region). GFP+ cell density (% of total GFP+ cells across bins). AAV-Cre vs. AAV-CCL2: ** P = 0.037 (Bin #1), *** P = 0.0009 (Bin #3); AAV-Cre vs. naive: # P = 0.0017 (Bin #3), # P = 0.0017 (Bin #4). All other P-values > 0.05. n = 3 for naive and AAV-Cre, n = 6 for AAV-CCL2, two-way ANOVA, P values corrected for multiple comparisons by Holm-Sidak test. Scale = 100 μm. (E) Immune cells (CD45, pseudocolored in green) breaching 4V ChP epithelium. In AAV-GFP condition, CD45+ cells were mostly seen in the 4V ChP stromal space. In AAV-CCL2 condition, Iba1+ and CD45+ cells were in stromal space, on the ChP surface, and between epithelial cells (arrowheads). Dashed lines: 4V ChP. Arrowheads: putative transmigrating immune cells at distal tip of 4V ChP villi. Arrow: transmigrating immune cell at trough between 4V ChP villi. Scale = 50 μm. (F) Schematic depicting a breaching immune cell. (G) Number of cells breaching the barrier per ChP villus: GFP = 0, n = 9; CCL2 = 0.57 ± 0.16, n = 6. * P = 0.018 Welch’s t-test. (H) Violin plots and individual cell data illustrating the distribution of relative distances of the “barrier breaching” CD45+/hi cells from the distal tip of the adjacent 4V ChP villus (‘0’: cell at tip; ‘1’: cell at trough). 65.5% of cells had a relative distance of ≤ 0.1 from the tip of the villus, 6.9% cells had relative distance ≥ 0.5, n = 29 cells from 6 AAV-CCL2 individuals. (I) Proliferating (Ki67, green pseudocolored) immune cells (Iba1, red) in 4V ChP of AAV-GFP or AAV-CCL2 individuals. Arrowheads in I, J: BrdU+;Iba1+ cells. Scale bars = 50 μm. (J) Top: schematic of BrdU labeling. Bottom: LV ChP BrdU+ and Iba1+ staining related to (K). (K) Iba1+ cells that are BrdU+ (% of total). ***P = 0.0006, unpaired t-test. Scale = 50 μm. All data presented as mean ± S.E.M. except in (H). See also Figure S5.
Indeed, we noticed that a subset of 4V ChP Iba1+;CD45+/hi cells were positioned between epithelial cells in AAV-CCL2 samples (Figure 5E-G), suggesting immune cell infiltration across the barrier. Consistent with this hypothesis, these CD45+/hi cells were more frequently located near the distal tip of ChP villi (Figure 5H). In addition, we observed increased expression of Ly6C, a marker of infiltrating monocytes, in AAV-CCL2 samples (Figure S5E). Together, these data indicate that elevated CSF-CCL2 driven by ChP CCL2 overexpression promotes immune cell infiltration across the ChP barrier at the distal tips of ChP villi “hotspots.”
CSF-CCL2 elevation also increased the population of proliferating Ki67+;Iba1+ cells in 4V ChP (Figure 5I). These data were corroborated by a larger percentage of BrdU+;Iba1+ cells in LV ChP (Figure S5F). Similarly, an increased BrdU+;Iba1+ cell population was captured in BrdU-pulsed MIA samples (Figure 5J, K). Taken together, these data demonstrate that both infiltration and proliferation from the circulation contribute to the accumulation of ChP immune cells following CSF-CCL2 augmentation.
Inflammation disrupts the ChP barrier by loosening tight junctions
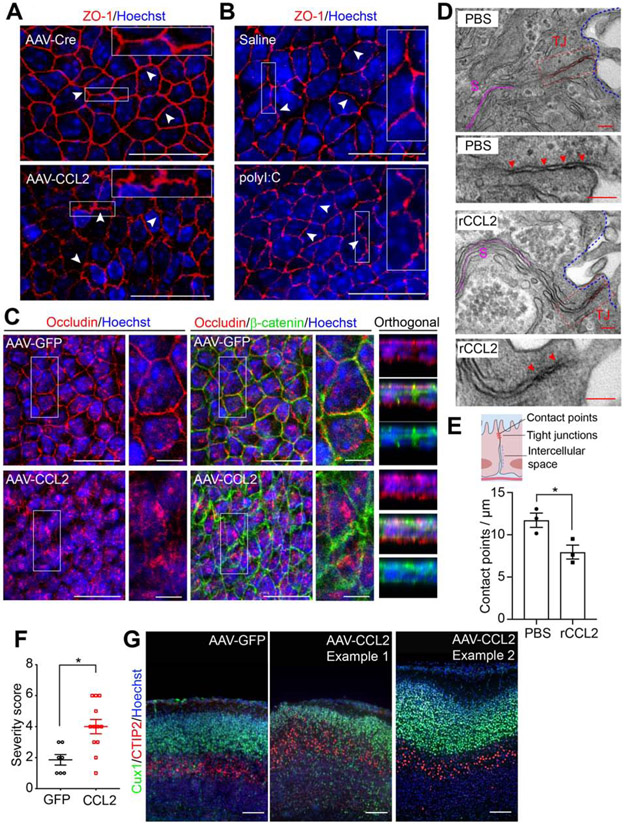
Tight junctions between ChP epithelial cells contribute to blood-CSF barrier functions (Ghersi-Egea et al., 2018; Saunders et al., 2018). Inflammation has been shown to induce redistribution of tight junction proteins in endothelial cells (Murakami et al., 2009; Stamatovic et al., 2003, 2005). To test if CSF-CCL2 augmentation and accumulation of activated immune cells at the ChP are accompanied by altered tight junctions, we first analyzed the tight junction marker ZO-1 in LV ChP explants. Both CSF-CCL2 augmentation and MIA disrupted the ZO-1 distribution between ChP epithelial cells (Figure 6A, B, S6A). We also found a prominent difference in the subcellular distribution of Occludin expression: in control explants, Occludin continuously localized along each cell, outlining the epithelial cell borders, similar to ZO-1 staining. However, in AAV-CCL2 explants, the Occludin staining pattern was disrupted and appeared diffuse (Figure 6C). Meanwhile, β-catenin, a component of adherens junctions that regulates cell adhesion and polarity (Brembeck et al., 2006), revealed a wider intracellular border in the CCL2 condition (Figure 6C). Additionally, Claudin-1 staining in LV ChP explants revealed intermediate changes (not shown). CSF-CCL2 elevation led to a modest down-regulation of Cldn1 and Cldn2 mRNA, but not Ocln or Cldn3 mRNA (Figure S6B-E). Electron microscopy (EM) revealed reduced numbers of tight junction contact points, wider junctions, and a wider intercellular space between epithelial cells in CCL2 condition (Figure 6D, E, Figure S6F, G). Taken together, these data demonstrate that MIA and CSF-CCL2-associated inflammation weakens ChP tight junctions, a core feature of compromised barrier integrity.
Figure 6. ChP barrier properties and embryonic brain development are altered by CSF-CCL2 augmentation.
(A-B) Mean z-projection images showing ZO-1 staining of LV ChP explants from (A) AAV-GFP or -CCL2 conditions, and (B) E14.5 control or MIA conditions. Arrowheads: representative staining patterns; scale = 20 μm. (C) Representative mean z-projection images, and orthogonal images of boxed areas showing Occludin and β-catenin staining. Scale = 20 μm, or 5 μm in close-up views. For each condition in (A) to (C), two regions of interest per explant were examined, and whole mount LV ChP explants of n ≥ 3 individuals from two litters were scanned and assessed. (D) EM of tight junctions (TJ) between ChP epithelial cells from PBS or recombinant CCL2 (rCCL2) conditions. Scale = 100 nm. TJ: red; intercellular space (S): magenta; arrowheads: contact points; blue dashed lines: CSF-facing apical surface of the LV ChP (E) Top: schematic showing TJ between ChP epithelial cells. Bottom: number of contact points per μm. LV ChP samples from n = 3 individuals, at least 10 TJ per sample were analyzed, *P = 0.033, unpaired t-test. (F) P0 cortical disorganization severity scores. n = 7 from two litters for AAV-GFP, and n = 12 from three litters for AAV-CCL2. ** P = 0.0048, unpaired t-test. (G) Representative zoomed-in images showing cortical disorganization patches of P0 samples from (F). Scale = 100 μm. See also Figure S6.
We hypothesized that CSF-CCL2 augmentation ultimately dysregulates fetal brain development. First, at P0, we found massive immune cell accumulation at the LV ChP, and prominent infiltration of myeloid cells along the CSF-contacting surface of the forebrain (Figure S6H). CSF-CCL2 augmentation disrupted brain development (Figure 6F) such that 8/12 AAV-CCL2 brains exhibited disorganized cortical patches along the dorso-lateral cerebral cortex, while these types of patches were not observed in the AAV-GFP group (Figure 6G, S6I). Together, these results demonstrate that ChP overexpression-driven CSF-CCL2 augmentation can ultimately dysregulate fetal brain development.
DISCUSSION
We demonstrate that the developing ChP barrier is capable of responding to external insults, and propagates maternal inflammation to the developing brain. Using the MIA model, we uncovered a broad inflammatory response in the developing brain that manifests as an accumulation of macrophages at the ChP and an increase in pro-inflammatory CSF cytokines. We developed live imaging and analysis approaches for monitoring ChP macrophage behaviors in vivo in the embryonic brain with subcellular resolution. Using these tools and histological analyses, we found that elevated CCL2 at the CSF-ChP interface was sufficient to drive ChP immune cell activation, recruitment, and proliferation. In turn, ChP immune cells abandoned their regular tiling pattern and relocated along the ChP free margin, where they breached the barrier that was weakened by CCL2-associated inflammation. Anatomically distinct “hotspots” for immune cell entry from ChP to CSF emerged at the distal tips of ChP villi, where immune cells appeared strategically positioned to enter the ventricles.
Microglia, ChP macrophages, meningeal macrophages, and perivascular macrophages represent the majority of CNS myeloid cells critical for the maintenance of CNS homeostasis. During brain development, microglia exhibit a phagocytic transcriptomic profile, and regulate neurogenesis and synapse refinement (Cunningham et al., 2013; Kracht et al., 2020; Schafer et al., 2012; Squarzoni et al., 2014; Wilton et al., 2019). Aberrant microglial activity during development has devastating, life-long consequences on brain function (Bolton et al., 2017; Nelson and Lenz, 2017; Williamson et al., 2011). Although distinct in ontology, microglia share a unique transcriptomic signature with the brain border macrophages at embryonic age E14.5 (Hammond et al., 2018; Li and Barres, 2018). Our findings complement this emerging field of neuroimmunology, where the pro-inflammatory signature of the embryonic CSF barrier in response to MIA reflects altered CNS immune cell function, and consequently impacts cerebral cortical development. Embryonic microglia regulate neural progenitor cell numbers, and the proliferation dynamics of neural progenitor cells is disrupted by MIA (Ben-Reuven and Reiner, 2019; Cunningham et al., 2013; Stolp et al., 2011). Thus, the aberrant macrophage accumulation we observed at the ventricular zone of the developing cerebral cortex (Figure S3B, Figure S6H) indicates that CCL2 in the CSF that originates from ChP may indirectly modulate neural progenitor proliferation by recruiting phagocytic macrophages into the brain parenchyma. Multiple cytokines and pathways from various sources contribute to the overall inflammatory response in the brain upon MIA stimulation, including IL17a, IL6 (Ben-Reuven and Reiner, 2019; Choi et al., 2016; Smith et al., 2012), and CCL2 as discovered in this study.
During mid-gestation in mouse, the blood-brain barrier (BBB) is still developing and remains permeable to small molecules (Ben-Zvi et al., 2014; Daneman et al., 2010). Similar features are conserved across different species during early life (Ek et al., 2001, 2003, 2006; Knott et al., 1997; Saunders et al., 2012). Inflammation during gestation and early life increases BBB permeability and white matter damage (Stolp et al., 2005a, 2005b, 2011). The embryonic ChP has functional tight junctions (Liddelow et al., 2012). However, little is known about how insults during development affect the ChP (Fame and Lehtinen, 2020). Our findings demonstrate that during early brain development, inflammation leads to ChP immune cell accumulation, and loosens the ChP barrier. These findings extend our view of the gatekeeper role of embryonic ChP for not only as a gatekeeper regulating CNS entry of peripheral nutrients, toxins, and xenobiotics, but also entry of peripheral immune cells. During early cortical development, the meninges are also maturing and secrete factors important for brain development (Engelhardt et al., 2017; Siegenthaler and Pleasure, 2011; Siegenthaler et al., 2009). Our data hint that the developing meninges may also contribute to the CSF inflammatory response to MIA (Figure 2E, Figure S2A). Thus, the collective vulnerability of all brain barriers to inflammation likely leads to the manifestation of neurodevelopmental deficits caused by MIA.
The emerging field of brain border macrophages relies largely on histological and FACS-based evaluation of cells (Kierdorf et al., 2019; Li and Barres, 2018). Remarkably, behaviors of these cells during development have never been visualized in real time. Our two-photon live imaging platform captures macrophage motility and mobility at the embryonic ChP barrier, thereby providing an opportunity to systematically investigate the early maturation of the brain’s immune system. Our findings that embryonic ChP macrophages are motile, mobile, and capable of responding to external stimuli complement recent findings that human fetal microglia are phagocytic and acquire immune-sensing properties early in development, at 9 to 18 gestational weeks (Kracht et al., 2020). Given the morphological differences between the embryonic ChP macrophages and ramified adult macrophages and microglia, the typical approaches to skeletonize and analyze cell branching as a measure of activity were not feasible (Davalos et al., 2005; Madry et al., 2018; Nimmerjahn et al., 2005; Shipley et al., 2020). We overcame this challenge by developing a method using the mean standard deviation of GFP fluorescence intensity adjacent to the cell body to quantitatively analyze process movements. One limitation of this approach is that the imaging time is constrained by the health of the preparation, averaging approximately one hour. While we did not observe an immediate additional immune response of 4V ChP in our surgical preparation, 4V ChP exposure is nevertheless invasive. Thus, we cannot exclude the possibility that an additional inflammatory response from the preparation of the embryo for imaging may further alter cell behaviors. Performing the imaging shortly following viral transduction may also introduce an unintended “altered baseline” that differs from truly naïve conditions (Figure S4J). Future advances in imaging the fully intact embryonic brain (Yuryev et al., 2016), including with three-photon imaging (Ouzounov et al., 2017), should facilitate longer-term tracking of individual cells.
CSF from ASD patients display hallmarks of neuroinflammation including increased CCL2 expression (Ashwood et al., 2006; Vargas et al., 2005). Neuropathology of ASD brain tissue has also revealed microglial activation and T cell infiltration at the brain-CSF interface (DiStasio et al., 2019; Morgan et al., 2010; Vargas et al., 2005). Our data suggest an additional potential route of infiltration at the ChP blood-CSF barrier. Future studies could examine this hypothesis in clinical samples. While CSF-CCL2 was upregulated in E14.5 mouse CSF following MIA, additional cytokines and chemokines (such as IL6 and CXCL10, Figure 1F, Figure S2E) were also elevated under these conditions. Thus, CCL2 represents one of multiple factors that together contribute to disease pathology. Together with recent advances in MRI diagnostics implicating increased extra-axial CSF as predictive of ASD (Shen, 2018), our findings of ChP-CSF inflammation should inspire the development of CSF-based biomarkers for the early diagnosis of ASD, and novel therapies aimed at barrier intervention for preventing neurodevelopmental disorders.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, maria.lehtinen@childrens.harvard.edu.
Materials Availability
This study did not generate mouse lines. The plasmid generated for this study is available from the corresponding author upon request.
Data and Code Availability
The imaging registration, cell motility and mobility analysis algorithms generated for this study are available at https://github.com/LehtinenLab/Cui2020-code. Original data is available from the corresponding author upon request.
EXPERIMENT MODEL AND SUBJECT DETAILS
Animals
All mouse work was performed in accordance with the Institutional Animal Care and Use Committees (IACUC) at Boston Children’s Hospital. Six to eight week old C57BL/6 and CD-1 (ICR) breeders were purchased from Taconic Farms or Charles River Laboratories (CRL), and gut SFB (Eubacteria as a control (Barman et al., 2008)) was confirmed after import into the facility. In-house bred timed-pregnant C57BL/6 mice (E14.5 or E15.5) were used for ChP histology, CSF, and AF analysis following MIA. Timed pregnant CD-1 dams were used for in utero ICV studies and were purchased (CRL). Cx3cr1GFP (B6.129P2(Cg)-Cx3cr1tm1Litt/J, Jax Stock #005582) and Ccr2RFP (B6.129(Cg)-Ccr2tm2.1Ifc/J, Jax Stock #017586) lines were maintained by breeding with C57BL/6. For both reporter lines, C57BL/6 females were bred with homozygous Cx3cr1GFP or Ccr2RFP males, such that all samples used for experiments were heterozygous. All animals were housed under 12hr/12hr day night cycle with access to standard chow and water ad libitum.
METHODS DETAILS
Maternal Immune Activation
For timed pregnancies, females were checked for the presence of plugs, and the date of the plug was noted as embryonic day 0.5 (E0.5). Bodyweight was monitored every 3-4 days, and ultrasound was available to help confirm gestational age. On E12.5, pregnant dams received a single dose (20 mg/kg; i.p.) of polyinosinic-polycytidylic acid (polyI:C, Sigma Aldrich) or saline as vehicle control. Maternal blood was collected by tail-nick three hours following the injections. Following coagulation and centrifugation, serum samples were analyzed by LEGENDplex™ Mouse Inflammation Panel FACS-ELISA (BioLegend) to confirm successful immune activation.
Embryonic body fluid collection and analysis
AF was collected by inserting a glass capillary into the intra-amniotic space close to the snout. CSF was collected by inserting a glass capillary into cisterna magna, and processed as described (Zappaterra et al., 2013). Samples were pooled across litters and their purity was erified under the dissection scope. Blood was collected by glass capillary following a neck artery nick. Serum samples were analyzed following coagulation and centrifugation.
For LegendPlex cytokine FACS-ELISA analysis, CSF and AF samples were diluted two-fold and processed according to the manufacturer’s instructions. Following resuspension, beads were run on a FACS Celesta (BD Biosciences) and results were analyzed by LegendPlex v7.1 software. For CCL2 sandwich ELISA (Abcam), CSF and serum samples were diluted for 10-fold and processed according to the manufacturer’s instructions.
Brain tissue processing for histology
Samples were fixed in 4% PFA at 4°C for 2-3 hrs. For cryosectioning, samples were sucrose protected as at 4°C as follows: 30% sucrose (overnight), 1:1 mixture of 30% sucrose and Optimal Cutting Temperature (OCT, overnight), and OCT (1 hr). Samples were then frozen in OCT and cryosectioned at 14 μm unless otherwise noted.
Immunostaining and image acquisition
Cryosections were rehydrated with PBS, then permeabilized and blocked with 5% goat serum in PBS with either 0.3% TritonX-100 (0.04% Tween-20 was used for CCL2 staining). Following overnight incubation with primary antibodies at 4°C, sections were washed, incubated with secondary antibodies at room temperature for one hour, and counterstained with Hoechst (ThermoFisher Scientific) before being mounted with Fluoromount-G (SouthernBiotech). E14.5-E15.5 ChP explants were micro-dissected in HyClone Hank's 1X Balanced Salt Solutions (HBSS, GE Healthcare) and drop-fixed in 4% PFA. After fixing for 15 min. at room temperature, ChP explants were stabilized on a Sylgard plate for immunostaining. For BrdU and Ki67 staining, an antigen retrieval step was included: A vegetable steamer (IHC World) was filled with water and preheated until the chamber temperature reached 100°C; sections were immersed in boiling citric acid buffer (10 mM citric acid, 0.05% Tween 20, pH 6) and steamed for 20 min. Sections were cooled to room temperature prior to immunostaining. RNAscope in situ hybridization to detect Ccl2 transcripts (ACD) was processed according to the manufacturer’s instructions. For Ccl2 transcript detection in MIA samples, due to low abundance of the transcript at baseline, a high-sensitivity DAB reaction protocol was used with 30 min incubation following the addition of DAB substrate mixture.
Epifluorescence images were acquired using a Nikon TiEclipse (Cellular Imaging Core, IDDRC, BCH). Confocal images (CCL2 staining and ZO-1 staining) were acquired with Zeiss LSM710 (IDDRC, Cellular Imaging Core, BCH). Image acquisition settings (wavelengths, exposure, gain, pinhole, etc.) were kept consistent for different samples.
RNA extraction, qRT-PCR
E14.5 LV ChP, 4V ChP, meninges and neuroepithelium were micro-dissected in HBSS. Tissue from three individuals were pooled together for RNA extraction (RecoverAll Total Nucleic Acid Isolation Kit, ThermoFisher Scientific). 100 ng RNA was used for reverse transcription (ImProm-II Reverse Transcription System, Promega), and 1 μl cDNA product was used for Taqman assay (Taqman Fast Universal PCR Master Mix, ThermoFisher Scientific).
Embryonic ChP single cell RNA Seq dataset
Mouse embryonic ChP single cell RNAseq dataset was acquired and analyzed by N.D., N.H., M.K.L., R.H.H., and A.R. in Dani et al., 2019. Full material and method details are available at: https://www.biorxiv.org/content/10.1101/627539v1.full. Briefly, whole embryonic ChP tissue from each ventricle was micro-dissected, digested, and live cells were FACS sorted. Single cells (~ 7,000 cells) were processed through the 10X Genomics Single Cell 3’ platform.
In utero intracerebroventricular (ICV) injections and BrdU pulses
Timed pregnant mice (E13.5) were anesthetized with isoflurane and placed on a heatpad. A laparotomy was performed and 1 μl of designated reagents (including fast green dye for easy visualization of the injected solution filling the ventricles) was introduced into one lateral ventricle. Mouse Ccl2 (NCBI reference sequence: NM_011333.3) was synthesized (IDT) and assembled into AAV2/5 vector (pAAV-MCS empty vector, Agilent) using EcoRI and HindIII (New England Biolabs) and produced by the Viral Core (Boston Children’s Hospital). For AAV transduction, the desired AAV was diluted in sterile PBS to 1×1013 gc/ml, and a final volume of 1 μl was introduced into one individual. Alternatively, 1 μl of 100 ng/ml CCL2 protein (Recombinant Mouse CCL2/JE/MCP-1 Protein, CF, R&D Systems, reconstituted in sterile PBS) was delivered per embryo and equal volume of sterile PBS was used as control. BrdU (50 mg/kg, Millipore Sigma; i.p.) was delivered maternally at designated time points.
Two-photon imaging of embryonic ChP
Timed-pregnant mice (E14.5) were anesthetized with isoflurane and placed on a heating pad. A laparotomy was performed, and embryos were gently exposed. Vasculature labeling was achieved by adapting a previous protocol (Ben-Zvi et al., 2014): 3.5 μl of 5 mg/ml Texas Red-Dextran (70 kD, ThermoFisher Scientific) was delivered into the developing liver using a fine-tip glass capillary tube. During the following five-minute circulation time, samples were kept moist using warm HBSS. The samples and the placenta were carefully transferred into an imaging chamber filled with continuously circulating artificial CSF at 37°C (Alzet formulation as provided online: https://www.alzet.com/guide-to-use/preparation-of-artificial-csf/, 150 mM Na, 3 mM K, 1.4 mM Ca, 0.8 mM Mg, 1 mM P, 155 mM Cl). The skin and skull over the 4th ventricle were removed, exposing the ventricular system.
3D volumetric images were acquired to robustly track the ChP across short and long timescales due to mouse motion, changes in posture, and occasional axial drift of ChP. Recordings were 30 to 45 min long on average. Two-photon imaging of immune cells was performed using a resonant-scanning two-photon microscope (Olympus MPE-RS Multiphoton Microscope; 12.8 frame/s; 512 × 512 pixels/frame; 0.16 volumes/s, 81 planes/volume; volume size: 254 × 254 × 400 μm3) and a 25X objective (Olympus XLPLN25XSVMP2, 1.0 NA, 4 mm W.D.) with ~1.3X zoom (~394 × 394 μm2). Volume scanning on the Olympus was achieved by using a piezoelectric microscope stage (nPFocus250). Laser power at 940 nm (Mai Tai DeepSee laser, Spectra Physics) measured below the objective was 30-40 mW.
Transmission electron microscopy
E14.5 LV ChP tissue was micro-dissected in HBSS (Thermo Fisher Scientific) and drop-fixed immediately in FGP fixative (5% Glutaraldehyde, 2.5% Paraformaldehyde and 0.06% picric acid in 0.2 M sodium cacodylate buffer, pH 7.4). After 2 hours of fixation at room temperature, the tissue was washed in 0.1 M cacodylate buffer and post-fixed with 1% Osmiumtetroxide (OsO4)/1.5% Potassium ferrocyanide (KFeCN6) for 1 hour, washed twice in water, and once in 50mM Maleate buffer pH 5.15 (MB) and incubated in 1% uranyl acetate in MB for 1hr followed by 1 wash in MB, 2 washes in water and subsequent dehydration in grades of alcohol (10 min each; 50%, 70%, 90%, 2×10 min 100%). Samples were then put in propyleneoxide for 1 hr and infiltrated ON in a 1:1 mixture of propyleneoxide and TAAB Epon (TAAB Laboratories Equipment Ltd, https://taab.co.uk). The following day, the samples were embedded in TAAB Epon and polymerized at 60°C for 48 hrs. Ultrathin sections (about 80 nm) were cut on a Reichert Ultracut-S microtome, picked up on to copper grids stained with lead citrate. Images were acquired with a JEOL 1200EX transmission electron microscope, and recorded with an AMT 2k CCD camera (Biological Electron Microscopy Facility, Harvard Medical School).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantifications of cell numbers and macrophage activation
For brain section staining and analysis, at least three sections from each individual were stained, imaged and analyzed. Sections from consistent anatomical locations in the ChP of at least three individuals (please refer to specific sample sizes in Figure Legends) were analyzed in each group. All cell number quantifications were performed in Fiji (ImageJ) automatically using the “Analyze particles” function in Fiji after adjusting brightness/contrast to a consistent setting and adjusting threshold (Otsu) to generate a binary image. If there were clearly two binary objects (e.g. cells in close contact to each other in the raw image), watershed was used to separate them. ChP tissue area was traced with “Polygon selections” according to Hoechst staining, measured and used to normalize cell numbers. Cells whose Iba1+ staining area appeared to be external to the traced tissue margin were considered “surface/epiplexus cells.” Circularity and solidity of Iba1+ macrophages were measured using “Shape descriptions” in combination with the “Analyze particles” function in Fiji. Iba1+ cell morphology was characterized based on previously published observations on developing microglia at similar developmental stages (Bolton et al., 2017; Wu et al., 1992). Using the “Analyze particles” function, the percentage of CD68+ signal within Iba1+ macrophages was quantified as (CD68+; Iba1+ / Iba1+) % to reflect ChP macrophage phagocytic activity.
Brain development severity score index
Disruption to brain development was scored based on CUX1, CTIP2, and Iba1 staining of four coronal sections per sample, and sections from consistent anatomical locations were analyzed. Point were assigned based on the presence of the following phenotypes: (1) alterations of gross brain morphology along the dorsal-lateral cerebral cortex; (2) cell distribution disruptions or clusters of cells occurring near the pial surface; (3) severe cortical thinning and ventricle enlargement; (4) severity of SVZ disruption (score = 1 if SVZ region thickness was irregular along the ventricle, score = 2 if massive macrophage infiltration was observed following Iba1 staining); (5) in the event of cortical patches, one point was given per patch. The total score from the above criteria provided the “Severity score” presented in Figure 6F.
Nearest neighbor analysis
The nearest neighbor analyses were carried out as previously described (Shipley et al., 2020). Specifically, locations of cell centers were selected using the Cell Counter plugin in Fiji. The tissue outline was drawn manually and the image region within this outline was converted to a binary mask using Fiji. Whole LV ChP explants were used to trace Cx3cr1GFP/+ cell locations. Areas where the tissue was pinned (to stabilize for staining Figure S4A, B) or folded were excluded from further analyses, to avoid bias in cell-cell distances introduced by tissue-pinning or tissue-folding. The nearest-neighbor distance was calculated for each individual cell, and the cumulative distribution function was plotted using the spatstat package in R.
Simulated distributions based on a 2D Poisson distribution of cell location, assuming random cell locations (vs. tiled spacing) were generated iteratively 100 times. The mean of these simulated distributions yielded the “Poisson” estimate, while their extrema yielded a p = 0.01 acceptance interval.
Studentized permutation test was used to compare differences of different groups (Baddeley et al., 2016). Basically, it does a random permutation of the point pattern list, and calculates the Monte Carlo P value based on measures of variances from the group means.
Analysis of CD45+/hi cells breaching the barrier near the tips or troughs of ChP villi
First, the user traced the border of the 3V ChP from a coronal IHC section. Next, the user manually selected the position of each immune cell that was putatively crossing the epithelium. The cell position on the border curve (outlining the epithelium of the ChP) was the point on the curve closest to the manually selected point. The border curve was reparameterized along its path such that each point on the curve had a unique path distance, D, starting from the leftmost edge of the ChP in the image. Crucially, D is monotonic, as opposed to the border curve, which can fold back on itself. The border curve “prominence” was plotted as a function of D, the border path distance from the leftmost edge. These values were high-pass filtered to account for the large curvature of the ChP, preserving only the prominence of the villi. Each cell position was also converted to a coordinate along the border curve. The local extrema of this curve represent the alternating tips and troughs of the ChP villi. The relative distance of each cell’s position from the nearest tip and trough is given as:
such that a point exactly on the tip would have a relative distance of 0, and a point in the trough would have a relative distance of 1. This relative distance score was calculated for each breaching cell across all AAV-CCL2 ChP samples.
Imaging registration
We used a previously described custom 3D registration pipeline in Matlab (Mathworks) to account for tissue motion and warping (Shipley et al., 2020). Due to the rapid motility of immune cells across seconds, 3D registration of individual volumes was necessary to properly account for ChP movement at these rapid timescales.
To account for fast intra-volume changes, within each individual volume, each plane was iteratively matched to its neighbor by cross-correlating the Fourier transformation of the two images to find the transverse X and Y shifts.
After intra-volume alignment, reference volumes were generated by averaging every 20 volumes. To account for inter-volume lateral and axial shifts, each volume was then registered to its respective reference volume by cross-correlating the 3D Fourier transformation of the two volumes to find the X, Y, and Z shifts. Each reference volume was registered to the first reference volume using the same method.
Axial projections, such as mean, median, and maximum projections, were then performed. Finally, the movie of these projected images was further stabilized in three successive steps: (i.) matching each frame to the average of the first 50 frames, (ii.) matching each frame of the resulting movie iteratively to its neighbor, (iii.) matching each frame of the resulting movie to the average of the first 50 frames.
Motility analysis
The aligned, 4D XYZT recording was mean projected over time to obtain an average XYZ volume. In this average volume, 3D XYZ coordinates of immune cell centroids were manually selected. True positions were defined as the weighted centroid within a 15x15x20 μm neighborhood centered on the manual selection.
For analyses of individual immune cells, the raw XYZT movie was cropped to a 40 x 40 x 50 μm3 area around a centroid. A 5-minute portion of this cropped 4D movie was then mean Z projected to obtain a 40 x 40 μm2 movie (XYT) video. A target image was created by averaging every frame of this movie. Local translational registration was performed by cross-correlating each movie frame with this target image in Fourier space (i.e. rigid-body translation correction, (Guizar-Sicairos and Fienup, 2008)). These XY translation corrections were cumulatively summed to calculate the total displacement of each cell in the XY plane.
The mean of the registered movie was taken as the mean fluorescence image. The mean fluorescence image was binarized using Otsu’s method in MATLAB. If there were two binary objects (e.g. from a neighboring cell), a watershed transform was applied, and the region ontaining the center of the field of view was considered the field of view, and the rest was masked out.
This masked, registered movie was bandpass filtered. A low-pass movie was made with a rolling mean of 3 frames; a high-pass movie was made with a rolling mean of 20 frames; the bandpass filtered movie was made by taking the absolute difference between the low-pass and high-pass movies. A standard deviation bandpass image was created by taking the standard deviation of the rectified, bandpass filtered movie; a mean image was created by taking the mean of the original movie over time. The standard deviation image was pixelwise divided by the mean image to make a normalized motility image.
The binarized fluorescent image was iteratively dilated to make 16 equidistant annuli, each with a width of 0.5 μm. The motility image was masked by each annulus mask, to make a 16-element array of average motility across all pixels at each distance from 0 to 8 μm beyond the outer edge of a cell (in 0.5 μm bins). This process was repeated for each individual cell centroid, and for each recorded movie.
Statistical Analyses
All experiments were repeated in a minimum of two litters of mice each group, from independent experiments, unless otherwise specified. Statistical analyses were performed using Prism 7 (GraphPad), R 3.5 and Python 2.7. Quantitative data from biological replicates are presented as the “mean ± S.E.M.”, unless otherwise noted. Appropriate statistical tests were selected based on sample size, distribution of data, and the homogeneity of variances. Outliers were excluded using the ROUT method (Q = 1%). For datasets with n > 8, D'Agostino-Pearson normality test was used to assess whether the dataset was normally distributed. Parametric tests were chosen for datasets with n < 8 and for larger datasets when the data were distributed normally. The Mann-Whitney test was used for non-parametric comparisons of two groups of datasets. The F test or Brown Forsythe’s test were used to assess variances between data sets from different groups. Differences between two different groups (subjected to different treatment) were analyzed by two-tailed unpaired t-test when variances between groups were similar, otherwise Welch’s t-test was selected. For the GLMM analysis of cell motility and mobility across conditions, data were analyzed in Python 2.7, and R 3.5; statistics were calculated using the lme4 and afex packages in R; P values were calculated using the Kenward-Roger approximation for degrees of freedom. For the nearest-neighbor analyses, studentized permutation test was used to compare differences of different groups. Please refer to figures and figure legends for statistical details. P values < 0.05 were considered significant (* P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001), P > 0.05 were not statistically significant (N.S.).
Supplementary Material
Video S1, Related to Figure 4. Live imaging of a 3D volume of ChP. 3D volume scan (following 3D registration, alignment and averaging of volumes) demonstrates 4V ChP vasculature (red, labeled using embryonic liver injection of Texas Red-conjugated dextran), and Cx3cr1+/GFP macrophages (green) in E14.5 Cx3cr1+/GFP. Scale bar: 50 μm. Same dataset as in Figure 4D. Note the curved shape of 4V ChP villi in lower half of each image, and the dark region at the perimeter of each villus (containing non-labeled ChP epithelial cells).
Video S2 Related to Figure 4. Time-lapse live imaging of Cx3cr1+/GFP ChP macrophages from control condition. This thirty-minute movie (each frame is a maximum intensity projections of a two-photon imaging volume) demonstrates tdTomato+ 4V ChP epithelial cells and adjacent Cx3cr1+/GFP ChP macrophages at E14.5, one day after ICV injection of AAV-Cre and AAV-tdTomato. Scale bar: 50 μm. Timestamps are in units of hours:minutes:seconds. See also still images in Figure 4E and Figure S4I.
Video S3 Related to Figure 4. An example movie showing interactions between processes of two Cx3cr1+/GFP ChP macrophages from control condition (AAV-Cre + AAV-tdTomato). Time-lapse video of maximum intensity projections of two-photon imaging volumes. Left: projection across Z, i.e. across the depth of the volume. Right: projection across X, i.e. across the horizontal width of the volume. Arrowhead indicates where processes from the two cells came into close proximity. Scale bar: 20 μm. Timestamps are in units of hours:minutes:seconds.
Video S4 Related to Figure 4. Time-lapse live imaging of ChP macrophages at E14.5 from CCL2 condition (AAV-CCL2 + AAV-tdTomato). Thirty-minute imaging dataset (maximum intensity projections of two-photon imaging volumes across Z planes) demonstrates tdTomato+ 4V ChP epithelial cells and adjacent Cx3cr1+/GFP macrophages. Scale bar in first frame: 50 μm. Timestamps are in units of hours:minutes:seconds. Note the large degree of motility of immune cell processes. See also Figure 4G and Figure S4I.
Video S5 Related to Figure 4. High-magnification movies of motility in individual ChP macrophages. Movies are from imaging sessions at E14.5 in the following conditions: (top) AAV-Cre + AAV-tdTomato, (bottom) AAV-CCL2 + AAV-tdTomato. Timestamps are in units of hours:minutes:seconds.
Video S6 Related to Figure 4. Example movie showing cell body mobility of a ChP macrophage in CCL2 condition (AAV-CCL2 + AAV-tdTomato). Time-lapse video of maximum intensity projections of two-photon imaging volumes. Left: projection across Z, i.e. across the depth of the volume. Right: projection across X, i.e. across the width of the volume. White arrowhead in all frames indicates the initial location of the cell body; green arrowhead in the last frame indicates the front edge of the cell at the end of the recording. Scale bar in first frame: 20 μm. Timestamps are in units of hours:minutes:seconds.
Video S7 Related to Figure 4. Montage of movies of cell process motility for individual Cx3cr1+/GFP ChP macrophages. Presented cells were from one AAV-Cre individual, or one CCL2 individual with greatest overall motility (see also Figure 4I). For each cell, two movies were generated: left movie: actual fluorescence signal over time; right: normalized motility (sliding estimate of standard deviation of band-passed fluorescence time series at each pixel) / (fluorescence intensity averaged across all frames in the movie). Time window is 5 min (the whole video). Timestamps are in units of minutes:seconds. Scale bars: 10 μm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-MCP1 (CCL2) | Abcam | Cat#ab7202 |
| Rabbit anti-Iba1 | Fujifilm Wako | Cat#019-19741 |
| Rat monoclonal [FA-11] anti-CD68 | Abcam | Cat#ab53444 |
| Chicken polyclonal to GFP | Abcam | Cat#ab13970 |
| Rabbit anti-RFP | Rockland | Cat#600-401-379 |
| Rat Anti-Mouse CD45, Clone 30-F11 | BD Pharmingen | Cat#550539 |
| Mouse Anti-BrdU (5-bromo-2’-deoxyuridine), Clone B44 | Becton Dickinson | Cat#347580 |
| Mouse monoclonal Anti-AQP1 (1/22) | Santa Cruz | cat#sc-32737 |
| Mouse Anti-Ki-67, Clone B56 | BD Pharmingen | cat#550609 |
| Rabbit polyclonal Anti-ZO-1 | ThermoFisher Scientific | cat#61-7300 |
| Rabbit Occludin Polyclonal Antibody | ThermoFisher Scientific | cat#71-1500 |
| Rabbit Claudin1 Polyclonal Antibody | ThermoFisher Scientific | cat#51-9000 |
| Mouse Anti-Clathrin Heavy Chain, Clone 23/Clathrin Heavy Chain | BD Transduction Laboratories | Cat#610449 |
| Rabbit polyclonal anti-CDP (CUX1) | Santa Cruz | Cat#sc-13024 |
| Rat monoclonal [25B6] to Ctip2 | Abcam | Cat#ab18465 |
| Rabbit polyclonal Cleaved Caspase-3 (Asp175) | Cell Signaling Technology | Cat#9661 |
| Rat monoclonal [ER-MP20] to Ly6c | Abcam | Cat#ab15627 |
| Bacterial and Virus Strains | ||
| AVV2/5-mCCL2-hGH | Boston Children’s Hospital viral core, IDDRC | N/A |
| AAV2/5-CAG-GFP-WPRE | Boston Children’s Hospital viral core, IDDRC | N/A |
| AAV2/5-CAG-Cre-WPRE | Boston Children’s Hospital viral core, IDDRC | N/A |
| AAV2/5-CAG-tdTomato-WPRE | Boston Children’s Hospital viral core, IDDRC | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| polyinosinic-polycytidylic acid (polyI:C) | Sigma | Cat#P9582, Lot#086M4045V, 077M4039V |
| Recombinant Mouse CCL2/JE/MCP-1 Protein, CF | R&D Systems | Cat#479-JE-050/CF |
| Critical Commercial Assays | ||
| LEGENDplex™ Mouse Inflammation Panel FACS-ELISA | BioLegend | Cat#740446 |
| MCP-1 ELISA kit | Abcam | Cat#ab208979 |
| RNAscope H202 & Protease Plus Reagents | ACD Bio | Cat#322330 |
| RNAscope 2.5 HD Dectection Reagent-BROWN | ACD Bio | Cat#322310 |
| RecoverAll Total Nucleic Acid Isolation Kit | ThermoFisher Scientific | Cat#AM1975 |
| ImProm-II Reverse Transcription System | Promega | Cat#A3800 |
| Taqman Fast Universal PCR Master Mix | ThermoFisher Scientific | Cat#4366073 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Taconic Farms, Charles River Laboratories | N/A |
| Mouse: CD-1 (ICR) | Charles River Laboratories | N/A |
| Mouse: Cx3cr1GFP (B6.129P2(Cg)-Cx3cr1tm1LItt/J) | The Jackson Laboratory | Stock# 005582 |
| Mouse: Ccl2 KO (B6.129S4-Ccl2tm1Rol/J) | The Jackson Laboratory | Stock# 004434 |
| Mouse: Ccr2RFP (B6.129(Cg)-Ccr2tm2.1Ifc/J) | The Jackson Laboratory | Stock# 017586 |
| Oligonucleotides | ||
| Mouse GAPD (GAPDH) Endogenous Control (VIC™/MGB probe) | ThermoFisher Scientific | Cat#4352339E |
| Mouse Ccl2 (FAM-MGB, Assay ID: Mm00441242_m1) | ThermoFisher Scientific | Cat#4453320 |
| Mouse Cxcl10 (FAM-MGB, Assay ID: Mm00445235_m1) | ThermoFisher Scientific | Cat#4453320 |
| Mouse Ccl3 (FAM-MGB, Assay ID: Mm00441259_g1) | ThermoFisher Scientific | Cat#4453320 |
| Mouse Ccl4 (FAM-MGB, Assay ID: Mm00443111_m1) | ThermoFisher Scientific | Cat#4453320 |
| Mouse Occln (FAM-MGB, Assay ID: Mm00500912_m1) | ThermoFisher Scientific | Cat# 4448892 |
| Mouse Cldn1 (FAM-MGB, Assay ID: Mm00516701_m1) | ThermoFisher Scientific | Cat# 4448892 |
| Mouse Cldn2 (FAM-MGB, Assay ID: Mm00516703_s1) | ThermoFisher Scientific | Cat# 4448892 |
| Mouse Cldn3 (FAM-MGB, Assay ID: Mm01196233_s1) | ThermoFisher Scientific | Cat#4448892 |
| Mouse GAPD (GAPDH) Endogenous Control (VIC-MGB) | ThermoFisher Scientific | Cat#4352339E |
| Mouse Ccl2 RNAscope probe (Probe-Mm-CCL2) | ACD Bio | Cat#311791 |
| Recombinant DNA | ||
| Mouse Ccl2 CDS (NCBI reference sequence: NM_011333.3) | IDT | gBlocks Gene Fragments |
| pAAV-MCS empty vector | Agilent | Cat#240071 |
| Software and Algorithms | ||
| LegendPlex v7.1 software | Biolegend | https://www.biolegend.com/enus/legendplex#software |
| GraphPad Prism 7.04 | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| FIJI (version 2.0.0-rc-69/1.52p) | ImageJ | https://imagej.net/Fiji |
| MATLAB 2018b | MathWorks | https://www.mathworks.com/?s_tid=gn_logo |
| Python 2.7 | Python Software Foundation | www.python.org/ |
| R 3.5 | The R Foundation | https://cran.r-project.org/ |
| Imaging registration and analysis algorithms | Shipley et al., 2020 and this study | https://github.com/LehtinenLab/Cui2020-code |
| Other | ||
| Hoechst 33342 | ThermoFisher Scientific | Cat#H3570 |
| 70kD Texas Red-Dextran | ThermoFisher Scientific | Cat#D1830 |
HIGHLIGHTS.
Embryonic choroid plexus (ChP) mounts inflammatory responses to maternal infection
Embryonic CSF contains key biomarkers of neurodevelopmental disorders
Two-photon imaging reveals macrophage motility and mobility at embryonic ChP
CCL2 weakens ChP barrier and recruits immune cells through ChP barrier hotspots
ACKNOWLEDGEMENTS
We thank the Lehtinen lab for helpful discussions; B. Stevens, A. Walker, T. Hammond, and M. Marin-Rodero for advice on MIA models; J. Reyes, V Lagomarsino, and K. Shaw for technical assistance; N. Chamberlin for manuscript advice; C. Chen and the IDDRC Cellular Imaging Core; M. Ericsson and HMS EM Facility; C. Wu and Y Zhang at BCH Viral Core. This work was supported by: NIH T32 HL110852 (J.C.), William Randolph Hearst Fellowship (J.C. and N.D.); Reagan Sloane Shanley Research Internship (N.D.); NSF Graduate Research Fellowship (F.B.S.); T32 5T32DK007516 (A.U.S.); David Mahoney Neuroimaging Grant Program - Dana Foundation, NIH New Innovator Award DP2 DK105570, R01 DK109930, McKnight Scholar Award, Pew Scholar Award (M.L.A.); Simons Foundation (SFARI [award #402089]), R01 NS088566 (M.K.L.), RF1DA048790 (M.K.L.); the New York Stem Cell Foundation (M.K.L.); BCH IDDRC 1U54HD090255; and Viral Core P30EY012196. M.K.L. is a New York Stem Cell Foundation - Robertson Investigator. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abdallah MW, Larsen N, Grove J, Nørgaard-Pedersen B, Thorsen P, Mortensen EL, and Hougaard DM (2012). Amniotic fluid chemokines and autism spectrum disorders: An exploratory study utilizing a Danish Historic Birth Cohort. Brain. Behav. Immun 26, 170–176. [DOI] [PubMed] [Google Scholar]
- Ashwood P, Wills S, and Van de Water J (2006). The immune response in autism: a new frontier for autism research. J. Leukoc. Biol 80, 1–15. [DOI] [PubMed] [Google Scholar]
- Baddeley A, Rubak E, and Turner R (2016). Spatial Point Patterns: Methodology and Applications with R (CRC Press Taylor & Francis Group, LLC; ). [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, and Salzman N (2008). Enteric Salmonellosis Disrupts the Microbial Ecology of the Murine Gastrointestinal Tract. 76, 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch K, Deczkowska A, David E, Castellano JM, Miller O, Kertser A, Berkutzki T, Barnett-Itzhaki Z, Bezalel D, Wyss-Coray T, et al. (2014). Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science (80-. ). 346, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Reuven L, and Reiner O (2019). Dynamics of cortical progenitors and production of subcerebral neurons are altered in embryos of a maternal inflammation model for autism. Mol. Psychiatry [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Lacoste B, Kur E, Andreone BJ, Mayshar Y, Yan H, and Gu C (2014). Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature 509, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbo SD, Block CL, Bolton JL, Hanamsagar R, and Tran PK (2018). Beyond infection - Maternal immune activation by environmental factors , microglial development , and relevance for autism spectrum disorders. 299, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton JL, Marinero S, Hassanzadeh T, Natesan D, Le D, Belliveau C, Mason SN, Auten RL, Bilbo SD, Ziats MN, et al. (2017). Gestational Exposure to Air Pollution Alters Cortical Volume , Microglial Morphology , and Microglia-Neuron Interactions in a Sex-Specific Manner. 9, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Rosário M, and Birchmeier W (2006). Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev 16, 51–59. [DOI] [PubMed] [Google Scholar]
- Chau KF, Springel MW, Broadbelt KG, Park H. yeon, Topal S, Lun MP, Mullan H, Maynard T, Steen H, LaMantia AS, et al. (2015). Progressive Differentiation and Instructive Capacities of Amniotic Fluid and Cerebrospinal Fluid Proteomes following Neural Tube Closure. Dev. Cell 35, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau KF, Shannon ML, Fame RM, Fonseca E, Mullan H, Johnson MB, Sendamarai AK, Springel MW, Laurent B, and Lehtinen MK (2018). Downregulation of ribosome biogenesis during early forebrain development. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, He Y, Tian Y, Wang Y, Wu Z, Lan T, Wang H, Cheng K, and Xie P (2020). Different Serotypes of Adeno-Associated Virus Vector- and Lentivirus-Mediated Tropism in Choroid Plexus by Intracerebroventricular Delivery. Hum. Gene Ther 31, 440–447. [DOI] [PubMed] [Google Scholar]
- Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, Hoeffer CA, Littman DR, and Huh JR (2016). The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science (80-. ). 351, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Martinez-Cerdeno V, and Noctor SC (2013). Microglia Regulate the Number of Neural Precursor Cells in the Developing Cerebral Cortex. J. Neurosci 33, 4216–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R, Zhou L, Kebede AA, and Barres BA (2010). Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani N, Herbst RH, Habib N, Head J, Dionne D, Nguyen L, McCabe C, Cui J, Shipley FB, Jang A, et al. (2019). A cellular and spatial map of the choroid plexus across brain ventricles and ages. BioRxiv 627539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, and Gan WB (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci 8, 752–758. [DOI] [PubMed] [Google Scholar]
- DiStasio MM, Nagakura I, Nadler MJ, and Anderson MP (2019). T lymphocytes and cytotoxic astrocyte blebs correlate across autism brains. Ann. Neurol 86, 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek CJ, Habgood MD, Dziegielewska KM, Potter A, and Saunders NR (2001). Permeability and route of entry for lipid-insoluble molecules across brain barriers in developing Monodelphis domestica. J. Physiol 536, 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek CJ, Habgood MD, Dziegielewska KM, and Saunders NR (2003). Structural characteristics and barrier properties of the choroid plexuses in developing brain of the opossum (Monodelphis domestica). J. Comp. Neurol 460, 451–464. [DOI] [PubMed] [Google Scholar]
- Ek CJ, Dziegielewska KM, Stolp H, and Saunders NR (2006). Functional effectiveness of the blood-brain barrier to small water-soluble molecules in developing and adult opossum (Monodelphis domestica). J. Comp. Neurol 496, 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt B, Vajkoczy P, and Weller RO (2017). The movers and shapers in immune privilege of the CNS. Nat. Immunol 18, 123–131. [DOI] [PubMed] [Google Scholar]
- Estes ML, and McAllister AK (2016). Maternal immune activation: Implications for neuropsychiatric disorders. Science (80-. ). 353, 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame RM, and Lehtinen MK (2020). Emergence and Developmental Roles of the Cerebrospinal Fluid System. Dev. Cell 52, 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame RM, Shannon ML, Chau KF, Head JP, and Lehtinen MK (2019). A concerted metabolic shift in early forebrain alters the CSF proteome and depends on MYC downregulation for mitochondrial maturation. Dev. 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia L, Kijima N, Morrissy AS, De Antonellis P, Guerreiro-Stucklin A, Holgado BL, Wu X, Wang X, Parsons M, Zayne K, et al. (2018). A Hematogenous Route for Medulloblastoma Leptomeningeal Metastases. Cell 172, 1050–1062.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghersi-Egea JF, Strazielle N, Catala M, Silva-Vargas V, Doetsch F, and Engelhardt B (2018). Molecular anatomy and functions of the choroidal blood-cerebrospinal fluid barrier in health and disease. Acta Neuropathol. 135, 337–361. [DOI] [PubMed] [Google Scholar]
- Guizar-Sicairos M, and Fienup JR (2008). Direct image reconstruction from a Fourier intensity pattern using HERALDO. Opt. Lett 33, 2668–2670. [DOI] [PubMed] [Google Scholar]
- Haddad MR, Donsante A, Zerfas P, and Kaler SG (2013). Fetal Brain-directed AAV Gene Therapy Results in Rapid , Robust , and Persistent Transduction of Mouse Choroid Plexus Epithelia. Mol. Ther. Acids 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. (2018). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe CL, LaFrance-Corey RG, Goddery EN, Johnson RK, and Mirchia K (2017). Neuronal CCL2 expression drives inflammatory monocyte infiltration into the brain during acute virus infection. J. Neuroinflammation 14, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, and Patterson PH (2011). Activation of the maternal immune system induces endocrine changes in the placenta via IL-6. Brain Behav. Immun 25, 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KL, Croen LA, Yoshida CK, Heuer L, Hansen R, Zerbo O, Delorenze GN, Kharrazi M, Yolken R, and Ashwood P (2016). Autism with intellectual disability is associated with increased levels of maternal cytokines and chemokines during gestation. 22, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser K, Jang A, Lun MP, Procházka J, Machon O, Procházková M, Laurent B, Gyllborg D, van Amerongen R, Kompaníková P, et al. (2020). MEIS-WNT5A axis regulates development of 4th ventricle choroid plexus. BioRxiv 2020.05.07.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K, Masuda T, Joana M, Jordão C, and Prinz M (2019). Macrophages at CNS interfaces : ontogeny and function in health and disease. Nat. Rev. Neurosci 20. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, Longman RS, Honda K, Littman DR, Choi GB, et al. (2017). Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott GW, Dziegielewska KM, Habgood MD, Li ZS, and Saunders NR (1997). Albumin transfer across the choroid plexus of South American opossum (Monodelphis domestica). J. Physiol 499, 179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel I, Chicha L, Britschgi M, Schobel SA, Bodmer M, Hellings JA, Toovey S, and Prinssen EP (2014). Maternal immune activation and abnormal brain development across CNS disorders. Nat. Rev. Neurol 10, 643–660. [DOI] [PubMed] [Google Scholar]
- Kowash HM, Potter HG, Edye ME, Prinssen EP, Bandinelli S, Neill JC, Hager R, and Glazier JD (2019). Poly(I:C) source, molecular weight and endotoxin contamination affect dam and prenatal outcomes, implications for models of maternal immune activation. Brain. Behav. Immun 82, 160–166. [DOI] [PubMed] [Google Scholar]
- Kracht L, Borggrewe M, Eskandar S, Brouwer N, Laman JD, Scherjon SA, Prins JR, Kooistra SM, and Eggen BJL (2020). Human fetal microglia acquire homeostatic immune-sensing properties early in development. 537, 530–537. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. (2011). The Cerebrospinal Fluid Provides a Proliferative Niche for Neural Progenitor Cells. Neuron 69, 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, and Barres BA (2018). Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol 18, 225–242. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Temple S, Møllgård K, Gehwolf R, Wagner A, Bauer H, Bauer HC, Phoenix TN, Dziegielewska KM, and Saunders NR (2012). Molecular characterisation of transport mechanisms at the developing mouse blood-CSF interface: A transcriptome approach. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Rutledge BJ, Gu L, Fiorillo J, Lukacs NW, Kunkel SL, North R, Gerard C, and Rollins BJ (1998). Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J. Exp. Med 187, 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Kyrargyri V, Arancibia-Cárcamo IL, Jolivet R, Kohsaka S, Bryan RM, and Attwell D (2018). Microglial Ramification, Surveillance, and Interleukin-1β Release Are Regulated by the Two-Pore Domain K + Channel THIK-1. Neuron 97, 299–312.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova NV, Yu CZ, Hsiao EY, Moore MJ, and Patterson PH (2012). Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain. Behav. Immun 26, 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada González F, Perrin P, et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670. [DOI] [PubMed] [Google Scholar]
- Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J, Courchesne E, and Everall IP (2010). Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol. Psychiatry 68, 368–376. [DOI] [PubMed] [Google Scholar]
- Murakami T, Felinski EA, and Antonetti DA (2009). Occludin phosphorylation and ubiquitination regulate tight junction trafficking and vascular endothelial growth factor-induced permeability. J. Biol. Chem 284, 21036–21046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LH, and Lenz KM (2017). Microglia depletion in early life programs persistent changes in social, mood-related, and locomotor behavior in male and female rats. Behav. Brain Res 316, 279–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, and Helmchen F (2005). Resting Microglial Cells Are Highly Dynamic Surveillants of Brain Parenchyma in Vivo. Science (80-. ). 308, 1314–1319. [DOI] [PubMed] [Google Scholar]
- Ouzounov DG, Wang T, Wang M, Feng DD, Horton NG, Cruz-Hernández JC, Cheng Y-T, Reimer J, Tolias AS, Nishimura N, et al. (2017). In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat. Methods 14, 388–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson PH (2011). Maternal infection and immune involvement in autism. Trends Mol. Med 17, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Coisne C, Baumjohann D, Benvenuto F, Bottinelli D, Lira S, Uccelli A, Lanzavecchia A, Engelhardt B, and Sallusto F (2009). C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat. Immunol 10, 514–523. [DOI] [PubMed] [Google Scholar]
- Rudolph MD, Graham AM, Feczko E, Miranda-Dominguez O, Rasmussen JM, Nardos R, Entringer S, Wadhwa PD, Buss C, and Fair DA (2018). Maternal IL-6 during pregnancy can be estimated from newborn brain connectivity and predicts future working memory in offspring. Nat. Neurosci 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders NR, Liddelow SA, and Dziegielewska KM (2012). Barrier mechanisms in the developing brain. Front. Pharmacol 3 March, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders NR, Dziegielewska KM, Møllg K, and Habgood MD (2018). Physiology and molecular biology of barrier mechanisms in the fetal and neonatal brain. 23, 5723–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B (2012). Microglia Sculpt Postnatal Neural Circuits in an Activity and Complement-Dependent Manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, and Baruch K (2014). The resolution of neuroinflammation in neurodegeneration: Leukocyte recruitment via the choroid plexus. EMBO J. 33, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MD (2018). Cerebrospinal fluid and the early brain development of autism. J. Neurodev. Disord 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Yim Y, Park A, Berrios J, Lafourcade M, Pascual LM, Soares N, Yeon Kim J, Kim S, Kim H, Waisman A, et al. (2017). Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipley FB, Dani N, Xu H, Deister C, Cui J, Head J, Klein E, Marsh S, Sadegh C, Fame RM, et al. (2020). Tracking calcium dynamics and immune surveillance at the choroid plexus blood-cerebrospinal fluid interface Neuron In press Available SSRN; Https//Ssrn.Com/Abstract=3554055 or Http//Dx.Doi.Org/10.2139/Ssrn.3554055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler JA, and Pleasure SJ (2011). We have got you “covered”: how the meninges control brain development. Curr. Opin. Genet. Dev 21, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA, Folias AE, Choe Y, May SR, Kume T, et al. (2009). Retinoic Acid from the Meninges Regulates Cortical Neuron Generation. Cell 139, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SEP, Li J, Garbett K, Mirnics K, and Patterson PH (2007). Maternal immune activation alters fetal brain development through interleukin-6. J. Neurosci 27, 10695–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SEP, Elliott RM, and Anderson MP (2012). Maternal immune activation increases neonatal mouse cortex thickness and cell density. J. Neuroimmune Pharmacol 7, 529–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, and Garel S (2014). Microglia Modulate Wiring of the Embryonic Forebrain. Cell Rep. 8, 1271–1279. [DOI] [PubMed] [Google Scholar]
- Stamatovic SM, Keep RF, Kunkel SL, and Andjelkovic AV (2003). Potential role of MCP-1 in endothelial cell tight junction “opening” signaling via Rho and Rho kinase. J. Cell Sci 116, 4615 LP–4628. [DOI] [PubMed] [Google Scholar]
- Stamatovic SM, Shakui P, Keep RF, Moore BB, Kunkel SL, Van Rooijen N, and Andjelkovic AV (2005). Monocyte chemoattractant protein-1 regulation of blood-brain barrier permeability. J. Cereb. Blood Flow Metab 25, 593–606. [DOI] [PubMed] [Google Scholar]
- Stolp HB, Dziegielewska KM, Ek CJ, Habgood MD, Lane MA, Potter AM, and Saunders NR (2005a). Breakdown of the blood-brain barrier to proteins in white matter of the developing brain following systemic inflammation. Cell Tissue Res. 320, 369–378. [DOI] [PubMed] [Google Scholar]
- Stolp HB, Dziegielewska KM, Ek CJ, Potter AM, and Saunders NR (2005b). Long-term changes in blood-brain barrier permeability and white matter following prolonged systemic inflammation in early development in the rat. Eur. J. Neurosci 22, 2805–2816. [DOI] [PubMed] [Google Scholar]
- Stolp HB, Turnquist C, Dziegielewska KM, Saunders NR, Anthony DC, and Molnár Z (2011). Reduced ventricular proliferation in the foetal cortex following maternal inflammation in the mouse. Brain 134, 3236–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, and Pardo CA (2005). Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann. Neurol 57, 67–81. [DOI] [PubMed] [Google Scholar]
- Williamson LL, Sholar PW, Mistry RS, Smith SH, and Bilbo SD (2011). Microglia and Memory : Modulation by Early-Life Infection. 31, 15511–15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton DK, Dissing-Olesen L, and Stevens B (2019). Neuron-Glia Signaling in Synapse Elimination. Annu. Rev. Neurosci 42, 107–127. [DOI] [PubMed] [Google Scholar]
- Wu CH, Wen CY, Shieh JY, and Ling EA (1992). A quantitative and morphometric study of the transformation of amoeboid microglia into ramified microglia in the developing corpus callosum in rats. J. Anat 181, 423–430. [PMC free article] [PubMed] [Google Scholar]
- Yuryev M, Pellegrino C, Jokinen V, Andriichuk L, Khirug S, Khiroug L, and Rivera C (2016). In vivo Calcium Imaging of Evoked Calcium Waves in the Embryonic Cortex. Front. Cell. Neurosci 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappaterra MW, Lamantia AS, Walsh CA, and Lehtinen MK (2013). Isolation of Cerebrospinal Fluid from Rodent Embryos for use with Dissected Cerebral Cortical Explants. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1, Related to Figure 4. Live imaging of a 3D volume of ChP. 3D volume scan (following 3D registration, alignment and averaging of volumes) demonstrates 4V ChP vasculature (red, labeled using embryonic liver injection of Texas Red-conjugated dextran), and Cx3cr1+/GFP macrophages (green) in E14.5 Cx3cr1+/GFP. Scale bar: 50 μm. Same dataset as in Figure 4D. Note the curved shape of 4V ChP villi in lower half of each image, and the dark region at the perimeter of each villus (containing non-labeled ChP epithelial cells).
Video S2 Related to Figure 4. Time-lapse live imaging of Cx3cr1+/GFP ChP macrophages from control condition. This thirty-minute movie (each frame is a maximum intensity projections of a two-photon imaging volume) demonstrates tdTomato+ 4V ChP epithelial cells and adjacent Cx3cr1+/GFP ChP macrophages at E14.5, one day after ICV injection of AAV-Cre and AAV-tdTomato. Scale bar: 50 μm. Timestamps are in units of hours:minutes:seconds. See also still images in Figure 4E and Figure S4I.
Video S3 Related to Figure 4. An example movie showing interactions between processes of two Cx3cr1+/GFP ChP macrophages from control condition (AAV-Cre + AAV-tdTomato). Time-lapse video of maximum intensity projections of two-photon imaging volumes. Left: projection across Z, i.e. across the depth of the volume. Right: projection across X, i.e. across the horizontal width of the volume. Arrowhead indicates where processes from the two cells came into close proximity. Scale bar: 20 μm. Timestamps are in units of hours:minutes:seconds.
Video S4 Related to Figure 4. Time-lapse live imaging of ChP macrophages at E14.5 from CCL2 condition (AAV-CCL2 + AAV-tdTomato). Thirty-minute imaging dataset (maximum intensity projections of two-photon imaging volumes across Z planes) demonstrates tdTomato+ 4V ChP epithelial cells and adjacent Cx3cr1+/GFP macrophages. Scale bar in first frame: 50 μm. Timestamps are in units of hours:minutes:seconds. Note the large degree of motility of immune cell processes. See also Figure 4G and Figure S4I.
Video S5 Related to Figure 4. High-magnification movies of motility in individual ChP macrophages. Movies are from imaging sessions at E14.5 in the following conditions: (top) AAV-Cre + AAV-tdTomato, (bottom) AAV-CCL2 + AAV-tdTomato. Timestamps are in units of hours:minutes:seconds.
Video S6 Related to Figure 4. Example movie showing cell body mobility of a ChP macrophage in CCL2 condition (AAV-CCL2 + AAV-tdTomato). Time-lapse video of maximum intensity projections of two-photon imaging volumes. Left: projection across Z, i.e. across the depth of the volume. Right: projection across X, i.e. across the width of the volume. White arrowhead in all frames indicates the initial location of the cell body; green arrowhead in the last frame indicates the front edge of the cell at the end of the recording. Scale bar in first frame: 20 μm. Timestamps are in units of hours:minutes:seconds.
Video S7 Related to Figure 4. Montage of movies of cell process motility for individual Cx3cr1+/GFP ChP macrophages. Presented cells were from one AAV-Cre individual, or one CCL2 individual with greatest overall motility (see also Figure 4I). For each cell, two movies were generated: left movie: actual fluorescence signal over time; right: normalized motility (sliding estimate of standard deviation of band-passed fluorescence time series at each pixel) / (fluorescence intensity averaged across all frames in the movie). Time window is 5 min (the whole video). Timestamps are in units of minutes:seconds. Scale bars: 10 μm.
Data Availability Statement
The imaging registration, cell motility and mobility analysis algorithms generated for this study are available at https://github.com/LehtinenLab/Cui2020-code. Original data is available from the corresponding author upon request.