Abstract
Machine learning models for radiology benefit from large-scale data sets with high quality labels for abnormalities. We curated and analyzed a chest computed tomography (CT) data set of 36,316 volumes from 19,993 unique patients. This is the largest multiply-annotated volumetric medical imaging data set reported. To annotate this data set, we developed a rule-based method for automatically extracting abnormality labels from free-text radiology reports with an average F-score of 0.976 (min 0.941, max 1.0). We also developed a model for multi-organ, multi-disease classification of chest CT volumes that uses a deep convolutional neural network (CNN). This model reached a classification performance of AUROC > 0.90 for 18 abnormalities, with an average AUROC of 0.773 for all 83 abnormalities, demonstrating the feasibility of learning from unfiltered whole volume CT data. We show that training on more labels improves performance significantly: for a subset of 9 labels – nodule, opacity, atelectasis, pleural effusion, consolidation, mass, pericardial effusion, cardiomegaly, and pneumothorax – the model’s average AUROC increased by 10% when the number of training labels was increased from 9 to all 83. All code for volume preprocessing, automated label extraction, and the volume abnormality prediction model is publicly available. The 36,316 CT volumes and labels will also be made publicly available pending institutional approval.
Keywords: chest computed tomography, multilabel classification, convolutional neural network, deep learning, machine learning
Graphical abstrtact
1. Introduction1
Automated interpretation of medical images using machine learning holds immense promise (Hosny et al., 2018; Kawooya, 2012; Schier, 2018). Machine learning models learn from data without being explicitly programmed and have demonstrated excellent performance across a variety of image interpretation tasks (Voulodimos et al., 2018). Possible applications of such models in radiology include human-computer interaction systems intended to further reduce the 3 – 5% real-time diagnostic error rate of radiologists (Lee et al., 2013) or automated triage systems that prioritize scans with urgent findings for earlier human assessment (Annarumma et al., 2019; Yates et al., 2018). Previous work applying machine learning to CT interpretation has focused on prediction of one abnormality at a time. Even when successful, such focused models have limited clinical applicability because radiologists are responsible for a multitude of findings in the images. To address this need, we investigate the simultaneous prediction of multiple abnormalities using a single model.
There has been substantial prior work on multiple-abnormality prediction in 2D projectional chest radiographs facilitated by the publicly available ChestX-ray14 (Wang et al., 2017), CheXpert (Irvin et al., 2019), and MIMIC-CXR (Johnson et al., 2019) datasets annotated with 14 abnormality labels. However, to the best of our knowledge, multilabel classification of whole 3D chest computed tomography (CT) volumes for a diverse range of abnormalities has not yet been reported. Prior work on CTs includes numerous models that evaluate one class of abnormalities at a time – e.g., lung nodules (Ardila et al., 2019; Armato et al., 2011; Pehrson et al., 2019; Shaukat et al., 2019; Zhang et al., 2018), pneumothorax (Li et al., 2019), emphysema (Humphries et al., 2019), interstitial lung disease (Anthimopoulos et al., 2016; Bermejo-Peláez et al., 2020; Christe et al., 2019; Christodoulidis et al., 2017; Depeursinge et al., 2012; Gao et al., 2018, 2016; Walsh et al., 2018; Wang et al., 2019), liver fibrosis (Choi et al., 2018), colon polyps (Nguyen et al., 2012), renal cancer (Linguraru et al., 2011), vertebral fractures (Burns et al., 2016), and intracranial hemorrhage (Kuo et al., 2019; Lee et al., 2019). The public DeepLesion dataset (Yan et al., 2018) has enabled multiple studies on detection of focal lesions (Khajuria et al., 2019; Shao et al., 2019). There are three obstacles to large-scale multilabel classification of whole CTs: acquiring sufficently large datasets, preparing labels for each volume, and the technical challenges of developing a large-scale multi-label machine learning model for the task. In this study, we address all of these challenges in order to present a fully automated algorithm for multi-organ and multi-disease diagnosis in chest CT.
Acquiring a large CT dataset appropriate for computational analysis is challenging. There is no standardized software for bulk downloading and preprocessing of CTs for machine learning purposes. Each CT scan is associated with multiple image sets (“series”), each comprising on the order of 100,000,000 voxels. These volumes need to be organized and undergo many pre-processing steps.
To train a multilabel classification model, each volume must be associated with structured labels indicating the presence or absence of abnormalities. Given the number of organs and diseases, manual abnormality labeling by radiologists for the thousands of cases required to train an accurate machine learning model is virtually impossible. Instead, methods that automatically extract accurate labels from radiology reports are necessary (Irvin et al., 2019; Johnson et al., 2019; Wang et al., 2017).
Prior work in automated label extraction from radiology reports can be divided into two primary categories: whole-report classifiers (Banerjee et al., 2017; Chen et al., 2018; Pham et al., 2014; Zech et al., 2018) that predict all labels of interest simultaneously from a numerical representation of the full text, and rule-based methods that rely on handcrafted rules to assign abnormality labels. Whole-report classifiers suffer two key drawbacks: they are typically uninterpretable and they require expensive, time-consuming manual labeling of training reports, where the number of manual labels scales linearly with the number of training reports and with the number of abnormalities. Rule-based systems (Chapman et al., 2001; Demner-Fushman et al., 2016; Irvin et al., 2019; Peng et al., 2018) are a surprisingly good alternative, as radiology language is rigid in subject matter, content, and spelling. We propose and validate a rule-based label extraction approach for chest CT reports designed to extract 83 abnormality labels.
Development of a multi-label classification model is challenging due to the complexity of multi-organ, multi-disease identification from CT scans. We will show that the frequency of particular abnormalities in CTs varies greatly, from nodules (78%) to hemothorax (<1%). There are hundreds of possible abnormalities; multiple abnormalities usually occur in the same scan (10±6); and the same abnormality can occur in multiple locations in one scan. Different abnormalities can appear visually similar, e.g., atelectasis and pneumonia (Edwards et al., 2016), and the same abnormality can look visually different depending on severity (e.g., pneumonia of one lobe vs. an entire lung) (Franquet, 2001), shape (e.g., smooth nodule vs. spiculated nodule), and texture (e.g., reticular vs. groundglass) (Dhara et al., 2016). Variation in itself is not necessarily pathologic – even among “normal” scans the body’s appearance differs based on age, gender, weight, and natural anatomical variants (Hansell, 2010; Terpenning and White, 2015). Furthermore, there are hardly any “normal” scans available to teach the model what “normality” is. We will show that <1% of chest CTs in our data are “normal” (i.e., lacking any of the 83 considered abnormalities). This low rate of normality is likely a reflection of requisite pre-test probabilities for disease that physicians consider before recommending CT and its associated exposure to ionizing radiation (Costello et al., 2013; Purysko et al., 2016; Smith-Bindman et al., 2009).
Previous single-abnormality CT classification studies have relied on time-intensive manual labeling of CT pixels (Kuo et al., 2019; Li et al., 2019; Walsh et al., 2018), patches (Anthimopoulos et al., 2016; Bermejo-Peláez et al., 2020; Christodoulidis et al., 2017; Gao et al., 2018), or slices (Gao et al., 2016; Lee et al., 2019) that typically limits the size of the data set to <1,000 CTs and restricts the total number of abnormalities that can be considered. Inspired by prior successes in the field of computer vision on identifying hundreds of classes in whole natural images (Deng et al., 2009; Rawat and Wang, 2017), we hypothesize that it should be possible to learn multi-organ, multi-disease diagnosis from whole CT data given sufficient training examples. We build a model that learns directly from whole CT volumes without any pixel, patch, or slice-level labels, and find that transfer learning and aggregation of features across the craniocaudal extent of the scan enables high performance on numerous abnormalities.
In this study we address the challenges of CT data preparation, automated label extraction from free-text radiology reports, and simultaneous multiple abnormality prediction from CT volumes using a deep convolutional neural network. We hope that this work will contribute to the long-term goal of automated radiology systems that assist radiologists, accelerate the medical workflow, and benefit patient care.
2. Methods
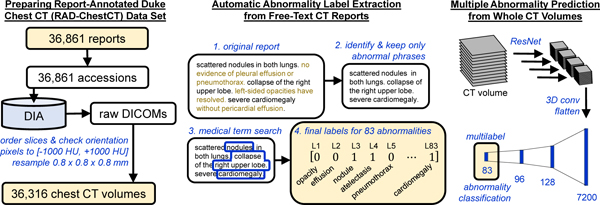
An overview of this study is shown in Figure 1.
Figure 1.
Study Overview. (a) Reports from chest CT scans performed without intravenous contrast material were acquired from the Duke Enterprise Data Unified Content Explorer (DEDUCE) search tool as well as the Epic electronic health record (EHR). Report accession numbers were used to download CT slices as DICOMs from the Duke Image Archive (DIA), which were processed into a final data set of 36,316 CT volumes. (b) We develop an approach for extracting binary labels for 83 different abnormalities from the free-text chest CT reports. (c) We train and evaluate a deep convolutional neural network model (shown here and detailed further in Figure 2) that takes as input a whole CT volume and predicts all 83 abnormality labels simultaneously.
2.1. Chest CT Data Set Preparation
The preparation of our retrospective chest CT data set included four stages: report download, report processing, volume download, and volume processing. The final dataset of 36,316 chest CT volumes obtained without intravenous contrast material and their associated reports from Duke University Health System spans January 2012 – April 2017 and was collected under IRB approval and in compliance with HIPAA. Informed consent was waived by the IRB.
In the first stage, 440,822 CT reports were obtained using the electronic health record and the Duke Enterprise Data Unified Content Explorer (DEDUCE) (Horvath et al., 2011) search tool. After filtering to remove duplicates, preliminary reports, un-addended versions, and empty reports, a dataset of 336,800 unique CT reports was obtained. This data set included head, chest, abdomen, and pelvis CTs, and include both intravenous contrast material enhanced and unenhanced scans. We then selected the 11% of reports (36,861) of chest CTs performed without intravenous contrast material based on the “protocol description” field.
Report text was prepared with standard natural language processing (NLP) preprocessing steps including lowercasing, replacing all whitespace with a single space, and removal of punctuation except for the periods inside decimal numbers which carry medical meaning (e.g., 1.2 cm mass versus 12 cm mass). We replaced all times with a “%time” token, dates with “%date”, and years with “%year.”
The chest CT scans were queried and downloaded using an application programming interface (API) developed for the Duke vendor neural archive. All scans were stored and preprocessed within the Duke Protected Analytics Computing Environment (PACE) which is a secure virtual network space that enables approved users to work with identifiable protected health information.
Using DICOM header information, the original series with the most slices was selected for subsequent analysis, thus rejecting secondary, derived, or reformatted series. In total 36,316 volumes were acquired out of 36,861 initially specified. Across the 36,316 scans, the slice thickness and reconstruction intervals were 0.625 mm for 54% of scans, 0.6 mm for 41% of scans, and other values ≤5 mm for the remaining 5% of scans. The scans came from two vendors, General Electric (57%) and Siemens (43%), and twelve different CT scanner models, representing the entire fleet across the Duke Health system. 53% of scans were reconstructed using filtered back projection (FBP), while the remaining 47% were produced with iterative reconstruction. Common scan indications include pulmonary nodules, cancer, and interstitial lung disease.
An end-to-end Python pipeline was developed to process the separate DICOM files corresponding to different slices of one CT into a single 3D numpy array (Van Der Walt et al., 2011) compatible with the major machine learning frameworks PyTorch (Paszke et al., 2019) and Tensorflow (Abadi et al., 2016). CT sections were ordered and verified to be in a consistent orientation to facilitate future work in abnormality localization. Raw pixel values in DICOMs have undergone a linear transformation to enable efficient disk storage; this transformation was reversed to obtain pixel values in Hounsfield units (HU), using the DICOM attributes RescaleSlope and RescaleIntercept. Pixel values were clipped to [−1000 HU,+1000 HU], which represent practical lower and upper limits of the HU scale, corresponding to the radiodensities of air and dense bone respectively (DenOtter and Schubert, 2019; Lamba et al., 2014). Each volume was resampled using SimpleITK (Lowekamp et al., 2013) to 0.8 × 0.8 × 0.8 mm to enable a consistent physical distance meaning of one pixel across all patients. To reduce storage requirements, the final 3D array was saved using lossless zip compression. The raw unprocessed DICOMs require 9.2 terabytes of storage. The final preprocessed arrays require 2.8 terabytes.
The data were randomly split into 70% volume training (25,355 volumes), 6% volume validation (2,085 volumes), 4% reserved for future studies (1,667), and 20% volume test (7,209 volumes) based on patient MRN so that no patient appears in more than one set. A subset of the volume training data was designated “report train” (639 reports) and “report test” (427 reports) and used to develop the automated label extraction method. Finally, we define a random subset of 2,000 training and 1,000 validation set scans that we use for architecture and ablation studies.
In total, 36,316 volumes paired with reports were successfully downloaded and prepared for analysis. We refer to the full data set as the Report-Annotated Duke Chest CTs (RAD-ChestCT) data set.
2.2. Automated image labeling through analysis of radiology reports
Manually recording the presence or absence of 83 different abnormalities for each of 36,316 volumes would require hand-coding over 3 million labels and is prohibitively time-consuming. To create a large data set of labeled CT volumes it is necessary to leverage automated label extraction approaches.
We follow the experimental setup of ChestX-Ray8 (Wang et al., 2017), CheXpert (Irvin et al., 2019), and MIMIC-CXR (Johnson et al., 2019), in which subsets of ~200 to 1,000 reports are used to develop and evaluate an automated label extraction method, and subsequently the final label extraction model is applied to the remaining tens of thousands of reports to obtain predicted labels. The predicted labels are then treated as ground truth in image-based experiments. We use 639 reports as a training set and 427 reports as a held-out test set.
The goal of radiology report label extraction is to analyze a free-text report and produce a binary vector of labels in which an entry is equal to one if the corresponding abnormality is present and is equal to zero otherwise. For example, if the predefined label order is [nodule, atelectasis, cardiomegaly] then the label [1,1,0] will be produced for a report in which nodule and atelectasis are present and cardiomegaly is absent.
A key challenge in radiology report label extraction is that the presence of a word, e.g., “nodule,” does not necessarily indicate presence of the abnormality in the scan, e.g., “the previously seen nodule is no longer visualized.” Furthermore, there are numerous phrasings for the same label, e.g., “enlarged heart,” “hypertrophic ventricles,” and “severe cardiomegaly” for the label “cardiomegaly.”
We propose two closely related approaches to extract 83 abnormalities labels from radiology reports, which we term Sentence Analysis for Radiology Label Extraction (SARLE). The first approach, SARLE-Hybrid, uses a machine learning sentence classifier followed by a rule-based term search, and is intended to be easier to adapt to text reports from other radiology modalities as it abstracts away abnormality detection rules. The second approach, SARLE-Rules, is fully rule-based and customized for chest CT reports. Both approaches are designed to be conceptually simple, to scale to a large number of abnormality labels, and to assign multiple abnormalities per scan when the report indicates that multiple abnormalities are present.
The language used in radiology reports varies by modality, anatomical region, and institution. We chose to create a custom lexicon targeted to the RAD-ChestCT dataset, rather than conforming a prospectively derived ontology to the dataset. Our 83 labels were chosen in an iterative manner with the goal of identifying all abnormalities mentioned in this dataset with at least ~2% frequency. First, we prepared an initial list of common abnormalities (e.g. nodule, cardiomegaly). Next, training set sentences that did not contain any of these abnormalities were examined in order to identify additional abnormalities. This process of abnormality sentence tagging followed by examination of untagged sentences was repeated until the final list of 83 abnormalities was obtained. The 83 extracted labels are shown in Table 1.
Table 1. List of 83 Abnormalities that SARLE Extracts from Radiology Reports.
Note that each abnormality is associated with a set of medical synonyms that are defined in a term search step. For example, the term search for cardiomegaly captures “cardiomegaly,” “dilated ventricles,” “enlarged right atrium,” and other synonyms. “Lung resection” captures pneumonectomy and lobectomy; “breast surgery” captures mastectomy and lumpectomy; pleural effusion captures “pleural effusion” and “pleural fluid” and so on. The term search for all abnormalities is available in Appendix B.
| Lung (22) | airspace disease, air trapping, aspiration, atelectasis, bronchial wall thickening, bronchiectasis, bronchiolectasis, bronchiolitis, bronchitis, consolidation, emphysema, hemothorax, interstitial lung disease, lung resection, mucous plugging, pleural effusion, infiltrate, pleural thickening, pneumonia, pneumonitis, pneumothorax, pulmonary edema, scattered nodules, septal thickening, tuberculosis |
| Lung Patterns (5) | bandlike or linear, groundglass, honeycombing, reticulation, tree in bud |
| Additional (47) | arthritis, atherosclerosis, aneurysm, breast implant, breast surgery, calcification, cancer, catheter or port, cavitation, clip, congestion, cyst, debris, deformity, density, dilation or ectasia, distention, fibrosis, fracture, granuloma, hardware, hernia, infection, inflammation, lesion, lucency, lymphadenopathy, mass, nodule, nodule > 1 cm, opacity, plaque, postsurgical, scarring, scattered calcifications, secretion, soft tissue, staple, stent, suture, transplant, chest tube, tracheal tube, GI tube (includes NG and GJ tubes) |
| Heart (9) | CABG (coronary artery bypass graft), cardiomegaly, coronary artery disease, heart failure, heart valve replacement, pacemaker or defibrillator, pericardial effusion, pericardial thickening, sternotomy |
The first approach, SARLE-Hybrid, is a hybrid machine learning and rule-based method. The motivation behind this approach is to use machine learning to eliminate the need for hand-crafted negation detection rules found in fully rule-based methods (Chapman et al., 2001; Peng et al., 2018), while enabling scaling to a large number of abnormality labels through a rule-based term search that leverages medical vocabulary. Note that wholly machine learning approaches, i.e., report classifiers (Chen et al., 2018; Pham et al., 2014; Zech et al., 2018) that take in a numerical representation of the full report text and output the entire vector of predicted abnormalities, require intensive manual labeling of all abnormalities of interest for all reports in the training set, which limits the size of the training data, limits the number of abnormalities that can be predicted, and is a suboptimal choice for rare labels that will have insufficient training examples.
Instead of performing abnormality-specific whole-report classification, the first step of SARLE-Hybrid performs binary sentence classification, distinguishing only between “normal” and “abnormal” sentences rather than particular abnormalities. We define a sentence as “normal” if it describes normal findings, e.g., “the lungs are clear,” or the lack of abnormal findings, e.g., “no masses.” We define a sentence as “abnormal” if it describes the presence of abnormal findings, e.g., “pneumonia in the right lung”; missing organs, e.g., “thyroid is absent”; or presence of devices, lines, or tubes. We train a Fasttext model (Bojanowski et al., 2017; Joulin et al., 2017; Mikolov et al., 2013) on manually labeled sentences from the 669 training reports. This system allows sentences to be subsequently classified as indicating one or multiple abnormalities.
After the sentence classification step, a rule-based term search using medical vocabulary for each abnormality is applied to the “abnormal” sentences to determine exactly which abnormal findings are present. We designed the term search to be easily modifiable in the code for customization to other abnormalities of interest. Examples of the term search are shown in Table 2. A full description of the entire term search for all 83 abnormalities is provided in Appendix B.
Table 2. Examples of the term search used in our radiology label extraction framework, from simple (e.g., mass) to complex (e.g., cardiomegaly).
The presence of any word in the “Any” column will result in considering the associated abnormality positive. The presence of any word in the “Term 1” column along with any word in the “Term 2” column will result in considering the associated abnormliaty positive. “Example Matches” shows example words and phrases that will result in a positive label for that abnormlaity based on the term search. Appendix B includes the full term search.
| Abnormality | Any | Term 1 | Term 2 | Example Matches |
|---|---|---|---|---|
| ‘mass’ | ‘mass’ | mass, masses | ||
| ‘nodule’ | ‘nodul’ | nodule, nodular, nodularity | ||
| ‘opacity’ | ‘opaci’ | opacity, opacities, opacification | ||
| ‘pericardial effusion’ | ‘pericardial effusion’,’pericardial fluid’ | ‘pericardi’ | ‘fluid’,’effusion’ | “pericardial effusion present”; “effusion in the pericardial sac”; “fluid also seen in the pericardial space” |
| ‘cardiomegaly’ | ‘cardiomegaly’ | ‘large’, ‘increase’, ‘prominent’,’dilat’ | ‘cardiac’, ‘heart’, ‘ventric’, ‘atria’, ‘atrium’ | “ventricular enlargement”; “the heart is enlarged”; “atrial dilation”; “increased heart size” |
The next variant, SARLE-Rules, is purely rule-based. It is identical to SARLE-Hybrid except that instead of a machine learning classifier in the first step, a rule-based system is used to identify phrases that are medically “normal” vs. “abnormal.” The advantages of SARLE-Rules are full interpretability and better handling of the minority of sentences that include both a normal and an abnormal statement (e.g., “the heart is enlarged without pericardial effusion”). The disadvantage is the extra work required to craft the rules. The rule-based phrase classifier differs from prior work in that it incorporates negation detection as well as “normality detection” based around words like “patent” (e.g., “the vessels are patent”). Negation scopes are defined directly on the sentence text, include a direction (forward/backward), and can be limited by other words (e.g., “and”, “with”) or the beginning/end of a sentence. The entirety of our “abnormality detection” including all negation detection requires fewer than 300 lines of Python code and does not have any dependencies on pretrained models (code is available at https://github.com/rachellea).
We report F-score, precision, recall, and accuracy of SARLE-Hybrid and SARLE-Rules on a held-out test set of 427 reports that were not used for classifier training or rule development. For the test reports we manually recorded abnormality-specific ground truth for 9 abnormalities commonly studied in the chest medical imaging literature: nodule, mass, opacity, consolidation, atelectasis, pleural effusion, pneumothorax, pericardial effusion, and cardiomegaly, for a total of 3,843 manual labels. The ground truth was obtained by a single observer (R.L.D., a 6th-year MD/PhD candidate). To ensure high label quality, a second observer (G.D.R, a fellowship-trained cardiothoracic radiologist) produced a random subset of 918 labels independently, resulting in 99% agreement.
We also report the F-score for the publicly available CheXpert labeler (Irvin et al., 2019) for the six CheXpert label categories that align with our label categories. We consider the CheXpert labeler’s “uncertain” outputs (e.g. “possible atelectasis”) as “positive” to match the way that our conservative ground truth was created.
Note that SARLE produces 83 abnormality labels per report, but due to the expense of obtaining abnormality-level ground truth, we only explicitly evaluate SARLE’s performance on this subset of 9 labels. We later demonstrate the value of the additional 74 labels by showing that they improve the performance of the downstream task of multilabel CT volume classification.
2.3. Development and evaluation of a whole CT volume multi-organ, multi-disease classifier
With the dataset and labels prepared, we train and evaluate a deep CNN to predict all abnormalities present in a CT volume. Following prior work on large-scale radiology datasets, we consider the automatically extracted labels ground truth (Irvin et al., 2019; Johnson et al., 2019).
Many architectures have been developed for image classification, including AlexNet (Krizhevsky et al., 2012), VGG (Simonyan and Zisserman, 2014), GoogLeNet (Szegedy et al., 2015), and ResNet (He et al., 2015). In medical imaging analysis, it is common to first pre-train one of these architectures on a large public data set of natural images (e.g., ImageNet (Deng et al., 2009)) and then refine the weights on the medical images specifically, a process called “transfer learning” (Raghu et al., 2019). ResNets are a particularly popular architecture for transfer learning. ResNets include “residual connections” (also called “skip connections”) which have been shown to smooth out the loss landscape and thereby facilitate training of deep networks (Li et al., 2018). Transfer learning has been shown to accelerate model convergence for medical imaging tasks (Raghu et al., 2019) including CT classification (Gao et al., 2018). However, the ResNet architecture is designed for two-dimensional images, and is thus not directly applicable to three-dimensional CT volumes.
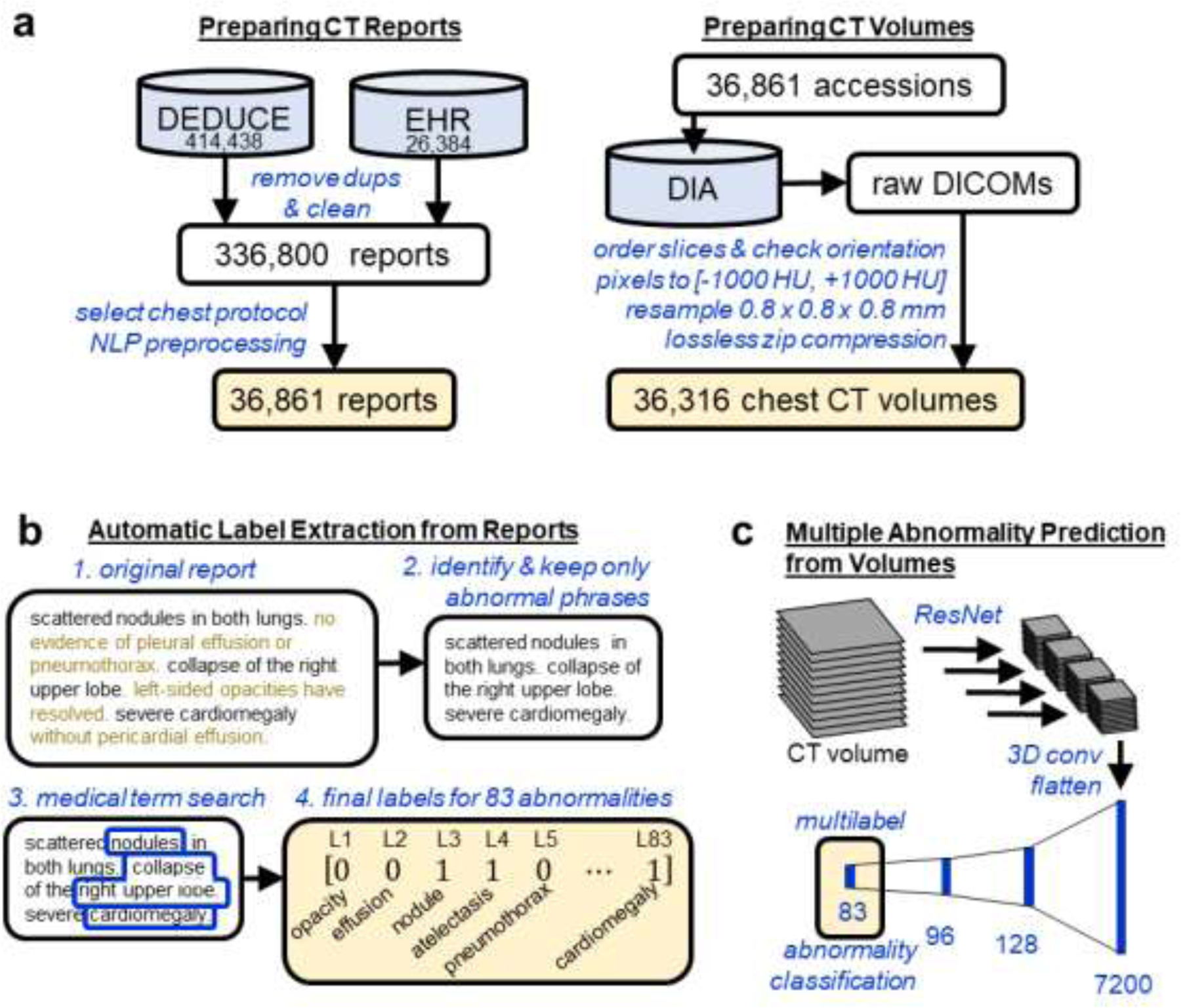
Our proposed network architecture, refered to as CT-Net, is shown in Figure 2. First we apply a ResNet-18 feature extractor to each stack of three adjacent grayscale axial slices, which have the same shape as RGB three-channel images and can therefore serve as ResNet input. The ResNet feature extractor is pretrained on ImageNet and its weights are refined on the CT classification task. In most applications of ResNets to medical images, the classification step occurs immediately after extracting the features, using a fully-connected layer. However, because the size of a whole CT volume is so large, a fully-connected layer applied directly to the ResNet output would require 1,116,114,944 parameters. Therefore, we reduce the size of the representation by orders of magnitude and aggregate features across the whole craniocaudal extent of the data by performing 3D convolutions. Once the representation is a reasonable size, we perform the final classification using fully connected layers. To provide more insights into the model we report results on two alternative architectures and perform an ablation study.
Figure 2. CT-Net volume classification architecture.

The CT volume is treated as a stack of three-channel images to enable use of a ResNet-18 (He et al., 2015) feature extractor pretrained on ImageNet (Deng et al., 2009). The ResNet-18 features for the stack of 134 three-channel images are concatenated and processed with several 3D convolutional layers to aggregate features across the craniocaudal extent of the scan and reduce the size of the representation. Then the representation is flattened and passed through three fully connected layers to produce predicted probabilities for the 83 abnormalities of interest.
To the best of our knowledge this is the first multilabel classification model that uses an entire CT volume as input. One prior study included a whole CT volume as a model input (Ardila et al., 2019), but the output was a lung cancer risk probability rather than predictions for multiple abnormalities. Most prior approaches have focused on 2D sections (Gao et al., 2016; Lee et al., 2019; Tang et al., 2019; Walsh et al., 2018) or patches (Anthimopoulos et al., 2016; Bermejo-Peláez et al., 2020; Christodoulidis et al., 2017; Gao et al., 2018; Kuo et al., 2019; Li et al., 2019; Wang et al., 2019), which either requires intensive manual labeling of CT subcomponents (infeasible for a dataset of 36,316 volumes), or accepting that labels will be extremely noisy (e.g., assigning the whole-volume label of “nodule” to all small patches in a CT is guaranteed to be wrong for most of the patches.)
The network is trained with a multilabel cross-entropy (CE) loss:
Where C is the number of abnormality labels and yi is a ground truth label for abnormality i. The predicted probability is calculated using the logistic function (a.k.a. sigmoid function): for score si, where score is the raw output of the last layer. This loss function enables the model to predict multiple abnormalities per scan simultaneously.
The CT volumes vary in shape between patients. To standardize the shape we pad or center crop all CTs to shape [402, 420, 420]. We clip pixel values to [−1000, 200] Hounsfield units, normalize to the range [−1,1], and center on the ImageNet mean. The model trained for 15 days with a batch size of 2 on two NVIDIA Titan XP GPUs with 11.9 GiB of memory each (a single CT scan requires all of the memory for a single GPU, as one CT is over 1,000 times larger than a typical 256 × 256 ImageNet example). We use a stochastic gradient descent optimizer with learning rate 10−3, momentum 0.99, and weight decay 10−7. Early stopping is performed on the validation loss with patience of 15 epochs. Data augmentation is performed through random jitters to the center crop, random flips, and random rotations of an input volume. The model is implemented in Pytorch (Paszke et al., 2019). All model code is publicly available on GitHub (https://github.com/rachellea).
We train two models using the same CNN architecture (up to the last fully-connected layer): (1) a multilabel CNN trained on all 83 labels simultaneously (CT-Net-83), and (2) a multilabel CNN trained on only the 9 labels for which report-level ground truth was obtained (CT-Net-9). The intent is to demonstrate the utility of extracting multiple labels by illustrating the change in performance when the number of labels is increased from 9 to 83. We only use the test set once, for the CT-Net-83 and CT-Net-9 simultaneously, after finishing all model development.
2.4. Architecture Comparison and Ablation Study
We compare the CT-Net architecture to two alternative architectures, BodyConv and 3DConv. BodyConv is a model similar to CT-Net-83, except instead of combining features across the whole craniocaudal extent of the scan at the beginning of the 3D convolution step, BodyConv only combines features across the craniocaudal extent at the fully connected layers. 3DConv is a model that uses only 3D convolutions and fully connected layers.
We perform an ablation study with models termed CT-Net-83 (Rand) and CT-Net-83 (Pool). The proposed CT-Net-83 architecture has three main components: the ResNet feature extractor, the 3D convolutions, and the final fully connected layers. CT-Net-83 (Rand) ablates the ResNet pretraining on ImageNet, by initializing the ResNet with random weights; this experiment was motivated by prior work (Raghu et al., 2019) suggesting that pretraining on ImageNet may not always be necessary for maximal performance on medical imaging tasks. CT-Net-83 (Pool) ablates the 3D convolution stage by replacing the 3D convolution layers with 3D max pooling layers of identical kernel size and stride. Note that we could not simply remove the 3D convolution stage entirely (i.e. proceed from the ResNet directly to a fully connected layer) due to the large size of the representation after the ResNet.
Performance in these experiments was obtained using a random subset of 2,000 training and 1,000 validation set scans. The full RAD-ChestCT data set of 36,316 scans was not used due to prohibitively long training/evaluation times. All these models are trained on 83 labels simultaneously.
2.5. Performance and Statistical Analysis
We report the area under the receiver operating characteristic (AUROC) and average precision for the CT-Net models. AUROC summarizes the sensitivity and specificity across different decision thresholds and ranges from 0.5 (random classifier) to 1.0 (perfect classifier.) Average precision is also known as the area under the precision-recall curve. While AUROC starts at a baseline of 0.5, average precision starts at a baseline equal to the frequency of positives for the particular abnormality being considered (Saito and Rehmsmeier, 2015). Therefore, an average precision of 0.4 would be high for a rare abnormality (e.g., frequency 0.02) and low for a common abnormality (e.g., frequency 0.8). Due to this frequency dependence we report the frequency for all abnormalities in our results.
We statistically compare the AUROCs of CT-Net-83 and CT-Net-9 using the DeLong test (DeLong et al., 1988), and obtain 95% AUROC confidence intervals using the DeLong method implemented in the pROC package in R version 3.6.2. The p-values for the DeLong test are corrected for multiple testing using the Benjamini and Hochberg method to control the false discovery rate (Benjamini and Hochberg, 1995).
3. Results
3.1. Automatic Label Extraction from Free-Text Reports
The performance of SARLE for automatic extraction of nine labels is shown in Table 3. The SARLE-Hybrid approach achieves an average F-score of 0.930 while the SARLE-Rules approach achieves an average F-score of 0.976, indicating that the automatically extracted labels are of high quality using both approaches. For the common labels, the Hybrid and Rules approaches perform equally well, e.g., atelectasis where both SARLE-Hybrid and SARLE-Rules achieve an F-score of 1.0. For the rarer findings – pericardial effusion, cardiomegaly, and pneumothorax – the SARLE-Rules approach outperforms SARLE-Hybrid. Because the SARLE-Rules approach achieved higher average performance and had better rare-abnormality performance, the labels produced by SARLE-Rules were used to train and evaluate the multilabel CNN on volumes.
Table 3: SARLE performance for the 427 chest CT test reports.
across the 9 labels with manually obtained ground truth. “# Pos” is the number of positive examples for that label in the report test set. F = equally weighted harmonic mean of precision and recall, P = Precision, R = Recall, Acc = Accuracy.
| Label | # Pos | SARLE-Hybrid | SARLE-Rules | ||||||
|---|---|---|---|---|---|---|---|---|---|
| F-score | P | R | Acc | F-score | P | R | Acc | ||
| nodule | 341 | 0.996 | 0.991 | 1 | 0.993 | 0.996 | 0.994 | 0.997 | 0.993 |
| opacity | 213 | 0.995 | 0.995 | 0.995 | 0.995 | 0.998 | 1 | 0.995 | 0.998 |
| atelectasis | 108 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| pleural effusion | 88 | 0.978 | 0.967 | 0.989 | 0.991 | 0.977 | 0.988 | 0.966 | 0.991 |
| consolidation | 78 | 0.969 | 0.951 | 0.987 | 0.988 | 0.975 | 0.963 | 0.987 | 0.991 |
| mass | 55 | 0.915 | 0.857 | 0.982 | 0.977 | 0.956 | 0.931 | 0.982 | 0.988 |
| pericardial effusion | 44 | 0.755 | 0.685 | 0.841 | 0.944 | 0.956 | 0.935 | 0.977 | 0.991 |
| cardiomegaly | 34 | 0.919 | 0.850 | 1 | 0.986 | 0.986 | 0.971 | 1 | 0.998 |
| pneumothorax | 8 | 0.842 | 0.727 | 1 | 0.993 | 0.941 | 0.889 | 1 | 0.998 |
SARLE-Rules outperforms CheXpert on our test set of 427 reports, for the six labels that overlap between the two approaches. SARLE-Rules has a better F-score on opacity (SARLE 0.998 vs. CheXpert 0.888), consolidation (SARLE 0.975 vs. CheXpert 0.969), cardiomegaly (SARLE 0.986 vs. CheXpert 0.449), and pneumothorax (SARLE 0.941 vs. CheXpert 0.727). Both models obtain perfect performance on atelectasis. CheXpert has a better F-score than SARLE-Rules on one label, pleural effusion (SARLE 0.977 vs. CheXpert 0.983).
The most common abnormalities in the 25,355 volume training CTs are nodule (19,567 examples which is 77% positive), calcification (17,228; 68%), opacity (13,833; 55%), coronary artery disease (12,585; 50%), postsurgical (10,900; 43%), and groundglass (8,401; 33%). Note that the “nodule” category refers to any nodule, including micronodules <3 mm in size; nodules greater than 1 cm are much less frequent, at 12%. The rarest abnormalities are all at frequency 1% or less, with the following counts: hardware (321), distention (306), bronchitis (175), hemothorax (137), heart failure (50), and congestion (37). Although these abnormalities are rare, the count of positive examples for many of these abnormalities still exceeds the size of many previously reported CT data sets which are on the order of 100 – 200 CT scans total (Anthimopoulos et al., 2016; Bermejo-Peláez et al., 2020; Christodoulidis et al., 2017; Gao et al., 2018; Li et al., 2019; Wang et al., 2019).
The median number of abnormality labels for a single scan in the volume training set is 10, with an interquartile range of 6. The full histogram of abnormalities per scan is available in Appendix C. Only 139 training set scans were negative for all 83 abnormalities (i.e., “normal”), which is less than 0.6% of the scans.
3.2. Multilabel CNN to Predict Abnormalities from CT Volumes
Table 4 compares the test set performance of the CT-Net-83 and CT-Net-9 models. The CT-Net-83 model outperforms the CT-Net-9 model on every abnormality, indicating the value of training on the additional 74 labels. This is an example of the benefit of transfer learning.
Table 4. CT volume test set AUROC for models trained on 9 vs. 83 labels.
The area under the receiver operating characteristic (AUROC) is shown for CT-Net-9 (trained only on the 9 labels shown) and CT-Net-83 (trained on the 9 labels shown plus 74 additional labels) for the test set of 7,209 examples. CT-Net-83 outperforms CT-Net-9 on all abnormalities, emphasizing the value of the additional 74 labels. Note that we also experimented with separate binary classifiers for each of the 9 labels independently, but these models did not converge (AUROC ~0.5). Positive Count and Positive Percent are for positive examples of the abnormality in the test set.
| Abnormality | Positive Count | Positive Percent | CT-Net-9 |
CT-Net-83 |
DeLong | ||
|---|---|---|---|---|---|---|---|
| AUROC | 95% CI | AUROC | 95% CI | p-value | |||
| nodule | 5,617 | 77.9 | 0.682 | 0.667–0.698 | 0.718 | 0.703–0.732 | 3.346×10−7 |
| opacity | 3,877 | 53.8 | 0.617 | 0.605–0.630 | 0.740 | 0.728–0.751 | <4.950×10−16 |
| atelectasis | 2,037 | 28.3 | 0.683 | 0.668–0.697 | 0.765 | 0.753–0.777 | <4.950×10−16 |
| pleural effusion | 1,404 | 19.5 | 0.945 | 0.937–0.952 | 0.951 | 0.945–0.958 | 1.882×10−2 |
| consolidation | 1,086 | 15.1 | 0.719 | 0.703–0.736 | 0.816 | 0.804–0.829 | <4.950×10−16 |
| mass | 863 | 12.0 | 0.624 | 0.604–0.644 | 0.773 | 0.755–0.791 | <4.950×10−16 |
| pericardial eff. | 1,078 | 15.0 | 0.659 | 0.640–0.677 | 0.697 | 0.679–0.714 | 8.315×10−8 |
| cardiomegaly | 649 | 9.0 | 0.791 | 0.774–0.807 | 0.851 | 0.836–0.867 | 7.000×10−13 |
| pneumothorax | 205 | 2.8 | 0.816 | 0.785–0.847 | 0.904 | 0.882–0.926 | 8.810×10−11 |
Table 5 shows the abnormality labels for which the CT-Net-83 model achieved the highest and lowest performance. Several of the highest-performing labels are abnormalities related to surgeries that affect a large area of the chest, including lung resection, sternotomy, CABG (coronary artery bypass graft), transplant, and “postsurgical” which encompasses a variety of descriptors of recent surgery. Other high-performing labels are human-made objects, including pacemaker or defibrillator, tracheal tube, catheter or port, heart valve replacement, chest tube, and GI tube. Finally, there are numerous common biological abnormalities that the model is able to identify with high AUROC, including pleural effusion, emphysema, pulmonary edema, fibrosis, interstitial lung disease, pneumothorax, and coronary artery disease.
Table 5. CT-Net-83 test set AUROC and Average Precision for abnormalities with the highest and lowest AUROCs.
Note that the baseline for average precision is equal to the frequency of the abnormality being considered; this frequency is provided in the Test Set Percent column. Thus, an average precision of 0.463 for honeycombing is high, given honeycombing’s baseline of only 0.027.
| Abnormality | AUROC | Average Precision | Test Set Percent | Test Set Count |
|---|---|---|---|---|
| pacemaker or defib | 0.975 | 0.699 | 0.039 | 279 |
| honeycombing | 0.972 | 0.463 | 0.027 | 193 |
| tracheal tube | 0.971 | 0.597 | 0.017 | 121 |
| lung resection | 0.967 | 0.876 | 0.194 | 1,398 |
| sternotomy | 0.965 | 0.598 | 0.071 | 514 |
| CABG | 0.965 | 0.527 | 0.040 | 288 |
| transplant | 0.963 | 0.751 | 0.057 | 414 |
| catheter or port | 0.954 | 0.716 | 0.105 | 755 |
| heart failure | 0.952 | 0.040 | 0.002 | 18 |
| pleural effusion | 0.951 | 0.869 | 0.195 | 1,404 |
| heart valve replacement | 0.949 | 0.219 | 0.018 | 133 |
| chest tube | 0.944 | 0.387 | 0.020 | 146 |
| GI tube | 0.940 | 0.492 | 0.022 | 162 |
| emphysema | 0.929 | 0.843 | 0.243 | 1,754 |
| pulmonary edema | 0.921 | 0.524 | 0.052 | 373 |
| fibrosis | 0.910 | 0.662 | 0.112 | 811 |
| interstitial lung disease | 0.906 | 0.764 | 0.153 | 1,102 |
| pneumothorax | 0.904 | 0.355 | 0.028 | 205 |
| postsurgical | 0.896 | 0.853 | 0.428 | 3,089 |
| hemothorax | 0.890 | 0.038 | 0.004 | 28 |
| coronary artery disease | 0.873 | 0.830 | 0.494 | 3,563 |
| cyst | 0.594 | 0.184 | 0.143 | 1,032 |
| granuloma | 0.588 | 0.116 | 0.083 | 595 |
| hardware | 0.577 | 0.020 | 0.017 | 120 |
| density | 0.560 | 0.115 | 0.090 | 647 |
| scattered nodules/nodes | 0.559 | 0.249 | 0.210 | 1,512 |
| infiltrate | 0.526 | 0.016 | 0.015 | 107 |
The model performs poorly on several labels, including cyst, density, and scattered nodules (Table 5). On further analysis we discovered that cysts most commonly appear in the kidneys, which are likely to be cropped out in the preprocessing due to appearing at the edge of the volume. “Density” is used in variable ways in radiology reports and may not correspond to a clear visual pattern. The “scattered nodules/nodes” category includes scattered micronodules which may affect only one or two pixels each and by definition are distributed over a wide area, which may be a difficult characteristic for the model to capture.
Overall, CT-Net-83 achieves an AUROC >0.9 on 18 abnormalities, 0.8 – 0.9 AUROC on 17 abnormalities, 0.7 – 0.8 AUROC on 24 abnormalities, 0.6 – 0.7 AUROC on 18 abnormalities, and <0.6 AUROC on 6 abnormalities. Performance of CT-Net-83 on all 83 abnormalities is provided in Appendix C.
In Figure 3 we present a boxplot summary of the AUROC across all 83 abnormalities for our proposed CT-Net-83 architecture and two alternative architectures on a validation set of 1,000 volumes for models trained on 2,000 volumes. CT-Net-83 in this figure shows the performance of the CT-Net-83 model on this smaller subset of data; the performance is lower than that shown in Tables 4 and 5, due to training on 12× less data. CT-Net-83 outperforms the two alternative architectures, BodyConv (which uses different 3D convolutions) and 3DConv (which uses all 3D convolutions instead of a pre-trained ResNet feature extractor).
Figure 3. Architecture Comparison and Ablation Study on Training/Validation Data Subset.
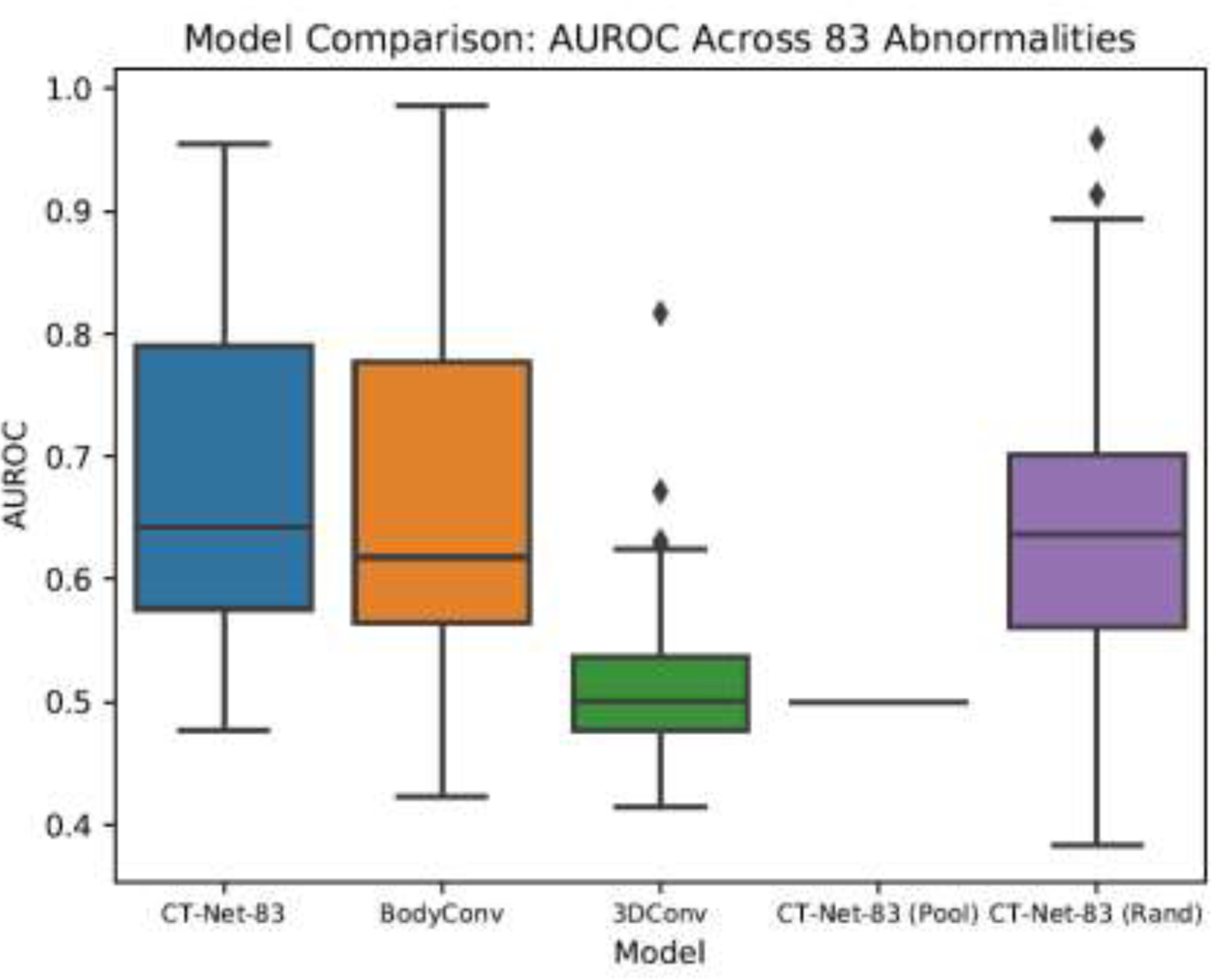
The AUROCs for each abnormality in this experiment were calculated on a random sample of 1,000 validation set scans, for models trained on a random subset of 2,000 training scans. CT-Net-83 is the proposed model. BodyConv and 3DConv are alternative architectures. CT-Net-83 (Pool) and CT-Net-83 (Rand) are ablated version of the CT-Net-83 model.
Figure 3 also demonstrates that the proposed CT-Net-83 outperforms the two ablated variants, (Rand) in which the ResNet feature extractor is randomly initialized instead of pretrained, and (Pool) in which the 3D convolutions have been replaced by max pooling operations. These results illustrate that for the task of multiple abnormality prediction in CT scans, transfer learning by pretraining the feature extractor on ImageNet does lead to better abnormality identification in spite of the differences between CT slices and natural images. Furthermore, the relationships learned in the 3D convolution stage are critical for the model’s performance. Without the 3D convolutions, the model does not converge.
4. Discussion
The three main contributions of this work are the preparation of the Report-Annotated Duke Chest CT data set (RAD-ChestCT) of 36,316 unenhanced chest CT volumes, the SARLE framework for automatic extraction of 83 labels from free-text radiology reports, and a deep CNN model for multiple abnormality prediction from chest CT volumes.
The RAD-ChestCT data set is the largest reported data set of multiply annotated chest CT volumes, with 36,316 whole volumes from 19,993 unique patients. We plan to make the CT volumes publicly available, pending deidentification and approval. Fewer than 6% of studies on deep learning in radiology use more than 10,000 cases (Soffer et al., 2019). Several previous studies on interstitial lung disease use between 120 and 1,157 chest CTs (Anthimopoulos et al., 2016; Bermejo-Peláez et al., 2020; Christodoulidis et al., 2017; Gao et al., 2018, 2016; Walsh et al., 2018; Wang et al., 2019). Two recent studies on acute intracranial hemorrhage used 1,300 (Lee et al., 2019) and 4,596 (Kuo et al., 2019) head CTs. The public LIDC-IRDI data set (Armato et al., 2011) of 1,018 chest CT volumes and the public DeepLesion data set (Yan et al., 2018) of 10,825 partial CT volumes are centered on focal lesions (e.g., nodules). Ardila et al. (Ardila et al., 2019) develop a lung cancer screening model on a National Lung Screening Trial (NLST) (Gatsonis et al., 2011) data set of 42,290 CT scans with cancer-related annotations, from 14,851 patients. This represents a greater total number of scans, but a smaller number of unique patients and unique annotations than RAD-ChestCT.
To the best of our knowledge RAD-ChestCT is the only chest CT data set with such a diverse range of abnormality annotations including both focal (nodule, mass, etc.). and diffuse (fibrosis, ILD, atelectasis, edema, pneumonia, etc.) abnormalities. In its present form it can be used for multilabel classification, weakly supervised abnormality localization, or exploratory research using unsupervised methods like clustering. One potential future direction would be to extend the RAD-ChestCT data set to include bounding box annotations to facilitate supervised abnormality localization. To accelerate other large-scale machine learning projects on CT data, we provide a detailed tutorial in Appendix A on how to transform raw CT DICOMs into 3D numpy arrays for analysis, and we have made our entire end-to-end Python CT preprocessing pipeline publicly available.
The SARLE framework for automatic label extraction from radiology reports is designed to be simple, scale to a large number of abnormality labels, and achieve high performance. It is the first approach to automatically extract numerous abnormality labels from chest CT reports. The general principle of first distinguishing between medically “normal” and “abnormal” phrases, and then performing an abnormality-specific vocabulary lookup, is applicable to any radiology modality. In Appendix B we present a detailed discussion of SARLE in the context of related work. Other studies in label extraction report F-scores between 0.52 – 1.0 for the extraction of anywhere between 3 and 55 abnormalities (Banerjee et al., 2017; Chen et al., 2018; Demner-Fushman et al., 2016; Irvin et al., 2019; Peng et al., 2018; Pham et al., 2014; Zech et al., 2018). Relative to these other approaches, SARLE achieves high F-score (average 0.976 across 9 abnormalities) and makes predictions on a large number of labels (83). We found that SARLE-Rules outperforms the CheXpert labeler on our test set of chest CT reports. This may be in part because the CheXpert labeler’s rules and vocabulary were originally designed for chest x-ray reports rather than chest CT reports. We also found that SARLE-Rules outperforms SARLE-Hybrid, and hypothesize that this may be because the Rules approach is phrase-based rather than whole-sentence based, which allows better handling of the small minority of sentences that contain both normal and abnormal medical findings. In the future, the SARLE-Hybrid approach could be extended by replacing the sentence classifier with a phrase classifier.
Our work was inspired by the ChestX-Ray8 study (Wang et al., 2017) and the CheXpert study (Irvin et al., 2019), which share the most similar overall experimental design. These studies as well as our present work involve preparation of a large database of radiology images, development and application of an automated label extraction approach to obtain structured disease annotations from radiology reports, and training and evaluation of a deep learning model on the radiology images using the automatically extracted labels as ground truth.
However, there are a few key differences between our work and the ChestX-Ray8/CheXpert studies. First, ChestX-Ray8/CheXpert are focused on projectional radiographs, which are two-dimensional images, while our work focuses on CT scans, which are three-dimensional and require different preprocessing steps and modeling considerations due to their volumetric nature. Our label extraction approaches also differ: the CheXpert work applies rules defined on a sentence graph to extract 14 abnormalities with F-scores ranging between 0.72 – 1.00 while our work uses rules defined directly on the sentence text to produce 83 labels per report, with F-scores for 9 abnormalities ranging between 0.941– 1.00.
The challenges in applying deep learning models to radiographs and CTs are different. A single CT is about 70x larger than a radiograph, which presents hardware and memory challenges and causes the abnormalities to be more spatially dispersed within the training example. Because radiographs are 2D projections of a 3D volume, radiographs are more ambiguous (de Hoop et al., 2010; Gibbs et al., 2007; Howarth and Tack, 2015; Self et al., 2013), which alters what abnormalities can be visualized and changes the implications of certain words in the reports. For example, the majority of nodules smaller than 1 centimeter are not visible on chest radiographs (MacMahon et al., 2017) whereas CT can detect nodules as small as 1 – 2 mm in diameter (Sánchez et al., 2018). That means our “nodule” category for CT includes nodules that from a volumetric perspective are up to 1,000× smaller than those visible on chest radiographs, and these tiny CT nodules are distributed across 1,000× more pixels. In spite of this, we obtain better performance on nodule and mass identification: nodule 0.718 ours vs. 0.716 ChestX-Ray8 (Wang et al., 2017), and mass 0.773 vs. 0.564 ChestX-Ray8.
Although the studies are on different kinds of data using different automatic labelers, for the purposes of placing our work in context, we note that our CT volume classifier’s performance is generally in the same range as that of prior chest x-ray work: pneumothorax 0.904 ours vs. 0.789 ChestX-Ray8 (Wang et al., 2017), pneumonia 0.816 ours vs. 0.633 ChestX-Ray8, infiltration 0.526 ours vs. 0.612 ChestX-Ray8. The CheXpert study (Irvin et al., 2019) reports performance on additional abnormalities: atelectasis 0.765 ours vs. 0.858 CheXpert, cardiomegaly 0.851 ours vs. 0.832, consolidation 0.816 ours vs. 0.899, edema 0.921 ours vs. 0.941, and pleural effusion 0.951 ours vs. 0.934.
Most prior research in CT scan classification has focused on a single category of diseases and relies on a fundamentally different modeling approach that requires manual slice-level, patch-level, or pixel-level labels. Several studies have focused on subtypes of interstitial lung disease (ILD) (Anthimopoulos et al., 2016; Bermejo-Peláez et al., 2020; Christodoulidis et al., 2017; Gao et al., 2018, 2016; Walsh et al., 2018) but all of these studies require manual labeling of regions of interest or manual pixel-level labeling, and all the models are trained on small patches (e.g., 32 × 32 pixels) or slices extracted from the CT scan. The advantage of patch or slice classification is that it provides some inherent localization. The disadvantage is that it limits the total number of CTs in the data set (all these studies use <1,200 CTs) and it limits the total number of abnormalities that can be considered (all consider <9 classes) due to the immense manual work required to obtain pixel-level, patch-level, or slice-level annotations.
A related study (Tang et al., 2019) performs binary classification of weakly-labeled slices for each of nodule, atelectasis, edema, and pneumonia separately versus normal scans, using an approximately 1:1 ratio between abnormal and normal scans. This is a different task from our multilabel setup, in which we use unfiltered hospital data where fewer than 1% of scans are normal, and we train one model on all abnormalities simultaneously. Two implications of our approach are noteworthy. Firstly, the application of multiple single label models does not provide a basis for accommodating interactions between co-existent diseases or imaging findings. Our multilabel approach provides a holistic image assessment paradigm that may better generalize across individual patients by accommodating for interactions across the clinical reality of multiple co-existent diseases and imaging findings. Secondly, the very low prevalence of true “normality” in an unfiltered sample of patients receiving unenhanced chest CT scans challenges the utility of interpretation workflows that are based upon patient-level prioritization of abnormal scans over those that are normal.
A substantial body of literature focuses on lung nodules and is the subject of multiple review articles (Pehrson et al., 2019; Shaukat et al., 2019; Zhang et al., 2018). For understandable reasons these methods typically rely on manually acquired nodule bounding boxes and can achieve AUROCs in the upper 0.90s. This is higher than our AUROC of 0.718 for nodules, but our model was trained using only whole-volume labels without any bounding box annotations. In the future it may be possible to improve our model’s performance on focal findings through combined training on RAD-ChestCT and DeepLesion or LIDC, through addition of bounding box annotations to RAD-ChestCT, or through active learning to improve a weakly-supervised lesion detector. Multi-scale architectures may facilitate improvement on both focal and diffuse findings simultaneously.
A critical contribution of our work is the demonstration that leveraging numerous automatically extracted abnormality labels enables learning from unfiltered hospital-scale CT data. A binary classifier trained on unfiltered hospital CT data does not converge (AUROC ~0.5) likely due to contamination of the “normal” class with other abnormalities, some of which may look similar to the target class. Training a multilabel classification model on 83 labels simultaneously instead of only 9 simultaneously boosts the average AUROC by over 10%, from 0.726 to 0.802. Furthermore, the model trained on all 83 labels achieves AUROCs over 0.90 for almost twenty different abnormalities including many medically significant abnormalities that have been the subject of prior work (Anthimopoulos et al., 2016; Christe et al., 2019; Humphries et al., 2019; Irvin et al., 2019; Li et al., 2019; Tang et al., 2019; Walsh et al., 2018) such as emphysema (0.929), pleural effusion (0.951), pulmonary edema (0.921), interstitial lung disease (0.906), honeycombing (0.972), pneumothorax (0.904), and fibrosis (0.910). We further show that our proposed model, CT-Net, is able to outperform two alternative models by leveraging transfer learning and 3D convolutions that combine abnormality features across the craniocaudal extent of the scan.
We hope this work will contribute to the long-term goal of augmented medical image interpretation systems that enhance the radiologists’ workflow, improve detection and monitoring of diseases, and advance patient care.
Supplementary Material
Highlights.
We create the RAD-ChestCT data set of 36,316 volumes from 19,993 unique patients
We develop a method to automatically extract 83 abnormality labels from CT reports
We develop a deep learning model for multi-abnormality classification of CT volumes
The CT volume model achieves mean AUROC of 0.773 for all 83 abnormalities
Training the CT volume model on more abnormality labels improves performance
Acknowledgements
We would like to thank Mark Martin, Justin Solomon, the Duke University Office of Information Technology (OIT), and the Duke Protected Analytics Computing Environment (PACE) team. We also thank the anonymous reviewers for providing insightful comments that improved the manuscript.
Funding Sources
This work was supported by NIH/NIBIB R01-EB025020, developmental funds of the Duke Cancer Institute from the NIH/NCI P30-CA014236 Cancer Center Support Grant, and GM-007171 the Duke Medical Scientist Training Program Training Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations: SARLE (Sentence Analysis for Radiology Label Extraction), RAD-ChestCT (Report-Annotated Duke Chest CT), AUROC (area under the receiver operating characteristic)
References
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng X, 2016. TensorFlow: A system for large-scale machine learning, in: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16) pp. 265–283. [Google Scholar]
- Annarumma M, Withey SJ, Bakewell RJ, Pesce E, Goh V, Montana G, 2019. Automated Triaging of Adult Chest Radiographs with Deep Artificial Neural Networks. Radiology 291, 196–202. 10.1148/radiol.2018180921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S, 2016. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans. Med. Imaging 35, 1207–1216. 10.1109/TMI.2016.2535865 [DOI] [PubMed] [Google Scholar]
- Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, Naidich DP, Shetty S, 2019. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med 10.1038/s41591-019-0447-x [DOI] [PubMed]
- Armato SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Van Beek EJR, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DPY, Roberts RY, Smith AR, Starkey A, Batra P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Vande Casteele A, Gupte S, Sallam M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY, Clarke LP, 2011. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys 38, 915–931. 10.1118/1.3528204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee I, Madhavan S, Goldman RE, Rubin DL, 2017. Intelligent Word Embeddings of Free-Text Radiology Reports. AMIA … Annu. Symp. proceedings. AMIA Symp 2017, 411–420. [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bermejo-Peláez D, Ash SY, Washko GR, San José Estépar R, Ledesma-Carbayo MJ, 2020. Classification of Interstitial Lung Abnormality Patterns with an Ensemble of Deep Convolutional Neural Networks. Sci. Rep 10, 338 10.1038/s41598-019-56989-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojanowski P, Grave E, Joulin A, Mikolov T, 2017. Enriching Word Vectors with Subword Information, in: Association for Computational Linguistics; pp. 135–146. [Google Scholar]
- Burns JE, Yao J, Muñoz H, Summers RM, 2016. Automated detection, localization, and classification of traumatic vertebral body fractures in the thoracic and lumbar spine at CT. Radiology 278, 64–73. 10.1148/radiol.2015142346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG, 2001. A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries. J. Biomed. Inform 34, 301–310. 10.1006/jbin.2001.1029 [DOI] [PubMed] [Google Scholar]
- Chen MC, Ball RL, Yang L, Moradzadeh N, Chapman BE, Larson DB, Langlotz CP, Amrhein TJ, Lungren MP, 2018. Deep Learning to Classify Radiology Free-Text Reports. Radiology 286, 845–852. 10.1148/radiol.2017171115 [DOI] [PubMed] [Google Scholar]
- Choi KJ, Jang JK, Lee SS, Sung YS, Shim WH, Kim HS, Yun J, Choi J-Y, Lee Y, Kang B-K, Kim JH, Kim SY, Yu ES, 2018. Development and Validation of a Deep Learning System for Staging Liver Fibrosis by Using Contrast Agent–enhanced CT Images in the Liver. Radiology 289, 688–697. 10.1148/radiol.2018180763 [DOI] [PubMed] [Google Scholar]
- Christe A, Peters AA, Drakopoulos D, Heverhagen JT, Geiser T, Stathopoulou T, Christodoulidis S, Anthimopoulos M, Mougiakakou SG, Ebner L, 2019. Computer-Aided Diagnosis of Pulmonary Fibrosis Using Deep Learning and CT Images. Invest. Radiol 54, 627–632. 10.1097/rli.0000000000000574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulidis S, Anthimopoulos M, Ebner L, Christe A, Mougiakakou S, 2017. Multisource Transfer Learning with Convolutional Neural Networks for Lung Pattern Analysis. IEEE J. Biomed. Heal. Informatics 21, 76–84. 10.1109/JBHI.2016.2636929 [DOI] [PubMed] [Google Scholar]
- Costello JE, Cecava ND, Tucker JE, Bau JL, 2013. CT radiation dose: Current controversies and dose reduction strategies. Am. J. Roentgenol 10.2214/AJR.12.9720 [DOI] [PubMed]
- de Hoop B, Schaefer-Prokop C, Gietema HA, de Jong PA, van Ginneken B, van Klaveren RJ, Prokop M, 2010. Screening for Lung Cancer with Digital Chest Radiography: Sensitivity and Number of Secondary Work-up CT Examinations. Radiology 255, 629–637. 10.1148/radiol.09091308 [DOI] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL, 1988. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 44, 837 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- Demner-Fushman D, Kohli MD, Rosenman MB, Shooshan SE, Rodriguez L, Antani S, Thoma GR, McDonald CJ, 2016. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Informatics Assoc 23, 304–310. 10.1093/jamia/ocv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L, 2009. ImageNet: A Large-Scale Hierarchical Image Database, in: CVPR. [Google Scholar]
- DenOtter TD, Schubert J, 2019. Hounsfield Unit, StatPearls. [PubMed] [Google Scholar]
- Depeursinge A, Vargas A, Platon A, Geissbuhler A, Poletti PA, Müller H, 2012. Building a reference multimedia database for interstitial lung diseases. Comput. Med. Imaging Graph 36, 227–238. 10.1016/j.compmedimag.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Dhara AK, Mukhopadhyay S, Dutta A, Garg M, Khandelwal N, 2016. A Combination of Shape and Texture Features for Classification of Pulmonary Nodules in Lung CT Images. J. Digit. Imaging 29, 466–475. 10.1007/s10278-015-9857-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RM, Godwin JD, Hippe DS, Kicska G, 2016. A Quantitative Approach to Distinguish Pneumonia from Atelectasis Using Computed Tomography Attenuation. J. Comput. Assist. Tomogr 40, 746–751. 10.1097/RCT.0000000000000438 [DOI] [PubMed] [Google Scholar]
- Franquet T, 2001. Imaging of pneumonia: trends and algorithms. Eur. Respir. J 18, 196–208. 10.1183/09031936.01.00213501 [DOI] [PubMed] [Google Scholar]
- Gao M, Bagci U, Lu L, Wu A, Buty M, Shin HC, Roth H, Papadakis GZ, Depeursinge A, Summers RM, Xu Z, Mollura DJ, 2018. Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Comput. Methods Biomech. Biomed. Eng. Imaging Vis 6, 1–6. 10.1080/21681163.2015.1124249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Xu Z, Lu L, Harrison AP, Summers RM, Mollura DJ, 2016. Multi-label deep regression and unordered pooling for holistic interstitial lung disease pattern detection, in: Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Springer Verlag, pp. 147–155. 10.1007/978-3-319-47157-0_18 [DOI] [Google Scholar]
- Gatsonis CA, Aberle DR, Berg CD, 2011. The national lung screening trial: Overview and study design. Radiology 258, 243–253. 10.1148/radiol.10091808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JM, Chandrasekhar CA, Ferguson EC, Oldham SAA, 2007. Lines and Stripes: Where Did They Go? —From Conventional Radiography to CT. RadioGraphics 27, 33–48. 10.1148/rg.271065073 [DOI] [PubMed] [Google Scholar]
- Hansell DM, 2010. Thin-Section CT of the Lungs: The Hinterland of Normal. Radiology 256, 695–711. 10.1148/radiol.10092307 [DOI] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2015. Deep Residual Learning for Image Recognition
- Horvath MM, Winfield S, Evans S, Slopek S, Shang H, Ferranti J, 2011. The DEDUCE Guided Query tool: providing simplified access to clinical data for research and quality improvement. J. Biomed. Inform 44, 266–76. 10.1016/j.jbi.2010.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL, 2018. Artificial intelligence in radiology. Nat. Rev. Cancer 18, 500–510. 10.1038/s41568-018-0016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth N, Tack D, 2015. Missed Lung Lesions: Side by Side Comparison of Chest Radiography with MDCT, in: Diseases of the Chest and Heart 2015–2018 Springer Milan, pp. 80–87. 10.1007/978-88-470-5752-4_10 [DOI] [PubMed] [Google Scholar]
- Humphries SM, Notary AM, Centeno JP, Strand MJ, Crapo JD, Silverman EK, Lynch DA, Genetic Epidemiology of COPD (COPDGene) Investigators, 2019. Deep Learning Enables Automatic Classification of Emphysema Pattern at CT. Radiology 191022 10.1148/radiol.2019191022 [DOI] [PMC free article] [PubMed]
- Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, Seekins J, Mong DA, Halabi SS, Sandberg JK, Jones R, Larson DB, Langlotz CP, Patel BN, Lungren MP, Ng AY, 2019. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison, AAAI. [Google Scholar]
- Johnson AEW, Pollard TJ, Berkowitz SJ, Greenbaum NR, Lungren MP, Deng C, Mark RG, Horng S, 2019. MIMIC-CXR: A large publicly available database of labeled chest radiographs [DOI] [PMC free article] [PubMed]
- Joulin A, Grave E, Bojanowski P, Mikolov T, 2017. Bag of Tricks for Efficient Text Classification, Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics Valencia, Spain. [Google Scholar]
- Kawooya M, 2012. Training for Rural Radiology and Imaging in Sub-Saharan Africa: Addressing the Mismatch Between Services and Population. J. Clin. Imaging Sci 2, 37 10.4103/2156-7514.97747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria T, Badr E, Al-Mallah M, Sakr S, 2019. LDLCT an instance-based framework for lesion detection on lung CT scans, in: Proceedings - IEEE Symposium on Computer-Based Medical Systems Institute of Electrical and Electronics Engineers Inc., pp. 523–526. 10.1109/CBMS.2019.00106 [DOI] [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE, 2012. ImageNet Classification with Deep Convolutional Neural Networks, in: Advances in Neural Information Processing Systems (NIPS) pp. 1097–1105. [Google Scholar]
- Kuo W, Hӓne C, Mukherjee P, Malik J, Yuh EL, 2019. Expert-level detection of acute intracranial hemorrhage on head computed tomography using deep learning. Proc. Natl. Acad. Sci. U. S. A 116, 22737–22745. 10.1073/pnas.1908021116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba R, McGahan JP, Corwin MT, Li CS, Tran T, Seibert JA, Boone JM, 2014. CT Hounsfield numbers of soft tissues on unenhanced abdominal CT scans: Variability between two different manufacturers’ MDCT scanners. Am. J. Roentgenol 203, 1013–1020. 10.2214/AJR.12.10037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Nagy PG, Weaver SJ, Newman-Toker DE, 2013. Cognitive and system factors contributing to diagnostic errors in radiology. Am. J. Roentgenol 10.2214/AJR.12.10375 [DOI] [PubMed]
- Lee H, Yune S, Mansouri M, Kim M, Tajmir SH, Guerrier CE, Ebert SA, Pomerantz SR, Romero JM, Kamalian S, Gonzalez RG, Lev MH, Do S, 2019. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat. Biomed. Eng 3, 173–182. 10.1038/s41551-018-0324-9 [DOI] [PubMed] [Google Scholar]
- Li H, Xu Z, Taylor G, Studer C, Goldstein T, 2018. Visualizing the Loss Landscape of Neural Nets, in: Advances in Neural Information Processing Systems 31. [Google Scholar]
- Li X, Thrall JH, Digumarthy SR, Kalra MK, Pandharipande PV, Zhang B, Nitiwarangkul C, Singh R, Khera RD, Li Q, 2019. Deep learning-enabled system for rapid pneumothorax screening on chest CT. Eur. J. Radiol 120 10.1016/j.ejrad.2019.108692 [DOI] [PubMed] [Google Scholar]
- Linguraru MG, Wang S, Shah F, Gautam R, Peterson J, Linehan WM, Summers RM, 2011. Automated noninvasive classification of renal cancer on multiphase CT. Med. Phys 38, 5738–5746. 10.1118/1.3633898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowekamp BC, Chen DT, Ibáñez L, Blezek D, 2013. The Design of SimpleITK. Front. Neuroinform 7 10.3389/fninf.2013.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, Mehta AC, Ohno Y, Powell CA, Prokop M, Rubin GD, Schaefer-Prokop CM, Travis WD, Van Schil PE, Bankier AA, 2017. Guidelines for management of incidental pulmonary nodules detected on CT images: From the Fleischner Society 2017. Radiology 10.1148/radiol.2017161659 [DOI] [PubMed]
- Mikolov T, Sutskever I, Chen K, Corrado G, Dean J, 2013. Distributed representations of words and phrases and their compositionality. Proc. 26th Int. Conf. Neural Inf. Process. Syst 2, 3111–3119. [Google Scholar]
- Nguyen TB, Wang S, Anugu V, Rose N, McKenna M, Petrick N, Burns JE, Summers RM, 2012. Distributed human intelligence for colonic polyp classification in computer-aided detection for CT colonography. Radiology 262, 824–833. 10.1148/radiol.11110938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszke A, Gross S, Massa F, Lerer A, Bradbury Google J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, Xamla AK, Yang E, Devito Z, Raison Nabla M, Tejani A, Chilamkurthy S, Ai Q, Steiner B, Facebook LF, Facebook JB, Chintala S, 2019. PyTorch: An Imperative Style, High-Performance Deep Learning Library, in: Advances in Neural Information Processing Systems 32 (NIPS 2019) Pre-Proceedings. [Google Scholar]
- Pehrson LM, Nielsen MB, Lauridsen CA, 2019. Automatic pulmonary nodule detection applying deep learning or machine learning algorithms to the LIDC-IDRI database: A systematic review. Diagnostics 10.3390/diagnostics9010029 [DOI] [PMC free article] [PubMed]
- Peng Y, Wang X, Lu L, Bagheri M, Summers R, Lu Z, 2018. NegBio: a high-performance tool for negation and uncertainty detection in radiology reports 2017, 188–196. [PMC free article] [PubMed] [Google Scholar]
- Pham A-D, Névéol A, Lavergne T, Yasunaga D, Clément O, Meyer G, Morello R, Burgun A, 2014. Natural language processing of radiology reports for the detection of thromboembolic diseases and clinically relevant incidental findings. BMC Bioinformatics 15, 266 10.1186/1471-2105-15-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purysko C, Renapurkar R, Bolen M, 2016. When does chest CT require contrast enhancement? Cleve. Clin. J. Med 83, 423–426. [DOI] [PubMed] [Google Scholar]
- Raghu M, Zhang C, Brain G, Kleinberg J, Bengio S, 2019. Transfusion: Understanding Transfer Learning for Medical Imaging, in: Advances in Neural Information Processing Systems 32. [Google Scholar]
- Rawat W, Wang Z, 2017. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput 29, 2352–2449. 10.1162/NECO_a_00990 [DOI] [PubMed] [Google Scholar]
- Saito T, Rehmsmeier M, 2015. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 10 10.1371/journal.pone.0118432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez M, Benegas M, Vollmer I, 2018. Management of incidental lung nodules less than 8 mm in diameter. J. Thorac. Dis 10, S2611–S2627. 10.21037/jtd.2018.05.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier R, 2018. Artificial Intelligence and the Practice of Radiology: An Alternative View. J. Am. Coll. Radiol 15, 1004–1007. 10.1016/j.jacr.2018.03.046 [DOI] [PubMed] [Google Scholar]
- Self WH, Courtney DM, McNaughton CD, Wunderink RG, Kline JA, 2013. High discordance of chest x-ray and computed tomography for detection of pulmonary opacities in ED patients: Implications for diagnosing pneumonia. Am. J. Emerg. Med 31, 401–405. 10.1016/j.ajem.2012.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q, Gong L, Ma K, Liu H, Zheng Y, 2019. Attentive CT Lesion Detection Using Deep Pyramid Inference with Multi-scale Booster, in: MICCAI. Springer, pp. 301–309. 10.1007/978-3-030-32226-7_34 [DOI] [Google Scholar]
- Shaukat F, Raja G, Frangi AF, 2019. Computer-aided detection of lung nodules: a review. J. Med. Imaging 6, 1 10.1117/1.jmi.6.2.020901 [DOI] [Google Scholar]
- Simonyan K, Zisserman A, 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition
- Smith-Bindman R, Lipson J, Marcus R, Kim KP, Mahesh M, Gould R, Berrington De González A, Miglioretti DL, 2009. Radiation dose associated with common computed tomography examinations and the associated lifetime attributable risk of cancer. Arch. Intern. Med 169, 2078–2086. 10.1001/archinternmed.2009.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E, 2019. Convolutional Neural Networks for Radiologic Images: A Radiologist’s Guide. Radiology 290, 590–606. 10.1148/radiol.2018180547 [DOI] [PubMed] [Google Scholar]
- Szegedy C, Wei Liu, Yangqing Jia, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A, 2015. Going deeper with convolutions, in: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) IEEE, pp. 1–9. 10.1109/CVPR.2015.7298594 [DOI] [Google Scholar]
- Tang R, Islam Tushar F, Han S, Hou R, Rubin GD, Lo JY, 2019. Classification of chest CT using case-level weak supervision, in: Hahn HK, Mori K (Eds.), Medical Imaging 2019: Computer-Aided Diagnosis SPIE, p. 42 10.1117/12.2513576 [DOI] [Google Scholar]
- Terpenning S, White CS, 2015. Imaging pitfalls, normal anatomy, and anatomical variants that can simulate disease on cardiac imaging as demonstrated on multidetector computed tomography. Acta Radiol. Short Reports 4, 204798161456244 10.1177/2047981614562443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Walt S, Colbert SC, Varoquaux G, 2011. The NumPy array: A structure for efficient numerical computation. Comput. Sci. Eng 13, 22–30. 10.1109/MCSE.2011.37 [DOI] [Google Scholar]
- Voulodimos A, Doulamis N, Doulamis A, Protopapadakis E, 2018. Deep Learning for Computer Vision: A Brief Review, Computational Intelligence and Neuroscience Hindawi Limited; 10.1155/2018/7068349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SLF, Calandriello L, Silva M, Sverzellati N, 2018. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet. Respir. Med 6, 837–845. 10.1016/S2213-2600(18)30286-8 [DOI] [PubMed] [Google Scholar]
- Wang C, Moriya T, Hayashi Y, Roth H, Lu L, Oda M, Ohkubo H, Mori K, 2019. Weakly-supervised deep learning of interstitial lung disease types on CT images. SPIE-Intl Soc Optical Eng, p. 53 10.1117/12.2512746 [DOI]
- Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM, 2017. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases, in: Computer Vision and Pattern Recognition IEEE; 10.1109/CVPR.2017.369 [DOI] [Google Scholar]
- Yan K, Wang X, Lu L, Summers RM, 2018. DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J. Med. Imaging 5, 1 10.1117/1.JMI.5.3.036501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates EJ, Yates LC, Harvey H, 2018. Machine learning “red dot”: open-source, cloud, deep convolutional neural networks in chest radiograph binary normality classification. Clin. Radiol 73, 827–831. 10.1016/j.crad.2018.05.015 [DOI] [PubMed] [Google Scholar]
- Zech J, Pain M, Titano J, Badgeley M, Schefflein J, Su A, Costa A, Bederson J, Lehar J, Oermann EK, 2018. Natural Language–based Machine Learning Models for the Annotation of Clinical Radiology Reports. Radiology 287, 570–580. 10.1148/radiol.2018171093 [DOI] [PubMed] [Google Scholar]
- Zhang G, Jiang S, Yang Z, Gong L, Ma X, Zhou Z, Bao C, Liu Q, 2018. Automatic nodule detection for lung cancer in CT images: A review. Comput. Biol. Med 10.1016/j.compbiomed.2018.10.033 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


