Abstract
Purpose
To automate the segmentation of retinal layers, we propose DeepRetina, a method based on deep neural networks.
Methods
DeepRetina uses the improved Xception65 to extract and learn the characteristics of retinal layers. The Xception65-extracted feature maps are inputted to an atrous spatial pyramid pooling module to obtain multiscale feature information. This information is then recovered to capture clearer retinal layer boundaries in the encoder-decoder module, thus completing retinal layer auto-segmentation of the retinal optical coherence tomography (OCT) images.
Results
We validated this method using a retinal OCT image database containing 280 volumes (40 B-scans per volume) to demonstrate its effectiveness. The results showed that the method exhibits excellent performance in terms of the mean intersection over union and sensitivity (Se), which are as high as 90.41 and 92.15%, respectively. The intersection over union and Se values of the nerve fiber layer, ganglion cell layer, inner plexiform layer, inner nuclear layer, outer plexiform layer, outer nuclear layer, outer limiting membrane, photoreceptor inner segment, photoreceptor outer segment, and pigment epithelium layer were found to be above 88%.
Conclusions
DeepRetina can automate the segmentation of retinal layers and has great potential for the early diagnosis of fundus retinal diseases. In addition, our approach will provide a segmentation model framework for other types of tissues and cells in clinical practice.
Translational Relevance
Automating the segmentation of retinal layers can help effectively diagnose and monitor clinical retinal diseases. In addition, it requires only a small amount of manual segmentation, significantly improving work efficiency.
Keywords: DeepRetina, retinal layer, auto-segmentation, OCT image, deep neural network
Introduction
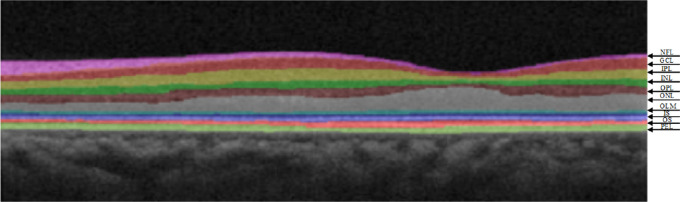
Fundus retinal diseases are very commonly diagnosed by ophthalmologists, and most fundus diseases are caused by retinopathy.1,2 It is estimated that more than 300 million people worldwide have fundus diseases such as age-related macular degeneration (AMD), diabetic retinopathy (DR), and central serous chorioretinopathy.3,4 Studies have shown that in most fundus diseases, retinal morphologic changes are observed earlier than visual field changes, and the analysis of retinal morphologic structure using specific and sensitive methods will contribute to the early detection of fundus retinal diseases. Therefore, performing a retinal layer analysis is essential for the early diagnosis and timely treatment of retinal diseases. In recent years, retinal fundus images have been widely used in the diagnosis, screening, and treatment of retinal diseases.5 Automating the segmentation of retinal layers in retinal optical coherence tomography (OCT) images can help effectively diagnose and monitor retinal diseases. In this study, the 10-layer structure of the retina, including the nerve fiber layer (NFL), ganglion cell layer (GCL), inner plexiform layer (IPL), inner nuclear layer (INL), outer plexiform layer (OPL), outer nuclear layer (ONL), outer limiting membrane (OLM), photoreceptor inner segment (IS), photoreceptor outer segment (OS), and pigment epithelium layer (PEL), is analyzed.6 Figure 1 shows the retinal OCT cross section of the macula region, including the different layers of the retina.
Figure 1.
Interlaminar structure diagram of the retinal OCT cross section of the macular area.
OCT is a noninvasive, real-time, micro-resolution medical imaging tool for micro-resolution volumetric scanning of biological tissues and is ideal for examining fundus nerve tissue.7 A quantitative analysis of retinal OCT images is critical for the diagnosis and treatment of retinal diseases; however, retinal OCT images are susceptible to speckle noise, and the contrast between adjacent faults is small, making it difficult to accurately segment the images. Therefore, automatic segmentation of the retinal layers in retinal OCT images is of great significance for the diagnosis and treatment of fundus retinal diseases.
In recent decades, computer-aided analysis methods for the segmentation of retinal OCT images have become increasingly popular, including automatic segmentation methods. In the development of computer-aided diagnostic systems for ophthalmic diseases, automatic segmentation of the retina has been considered a critical and challenging step.8 First, it is difficult to accurately segment the retinal layers because of the complexity of retinal OCT images and the limited resolution of the OCT scanning system.9 Second, affected by retinal diseases, the retinal layer may deteriorate and cause severe deformation.10,11 Although many algorithms for segmenting the retinal layer have been developed,12–22 these algorithms have a problem in that some layers are not segmented, which remains a daunting task. Existing retinal OCT image segmentation methods are mainly based on the active contour, classifiers, three-dimensional map search, and deep learning.
Gawish et al.23 proposed the use of active contours for the stratification of fundus retinal OCT images. They used continuous curves and energy function to transform the process of segmentation into a process of solving the minimum value of the energy function. The position of the curve corresponding to minimum energy is the contour of the layer. Although this method improves the antinoise performance and accuracy to some extent, the time complexity of this algorithm is high and sensitive to the initial position of the contour.
Abràmoff et al.24 proposed a classifier-based method based on support vector machine (SVM)25 and fuzzy c-means clustering.26 The SVM-based segmentation classifies image pixels in the feature space of the image, thus realizing image segmentation. The fuzzy c-means clustering-based segmentation determines the region to which the image pixels belong, thus realizing automatic segmentation of the fundus OCT image. With the gradual optimization of algorithms,27 the segmentation accuracy of such methods has been improved to within 2 pixels.
Hussain et al.28 used the graph theory method to divide a fundus OCT image into nine layers. This method involves dividing the graph into several subgraphs by continuously removing specific edges in the graph for an accurate image segmentation. Graph-based methods include GraphCut,29 GrabCut,30 and RandomWalk.31 Although the accuracy of this type of method can reach 1 pixel, it takes pixels as nodes and is easily affected by noise and image degradation.32–37
The segmentation algorithms have certain limitations in automatically segmenting the retinal layers in retinal OCT images. In the field of deep learning–based segmentation, Fang et al.38 proposed a convolutional neural network (CNN) and graph search–based method. They used a CNN to extract effective features of specific retinal layer boundaries and applied graph search methods on the probability maps generated by the CNN to obtain the final boundaries. Although this method can automate the segmentation of nine-layer boundaries of retina to some extent, the computational burden of this method is high and sensitive to the position of the boundaries. Roy et al.39 proposed the ReLayNet with fully convolutional deep architecture, for end-to-end segmentation of retinal layers. They used encoders to learn a hierarchy of contextual features, followed by decoders for semantic segmentation. Although ReLayNet exhibits well performance in segmentation of retina, it is easily affected by image degradation and poor spatial resolution. Apostolopoulos et al.40 proposed the fully convolutional neural network architecture, which combines dilated residual blocks in an asymmetric U-shape configuration. Overall, it demonstrates low computational costs and high performance, but it faces difficulties in segmenting all retinal layers accurately. Pekala et al.41 proposed automated approach segment images using fully convolutional networks (FCNs) together with Gaussian process–based regression. In aggregate, the proposed methods perform favorably, but its segmentation accuracy is not high. He et al.42 proposed a cascaded FCN framework and transformed the layer segmentation problem from pixel labeling into a boundary position regression problem. Although this method takes the topologically unconstrained results, there is some deviation in the segmentation of retina layer. Sedai et al.43 proposed a method for retinal layer segmentation and quantification of uncertainty based on Bayesian deep learning, and its average execution time for the retinal layer segmentation per image is somewhat slow. In addition, Shah et al.44 proposed a CNN-based framework to segment multiple surfaces simultaneously. Guo et al.45 proposed an automated method to segment seven retinal layer boundaries and two retinal plexuses in wide-field OCTA images. Hamwood et al.46 proposed a convolutional neural network approach for automatic segmentation of OCT retinal layers; the method is applied to detect the probability of retinal boundary locations in OCT images, which are subsequently used to segment the OCT image using a graph-search approach.
Chen et al.47 proposed an encoder-decoder model with separable convolution for image semantic segmentation. Based on this model, we carried out experiments and proposed a new DeepRetina method for the automatic segmentation of retinal layers in retinal OCT images. DeepRetina is based on a deep neural network; we improved Xception48 and used the improved Xception65 as the backbone network to extract and learn the characteristics of retinal layers. Xception65-extracted feature maps are inputted to an atrous spatial pyramid pooling (ASPP) module,49 and multiscale feature information is obtained. The encoder-decoder module47,50 is then used to recover the retinal information to capture a clearer retinal layer boundary. The segmentation result is optimized, and the automatic segmentation of the retinal OCT image is completed. This study is expected to promote the application of deep learning–based retinal segmentation methods for ophthalmic imaging. The research results have important practical significance for the early diagnosis and therapeutic monitoring of retinal diseases.
The rest of this article is organized as follows. The research method is described in the second section, including the details of DeepRetina and its application to the automatic segmentation of retinal OCT images. In the third section, the segmentation results of the retinal OCT image are given. The fourth and fifth sections present the discussions and conclusions, respectively.
Materials and Methods
Database Experiment
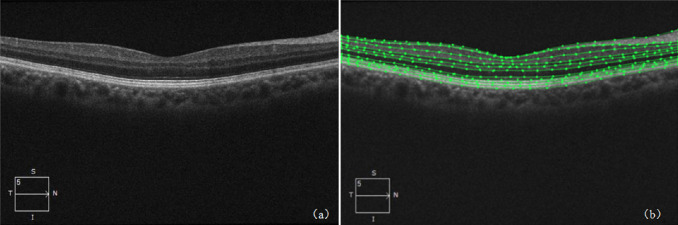
The database used in this study was a retinal OCT image database jointly maintained by the Shenzhen Eye Hospital affiliated with Jinan University and the Shenzhen University School of Medicine. The image database was scanned using a standard Cirrus HD-OCT (Carl Zeiss AG, Jena, Germany) device at 564 × 375 pixels and contained 280 volumes for a total of 11,200 images (40 B-scans per volume). The 40 B-scans per volume were scan centered at the fovea and 20 frames on either side of the fovea (fovea slice and scans laterally acquired at ±1, ±2, ±3,…, ±20 from the fovea slice). Figure 2a shows an example of the obtained retinal OCT image, which is an RGB image of 564 × 375 pixels.
Figure 2.
(a) Retinal OCT image and its annotation example. (b) The green line indicates the retinal layer, which forms a closed curve. Each closed curve represents a layer of retina containing 10 closed curves whose estimates do not cover the entire image.
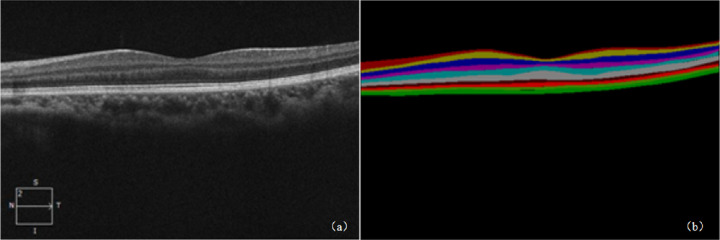
We randomly selected 7840 images from 196 volumes as the training set and the remaining 84 volumes, for a total of 3360 images, as the testing set (280 volumes were divided into 196 volumes and 84 volumes; each volume was different, and the randomization was performed at the patient level). The ratio of the training set to the testing set was 7:3. Our data set included control subjects and patients with age-related macular degeneration and diabetic retinopathy, and the ratio of controls to patients was 9:1. We invited a clinical expert (Shenzhen Institute of Ophthalmology Prevention, Shenzhen Key Laboratory of Ophthalmology, deputy chief physician of the Medical Imaging Department of Shenzhen Eye Hospital Affiliated to Jinan University) who has performed retinal segmentation for more than 10 years to label the data using the deep learning annotation tool Labelme (for manually drawing boundaries on the target to generate network-trainable JSON files, including the target location area and tag name), as shown in Figure 2b. Each layer of the retina after labeling had its own unique regional color (i.e., pixel value) and label name. The pixel values and label names of the same type of retinal layers were the same but different for the different retinal layers. Each image calibrated by the expert was marked with 10 layers of the retinal structure. Figure 3 shows the original image of the retina and its corresponding label. A complete retinal OCT image database was obtained after the data labeling was completed.
Figure 3.
Example of the original image of the retina and its corresponding label. (a) Original image of the retina with 564 × 375 pixels. (b) Label diagram labeled and converted using Labelme, with 10 different colors representing the 10 layers of the retinal structure.
For further performance validation on diseased retinas, we used the publicly available Duke data set.51 The data set includes 110 OCT B-scan images spanning 10 different patients with diabetic macular edema, with 11 B-scans per patient, and each image consisted of 740 × 512 pixels.
Method Overview
DeepRetina uses the expert-labeled retinal OCT image database as the training sample. Before inputting to the depth network model, the training data are preprocessed to overcome the low resolution of the retinal OCT images, low contrast between the layers, and effect of speckle noise. The overall network model of DeepRetina comprises the following operations: extracting and learning the retinal layer characteristics using the improved Xception65 as the backbone network, combining ASPP for reinforcement learning and generating retinal multiscale feature information, and using the encoder-decoder module to recover retinal information to capture clearer retinal layer boundaries and further optimize the segmentation results. Finally, the automatic segmentation of the retinal OCT image is completed using the TensorFlow deep learning framework.
Overall Network Model Structure
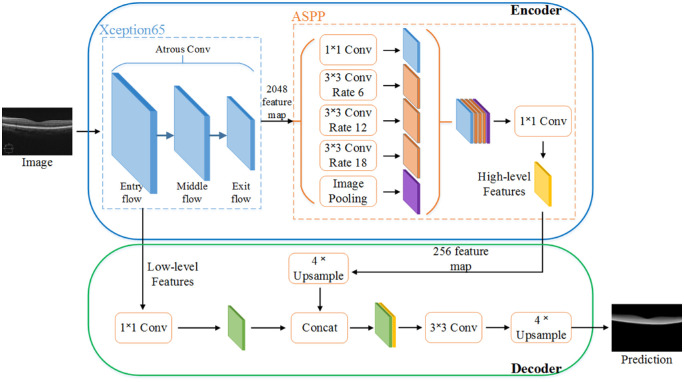
Figure 4 shows the overall neural network structure of DeepRetina, including Xception65 as the backbone network and ASPP for strengthening the learning of the feature map generated by Xception65 and generating retinal multiscale feature information. There is no strict limit on the size of the input image to obtain multiscale feature information. The encoder-decoder module is used to recover the retinal information and capture clearer retinal layer boundaries while further optimizing the retinal segmentation results.
Figure 4.
Overall neural network structure diagram of DeepRetina. The original retina image passes through Xception65, ASPP, and encoder-decoder; the predicted image is then outputted.
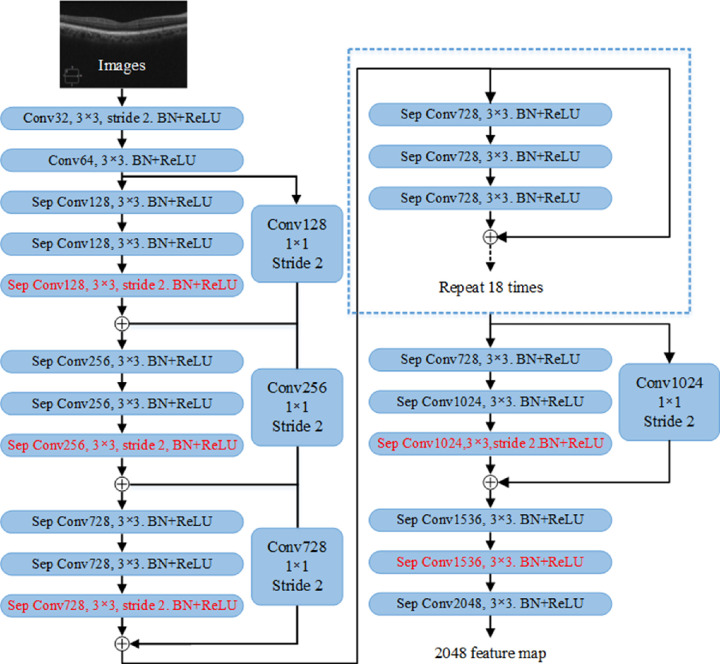
Backbone Network
Figure 5 shows the network structure of the improved Xception65. To optimize the network model, a deep separable convolution48 was introduced in Xception65, which allows extracting feature maps at any resolution, so there is no strict requirement on the size of the input image to obtain deep semantic information and shallow image detail. The improved Xception65 network has more layers and a deeper network depth. Moreover, all the pooling layers are replaced with convolution layers and merged into the existing convolution layer. In addition, batch normalization and ReLU layers are added after each 3 × 3 depth-wise convolution.
Figure 5.
Improved Xception network structure diagram. More network layers are added, depth separable convolutions are introduced, and BN (Batch Normalization) and ReLU layers are added after each 3 × 3 depth convolution.
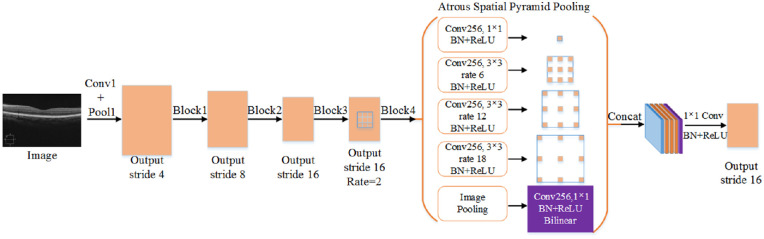
Improved Atrous Spatial Pyramid Pooling
ASPP is based on a spatial pyramid pool,52,53 with the introduction of an atrous convolution47 for capturing retinal multirange feature information. Figure 6 shows the network structure. In the experiment, we found that ASPP with different atrous rates can effectively capture retinal multiscale feature information. With the increase in the rate, the 3 × 3 convolution degenerates into a 1 × 1 convolution; therefore, we used four atrous convolutions with an atrous rate = {2,6,12,18} and added a 1 × 1 convolution. In addition, to obtain global information, an image pooling branch was added for the global pooling of the previously outputted feature map. Bilinear interpolation was employed to ensure that its resolution was the same as those of other branches. Finally, all the branches were connected, and a 1 × 1 convolution was performed.
Figure 6.
Improved ASPP network structure diagram. Atrous convolution is introduced, and the image pooling branches are combined.
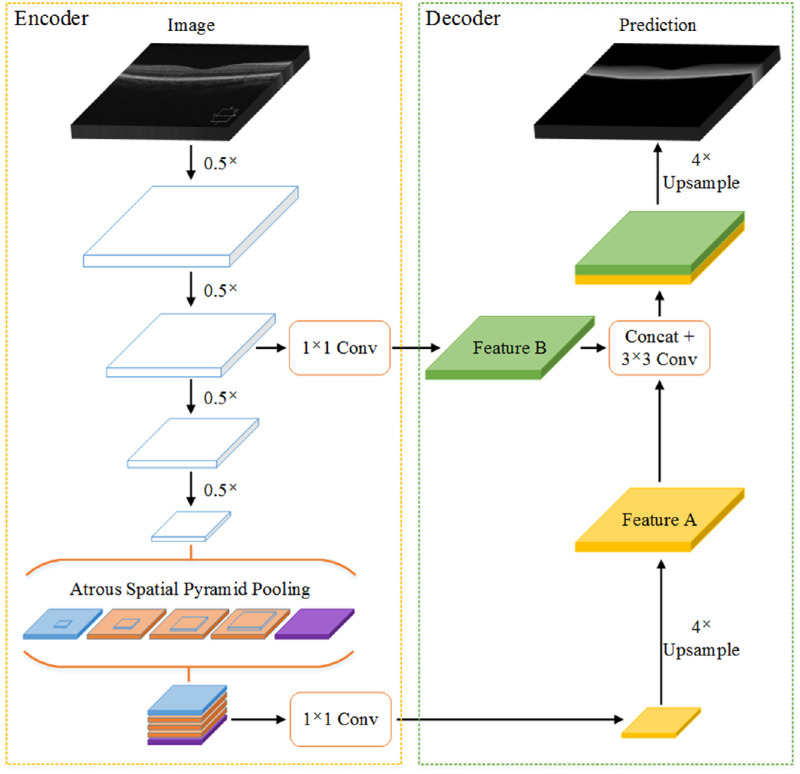
Encoder-Decoder Module
In general, an encoder-decoder module consists of an encoder with rich multiscale feature information and a valid decoder that restores the boundary of the object. The encoder obtains higher retinal feature information by reducing the feature map, and the decoder gradually recovers the retinal information to obtain a clearer retinal layer boundary. Figure 7 shows the network structure of the encoder-decoder. The encoder enhances the ASPP module, which uses atrous convolution with different rates to detect multiscale convolution features and extract features at any resolution. The decoder performs a fourfold bilinear upsampling of the feature outputted from the encoder to obtain a 256-dimensional feature A and then concatenates the 256-dimensional feature B obtained using the 1 × 1 convolution in the encoder with feature A and passes through a 3 × 3 convolution to refine the feature. Finally, a fourfold bilinear upsampling is performed to obtain the prediction results.
Figure 7.
Encoder-decoder block diagram. The encoder obtains rich multiscale feature information of the retina, and the decoder gradually recovers the retinal information to obtain a clearer retinal layer boundary.
Focal-Loss Function
The focal-loss function is used to solve the imbalance problem of training samples. To better solve the imbalance problem of training samples, we introduce a weighting factor, α, α ∈ [0,1]. As α increases, the influence of the background category on training is greatly reduced. In addition, to better control the weights of positive and negative samples, a modulation factor, (1 – Pt)γ, is introduced, where γ ≥ 0, thereby obtaining the focal-loss function:
| (1) |
Pt is defined as
| (2) |
where p is the probability of y = 1, p ∈ [0,1].
We found that (α, γ) = (0.25, 2) works best in experiments. In this way, the focal-loss function solves both the imbalance problem of training samples and the weight assignment problem of the positive and negative samples. This improves greatly the precision and speed of segmentation.
Overall Experiment
Data Preprocessing
To complete the layer segmentation of the retinal OCT image, we first preprocessed the training set images. To meet the requirements of deep learning neural network training and enhance the generalization ability of the network model, we carried out data augmentation processing on the images and their labels. Data augmentation methods include geometric transformation (translation, scaling, rotation, and flipping), sharpening, contrast change, brightness change, and HSV (Hue, Saturation, Value) color space change. The images of the training set are used for network training, and the images of the test set are used for performance evaluation. In our data set, there is no overlap between the training and test data to ensure the accuracy of network training.
Network Training
We trained the network in the experiment, and the database images used are given in Database Experiment. For the training parameters, we set the learning rate to 1e-4, the learning rate decay to 0.1, the momentum to 0.9, and the weight attenuation to 0.00004, and the input image was clipped to 512 × 512 pixels each time during network training. Based on the TensorFlow deep learning framework, the training was performed on a server with 4 NVIDIA GP102 TITAN Xp graphics cards and Intel Xeon Gold 6148 CPU (AMAX Information Technologies Inc, Suzhou China) @ 2.40 GHz × 51. The batch size was set to 32, and 130,000 iterations were performed. The entire training process took approximately 72 hours.
Performance Evaluation
To further assess the accuracy of the method, the following criteria were employed to evaluate the network model trained in the experiment: mean intersection over union (MIoU), sensitivity (Se), and CPU and GPU average calculation times. If both MIoU and Se remained high and the average computation time of the CPU or GPU was low, the network model was said to have excellent performance.
All the targets in the test set image already contained specific pixel values and tag names (i.e., ground truth; see Database Experiment for details), and a new predicted segmentation would be generated during the network test. MIoU is a standard measure for image semantic segmentation. It can be used to calculate the intersection and union ratio of the ground truth and predicted segmentation, thus determining the accuracy of the segmentation prediction. k + 1 categories (including background category) are assumed, and intersection over union (IoU) is calculated in each category separately, referring to Equation (3). The calculation method of MIoU is shown in Equation (4). For simplicity, Equation (3) is transformed into Equation (5). Pii represents the number of pixels correctly predicted as category i, Pij represents the number of pixels belonging to category i but predicted as category j, and Pji represents the number of pixels belonging to category j but predicted as category i.
| (3) |
| (4) |
| (5) |
Here, the true positive (TP) indicates correctly segmented retinal pixels, false positive (FP) indicates correctly segmented nonretinal pixels, and false negative (FN) indicates incorrectly segmented retinal pixels. The Se is calculated using Equation (6).
| (6) |
For further performance evaluation, besides the IoU and sensitivity, we calculated the mean thickness difference (in pixels) between the automated and manual segmentations. First, for each B-scan in the test set, we calculated the mean thickness difference of each layer between automated segmentations and manual segmentations. Next, after taking the absolute value of these differences, the mean and standard deviation across all B-scans of the test set were calculated. The smaller the mean and standard deviation, the better the retinal layer segmentation.
Results
Retinal Segmentation Results
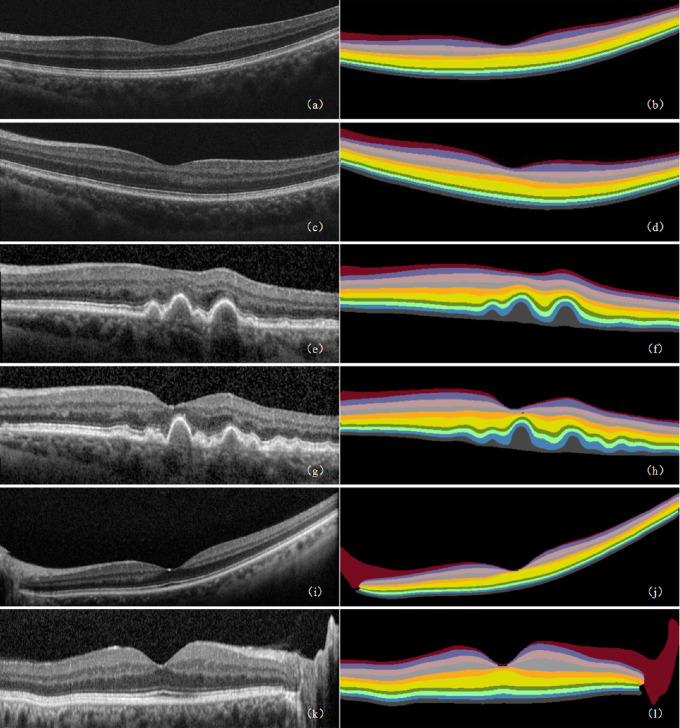
The MIoU of the DeepRetina segmentation model was up to 90.41%, and its Se was up to 92.15% for the testing set. The IoU and Se of values of the NFL, GCL, IPL, INL, OPL, ONL, OLM, IS, OS, and PEL were 91.82 and 95.02%, 92.79 and 94.36%, 90.71 and 92.31%, 90.95 and 92.76%, 89.90 and 91.74%, 93.23 and 95.57%, 88.52 and 89.12%, 89.23 and 90.49%, 88.93 and 90.51%, and 88.02 and 89.64%, respectively. The GPU average time required to automatically segment a retinal OCT image was approximately 0.52 seconds, and the average CPU average time was approximately 1.43 seconds, which met the clinical requirements for retinal segmentation. For the retinal OCT images in the testing set, we used the segmentation model to perform a 10-layer segmentation and obtained the predictions (see Fig. 8). Table 1 lists the segmentation performance results for the retinal OCT image test set obtained using our segmentation model. Table 2 lists the segmentation performance results of each of the specific categories (control, AMD, and DR).
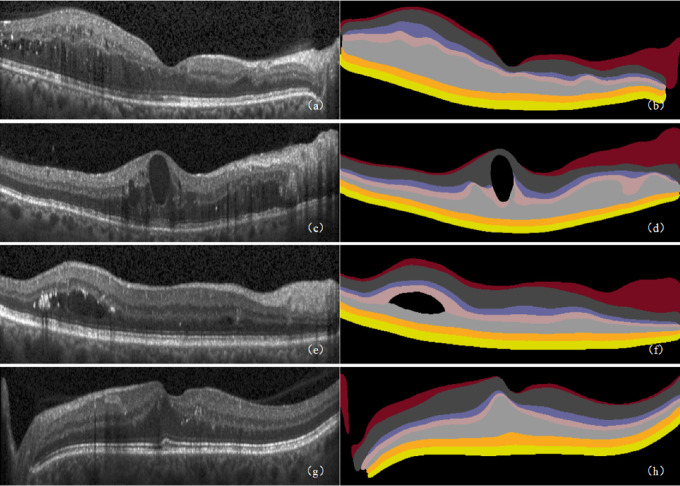
Figure 8.
Retinal 10-layer segmentation prediction results. (a, c, e, g, i, k) Original images. (b, d, f, h, j, l) Corresponding segmentation results. In the segmentation result graph, the pixel values of the different retinal layers are different, thereby showing the 10 layers of the retinal interlayer structure.
Table 1.
Test Performance Results for 10 Layers of Retina Segmentation
| Automated Versus Manual Segmentation | |||||||
|---|---|---|---|---|---|---|---|
| Retinal Layers | Total Layers | Complete Segmentation | Incomplete Segmentation | IoU (%) | Se (%) | Mean Deviation | Standard Deviation |
| NFL | 3360 | 3087 | 273 | 91.82 | 95.02 | 0.26 | 0.37 |
| GCL | 3360 | 3119 | 241 | 92.79 | 94.36 | 0.20 | 0.29 |
| IPL | 3360 | 3050 | 310 | 90.71 | 92.31 | 0.41 | 0.50 |
| INL | 3360 | 3058 | 302 | 90.95 | 92.76 | 0.21 | 0.33 |
| OPL | 3360 | 2998 | 362 | 89.90 | 91.74 | 0.62 | 0.74 |
| ONL | 3360 | 3135 | 225 | 93.23 | 95.57 | 0.12 | 0.26 |
| OLM | 3360 | 2979 | 381 | 88.52 | 89.12 | 0.82 | 0.84 |
| IS | 3360 | 2996 | 364 | 89.23 | 90.49 | 0.59 | 0.63 |
| OS | 3360 | 2988 | 372 | 88.93 | 90.51 | 0.78 | 0.76 |
| PEL | 3360 | 2962 | 398 | 88.02 | 89.64 | 0.88 | 0.87 |
Table 2.
Segmentation Performance Results of Control, AMD, and DR
| Control | AMD | DR | ||||
|---|---|---|---|---|---|---|
| Retinal Layers | IoU (%) | Se (%) | IoU (%) | Se (%) | IoU (%) | Se (%) |
| NFL | 91.82 | 95.02 | 90.29 | 92.98 | 90.06 | 91.99 |
| GCL | 92.79 | 94.36 | 91.46 | 92.68 | 90.57 | 92.26 |
| IPL | 90.71 | 92.31 | 89.87 | 91.22 | 89.31 | 90.66 |
| INL | 90.95 | 92.76 | 89.96 | 91.58 | 89.28 | 90.79 |
| OPL | 89.90 | 91.74 | 89.07 | 91.23 | 88.64 | 89.97 |
| ONL | 93.23 | 95.57 | 92.11 | 93.37 | 91.61 | 92.73 |
| OLM | 88.52 | 89.12 | 87.36 | 89.06 | 86.79 | 89.00 |
| IS | 89.23 | 90.49 | 88.79 | 89.79 | 88.49 | 89.34 |
| OS | 88.93 | 90.51 | 87.89 | 90.00 | 87.43 | 90.11 |
| PEL | 88.02 | 89.64 | 87.65 | 88.94 | 86.92 | 88.35 |
“Total layers” is the total number of images containing those layers. “Complete segmentation” means that the retinal layer is exactly segmented. “Incomplete segmentation” means that the retinal layer is not completely segmented. Please note that, because of the low resolution of the retinal OCT image and noise-induced interference, this led to the retinal layer not being completely segmented, and it is difficult to achieve accurate segmentation of the retinal layer.
The MIoU of the DeepRetina segmentation model was up to 90.55%, and its Se was up to 92.29% for the Duke data set. The IoU and Se of values of the NFL, GCL+IPL, INL, OPL, ONL+OLM, IS, and OS+PEL are listed in Table 3. Figure 9 shows the prediction results obtained on the Duke data set.
Table 3.
Test Results for Seven Layers of Retina Segmentation on the Duke Data Set
| Characteristic | NFL | GCL+IPL | INL | OPL | ONL+OLM | IS | OS+PEL |
|---|---|---|---|---|---|---|---|
| IoU (%) | 90.64 | 92.36 | 90.79 | 89.22 | 92.54 | 89.47 | 88.81 |
| Se (%) | 93.77 | 93.89 | 92.31 | 90.71 | 94.10 | 91.02 | 90.21 |
Figure 9.
Retinal seven-layer segmentation prediction results on the Duke data set. (a, c, e, g) Original images. (b, d, f, h) Corresponding segmentation results. In the segmentation result graph, the pixel values of the different retinal layers are different, thereby showing the seven layers of the retinal interlayer structure.
Results of Optimization Experiments
We used ResNet101, Inception-v3, and Xception65 as the backbone networks of the segmentation model for comparison. Table 4 lists their effects on the performance of the segmentation model under the same network parameters. The MIoU and Se of Xception65 were the highest, while those of ResNet101 and Inception-v3 were lower. Figure 10 shows their prediction results on retinal segmentation. Therefore, we chose Xception65 as the backbone network.
Table 4.
Experimental Results for Different Optimization Methods
| Optimization Method | MIoU (%) | Se (%) | Test Time (s/Image) | |
|---|---|---|---|---|
| backbone | ResNet101 | 85.91 | 86.63 | 0.48 |
| Inception-v3 | 88.03 | 89.75 | 0.54 | |
| Xception65 | 90.12 | 92.06 | 0.53 | |
| Atrous rates | {18, 36, 54} | 86.45 | 86.64 | 0.69 |
| {12, 24, 36} | 88.93 | 89.42 | 0.58 | |
| {6, 12, 18} | 90.11 | 91.79 | 0.51 | |
The bold values represent the bset result.
Figure 10.
Prediction results of different backbone networks on retinal segmentation. (a, b) ResNet101 prediction results. (c, d) Inception-v3 prediction results. (e, f) Xception65 prediction results.
In the experiment, we found that atrous convolution with different rates can help effectively capture retinal multiscale feature information. At higher rates, the 3 × 3 convolution cannot capture the feature information of the full image but degenerates into a simple 1 × 1 convolution. To this end, we used atrous convolution rates of {6, 12, 18}, {12, 24, 36}, and {18, 36, 54} for comparison. Table 4 lists the performance results of the segmentation model. When the rate is {6, 12, 18}, the segmentation effect is the best, and MIoU and Se are higher. Figure 11 shows their prediction results on retinal segmentation. Therefore, we selected the atrous rate as {6, 12, 18}.
Figure 11.
Prediction results of different atrous rates on retinal segmentation. (a, b) Atrous rate of {18, 36, 54} prediction results. (c, d) Atrous rate of {12, 24, 36} prediction results. (e, f) Atrous rate of {6, 12, 18} prediction results.
Folded Cross-Validation
To provide a better gauge of the algorithm's overall performance, we split our data set (280 volumes for a total of 11,200 images, 40 B-scans per volume) into five equally sized nonoverlapping subsets, and we used four subsets for training (224 volumes) and one subset for testing (56 volumes) and rotated the subset used for testing. We repeated this process for a fivefold cross-validation, resulting in testing performed on each subset. The average results of fivefold cross-validation on 10 layers of retina segmentation are tabulated in Table 5.
Table 5.
Average Results of Fivefold Cross-Validation on 10 Layers of Retina Segmentation
| Characteristic | NFL | GCL | IPL | INL | OPL | ONL | OLM | IS | OS | PEL |
|---|---|---|---|---|---|---|---|---|---|---|
| MIoU (%) | 92.08 | 92.56 | 90.78 | 91.16 | 90.56 | 92.89 | 88.78 | 89.08 | 89.49 | 88.69 |
| Se (%) | 95.03 | 94.29 | 91.93 | 92.98 | 91.47 | 94.77 | 89.91 | 90.11 | 90.53 | 89.61 |
Comparison with Existing State-of-the-Art Methods
We compared the proposed methods with existing advanced methods, as listed in Table 6. For proper and fair comparison, we implemented these methods and followed what was described in the original publications; all methods were tested on the same test set. The results show that the proposed method achieves better performance for retinal segmentation, with an MIoU and Se obtained for DeepRetina of 0.9041 and 0.9215, respectively. Our method achieved satisfactory results compared with existing methods.
Table 6.
Results Compared with Existing Methods
| Method | Year | MIoU | Se |
|---|---|---|---|
| Fang et al.38 | 2017 | 0.8709 | 0.8691 |
| Roy et al.39 | 2017 | 0.8823 | 0.8911 |
| Apostolopoulos et al.40 | 2017 | 0.8629 | 0.8723 |
| He et al.42 | 2018 | 0.8812 | 0.8924 |
| Sedai et al.43 | 2018 | 0.8845 | 0.9087 |
| Shah et al.44 | 2018 | 0.8832 | 0.8992 |
| Guo et al.45 | 2018 | 0.8854 | 0.9061 |
| Hamwood et al.46 | 2018 | 0.8768 | 0.8829 |
| Gholami et al.54 | 2018 | 0.8903 | 0.9194 |
| Pekala et al.41 | 2019 | 0.8915 | 0.9078 |
| Proposed method | 2020 | 0.9041 | 0.9215 |
For better evaluating the performance on Duke data set, we put our results in correspondence with other methods evaluated on Duke data set, as listed in Table 7. For fair comparison, we implemented these methods and followed what was described in the original publications, and all methods were trained and tested on the Duke data set. The results show that the performance of the proposed method was improved over the state of the art evaluated on the Duke data set.
Table 7.
Results Compared with Other Methods Evaluated on the Duke Data Set
Discussion
The proposed retinal layer automatic segmentation method—namely, DeepRetina—provided satisfactory segmentation results. The method effectively performed automatic segmentation of the retinal layer, with the continuous optimization of the network model through a series of optimization experiments to improve its performance.
The setting of the network parameters and the training parameters during training is critical to improve the performance of the network model. To improve the convergence speed of the network model, we froze the logit layer of the last layer in the network, and all trained weights were recalled, with weight attenuation set to 0.00004. During the training, the learning rate was set to 0.007, so that the loss could be quickly reduced to a lower value, and then the learning rate was reduced to 0.0001, so that the loss could be steadily decreased. Figure 12 shows the whole convergence process. In addition, the network model was trained on a server with 4 NVIDIA GP102 TITAN Xp graphics cards and Intel Xeon Gold 6148 CPU @ 2.40 GHz × 51. The batch size was set to 32, and compared with the smaller batch size, the larger batch size could improve the performance of the network model.
Figure 12.
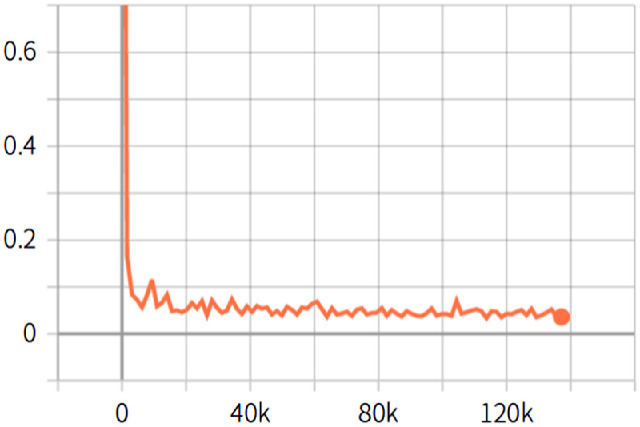
Loss convergence graph. The abscissa indicates the number of iterations, the ordinate indicates the loss value, and the orange curve indicates the entire convergence process of the training.
In this work, the average time required to segment a retinal OCT image was approximately 1.43 seconds with an Intel Xeon Gold 6148 CPU running at 2.40 GHz with 4 GB of RAM. Nevertheless, the testing procedure can be performed offline and accelerated by GPU programming, with an average time required to segment a retinal OCT image approximately 0.52 seconds with a GeForce GTX 1080 graphics card. This method is currently implemented using TensorFlow, and its computational performance can be further improved in future research. For example, using the PyTorch, a deep learning framework supports dynamic computational graphs and more efficient memory usage, and this can further improve the speed of retinal segmentation.
The conventional image segmentation model requires extracting relevant information from a manually designed feature extractor. Our DeepRetina segmentation model relies on the heuristic method of autonomous learning, does not require artificial design features, and is more similar to a biological neural network, thus reducing the complexity of the network model.52,53 This study is expected to promote the application of deep learning–based segmentation methods for the analysis of retinal layers.
As mentioned earlier, the proposed segmentation model provides excellent performance for retinal layer segmentation applications and yields good results. However, because of the low resolution of the retinal OCT image and noise-induced interference, it is difficult to achieve accurate segmentation of the retinal layer. Our method can be further improved in the future. First, more retinal OCT images should be collected to train a better model. In particular, high-resolution retinal OCT images can be employed, as they have better pixel contrast and can improve network model performance. Second, better image enhancement processing can be performed to improve the generalization ability of the network model. Finally, in training and testing, attempts should be made to transform the retinal OCT image to a three-dimensional image, improve the network structure, and adjust the training parameters to obtain better segmentation results. To successfully apply our method for the diagnosis and treatment of retinal diseases, more retinal layer analyses should be conducted, including on relevant retinal diseases.
Conclusions
In this article, we proposed a segmentation model called DeepRetina for the auto-segmentation of retinal layers in retinal OCT images. Only a small amount of manual segmentation is required for training the network, and the trained model can automatically perform segmentation of retinal layers, significantly increasing the segmentation rate and improving work efficiency. The method achieves good results in terms of the segmentation precision and sensitivity and is very robust. It has great potential for the diagnosis of ophthalmic diseases through existing computer-aided diagnostic systems. Our research can aid in the early diagnosis and treatment of clinical retinal diseases and is expected to promote the application of deep learning–based segmentation methods for the analysis of retinal layers. In addition, our method can provide a segmentation model framework for other types of tissues and cells in clinical practice, with good application prospects.
Acknowledgments
Supported by the Shenzhen Science and Technology Project (JCYJ20170302152605463, JCYJ20170306123423907, and JCYJ20180507182025817), the National Natural Science Foundation of China–Shenzhen Joint Fund Project (U1713220) and the Research Startup Fund of Shenzhen University General Hospital (0000030502).
Disclosure: Q. Li, None; S. Li, None; Z. He, None; H. Guan, None; R. Chen, None; Y. Xu, None; T. Wang, None; S. Qi, None; J. Mei, None; W. Wang, None
References
- 1. Hayreh SS, Zimmerman MB.. Fundus changes in branch retinal arteriolar occlusion. Retina. 2015; 35(10): 2060–2066. [DOI] [PubMed] [Google Scholar]
- 2. Figueiredo IN, Kumar S, Oliveira CM, Ramos JD, Engquist B. Automated lesion detectors in retinal fundus images. Comput Biol Med. 2015; 66: 47–65. [DOI] [PubMed] [Google Scholar]
- 3. Abramoff MD, Garvin MK, Sonka M. Retinal imaging and image analysis. IEEE Rev Biomed Eng. 2010; 3: 169–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morgan JIW. The fundus photo has met its match: optical coherence tomography and adaptive optics ophthalmoscopy are here to stay. Ophthalmic Physiol Opt. 2016; 36(3): 218–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fraz MM, Remagnino P, Hoppe A, et al.. Blood vessel segmentation methodologies in retinal images—a survey. Comput Methods Programs Biomed. 2012; 108(1): 407–433. [DOI] [PubMed] [Google Scholar]
- 6. Kim EK, Park HYL, Park CK. Relationship between retinal inner nuclear layer thickness and severity of visual field loss in glaucoma. Nature. 2017; 7(1): 5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang D, Swanson EA.. Optical coherence tomography. Science. 1991; 254: 1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirbas C, Quek F.. A review of vessel extraction techniques and algorithms. ACM Computing Surveys (CSUR). 2004; 36(2): 81–121. [Google Scholar]
- 9. Spaide RF, Curcio CA.. Anatomical correlates to the bands seen in the outer retina by optical coherence tomography: literature review and model. Retina. 2011; 31(8): 1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hagiwara A, Mitamura Y, Kumagai K, Baba T, Yamamoto S. Photoreceptor impairment on optical coherence tomographic images in patients with retinitis pigmentosa. Br J Ophthalmol. 2013; 97(2): 237–238. [DOI] [PubMed] [Google Scholar]
- 11. Burke TR, Rhee DW, Smith RT, Tsang SH, Allikmets R, Chang S. Quantification of peripapillary sparing and macular involvement in Stargardt disease (STGD1). Invest Ophthalmol Vis Sci. 2011; 52(11): 8006–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kafieh R, Rabbani H, Kermani H. A review of algorithms for segmentation of optical coherence tomography from retina. J Med Signals Sensors. 2013; 3(1): 45–60. [PMC free article] [PubMed] [Google Scholar]
- 13. DeBuc DC. A review of algorithms for segmentation of retinal image data using optical coherence tomography. Image Segmentation. 2011; 1: 15–54. [Google Scholar]
- 14. Vermeer KA, van der Schoot J, Lemij HG, de Boer JF. Automated segmentation by pixel classification of retinal layers in ophthalmic OCT images. Biomed Opt Express. 2011; 2(6): 1743–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mujat M, Chan RC, Cense B, Park BH, Boer JF.. Retinal nerve fiber layer thickness map determined from optical coherence tomography images. Opt Express. 2015; 13(23): 9480–9491. [DOI] [PubMed] [Google Scholar]
- 16. Mayer MA, Hornegger J, Mardin CY, Tornow RP. Retinal nerve fiber layer segmentation on FD-OCT scans of normal subjects and glaucoma patients. Biomed Opt Express. 2010; 1(5): 1358–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Novosel J, Thepass G, Lemij HG, de Boer JF, Vermeer KA, van Vliet LJ. Loosely coupled level sets for simultaneous 3D retinal layer segmentation in optical coherence tomography. Med Image Anal. 2015; 26(1): 146–158. [DOI] [PubMed] [Google Scholar]
- 18. Ishikawa H, Stein DM, Wollstein G, Beaton S, Fujimoto JG, Schuman JS. Macular segmentation with optical coherence tomography. Invest Ophthalmol Vis Sci. 2005; 46(6): 2012–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dufour PA, Lou L, Ceklic L, Abdillahi H, et al. Graph-based multi-surface segmentation of OCT data using trained hard and soft constraints. IEEE Trans Med Imaging. 2013; 32(3): 531–543. [DOI] [PubMed] [Google Scholar]
- 20. Carass A, Lang A, Hauser M, Calabresi PA, Ying HS, Prince JL. Multiple-object geometric deformable model for segmentation of macular OCT. Biomed Opt Express. 2014; 5(4): 1062–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiu SJ, Li XT, Nicholas P, Toth CA, Izatt JA, Farsiu S. Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation. Opt Express. 2010; 18(18): 19413–19428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garvin MK, Abramoff MD, Kardon R, Russell SR, Wu X, Sonka M. Intraretinal layer segmentation of macular optical coherence tomography images using optimal 3-D graph search. IEEE Trans Med Imaging. 2008; 27(10): 1495–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gawish A, Fieguth P, Marschall S, Bizheva K. Undecimated hierarchical active contours for oct image segmentation. In: 2014 IEEE International Conference on Image Processing (ICIP). IEEE: Paris, FRANCE; 2014: 882–886. [Google Scholar]
- 24. Abràmoff MD, Lee k, Niemeijer M, et al. Automated segmentation of the cup and rim from spectral domain OCT of the optic nerve head. Invest Ophthalmol Vis Sci. 2009; 50(12): 5778–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Giri EP, Arymurthy AM.. Quantitative evaluation for simple segmentation SVM in landscape image. In: 2014 International Conference on Advanced Computer Science and Information Systems (ICACSIS). IEEE: Jakarta, INDONESIA; 2014: 369–374. [Google Scholar]
- 26. Li C, Li Y.. Novel fuzzy c-means segmentation algorithm for image with the spatial neighborhoods. In: 2012 2nd International Conference on Remote Sensing, Environment and Transportation Engineering (RSETE). IEEE: Nanjing, China; 2012: 1–4. [Google Scholar]
- 27. Agarwal P, Kumar S.. A combination of bias-field corrected fuzzy c-means and level set approach for brain MRI image segmentation. In: 2015 Second International Conference on Soft Computing and Machine Intelligence (ISCMI). IEEE: Hong Kong, PEOPLES R CHINA; 2015: 84–87. [Google Scholar]
- 28. Hussain MA, Bhuiyan A, Ramamohanarao K. Disc segmentation and BMO-MRW measurement from SD-OCT image using graph search and tracing of three bench mark reference layers of retina. In: 2015 IEEE International Conference on Image Processing (ICIP). IEEE: Quebec City, CANADA; 2015: 4087–4091. [Google Scholar]
- 29. Boykov YY, Jolly MP.. Interactive graph cuts for optimal boundary & region segmentation of objects in N-D images. In: IEEE International Conference on Computer Vision. IEEE: VANCOUVER, CANADA; Vol. 1; 2001: 105–112. [Google Scholar]
- 30. Rother C, Kolmogorov V, Blake A. GrabCut: interactive foreground extraction using iterated graph cuts. ACM Trans Graphics. 2004; 23: 309–314. [Google Scholar]
- 31. Shang S, Kulkarni SR, Cuff PW, Hui P. A randomwalk based model incorporating social information for recommendations. In: 2012 IEEE International Workshop on Machine Learning for Signal Processing. IEEE: Santander, Spain; 2012: 1–6. [Google Scholar]
- 32. Sun S, Sonka M, Beichel RR. Graph-based 4D lung segmentation in CT images with expert-guided computer-aided refinement. In: 2013 IEEE 10th International Symposium on Biomedical Imaging. IEEE: San Francisco, CA; 2013: 1312–1315. [Google Scholar]
- 33. Çığla C, Alatan AA. Efficient graph-based image segmentation via speeded-up turbo pixels. In: 2010 IEEE International Conference on Image Processing. IEEE: Hong Kong, PEOPLES R CHINA; 2010: 3013–3016. [Google Scholar]
- 34. Cui H, Wang X, Zhou J, Fulham M, Eberl S, Feng D. Topology constraint graph-based model for non-small-cell lung tumor segmentation from PET volumes. In: 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI). IEEE: Beijing, PEOPLES R CHINA; 2014: 1243–1246. [Google Scholar]
- 35. Hickson S, Birchfield S, Essa I, Christensen H. Efficient hierarchical graph-based segmentation of RGBD videos. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition. IEEE: Columbus, OH; 2014: 344–351. [Google Scholar]
- 36. Zhang H, Essa E, Xie X. Graph based segmentation with minimal user interaction. In: 2013 IEEE International Conference on Image Processing. IEEE: Melbourne, AUSTRALIA; 2013: 4074–4078. [Google Scholar]
- 37. Li X, Jin L, Song E, Li L. Full-range affinities for graph-based segmentation. In: 2013 IEEE International Conference on Image Processing. IEEE: Melbourne, AUSTRALIA; 2013: 4084–4087. [Google Scholar]
- 38. Fang L, Cunefare D, Wang C, Robyn H, Li S, Farsiu S. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed Opt Express. 2017; 8(5): 2732–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roy AG, Conjeti S, Karri SPK, et al.. ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks. Biomed Opt Express. 2017; 8(8): 3627–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Apostolopoulos S, Zanet SD, Ciller C, Wolf S, Sznitman R. Pathological OCT retinal layer segmentation using branch residual u-shape networks. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer International Publishing, Cham, Switzerland: Quebec City, QC, Canada; 2017. [Google Scholar]
- 41. Pekala M, Joshi N, Freund DE, Bressler NM, Debuc DC, Burlina PM. Deep learning based retinal OCT segmentation. Comput Biol Med. 2019; 144: 103445. [DOI] [PubMed] [Google Scholar]
- 42. He Y, Aaron C, Bruno J, et al.. Topology guaranteed segmentation of the human retina from OCT using convolutional neural networks. arXiv preprint. 2018; 1803: 05120. [Google Scholar]
- 43. Sedai S, Antony B, Mahapatra D, Garnavi R. Joint segmentation and uncertainty visualization of retinal layers in optical coherence tomography images using Bayesian deep learning. In: Computational Pathology and Ophthalmic Medical Image Analysis. SPRINGER INTERNATIONAL PUBLISHING AG: Granada, SPAIN; 2018: 219–227. [Google Scholar]
- 44. Shah A, Zhou L, Abrámoff MD, Wu X. Multiple surface segmentation using convolution neural nets: application to retinal layer segmentation in OCT images. Biomed Opt Express. 2018; 9: 4509–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guo Y, Camino A, Zhang M, et al.. Automated segmentation of retinal layer boundaries and capillary plexuses in wide-field optical coherence tomographic angiography. Biomed Opt Express. 2018; 9: 4429–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hamwood J, David AC, Read SA, Vincent SJ, Collins MJ. Effect of patch size and network architecture on a convolutional neural network approach for automatic segmentation of OCT retinal layers. Biomed Opt Express. 2018; 9: 3049–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In: The European Conference on Computer Vision (ECCV). Springer International Publishing, Cham, Switzerland: Munich, Germany; 2018: 801–818. [Google Scholar]
- 48. Chollet F. Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE: Honolulu, HI; 2017: 1251–1258. [Google Scholar]
- 49. Chen LC, Papandreou G, Schroff F, Adam H. Rethinking atrous convolution for semantic image segmentation. 2017. arXiv:1706.05587.
- 50. Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Machine Intell. 2017; 39(12): 2481–2495. [DOI] [PubMed] [Google Scholar]
- 51. Chiu SJ, Allingham MJ, Mettu PS, Cousins SW, Izatt JA, and Farsiu S. Kernel regression based segmentation of optical coherence tomography images with diabetic macular edema. Biomed Opt Express. 2015; 6(4): 1172–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lazebnik S, Schmid C, Ponce J. Beyond bags of features: Spatial pyramid matching for recognizing natural scene categories. In: 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'06). ZHEJIANG UNIV, EDITORIAL BOARD, 20 YUGU RD: HANGZHOU, CHINA; Vol. 2; 2006: 2169–2178. [Google Scholar]
- 53. Grauman K, Darrell T.. The pyramid match kernel: Discriminative classification with sets of image features. In: Tenth IEEE International Conference on Computer Vision (ICCV'05). IEEE: Beijing, PEOPLES R CHINA; Vol. 2; 2005: 1458–1465. [Google Scholar]
- 54. Gholami P, Roy P, Parthasarathy MK, Ommani A, Zelek J, Lakshminarayanan V. Intra-retinal segmentation of optical coherence tomography images using active contours with a dynamic programming initialization and an adaptive weighting strategy. In: Optical Coherence Tomography and Coherence Domain Optical Methods in Biomedicine XXII , International Society for Optics and Photonics. SPIE-INT SOC OPTICAL ENGINEERING: San Francisco, CA; 2018: 104832M. [Google Scholar]
- 55. Kepp T, Ehrhardt J, Heinrich MP, Hüttmann G, Handels H. Topology-preserving shape-based regression of retinal layers in OCT image data using convolutional neural networks. In: 2019 IEEE 16th International Symposium on Biomedical Imaging. IEEE: Venice, ITALY; 2019: 1437–1440. [Google Scholar]
- 56. Wei H, Peng P.. The segmentation of retinal layer and fluid in SD-OCT images using mutex dice loss based fully convolutional networks. IEEE Access. 2020; 8: 60929–60939. [Google Scholar]