Abstract
The newly emerged SARS-CoV-2 strains from the coronavirus (CoV) family is causing one of the most disruptive pandemics of the past century. Developing antiviral drugs is a challenge for the scientific community and pharmaceutical industry. Given the health emergency, repurposing of existing antiviral, antiinflammatory or antimalarial drugs is an attractive option for controlling SARS-CoV-2 with drugs. However, phytochemicals selected based on ethnomedicinal information as well as in vitro antiviral studies could be promising as well. Here, we summarise the phytochemicals with reported anti-CoV activity, and further analyzed them computationally to accelerate validation for drug development against SARS-CoV-2. This systematic review started from the most potent phytocompounds (IC50 in μM) against SARS-CoV, followed by a cluster analysis to locate the most suitable lead(s). The advanced molecular docking used the crystallography structure of SARS-CoV-2-cysteine-like protease (SARS-CoV-2-3CLpro) as a target. In total, seventy-eight phytochemicals with anti-CoV activity against different strains in cellular assays, were selected for this computational study, and compared with two existing repurposed FDA-approved drugs: lopinavir and ritonavir. This review brings insights in the potential application of phytochemicals and their derivatives, which could guide researchers to develop safe drugs against SARS-CoV-2.
Keywords: Coronavirus, Herbal medicine, MERS, Molecular docking, SARS-CoV-2, Natural products
The newly emerged Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), also called novel coronavirus-2019 (nCoV-2019), which causes coronavirus disease-2019 (COVID-19), is one of the most serious pandemics, with widespread morbidity and mortality globally [1,2]. Genomic analysis quickly confirmed that the newly emerged SARS-CoV-2 is related to two known CoV-strains, the Middle East Respiratory Syndrome coronavirus (MERS-CoV) and the Severe Acute Respiratory Syndrome coronavirus (SARS)-CoV [3]. Several genomic sequences also confirmed that the emerged SARS-CoV-2 genome is 75–85% identical with previously reported CoV strains, but with significant genomic variations in different geographic regions and continents [[4], [5], [6]].
Furthermore, patients with pre-existing pulmonary disease, kidney dysfunction, diabetes, or immune-compromised persons are the most susceptible community with higher mortality rates from SARS-CoV-2 infection. Proper techniques for early detection to control the pandemic is another challenge for medical health procedures and policy. According to the World Health Organization (WHO) dashboard report, till 30th November 2020, 63, 212, 447 positive cases have been reported from 216 countries and territories, from which 1,467,624 death cases have been confirmed. Recent data show that >80% of patients have no clinical signs or symptoms [1,3]. A cohort study by Yanez et al. revealed that the mortality from COVID-19 is higher in patients above 65 years, and the mortality is higher in the American compared to Chinese community [7]. Similarly, a study taken by Harrison et al. confirmed that comorbidities like chronic pulmonary disease and diabetes mellitus increase the mortality rate from COVID-19 [8]. Overall, the fatality rate for comorbidity and age groups varies from region to region and nation to nation. As a health emergency, the development of effective and safe treatments for SARS-CoV-2 is a challenge; it can be based on the current genomic information and previous knowledge on CoV. Some compounds have already been tested on other coronaviruses, and may constitute promising leads. Moreover, we focused on phytochemicals because they (or medicinal plants containing them) often have a documented history of safe use.
A PubMed search with the term “SARS-CoV-2" on 12th November 2020, resulted in 43,623 articles, 5909 of which were reviews (https://pubmed.ncbi.nlm.nih.gov/?term=SARS-CoV-2&size=200&filter=pubt.review). Further, the term “SARS-CoV-2 AND 3CLpro” yielded 116 articles, including 15 reviews (https://pubmed.ncbi.nlm.nih.gov/?term=SARS-CoV-2+AND+3CLpro&size=200). Combining the terms “SARS-CoV-2” and “phytochemicals” retrieved only 17 reviews (https://pubmed.ncbi.nlm.nih.gov/?term=SARS-CoV-2+AND+Phytochemicals&filter=pubt.review&size=200). So, this review focuses on identifying potential phytochemical(s) to combat SARS-CoV based on the published literature, in combination with a computational intelligence platform.
SARS-CoV-2: insights into various drug targets and their potential inhibitors
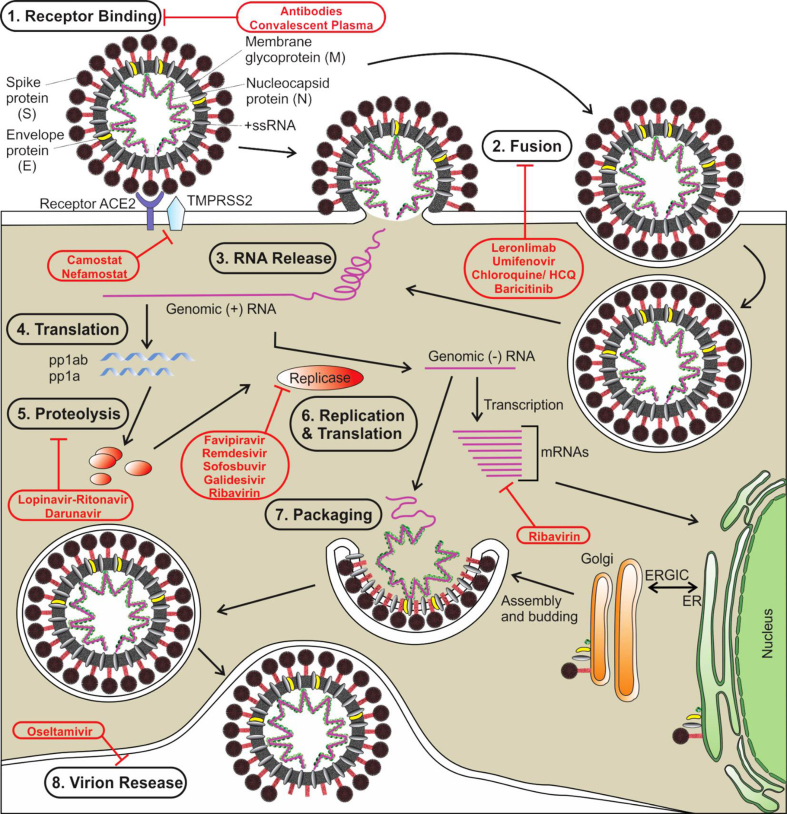
SARS-CoV-2 recognizes angiotensin-converting enzyme 2 (ACE2) on the host cell surface membrane binds to this via a viral envelope protein [9]. That spike proteins (S) accounts for viral entry by interacting with the host cell ACE2 receptor. Once the viral genome is introduced into the host cell, it is subsequently transcribed and translated, and the resulting polyprotein is processed by a protease (3CLpro) into virus-derived structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, as well as several non-structural proteins, used for building the viral particles [10]. During the fusion of spike proteins (S) with a specific receptor (like ACE2), a critical conformational change occurs in the S glycoprotein, which is an important drug target [11]. Several natural products have shown anti-SARS-CoV activity against this protein. For details see Fig. 1, adopted from Saha and co-worker [12].
Fig. 1.
The life cycle of SARS-CoV-2, and probable targets of different antiviral drugs being repurposed or investigated for COVID-19. Abbreviations used: ACE2: angiotensin-converting enzyme 2; HCQ: hydroxychloroquine; TMPRSS2: transmembrane protease serine 2; mRNA: messenger RNA; +ssRNA: positive-strand RNA; pp1ab: polyprotein 1 ab; ER: endoplasmic reticulum; ERGIC: ER-Golgi intermediate compartment (Adopted from Saha et al. [12]).
SARS-CoV chymotrypsin like protease (3CLpro)
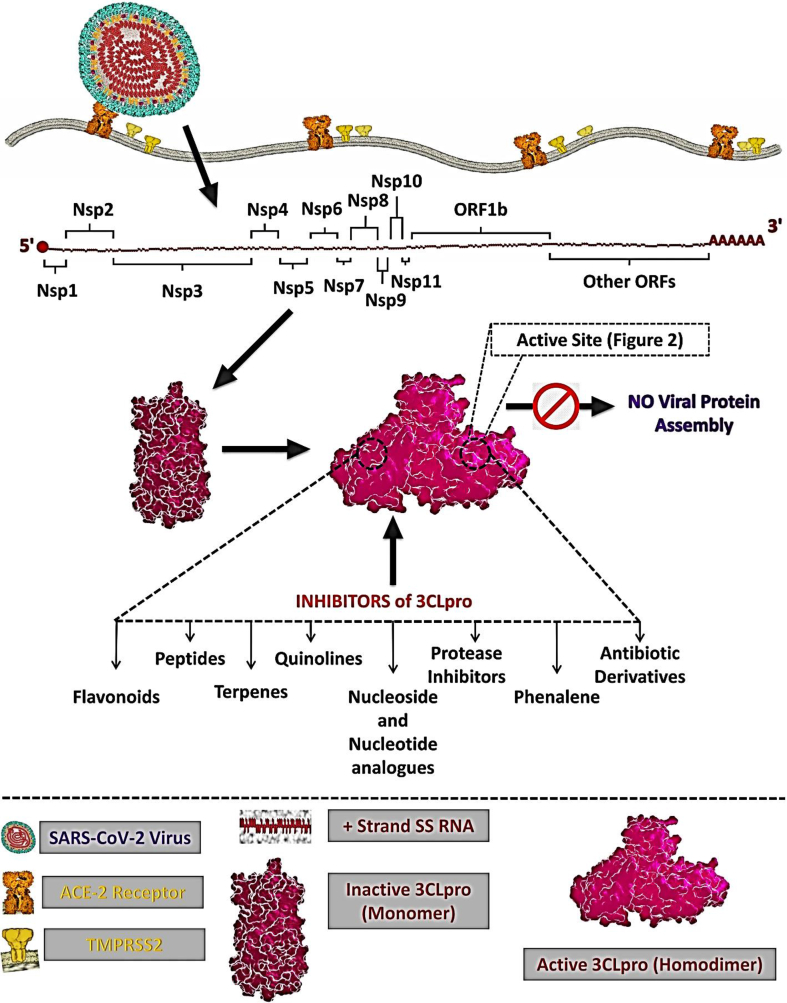
Predominantly, the SARS-CoV produces several functional proteins in host cells from two overlapping polyproteins (PP1a of 486 kDa and PP1ab of 790 kDa). Polyproteins (PPs) release the mature 16 nonstructural proteins (NSPs) through two types of cysteine proteases during viral replication [13,14]. The C-terminal end of these PPs is cleaved (NSP4-NSP16) by chymotrypsin-like cysteine protease or 3CLpro and the N-terminal end is cleaved (NSP1-NSP3) by papain-like protease or PLpro [[15], [16], [17]]. In brief, 3CLpro plays an important role to cleave 16 NSPs to produce most of the viral individual functional proteins, together with the RNA-dependent RNA polymerase (RdRp), helicase, single-stranded RNA-binding protein (ssRBP), exoribonuclease, endoribonuclease, ribose methyltransferase etc. Thus, the 3CLpro is critical for the entire RNA replication, transcription and further viral proliferation [15,16]. As an ideal target enzyme, the 3CLpro is a cysteine protease of 306 amino acids residues (the N-terminal part has two domain (I and II) within amino acid residues 1–184, while the C-terminal part (domain III) consists of amino acid residues 201–303). Domain III isconnected to domain II via a long loop (a globular cluster of five alpha-helices), which is vital for the active dimer-structure of the protease. The entire protein architecture and function are affected by individual classes of inhibitors (both synthetic and natural constituents) as shown in Fig. 2 [12]. Flavonoids, peptides, terpenes, quinolines, and nucleoside analogues have shown significant activity against SARS-CoV-2. Notably, humans do not have a 3CLpro homologue. Thus, 3CLpro is an attractive target for COVID-19 antiviral drug development [[17], [18], [19]].
Fig. 2.
Schematic representation for the localization and function of possible inhibitor classes against 3CLpro from SARS-CoV-2 (Adopted from Singh et al. [19]).
Several natural products have demonstrated anti-SARS-CoV activity via SARS-CoV 3CLpro. The docking approach on SARS-CoV 3CLpro is supporting for the identification of potential candidates for repurposing against SARS-CoV-2 [20].
Phytochemicals with anti-CoV activity
Previously, several Ayurvedic formulations and western natural regimens have been recorded against a range of viral diseases: chikungunya, dengue, human immunodeficiency virus, Ebola, etc., and those could also be an alternative option against SARS-CoV-2 [21,22]. The Ministry of Ayush (India) also started clinical trials for COVID-19 with four Ayurvedic herbs: ashwagandha (Withania somnifera (L.) Dunal), guduchi (Tinospora cordifolia (Willd.) Miers), yasthimadhu (Glycyrrhiza glabra L.), and pipli (Piper longum L.) [23]. The WHO encourages developing natural product-based medicines against any human ailments/diseases [24,25]. Approximately 70–80% of mainstream medicines originate from natural products [[7], [8], [9]]. Thus, it is worth searching for bioactive natural products with reported antiviral efficacy against CoV [[21], [22], [23], [24], [25]].
Primarily, we collected literature from scientific journals via a library and electronic search (using databases viz. PubMed, ScienceDirect, Wiley library, SCI hub, and Google Scholar). The specific terms ‘natural products for coronavirus treatment’ and ‘anti-coronavirus phytochemicals', were used for this literature search. From these, seventy-eight phytoconstituents with an exclusive anti-CoV activity (IC50 in μM), plant source and possible inhibition mechanisms were extracted [Table 1]. All these phytochemicals originate from thirty-one plant species, within twenty-two taxonomic families [Table 1]. Plant families with the highest number of species include Cupressaceae, Lamiaceae, Taxaceae (3 species each), followed by Apocyanaceae, Euphorbiaceae, Fabaceae (2 species each).
Table 1.
Phytochemicals (n = 78) with anti-CoV potency, plant source and mode of inhibition.
| Sl. No | Phytochemical (chemical class) | Taxonomic name (family) | Concentration (IC50/EC50) (μM) | Mode/target of inhibition | Tested against type of CoV | References |
|---|---|---|---|---|---|---|
| 1. | Abietane (T) | Lycopus europaeus L. (Lamiaceae) | 0.1–10 (1.39) | 3CLpro | SARS-CoV | [26] |
| 2. | Aloe emodin (AQ) | Isatis tinctoria L. (Brassicaceae) | 3.70–370.3 (8.3) | 3CLpro | SARS-CoV | [27] |
| 3. | Amentoflavone (P) |
Torreya nucifera (L.) Siebold & Zucc. (Taxaceae) |
1-1000 (8.3) | 3CLpro | SARS-CoV | [28] |
| 4. | Apigenin (P) |
Torreya nucifera (L.) Siebold & Zucc. (Taxaceae) |
1-1000 (280.8) | 3CLpro | SARS-CoV | [28] |
| 5. | Bavachinin (P) | Cullen corylifolium (L.) Medik. (Leguminosae) | 1-150 (38.4) | PLpro | SARS-CoV | [29] |
| 6. | Berbamine (A) | Berberis amurensis Rupr. (Berberidaceae) | 0.01–20 (1.48) | Restrict viral entry | HCoV-NL63 | [30] |
| 7. | β-sitosterol (S) | Isatis tinctoria L. (Brassicaceae) | 3.70–370.3 (12.10) | 3CLpro | SARS-CoV | [27] |
| 8. | Betulonic acid (T) | Juniperus formosana Hayata (Cupressaceae) | 8-80 (10) | 3CLpro | SARS-CoV | [26] |
| 9. | Broussochalcone A (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (88.1 for 3CLpro and 9.2 for PLpro) | 3CLpro and PLpro | SARS-CoV-3CLpro and PLPro enzyme assay | [31] |
| 10. | Broussochalcone B (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (57.8 for 3CLpro and 11.6 for PLpro) | 3CLpro and PLpro | [31] | |
| 11. | Broussoflavan A (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (92.4 for 3CLpro and 30.4 for PLpro) | 3CLpro and PLpro | SARS-CoV-3CLpro and PLPro enzyme assay | [31] |
| 12. | Catechin gallate (P) | Camellia sinensis (L.) Kuntze (Theaceae) | 0.22–2.26 (1.39) | Viral replication | SARS-CoV | [32] |
| 13. | Cepharanthine (A) |
Catharanthus roseus (L.) G.Don (Apocynaceae) |
0.82–16.5 (9.9–15.67) | 3CLpro | SARS-CoV | [33] |
| 14. | Cinnamtannin B (P) | Cinnamomum verum J.Presl (Lauraceae) | 0-500 (32.9) | Pseudovirus infection | SARS-CoV | [34] |
| 15. | Chrysin (P) | Scutellaria baicalensis Georgi (Lamiaceae) | 0-400 (200) | Restrict viral entry | SARS-CoV | [[21], [35]] |
| 16. | Corylifol A (P) | Cullen corylifolium (L.) Medik. (Fabaceae) | 1-150 (32.3) | PLpro | SARS-CoV | [29] |
| 17. | Curcumin (T) | Curcuma longa L. (Zingiberaceae) | 8-80 (40) | 3CLpro | SARS-CoV | [26] |
| 18. | Diplacone (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (10.4) | PLpro | SARS-CoV | [13] |
| 19. | Emetine (T) |
Carapichea ipecacuanha (Brot.) L. Andersson (Rubiaceae) |
0.1–5 (0.30, 1.43, 0.34, 0.12) | RNA, DNA and protein synthesis | HCoV-OC43, HCoV-NL63, MERS-CoV. | [30] |
| 20. | Emodin (AQ) | Rheum officinale Baill. (Polygonaceae) | 0.1–400 (200) | Restrict viral entry | SARS-CoV | [35] |
| 21. | Fangchinoline (A) | Panax ginseng C.A.Mey. (Araliaceae) | 2-20 (1.01 ± 0.07) | 3CLpro | HCoV-OC43 | [14] |
| 22. | Ferruginol (T) |
Chamaecyparis obtuse var. formosana Hayata (Cupressaceae) |
0.01–10 (1.39) | CoV-replication | SARS-CoV | [26] |
| 23. | Gallocatechin gallate (P) | Camellia sinensis (L.) Kuntze (Theaceae) | 0.002–2.183 (1.39) | Viral replication | SARS-CoV | [32] |
| 24. | Glycyrrhizic acid (T) | Glycyrrhiza Radix (Glycyrrhiza uralensis, G. glabra and G. inflata) (Fabaceae) | 0.1–1000 (365) | Viral replication | SARS-CoV | [36] |
| 25. | Hesperetin (P) | Isatis tinctoria L. (Cruciferae) | 3.31–331.1 (36.50) | 3CLpro | SARS-CoV | [27] |
| 26. | Hinokinin (L) | Phyllanthus amarus Schumach. & Thonn. (Phyllanthaceae) | 8-80 (>100) | 3CLpro | SARS-CoV | [26] |
| 27. | Homoharringtonine (A) | Cephalotaxus fortunei Hook. (Taxaceae) | 0.01–0.07 (0.012) | 3CLpro | Murine CoV | [37] |
| 28. | 4-Hydroxy-isolonchocarpin (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (35.4) | PLpro | SARS-CoV | [31] |
| 29. | Indigo (OC) | Isatis tinctoria L. (Brassicaceae) | 0.1–1000 (752) | 3CLpro | SARS-CoV | [27] |
| 30. | Isobavachalcone (P) | Cullen corylifolium (L.) Medik. (Fabaceae) | 1-150 (18.3) | PLpro | SARS-CoV | [29] |
| 31. | Juglanin (P) | Taxus caespitosa Nakai (Taxaceae) | 10-40 (2.3) | Blocks the 3a channel | SARS-CoV | [38] |
| 32. | Kazinol A (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (66.20) | PLpro | SARS-CoV PLpro enzyme assay | [31] |
| 33. | Kazinol B (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (31.40) | PLpro | SARS-CoV PLpro enzyme assay | [31] |
| 34. | Kazinol F (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (27.8) | PLpro | SARS-CoV PLpro enzyme assay | [31] |
| 35. | Kazinol J (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (15.20) | PLpro | SARS-CoV PLpro enzyme assay | [31] |
| 36. | Luteolin (P) |
Torreya nucifera (L.) Siebold & Zucc. (Taxaceae) |
1-1000 (20.2) | 3CLpro | SARS-CoV | [25] |
| 37. | Lycorine (A) | Lycoris aurea (L’Hér.) Herb. (Amaryllidaceae) | 0.01–5 (0.15, 0.47, 1.63, 0.31) | CoV-replication | HCoV-OC43, HCoV-NL63, MERS-CoV | [30] |
| 38. | Mimulone (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.01–100 (14.4 ± 0.27) | PLpro | SARS-CoV | [13] |
| 39. | Myricetin (P) | Isatis tinctoria L. (Brassicaceae) | 0.01–10 (2.74) | 3CLpro | SARS-CoV | [39] |
| 40. | Neobavaisoflavone (P) | Cullen corylifolium (L.) Medik. (Fabaceae) | 1-150 (18.3 ± 1.1) | PLpro | SARS-CoV | [29] |
| 41. | Papyriflavonol A (P) | Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) | 0.1–200 (3.7) | PLpro | SARS-CoV PLpro enzyme assay | [31] |
| 42. | Procyanidin A2 (P) | Cinnamomum verum J.Presl (Lauraceae) | 0.1–500 (29.9 ± 3.3) | Pseudovirus infection | SARS-CoV | [34] |
| 43. | Procyanidin B1 (P) | Cinnamomum verum J.Presl (Lauraceae) | 0.1–500 (41.3) | Pseudovirus infection | SARS-CoV | [34] |
| 44. | Psoralidin (P) | Cullen corylifolium (L.) Medik. (Fabaceae) | 1-150 (4.2) | PLpro | SARS-CoV | [29] |
| 45. | Quercetin (P) | Torreya nucifera (L.) Siebold & Zucc. (Taxaceae) | 1-200 (8.6) | PLpro | SARS-CoV-PLpro enzyme assay | [31] |
| 46. | Rosmariquinone (T) | Salvia miltiorrhiza Bunge (Lamiaceae) | 1-1000 (88.0) | CoV-replication | SARS-CoV | [40] |
| 47. | Rhein (G) | Houttuynia cordata Thunb (Saururaceae) | 0.1–400 (200) | Restrict viral entry | SARS-CoV | [35] |
| 48. | Saikosaponin A (T) | Bupleurum chinense DC (Apiaceae) | 5-25 (8.6) | 3CLpro | HCoV-22E9 | [41] |
| 49. | Saikosaponin B2 (T) | Bupleurum chinense DC (Apiaceae) | 5-25 (1.7) | Restrict viral entry | HCoV-22E9 | [41] |
| 50. | Saikosaponin C (T) | Bupleurum chinense DC (Apiaceae) | 5-25 (19.9) | CoV-replication | HCoV-22E9 | [41] |
| 51. | Saikosaponin D (T) | Bupleurum chinense DC (Apiaceae) | 5-25 (13.2) | CoV-replication | HCoV-22E9 | [41] |
| 52. | Savinin (L) | Chamaecyparis obtusa var. formosana (Hayata) Hayata (Cupressaceae) | 1-10 (9.1) | 3CLpro | SARS-CoV | [26] |
| 53. | Scutellarein (P) | Scutellaria baicalensis Georgi (Lamiaceae) | 0.01–10 (0.08) | 3CLpro | SARS-CoV | [39] |
| 54. | Silvestrol (OC) | Aglaia foveolata Pannell (Meliaceae) | 0.6–20 (40) | Cap-mRNA translation | HCoV-229 E | [42] |
| 55. | Sinigrin (G) | Isatis tinctoria L. (Brassicaceae) | 2.52–25.8 (217) | 3CLpro | SARS-CoV | [27] |
| 56. | Tanshinone I (T) | Salvia miltiorrhiza Bunge (Labiatae) | 1-1000 (1.6) | PLpro | SARS-CoV PLpro enzyme assay | [40] |
| 57. | Tetrandrine (A) | Stephania tetrandra (Menispermaceae) | 2-20 (0.33) | CoV-replication | HCoV-OC43-MRC-5 cells | [14] |
| 58. | Tetra-O-galloyl-β-d-glucose (P) | Phyllanthus emblica L. (Euphorbiaceae) | 0.1-104 (2.86) | Restrict viral entry | SARS-CoV | [43] |
| 59. | Theaflavin-3,3′-digallate (P) | Camellia sinensis (L.) Kuntze (Theaceae) | 4-20 (9.5) | 3CLpro | SARS-CoV | [44] |
| 60. | Tomentin A (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (6.2) | 3CLpro | SARS-CoV | [13] |
| 61. | Tomentin B (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (6.1) | 3CLpro | SARS-CoV | [44] |
| 62. | Tomentin C (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (11.6) | 3CLpro | SARS-CoV | [44] |
| 63. | Tomentin D (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (12.5) | 3CLpro | SARS-CoV | [44] |
| 64. | Tomentin E (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (5.0) | 3CLpro | SARS-CoV | [44] |
| 65. | Tylophorine (A) | Tylophora indica (Burm. f.) Merr. (Apocynaceae) | (0.008–1.47) (0.018) |
3CLpro | SARS-CoV | [45] |
| 66. | 3β-Friedelanol (T) | Euphorbia neriifolia L. (Euphorbiaceae) | 11.68 (132.4%) | 3CLpro | HCoV | [46] |
| 67. | 3 β,12-diacetoxy abieta-6,8,11,13-tetraene (T) | Juniperus formosana Hayata (Cupressaceae) | 365 (193) | 3CLpro | SARS-CoV | [26] |
| 68. | 3-Isotheaflavin-3-gallate (P) | Camellia sinensis (L.) Kuntze (Theaceae) | 4-20 (7) | 3CLpro | SARS-CoV | [44] |
| 69. | 3′-(3-methyl but-2-enyl)-3′,47-tri-hydroxy flavane (P) |
Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae) |
0-200 (35.8) | PLpro | SARS-CoV PLpro enzyme assay | [31] |
| 70. | 3′-O-methyl diplacol (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (9.5) | PLpro | SARS-CoV | [13] |
| 71. | 3′-O-methyl diplacone (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (13.2) | PLpro | SARS-CoV | [13] |
| 72. | 4′-O-methyl diplacol (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (9.2) | PLpro | SARS-CoV | [13] |
| 73. | 4′-O-methyl diplacone (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (12.7) | PLpro | SARS-CoV | [13] |
| 74. | 4′-O-methyl-bavachalcone (P) | Cullen corylifolium (L.) Medik. (Paulowniaceae) | 1-150 (10.1) | PLpro | SARS-CoV | [29] |
| 75. | 6-geranyl-4′,5,7-tri-hydroxy-3′,5′-di-methoxy flavanone (P) | Paulownia tomentosa Steud. (Paulowniaceae) | 0.1–100 (13.9) | PLpro | SARS-CoV | [13] |
| 76. | 7-β-Hydroxydeoxy cryptojaponol (T) |
Cryptomeria japonica (Thunb. ex L.f.) D.Don (Cupressaceae) |
365 (111) | 3CLpro | SARS-CoV | [34] |
| 77. | 7-Methoxy-cryptopleurine (A) | Tylophora indica (Burm. f.) Merr. (Apocynaceae) | (0.1–100) (20) |
3CLpro | SARS-CoV | [45] |
| 78. | 8β-hydroxy abieta-9 (11),13-dien-12-one (T) | Chamaecyparis obtusa var. formosana (Hayata) Hayata (Cupressaceae) | 0.1–10 (1.47) | CoV-replication | SARS-CoV | [46] |
Abbreviations: MRC-5: human embryo lung fibroblast; STCI-CoV: Swine testicular cells infected with CoV; 3CLpro: cysteine-like protease; PLpro: papain-like protease; PED-CoV: porcine epidemic diarrhea coronavirus; A: alkaloid; AQ: anthraquinone; G: glucoside/glycoside; L: lignin; P: polyphenol; Q: quinolone; T: terpene/terpenoid; S: sterol; MERS-CoV: Middle East respiratory syndrome coronavirus; HCoV-OC43: human coronavirus OC43; HCoV-NL63: human coronavirus NL63.
Documented most phytochemicals in Table 1 are poly-phenolic or flavonoid classes, but based on IC50/EC50 values, alkaloids and terpenoids are comparatively potential against different CoV strains. Briefly, the actib = ve most alkaloids against CoV are (presented in ascending order of IC50/EC50 values in round brackets): homoharringtonine (0.012 μM), lycorine (0.15–1.63 μM), tetrandrine (0.33), fangchinoline (1.01 μM), berbamine (1.48 μM), the potential flavonoids are scutellarein (0.08 μM), catechin gallate and gallocatechin gallate (1.39 μM), juglanin (2.3 μM), myricetin (2.7 μM), tetra-O-galloyl-β-d-glucose (2.86 μM), and potential terpenoids are emetine (0.12–1.43 μM), abietane and ferruginol (1.39 μM), tanshinone I (I.6 μM), saikosaponin B2 (1.7 μM), were recorded in several in vitro assays [Table 1]. In general, according to existing information, alkaloids are comparatively toxic than terpenoids and flavonoids. Thus, the primary aim of this review is to highlighted and structural analysis of such potential anti-CoV phytochemical(s) to provide further guidance for developing new therapeutic agents against SARS-CoV-2 through a computational intelligence platform.
Computational-intelligence analysis of select phytochemicals with anti-CoV activity
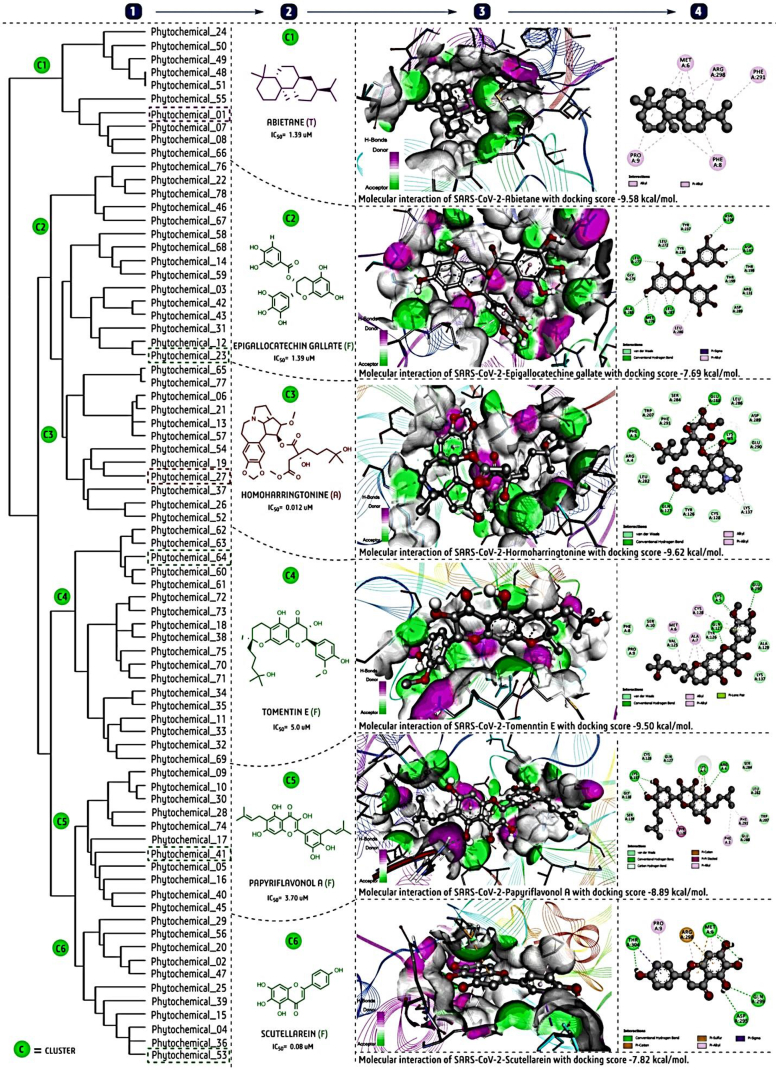
After that, taking all selected phytochemicals, we performed hierarchical clustering analysis using a ChemMine tool (https://chemminetools.ucr.edu/), to find out the structural characteristics of each cluster. Notably, each chemical simplified molecular-input line-entry system (SMILE) notation was retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and ChemSpider (http://www.chemspider.com/), and used for this clustering analysis. Furthermore, we carried out molecular docking against the SARS-CoV-2-3CLpro with six potent phytochemical(s) each selected from one of the six clusters [Fig. 3], and two repurposed drugs: lopinavir and ritonavir as reference [Fig. 4]. The three-dimensional crystallographic protein structure of 3CLpro was retrieved from the protein data bank (PDB ID: 6Y2E, https://www.rcsb.org/). The AutoDock 4.1 software for docking and the BIOVAI-Discovery Studio Visualizer-2019 client version (BIOVIA-Dassault Systèmes, San Diego, CA) software for protein-ligand interaction were used [47]. The default sets of docking parameters of AutoDock tools, like Kollman charges with polar hydrogen bonds were used for the target, while Geister partial charges for ligands were added for a blind docking study. A full grid size 126 × 126 × 126 and 5.5 Å spacing setting was used during each ligand-docking study. The lowest binding energy (kcal/mol) or interactive pose was selected out of ten generated poses for each ligand for further interaction analysis [47].
Fig. 3.
Cluster analysis and molecular docking study of phytochemicals with reported anti-CoV activity for potential use against SARS-CoV-2; (1). Cluster analysis of 78 phytochemicals using the ChemMine; tool, (2). Selection of active phytochemicals representing each cluster based on the lowest reported IC50 (μM); (3). Three-dimensional molecular interaction of SARS-CoV-2-3CLpro (Protein Data Bank ID: 6Y2E) with the most potent phytochemicals from each cluster by BIOVIA-Discovery Studio Visualizer 2.5 software: the AutoDock 4.1 software was used for molecular docking; (4). Two-dimensional interaction visualization by the same BIOVIA-Discovery Studio Visualizer 2.5 software with more clarification on the types of bond formation and interacting amino acids. The light pink dotted line represents the alkyl/pi–alkyl interactions, green the H–bond interaction, light-green for van der Waals and carbon-hydrogen interactions and brick-red color indicates pi-carbon or pi-sulfur, parrot-green shown a pi–loan pair, violet is used for pi-sigma, and dark pink for pi–pi stacked interactions in protein-ligand interaction for each docking complex.
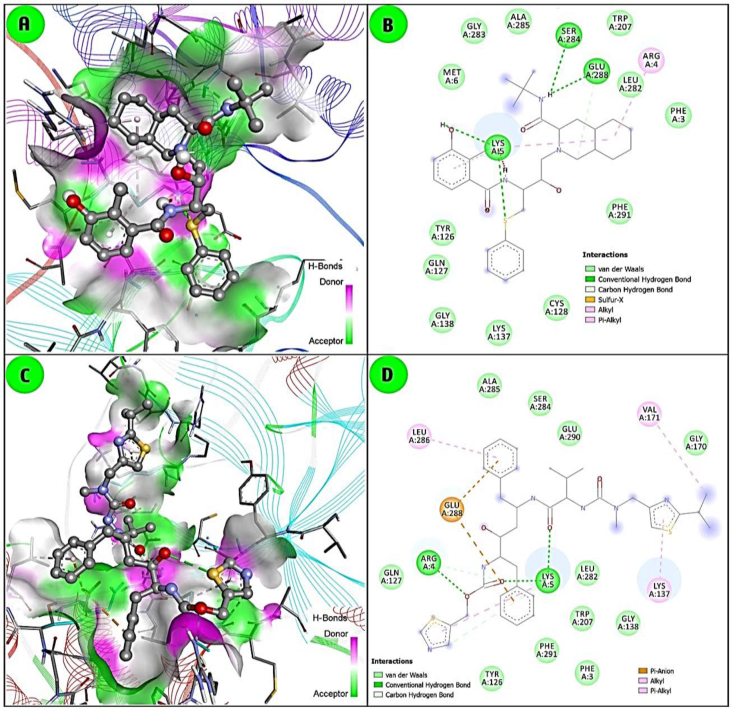
Fig. 4.
Molecular docking interaction of 3CLpro with two repurposed antiviral drugs, lopinavir (A and B) and ritonavir (C and D). Similarly, AutoDock 4.1 and BIOVIA-Discovery Studio Visualizer softwares were used.
The docking study was carried out with six phytochemicals, abietane, epigallocatechin gallate/EGCG, homoharringtonine, tomentine E, papyriflavonol A and scutellarein), representing each cluster, and that indicated potential activity based on binding affinity or docking score (kcal/mol) with SARS-CoV-2-3CLpro [Fig. 3]. The alkaloid homoharringtonine form cluster-3, had the best docking score (−9.62 kcal/mol), followed by the terpenoid abietane (−9.58) from cluster-1, the flavonoid tomentine E (−9.50) form cluster 4, papyriflavonol A (−8.89) from cluster-5, a scutellarein (−7.82) form cluster-6 and EGCG (−7.69) from cluster 2 [Fig. 3]. By comparison, the repurposed drug lopinavir (−9.00) and ritonavir (−8.75) are shown in Fig. 4. Overall, three to four H-bond interactions are found in both phytochemicals and antiviral drugs. In general, H-bond interactions are stronger than other interactions like van der Waals, carbon–hydrogen bond, pi-sigma, pi-alkyl, pi–lone pair etc. Indeed, both antiviral drugs and the flavonoid phytochemicals are more strongly bound by H-bonds than terpenoids and alkaloids [Fig. 4]. The interactions depend on the chemical structure, as flavonoids bear more side-chain hydroxyl (-OH) functional groups, which readily form H-bonds with the target enzyme. Based on the docking analysis, all modelled phytochemicals are predicted to have comparatively potent inhibition activity.
Within each cluster, a more detailed analysis was pursued.
Cluster 1- There are two majors structural subclusters: the abietane-like molecules (C1, C7, C8, C66), and the saikosaponins (with the related glycyrrhizic acid). The former are probably 3CLpro inhibitors, the latter interfere with viral entry. This suggests that the (so far unknown) mechanism of 3-β-friedelanol is 3CLpro inhibition. It may also permit explaining the structure–activity relationship observed [46]. The most potent abietane terpenoid showed the highest ligand efficiency, with pi-alkyl/alkyl interactions with 3CLpro. Both in experimental and in silico analysis, the abietane has shown potential activity; however, the exact relationship of structure with biological activity is still poorly understood.
Cluster-2: This cluster contain two major chemical classes; (i) terpenoids (C76, C22, C78, C46, C67) and (ii) (poly)phenolics (C58, C68, C14, C59, C3, C42, C43, C31, C12, C23). In this cluster, most compounds appear to act on 3CLpro. In the terpenoid subcluster, C76 and C67 are demonstrated 3CLpro inhibitors, whereas the other three have shown inhibition in a whole cell viral replication assay, where the molecular target is unclear. It is therefore tempting to assume that C22, C78 and C46 would as well exert their antiviral effect via inhibition of 3CLpro. The stronger potency of C22 and C78 compared to C76 and C46 may be due to a second substituent on the phenyl ring for the latter, which may provide steric hindrance. By the same reasoning for the second subcluster, all compounds active in cellular antiviral assays may well exert their activity via 3CLpro inhibition. An exception may be C31, which blocks the 3a channel. It is the only flavonone in the cluster, and this moiety seems important for channel-blocking activity [38], although similar flavonones also inhibit 3CLpro [15].
Cluster-3: This cluster contains of a range of alkaloids; whose reported mechanism of action is 3CLpro (also called Mainpro) inhibition. Among the six alkaloids of the first cluster, C65 is 100-fold more potent than C77, probably because the larger 6-membered ring of C77 is bulkier than the corresponding 5-membered ring of C56. The four bis-benzylisoquinoline alkaloids (C6, C21, C13 and C58) comprise cepharanthine, a compound with broad-spectrum antiviral activity, that may also inhibit 3CLpro, as its molecular target is unclear. However, apart from protease inhibition, other molecular targets have also been proposed [16].
Cluster-4: This cluster is the largest, and consists of flavonoids that seem to inhibit either via 3CLpro or PLpro. Modelling of flavonoids with 3CLpro has been reported [15]. The five tomentins (C60–C64) inhibit via 3CLpro; they are very close analogues, and their IC50 values differ at most two-fold, so that few conclusions can be drawn from this regarding structure–activity relationship. The other two subclusters fall into two chemical classes: diprenylated flavanones (C72, C73, C18, C38, C75, C70, C71) and a mixed group containing diphenylpropane derivatives (C34, C35) and flavanes (C11, C33, C32, C69. The first group inhibits mostly PLpro, the latter only 3CLpro tested both enzymes [31], and several compounds inhibit both, although the potency does not appear to correlate well [13]. Moreover, “the isolated compounds exerted significant SARS-CoV PLpro inhibitory activity through noncompetitive inhibition.” This implies that they do not interact directly with the ligand binding site.
Cluster-5: With the exception of curcumin (diarylheptanoid), this cluster falls into two chemical classes: flavanones (C41, C5, C16, C40, C45) and chalcones (C9, C10, C30, C74, C28). Both groups appear to contain inhibitors of PLpro and of 3CLpro. As noted for the previous cluster, Park et al. examined similar compounds, some of which inhibited both enzymes [32].
Cluster-6: This cluster consists of a range of flavonoids (C25, C39, C4, C36, C53), whose reported mechanisms of action are 3CLpro inhibition, except C15 (chrysin acts by restricting viral entry). The anthraquinones likely inhibit via interference with the binding of the spike protein with angiotensin-converting enzymes-2 (ACE2) [35]. The apparent exception (Aloe emodin) is, however, reported by Lin et al., as an inhibitor of the 3CLpro [27]. Conversely, the flavonoid chrysin would restrict viral entry. Two compounds (tanshinone I and indigo) do not fit into either of the aforementioned chemical classes and are outliers in the clusters.
Summary and concluding remarks
From a set of 78 phytochemicals with reported CoV activity, we could discern 6 clusters. Docking studies with SARS-CoV-2-3CLpro suggested that the most potent compound from each cluster could bind to the protease, and some did so with similar calculated free energy scores as lopinavir and ritonavir, which show in vitro activity against SARS-CoV-1 and are being repurposed for COVID-19 treatment. In several clusters, compounds with reported in vitro activity against SARS-CoV-2 in cellular assays were found, suggesting that they act via 3CLpro. Also for some other compounds the presumptive target may have to be reconsidered as 3CLpro. The docking studies provided explanations in several clusters for differences in potency, and can serve as the basis for further target-based lead optimsiation.
In the absence of effective treatments, several non-targeting alternative therapies such as natural killer (NK) cells, nitric oxide (NO), along with complementary and alternative medicine (CAM), including traditional Chinese medicine (TCM), Indian Ayurveda and Western herbal medicine (WHM) are used in different regions around the world to control the SARS-CoV-2 pandemic [[21], [22], [23], [24], [25],48,49]. Overall, 75–80% of aboriginal communities still depend on plant-based treatment regimens. Several herbal-based compounds and their derivatives have proven effective as antivirals against several viruses. Only limited therapeutic opportunities are presently available against SARS-CoV-2. As there is an urgent need for innovative drug development strategies, phytochemicals can potentially combat several complicated ailments, including CoV [[21], [22], [23], [24], [25]].
Plant secondary metabolites are preferable to other natural products for multi-target activities, lower–toxicity profiles and cost-effectiveness. For example, several TCM regimens showed potential activity in preliminary screening against SARS-CoV-2, and some are even at the clinical trial stage. An empirical study conducted by Luo et al. in Wuhan, Hubei Province, China, has evidenced faster recovery among patients (n = 54) treated with TCM. E.g. Qingfei Paidu decoction was used for the treatment of SARS-CoV-2 (n = 214) at 4 provincial hospitals in China, and a significant improvement (60%) was observed after three days of treatment, while 30% of patients had stable symptoms without exacerbation [[48], [49], [50]]. Zhang et al. executed a rational screen to find TCM that are frequently used in treating viral respiratory infections, as well as compounds that might directly inhibit SARS-CoV-2, an ongoing novel coronavirus that causes pneumonia, and found that 13 reported MERS-CoV/SARS–CoV compounds from TCM also have anti-SARS-CoV-2 activity [49].
Although since the COVID-19 outbreak more than 5000 review papers have been reported in PubMed, the importance of natural products against SARS-CoV-2 has not been explored much. But there is literature on various nature-derived compounds with potent anti-SARS-CoV and anti-MERS-CoV activity. It is clear that there is a close sequence similarity between SARS CoV-2 and SARS-CoV or MERS-CoV [50]. Using computational approaches to repurpose these anti-SARS-CoV or anti-MERS-CoV natural products could lead to candidates for developing a safe and cost-effective COVID-19 drug.
No doubt the potential of phytochemicals is large. At the same time, the challenges are considerable. More research will be necessary to study the mechanism of action underlying protection against the cytopathic effect of CoV, and crystallography to confirm the interaction of the phytocompounds with viral target proteins. In addition, advanced computational program and machine learning may also play a vital role in finding the most effective potential anti- COVID candidates [[51], [52], [53]].
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors largely supported themselves.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.WHO Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [accessed 25 September 2020]
- 2.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir Res. 2020;21:224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. J Am Med Assoc. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 6.Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanez N.D., Weiss N.S., Romand J.A., Treggiari M.M. COVID-19 mortality risk for older men and women. BMC Publ Health. 2020;20:1742. doi: 10.1186/s12889-020-09826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison S.L., Fazio-Eynullayeva E., Lane D.A., Underhill P., Lip G.Y.H. Comorbidities associated with mortality in 31,461 adults with COVID-19 in the United States: a federated electronic medical record analysis. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24:422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akaji K., Konno H. Design and evaluation of anti-SARS-coronavirus agents based on molecular interactions with the viral protease. Molecules. 2020;25:3920. doi: 10.3390/molecules25173920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha R.P., Sharma A.R., Singh M.K., Samanta S., Bhakta S., Mandal S. Repurposing drugs, ongoing vaccine, and new therapeutic development initiatives against COVID-19. Front Pharmacol. 2020;11:1258. doi: 10.3389/fphar.2020.01258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg Med Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D.E., Min J.S., Jang M.S., Lee J.Y., Shin Y.S., Park C.M. Natural Bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus oc43 infection of MRC-5 human lung cells. Biomolecules. 2019;9:696. doi: 10.3390/biom9110696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzym Inhib Med Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh E., Khan R.J., Jha R.K., Amera G.M., Jain M., Singh R.P. A comprehensive review on promising anti-viral therapeutic candidates identified against main protease from SARS-CoV-2 through various computational methods. J Genet Eng Biotechnol. 2020;18:69. doi: 10.1186/s43141-020-00085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koulgi S., Jani V., Uppuladinne M., Sonavane U., Nath A.K., Darbari H. Drug repurposing studies targeting SARS-CoV-2: an ensemble docking approach on drug target 3C-like protease (3CLpro) J Biomol Struct Dyn. 2020:1–21. doi: 10.1080/07391102.2020.1792344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhuiyan F.R., Howlader S., Raihan T., Hasan M. Plants Metabolites: possibility of natural therapeutics against the COVID-19 pandemic. Front Med. 2020;7:444. doi: 10.3389/fmed.2020.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boukhatem M.N., Setzer W.N. Aromatic herbs, medicinal plant-derived essential oils, and phytochemical extracts as potential therapies for Coronaviruses: future perspectives. Plants. 2020;9:800. doi: 10.3390/plants9060800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ministry of AYUSH First Report and Recommendations-Interdisciplinary Committee for integration of Ayurveda and Yoga Interventions in the National Clinical Management Protocol COVID 19. https://main.ayush.gov.in/event/first-report-and-recommendations-interdisciplinary-committee-integration-ayurveda-and-yoga [accessed 2 October 2020]
- 24.Yuan H., Ma Q., Ye L., Piao G. The traditional medicine and modern medicine from natural products. Molecules. 2016;21:559. doi: 10.3390/molecules21050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization WHO global report on traditional and complementary medicine 2019. https://apps.who.int/iris/handle/10665/312342 [accessed 2 October 2020]
- 26.Wen C.C., Kuo Y.H., Jan J.T., Liang P.H., Wang S.Y., Liu H.G. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem. 2007;50:4087–4095. doi: 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- 27.Lin C.W., Tsai F.J., Tsai C.H., Lai C.C., Wan L., Ho T.Y. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir Res. 2005;68:36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL (pro) inhibition. Bioorg Med Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D.W., Seo K.H., Curtis-Long M.J., Oh K.Y., Oh J.W., Cho J.K. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J Enzym Inhib Med Chem. 2014;29:59–63. doi: 10.3109/14756366.2012.753591. [DOI] [PubMed] [Google Scholar]
- 30.Shen L., Niu J., Wang C., Huang B., Wang W., Zhu N. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J Virol. 2019;93 doi: 10.1128/JVI.00023-19. e00023-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J.Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J Enzym Inhib Med Chem. 2017;32:504–515. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roh C. A facile inhibitor screening of SARS coronavirus N-protein using nanoparticle-based RNA oligonucleotide. Int J Nanomed. 2012;7:2173. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C.H., Wang Y.F., Liu X.J., Lu J.H., Qian C.W., Wan Z.Y. Antiviral activity of cepharanthine against severe acute respiratory syndrome coronavirus in vitro. Chin Med J (Engl) 2005;118:493–496. [PubMed] [Google Scholar]
- 34.Zhuang M., Jiang H., Suzuki Y., Li X., Xiao P., Tanaka T. Procyanidins and butanol extract of Cinnamomi cortex inhibit SARS-CoV infection. Antivir Res. 2009;82:73–81. doi: 10.1016/j.antiviral.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho T.Y., Wu S.L., Chen J.C., Li C.C., Hsiang C.Y. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antivir Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoever G., Baltina L., Michaelis M., Kondratenko R., Baltina L., Tolstikov G.A. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J Med Chem. 2005;48:1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- 37.Cao J., Forrest J.C., Zhang X. A screen of the NIH clinical collection small molecule library identifies potential anti-coronavirus drugs. Antivir Res. 2015;114:1–10. doi: 10.1016/j.antiviral.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz S., Sauter D., Wang K., Zhang R., Sun B., Karioti A. Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med. 2014;80:177–182. doi: 10.1055/s-0033-1360277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu M.S., Lee J., Lee J.M., Kim Y., Chin Y.W., Jee J.G. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg Med Chem Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J.Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg Med Chem. 2012;20:5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng P.W., Ng L.T., Chiang L.C., Lin C.C. Antiviral effects of saikosaponins on human coronavirus 229E in vitro. Clin Exp Pharmacol Physiol. 2006;33:612–616. doi: 10.1111/j.1440-1681.2006.04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller C., Schulte F.W., LangeGrünweller K., Obermann W., Madhugiri R., Pleschka S. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antivir Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C.N., Lin C.P., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3'-digallate (TF3) Evid Based Complement Alternat Med. 2005;2:209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang C.W., Lee Y.Z., Kang I.J., Barnard D.L., Jan J.T., Lin D. Identification of phenanthroindolizines and phenanthroquinolizidines as novel potent anti-coronaviral agents for porcine enteropathogenic coronavirus transmissible gastroenteritis virus and human severe acute respiratory syndrome coronavirus. Antivir Res. 2010;88:160–168. doi: 10.1016/j.antiviral.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang F.R., Yen C.T., Ei-Shazly M., Lin W.H., Yen M.H., Lin K.H. Anti-human coronavirus (anti-HCoV) triterpenoids from the leaves of Euphorbia neriifolia. Nat Prod Commun. 2012;7:1415–1457. [PubMed] [Google Scholar]
- 47.Swain S.S., Paidesetty S.K., Dehury B., Das M., Vedithi S.C., Padhy R.N. Computer-aided synthesis of dapsone-phytochemical conjugates against dapsone-resistant Mycobacterium leprae. Sci Rep. 2020;10:6839. doi: 10.1038/s41598-020-63913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo E., Zhang D., Luo H., Liu B., Zhao K., Zhao Y. Treatment efficacy analysis of traditional Chinese medicine for novel coronavirus pneumonia (COVID-19): an empirical study from Wuhan, Hubei Province, China. Chin Med. 2020;15:34. doi: 10.1186/s13020-020-00317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K. Is traditional Chinese medicine useful in the treatment of COVID-19? Am J Emerg Med. 2020;38:2238. doi: 10.1016/j.ajem.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat Biotechnol. 2020;38:379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 52.Zhang D.H., Wu K.L., Zhang X., Deng S.Q., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med. 2020;18:152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dey L., Chakraborty S., Mukhopadhyay A. Machine learning techniques for sequence-based prediction of viral-host interactions between SARS-CoV-2 and human proteins. Biomed J. 2020;43:438–450. doi: 10.1016/j.bj.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]