Endoplasmic reticulum (ER) stress signaling has long been associated with various pathological states in particular with the development of diseases with an underlying inflammation, such as diabetes, liver or cardiovascular dysfunctions, and cancer. ER stress signaling is mediated by three stress sensors. The most evolutionarily conserved one, the inositol‐requiring enzyme 1 alpha (IRE1), transduces most of the signals through an endoribonuclease (RNase) activity toward RNAs including mRNAs and microRNAs (miRNAs). By exploring phosphoinositide signaling in human macrophages, Hamid and colleagues discovered a novel function of IRE1 RNase that through the cleavage of pre‐miR‐2317 generates a mature miR‐2317 independently of the canonical Dicer endonuclease to yield specific biological outcomes (Hamid et al, 2020).
Subject Categories: Metabolism, RNA Biology
IRE1’s endoribonuclease activity is known to promote the unconventional splicing of XBP1 and the degradation of miRNAs. A study in this issue of EMBO Reports shows that IRE1 directly promotes the maturation of miR‐2137 to regulate lipid metabolism in macrophages.
ER stress signaling is involved in cell and organ homeostasis but also in their pathophysiology. Under specific physiological and pathological contexts, cells are exposed to intrinsic or extrinsic stresses resulting in ER homeostasis imbalance and subsequently in the activation of the ER stress sensors. These proteins trigger an adaptive pathway, the unfolded protein response (UPR), aiming to restore ER proteostasis. In case of acute/prolonged stress that cannot be resolved by the cell, the UPR leads to cell death (Almanza et al, 2019). The UPR is mediated by three sensors, namely inositol‐requiring enzyme 1 alpha (referred to as IRE1 hereafter), PKR‐like endoplasmic reticulum kinase (PERK), and activating transcription factor 6 (ATF6). IRE1 is the most conserved of these three sensors and contributes to the control of ER homeostasis (Almanza et al, 2019). One of IRE1 unique features lies in its RNase activity that contributes to (i) the nonconventional splicing of the X box‐binding protein‐1 (XBP1) mRNA leading to the production of the potent transcription factor XBP1s, and (ii) the degradation of cytoplasmic RNAs in a process called regulated‐IRE1 dependent decay (RIDD) of RNA (Hollien & Weissman, 2006; Maurel et al, 2014).
IRE1‐mediated control of ER homeostasis is exerted at many levels including protein, calcium, and lipid homeostasis. For instance, regarding protein homeostasis, IRE1 can sense protein misfolding (either directly or indirectly) and impact on this through activation of XBP1 mRNA splicing and RIDD (Almanza et al, 2019). In addition, IRE1 activity can be directly controlled by membrane lipid composition (Covino et al, 2018) and has been described to affect lipid metabolism through XBP1s and RIDD activation. Of note, protein and lipid stresses lead to different IRE1‐mediated transcriptional programs (Ho et al, 2020). Exploring and dissecting the precise mechanisms by which IRE1 senses and controls both protein and lipid homeostasis in physiology and pathology represents an extremely important challenge for the coming years.
In this issue of EMBO Reports, Hamid et al (2020) uncover a novel role of IRE1’s RNase activity in the maturation of pre‐miR2137, which in turn controls phosphoinositide levels in macrophages. miRNAs are small noncoding RNAs that act as regulators of gene expression in many physiological and pathological processes (Bartel, 2018). The generation of a functional mature miRNA from its synthesis to its maturation occurs through multiple molecular mechanisms that are still under investigation. In animals, the canonical miRNA biogenesis is initiated in the nucleus with the miRNA’s gene expression, under control of specific transcription factors. The primary miRNAs (pri‐miRNAs) are processed by the Drosha endonuclease/DGCR8 complex to hairpin pre‐miRNAs, which are then exported to the cytoplasm. Pre‐miRNAs are next cleaved by the Dicer endonuclease complexed with TRBP to generate miRNA duplexes. One of the strands of the duplex is loaded into Argonaute (Ago), a component of the RNA‐induced silencing complex (RISC). This now mature miRNA guides the complex to its mRNA targets, leading to their decay or to the inhibition of their translation (Bartel, 2018). Beside this mechanism, other so‐called noncanonical miRNA biogenesis pathways were also described, where a pre‐miRNA originates from intronic regions, from endogenous short hairpin RNA or chimeric hairpins from specific genome regions or where it bypasses the microprocessor steps mediated by both Drosha/DGCR8 or Dicer/TRBP complexes (Bartel, 2018; Fig 1).
Figure 1.
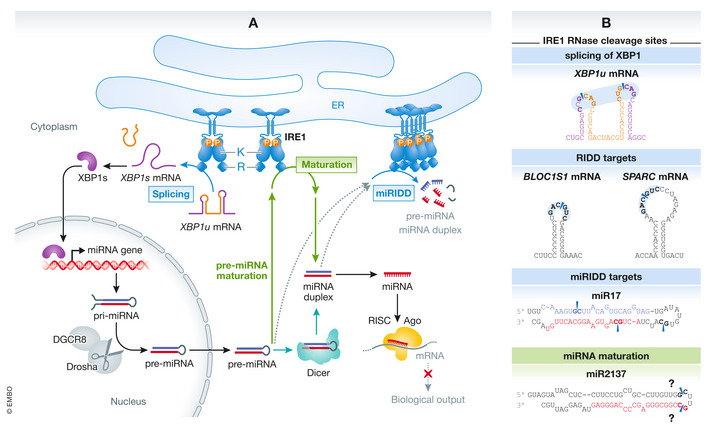
Mechanisms of IRE1‐dependent control of miRNA expression, maturation, and degradation. (A) Left: IRE1 promotes the production of the transcription factor XBP1s through a nonconventional mRNA splicing mechanism. XBP1s induces the expression of miRNA genes in the nucleus. Right: IRE1 degrades miRNAs through its endonuclease activity in a process called regulated IRE1‐dependent decay of miRNAs (miRIDD). Middle: Hamid et al provide evidence that IRE1 directly promotes the maturation of miRNAs (Hamid et al, 2020). Bottom: miRNA biogenesis cycle comprising production of pre‐miRNAs in the nucleus by DGCR8 and Drosha, Dicer‐mediated maturation, and RISC and Ago‐mediated action in the cytoplasm. (B) IRE1 cleavage sites on XBP1u mRNA (top), on RIDD mRNA and miRNA substrates (middle) and on miR2137 (bottom). Abbreviations: Ago, Argonaute; IRE1, Inositol‐requiring enzyme 1; K, kinase domain; miRIDD, regulated IRE1‐dependent decay of miRNAs; RISC, RNA‐induced silencing complex; R, RNAse domain; XBP1u, XBP1 unspliced; XBP1s, XBP1 spliced.
There is prior evidence for IRE1’s involvement in direct or indirect miRNA regulation (Maurel & Chevet, 2013; McMahon et al, 2017). First, several miRNAs such as miR‐17 or miR‐96 were described as direct targets of IRE1’s RIDD activity (Upton et al, 2012; Maurel & Chevet, 2013; McMahon et al, 2017). More recently, XBP1s was found to induce the expression of miRNA (e.g., miR‐148a and miR‐316) through transcription regulation in different cellular contexts. Together, these findings highlight the complexity of miRNA regulation by IRE1, which can either increase or decrease miRNA expression. In this issue, Hamid and colleagues add a novel layer of IRE1‐dependent regulation of the miRNA circuits showing that IRE1 can directly control the maturation of miR‐2137 in a Dicer‐independent manner (Hamid et al, 2020). The authors further provide evidence for a physiological role of this novel mechanism in the regulation of phosphoinositide levels in macrophages (Fig 1).
This study raises several questions about the mechanism of action through which IRE1 activity leads to pre‐miRNAs maturation. First, hairpin structures and nucleotide motifs of pri‐ and pre‐miRNAs are critical for their recognition and cleavage by endonucleases of the Drosha/DGCR8 microprocessor or the Dicer complex (Bartel, 2018). In comparison, IRE1 RNase requires two internal loops in the mRNA structure including a consensus CUGCAG site to initiate the unconventional splicing of XBP1 mRNA (Maurel et al, 2014). These mRNA hairpin structures with CUGCAG consensus sites were also shown to be important for RIDD activity although the exact targeted RNA structures and sequences still need to be clarified. Intriguingly, Hamid and colleagues did not identify any canonical IRE1 cleavage site in the pre‐miR‐2137 sequence. The recognition sequence cleaved by IRE1 in this particular context remains thus an outstanding question.
A second question is whether IRE1’s role in maturation is through an interaction with molecular actors involved in the canonical miRNA biogenesis, such as partners of Dicer or Ago that could lead to pre‐miRNA maturation. As discussed by Hamid and colleagues, although no direct interaction with Dicer and Ago2 molecules has been described so far, IRE1 is known to be associated with proteins found in the RISC. The importance of such interactions for its role in pre‐miRNA maturation needs to be explored. In this context, it would be highly relevant to re‐evaluate the impact of IRE1 activity on global miRNA circuits, integrating (i) XBP1s activity as a source of pri‐miRNA expression; (ii) RIDD pathway as a repressor of miRNA functions (miRIDD); and (iii) the novel Dicer‐independent IRE1 RNase function described by Hamid et al (2020) as part of the maturation process of pre‐miRNAs. Of note, IRE1 itself and XBP1s could also be targets of miRNAs, adding another layer of complexity (McMahon et al, 2017).
These different levels of IRE1‐mediated regulation of miRNA circuits will act together to impact the RNA targets of the miRNAs and thereby change protein expression patterns and cellular functions. Positioning such an integrated regulation (i.e., miRNA generation and degradation) in the physiological and pathological contexts will lead to a better understanding of their impact on biological processes, such as proliferation, cell death, lipid and protein homeostasis, and the roles played in metabolic, degenerative, or neoplastic diseases.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by grants from Institut National du Cancer (INCa PLBIO), Agence Nationale de la Recherche (ANR, IRE1inNASH and eRANET eRARE—ERAAT), Fondation pour la Recherche Médicale (FRM, équipe labellisée 2018) to EC ,and Institut des Neurosciences Cliniques de Rennes (INCR), la Ligue Contre le Cancer (comités 35, 56 & 85—AAP2019) to TA.
EMBO Reports (2020) 21: e51929.
See also: SM Hamid et al (December 2020)
Contributor Information
Tony Avril, Email: t.avril@rennes.unicancer.fr.
Eric Chevet, Email: eric.chevet@inserm.fr.
References
- Almanza A, Carlesso A, Chintha C, Creedican S, Doultsinos D, Leuzzi B, Luis A, McCarthy N, Montibeller L, More S et al (2019) Endoplasmic reticulum stress signalling – from basic mechanisms to clinical applications. FEBS J 286: 241–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018) Metazoan microRNAs. Cell 173: 20–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covino R, Hummer G, Ernst R (2018) Integrated functions of membrane property sensors and a hidden side of the unfolded protein response. Mol Cell 71: 458–467 [DOI] [PubMed] [Google Scholar]
- Hamid SM, Citir M, Terzi EM, Cimen I, Yildirim Z, Dogan AE, Kocaturk B, Onat UI, Arditi M, Weber C et al (2020) Inositol‐requiring enzyme‐1 regulates 1 phosphoinositide signaling lipids and macrophage growth. EMBO Rep 21: e51462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N, Yap WS, Xu J, Wu H, Koh JH, Goh WWB, George B, Chong SC, Taubert S, Thibault G (2020) Stress sensor Ire1 deploys a divergent transcriptional program in response to lipid bilayer stress. J Cell Biol 219: e201909165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS (2006) Decay of endoplasmic reticulum‐localized mRNAs during the unfolded protein response. Science 313: 104–107 [DOI] [PubMed] [Google Scholar]
- Maurel M, Chevet E (2013) Endoplasmic Reticulum stress signaling: the microRNA connection. Am J Physiol Cell Physiol 304: C1117–C1126 [DOI] [PubMed] [Google Scholar]
- Maurel M, Chevet E, Tavernier J, Gerlo S (2014) Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci 39: 245–254 [DOI] [PubMed] [Google Scholar]
- McMahon M, Samali A, Chevet E (2017) Regulation of the unfolded protein response by noncoding RNA. Am J Physiol Cell Physiol 313: C243–C254 [DOI] [PubMed] [Google Scholar]
- Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A et al (2012) IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase‐2. Science 338: 818–822 [DOI] [PMC free article] [PubMed] [Google Scholar]


