Abstract
Mammalian body temperature oscillates with the time of the day and is altered in diverse pathological conditions. We recently identified a body temperature‐sensitive thermometer‐like kinase, which alters SR protein phosphorylation and thereby globally controls alternative splicing (AS). AS can generate unproductive variants which are recognized and degraded by diverse mRNA decay pathways—including nonsense‐mediated decay (NMD). Here we show extensive coupling of body temperature‐controlled AS to mRNA decay, leading to global control of temperature‐dependent gene expression (GE). Temperature‐controlled, decay‐inducing splicing events are evolutionarily conserved and pervasively found within RNA‐binding proteins, including most SR proteins. AS‐coupled poison exon inclusion is essential for rhythmic GE of SR proteins and has a global role in establishing temperature‐dependent rhythmic GE profiles, both in mammals under circadian body temperature cycles and in plants in response to ambient temperature changes. Together, these data identify body temperature‐driven AS‐coupled mRNA decay as an evolutionary ancient, core clock‐independent mechanism to generate rhythmic GE.
Keywords: alternative splicing, circadian clock, mRNA decay, NMD, SR proteins, temperature
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; RNA Biology
This study identifies temperature‐controlled splicing variants triggering nonsense‐mediated decay and evolutionary conserved exons mediating temperature‐controlled gene expression levels, indicating that circadian and seasonal temperature signals feed into global gene expression control.

Introduction
Circadian clocks act as cell autonomous time‐measuring devices, which anticipate active and resting phases and coordinate behavior and physiology accordingly. The mammalian circadian timing system is organized in a hierarchical manner: The central pacemaker in the brain's suprachiasmatic nucleus (SCN) serves as a master regulator to coordinate circadian clocks throughout the body (Dibner et al, 2010). Therefore, information on the geophysical time (light‐dark) is transferred from the retina to the SCN, which then uses diverse neuronal and humoral cues to synchronize circadian clocks in other “light‐blind” parts of the brain and in other organs of the body (Gerber et al, 2015). Although the mechanisms of synchronization are different in the various organs, the conventional perception is that the majority of 24‐h rhythms depends on an identical transcription‐translation feedback loop in each cell of the body (Ko & Takahashi, 2006). However, only ~ 50% of circadian mRNA rhythms depend on de novo transcription (Koike et al, 2012; Menet et al, 2012), strongly arguing for an involvement of other post‐transcriptional mechanisms in generating rhythms (Preussner & Heyd, 2016; Shakhmantsir & Sehgal, 2019). Additionally, there is evidence for 24‐h rhythms which is cycle independent of the central oscillator (Kornmann et al, 2007). Core clock‐independent 24‐h rhythms in GE may be difficult to discriminate from real circadian oscillations in vivo (reviewed in (Preussner & Heyd, 2018)), and our mechanistic understanding of these rhythms is limited.
Systemic body temperature cycles (Refinetti & Menaker, 1992) can synchronize the central oscillator in peripheral clocks (Brown et al, 2002; Buhr et al, 2010; Saini et al, 2012), but can also directly result in rhythmic changes in GE. Examples include rhythmic expression of heat shock factors (Liu et al, 2019), of cold‐induced RNA‐binding proteins (RBPs), such as Cirbp (Morf et al, 2012; Liu et al, 2013), or rhythmic changes in the ratio of AS isoforms of U2af26 (U2af1l4) and many other related genes (Preussner et al, 2014; Preussner et al, 2017; Goldammer et al, 2018). All of these examples must be driven by highly sensitive mechanisms that are able to respond to physiological body temperature changes of 1–2°C (Gotic et al, 2016; Preussner et al., 2017), but the global impact of these mechanisms (and more generally of body temperature) on GE patterns remains enigmatic so far. In our previous work, we identified key components regulating temperature‐dependent AS. Essential regulators are members of the family of SR proteins (13 canonical members in human), which share a domain rich in serine and arginine residues, known as the RS domain (Manley & Krainer, 2010). The phosphorylation level of SR proteins controls their activity and serves as a fast and extremely sensitive molecular thermometer transferring the signal “body temperature” into global changes in AS (Haltenhof et al, 2020). SR proteins contain ultraconserved elements, allowing autoregulation of their own expression level (Lareau et al, 2007). Autoregulatory mechanisms can involve regulated inclusion of a poison exon containing a premature translation termination codon (PTC) or AS such that the normal stop codon becomes a PTC (Sureau et al, 2001; Lareau et al., 2007; Goncalves & Jordan, 2015). PTC‐containing mRNAs are recognized and degraded, usually by the nonsense‐mediated decay (NMD) surveillance pathway (Hug et al, 2016), and for SR proteins, this is understood as a mechanism associated with homeostatic control (Ni et al, 2007). However, the extreme evolutionary conservation is a strong indicator for further biological functions. AS‐inducing mRNA decay (AS‐decay) can also encompass other translation‐coupled pathways such as no‐go and non‐stop decay (Shoemaker & Green, 2012) and is not restricted to SR proteins. This mechanism thus allows a dynamic and cell‐wide control of GE (Braunschweig et al, 2013; Lykke‐Andersen & Jensen, 2015). While several thousand genes are affected by inhibition of the NMD pathway (Hurt et al, 2013), the role of mRNA decay in dynamically and actively controlling GE in response to changing cellular conditions has been described only in isolated cases (Wong et al, 2013; Tabrez et al, 2017). In flies and fungi, NMD mutants with altered circadian rhythms were described (Wu et al, 2017; Ri et al, 2019), thus suggesting a direct function of the NMD pathway in modulating the circadian clock (Mateos et al, 2018).
Here, we investigated to which extent AS‐decay is regulated by temperature and how this mechanism globally regulates body temperature‐dependent mRNA expression. We find that poison isoforms of different RBPs, especially SR proteins, are highly responsive to changes in the physiological temperature range and that temperature‐controlled AS‐NMD of SR proteins is sufficient to de novo generate 24‐h rhythms in GE. Temperature‐dependent AS‐decay is conserved from plants to human, controls thousands of mRNAs and represents a global mechanism for the generation of rhythmic 24‐h GE in vivo. The temperature‐sensing feedback loop inducing AS‐decay bears striking similarity to the classical circadian transcription‐translation feedback loop, both relying on rhythmic expression of a core machinery that controls itself and many output genes, but represents an independent and widespread mechanism to generate rhythmic GE. Furthermore, this mechanism shows wider evolutionary conservation than the classical core clock, pointing to an essential role in integrating temperature signals beyond the classical day–night cycle into cellular GE programs.
Results
Temperature‐dependent AS‐decay occurs frequently in primary mouse hepatocytes
The detection of temperature‐dependent poison isoforms requires depletion or pharmacological inhibition of their degradation pathways (Rehwinkel et al, 2006). NMD, non‐stop decay, and no‐go decay occur co‐translationally (Shoemaker & Green, 2012), and therefore blocking translation (e.g., via cycloheximide (CHX)) represents a fast and reliable way of blocking these pathways (Hurt et al., 2013). To identify temperature‐controlled splicing isoforms inducing mRNA decay, we performed RNA‐Seq on freshly isolated primary mouse hepatocytes, incubated either at 34 or 38°C, and treated either with DMSO or CHX (Fig 1A). A principal component analysis revealed clear clustering of the biological triplicates and comparable effects of both temperature and CHX on AS (Fig EV1A). Using Whippet (Sterne‐Weiler et al, 2018), we initially identified over 3,300 genes with CHX‐sensitive splicing events at either 34 or 38°C, over a fifth originating from cassette exon events. The events consist of all commonly recognized splicing patterns (Fig EV1B), and a quarter of them lies within genes previously associated with NMD (Fig EV1C), validating our approach for the identification of poison isoforms. To quantitatively estimate the percentage of potential NMD targets, we tested for the generation of PTCs. Stop codons are only rarely present in exons whose exclusion form is stabilized by CHX treatment or which exhibit no CHX‐dependent change in AS (6.3 and 8.9% of exons, respectively). Notably, the rate of stop codons in exons with a CHX‐stabilized inclusion isoform is strongly increased (to 27.7%, Fig EV1D), suggesting the NMD to be a prominent—but not the only—pathway through which these isoforms can be degraded. This number likely underestimates the true impact of the NMD pathway, as it does not account for PTCs that occur in later exons due to generation of frameshifts. Overall, these numbers are consistent with published data showing that about one‐third of all genes regulated upon CHX treatment are also regulated upon NMD factor knock‐down and we expect these isoforms to be degraded by the NMD pathway (Hurt et al., 2013). Finally, we find that about 30% of the CHX‐sensitive events were also SMG6/SMG7‐dependent using RNA‐Seq data from HeLa cells where these NMD‐specific factors (Chakrabarti et al, 2014) were knocked down (see Table EV1 and Materials and Methods section; data from (Colombo et al, 2017)). These data further validate the impact of NMD in degrading temperature‐controlled isoforms and highlight evolutionary conservation of this mechanism. However, despite the important impact of NMD we are not able to clearly distinguish between different degradation pathways in our data and we therefore refer more generally to mRNA decay in the following. Next, we identified 4,740 temperature‐controlled (comparing 34 and 38°C) splicing events in the CHX‐treated cells (Fig EV1E and Table EV1). Only 1/3 of the strongest 1,000 events and about 60% of all events respond in a similar manner in the DMSO control, indicating that these events do not lead to mRNA decay (Figs 1B and EV1F, heat‐skipped or cold‐skipped). All other isoforms, almost 2,000 in total (Table EV1), are only temperature‐dependent in CHX‐treated samples, indicating translation‐coupled degradation of one isoform, as it only accumulates in CHX‐treated cells. These cases can be categorized into four groups: (i) cold‐induced decay via inclusion; (ii) heat‐induced decay via inclusion; (iii) cold‐induced decay via exclusion; and (iv) heat‐induced decay via exclusion. Examples for each of these categories were confirmed by radioactive RT–PCRs with RNAs from independently generated hepatocytes (Figs 1C and EV1G) as follows: In Mettl16, exon 6 encodes a PTC, and the full length (fl) isoform—containing exon 6—is therefore stabilized via CHX. Exon 6 inclusion is strongly promoted at 34°C, representing a cold‐induced decay event. In Hnrnpdl, exon 8 inclusion generates a second exon junction complex after the canonical stop in exon 7, turning this stop into a PTC (Lindeboom et al, 2016). The fl isoform is stabilized by CHX and, as it predominates at 38°C, represents a heat‐induced decay event. In Hnrnph1 and Slc9a8, skipping of an exon (length not divisible by 3) generates a frameshift and a PTC in a downstream exon.
Figure 1. Characterization of temperature‐controlled AS‐decay in mammalian cells.
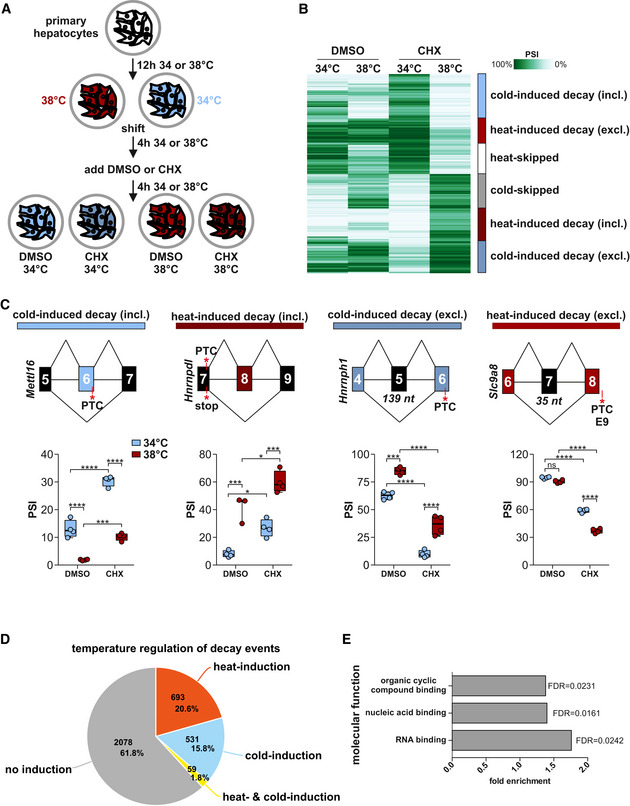
- Generation of RNA samples from mouse primary hepatocytes for analysis of temperature‐dependent AS‐decay.
- Automatic cluster mapping of the temperature‐dependent splicing changes in CHX, based on changes in PSI (percent spliced in). Categories (right) were manually assigned. Here the top 1,000 events are shown, see also Fig EV1E for all events.
- Examples for cold‐induced (blue) or heat‐induced (red) AS‐decay via exon inclusion or exon skipping. In each example, on top a simplified exon–intron structure is given, PTCs and the length of frameshift inducing exons are highlighted. Predicted splicing changes were validated by radioactive RT–PCR with primers binding to the surrounding constitutive exons. Box plots show median PSI of validation PCRs of four different samples from at least two different mice, whiskers show min to max. Individual data points are shown as circles. Statistical significance was determined by two‐way ANOVA and is indicated by asterisks: adjusted P values: not significant (ns), *P < 0.05, ***P < 0.001, ****P < 0.0001. See Fig EV1F and G for further examples.
- Classification of temperature dependence in genes with CHX‐dependent splicing events. Number of genes and percentages are indicated.
- GO‐term analysis of temperature‐controlled AS‐decay genes. All expressed genes (mean tpm > 0) served as background.
Data information: See also Fig EV1.
Figure EV1. Validation of temperature‐dependent AS in mammalian cells.

-
A.Principle component analysis of the investigated triplicate samples by RNA sequencing.
-
B.Distribution of splicing types in events identified to change after CHX treatment. AA alternative acceptor, AD alternative donor, AF alternative first exon, AL alternative last exon, CE cassette exon, RI retained intron, TE transcription end, TS transcription start.
-
C.Overlap of genes with CHX‐dependent events identified in our study in primary mouse hepatocytes (34 and 38°C) and by Hurt et al., 2013 in embryonic stem cells (37°C).
-
D.Number of alternative cassette exons that have a stop codon included, grouped by whether and how the exons are alternatively spliced upon CHX treatment. ****P < 0.0001, Chi‐square test.
-
E.Automatic cluster mapping of temperature‐dependent splicing changes in CHX, based on PSI (percent spliced in). All events with |ΔPSI| > 0.15 and P > 0.85 between CHX 34 and 38°C are shown. N = 4,740.
-
F, G.In F, examples for heat‐skipped (white) or cold‐skipped (gray) splicing events. Note that these splicing events are barely affected by CHX. In G, further examples for cold‐induced (blue) or heat‐induced (red) AS‐decay events. Box plots show median PSI of validation PCRs of four different samples from at least two different mice, whiskers show min to max. Individual data points are shown as circles. Statistical significance was determined by two‐way ANOVA and is indicated by asterisks: adjusted P values: not significant (ns), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
-
H.Comparison of temperature‐regulated AS events predicted by Whippet RNA‐Seq analysis and validated by RT–PCR from independent samples. The line depicts linear regression; goodness of fit is indicated by R 2.
-
I.Comparison of splicing changes in CHX (x‐axis) and DMSO (y‐axis) for all events presented in EV1E. The line depicts linear regression, goodness of fit is indicated by R 2, N = 4,740. The P value represents statistical probability of the slope being different from 0. Splicing changes in DMSO and CHX have the same directionality but ΔPSI values are generally larger in CHX.
-
J.Temperature dependence of the strongest decay events (top 500, largest ΔPSI in either 34 or 38°C comparison of DMSO vs CHX). Categories (right) were manually assigned. We define four categories: (i) cold‐induced decay, (ii) heat‐induced decay, (iii) temperature‐independent decay, and (iv) CHX‐sensitive temperature‐dependent splicing events. The generation of these isoforms could depend on de novo protein synthesis.
Overall, inclusion levels (percent spliced in, PSI) predicted by Whippet and validated by RT–PCR from independent biological samples show strong correlation, allowing us to draw reliable conclusions from our bioinformatics analysis (Fig EV1H). In all presented examples, as well as globally, the direction of PSI changes in DMSO correlates with PSI changes in CHX, confirming that these variants are present also in DMSO but are stabilized after CHX treatment (Fig EV1I). When analyzing all genes with potential poison events (all isoforms stabilized by CHX at either 34 or 38°C), we find that almost 40% of these genes contain temperature‐regulated splicing isoforms (Figs 1D and EV1J), showing that these isoforms are pervasively responsive to temperature changes, which is consistent with a global role in shaping temperature‐controlled GE. Interestingly, we find temperature‐ and CHX‐controlled events enriched in RBPs (Fig 1E and below), which could represent a temperature‐controlled core machinery that is able to amplify the temperature signal to control further downstream output events. In summary, temperature‐controlled AS coupled to mRNA decay represents a frequent mode of post‐transcriptional regulation, with wide implications for temperature‐controlled GE levels.
RBP expression is controlled through temperature‐dependent AS‐decay
Our bioinformatics analysis identified over 60 RBPs with temperature‐regulated poison exons of different types (Fig 2A, see also Table EV1). Most of these AS‐decay events are alternative cassette exons (e.g., Hnrnpdl, Hnrnph3), but there are also different types such as alternative last exons (e.g., Cirbp). In the presented examples, a temperature change of 4°C is sufficient to change AS by 20–30%, e.g., Hnrnpdl from 60 to 95%, indicating a strong regulatory impact on the expression of the respective RBPs (Fig 2A). We have recently shown this particular temperature‐dependent AS event in Cirbp to be regulated by CLK activity and found that E7b inclusion reduces Cirbp GE (Haltenhof et al., 2020). In line with these findings, we find the E7b isoform to be strongly stabilized by CHX and observe a correlation of higher abundance of the poison isoforms (particularly in the CHX condition) with decreased overall GE in the DMSO control in Cirbp and all other cases analyzed (Fig 2B). For example, in Hnrnpdl the poison isoform contributes more than 90% at 38°C, correlating with a 4‐fold reduction in GE. Note that although the change in Cirbp exon 7b inclusion between the two CHX conditions is visible as a drastic change in the Sashimi plot (Fig 2A, only 10% of reads from exon 6 span to exon 7b at 34°C, while this is the case for almost 70% of reads at 38°C), Whippet quantifies this as a difference of only about 15% (Fig 2B). For global correlations, we therefore include only skipped exon events, which seem more robust in the quantification. When considering all candidate RBPs with temperature‐dependent AS mRNA decay skipped exon events, we observe a highly significant global correlation of temperature‐dependent poison isoform generation and reduced GE levels in DMSO control (Fig 2C, top). Consistent with CHX preventing degradation of decay‐inducing isoforms, this correlation is completely abolished in the CHX samples (Fig 2C, bottom), strongly arguing for AS‐decay being the primary cause of temperature‐dependent changes in GE levels. Looking at all genes with temperature‐controlled poison cassette exons, the global trend is still present, although to a lesser extent (Fig EV2A), indicating that temperature‐dependent GE requires AS‐decay but is additionally controlled by other mechanisms. To provide evidence that temperature‐regulated AS‐decay can also result in altered protein levels, we generated a splicing sensitive GFP reporter. Here, exon inclusion results in an interrupted or non‐functional GFP variant while exon skipping results in functional GFP (Figs 2D and EV2B). Both an exon encoding a stop codon from Ythdf3 and an exon that interrupts the GFP‐coding frame from human Synerg (Haltenhof et al., 2020) show exon inclusion almost exclusively in cold (Fig 2E). In both examples, exon skipping in warm clearly correlates with higher GFP expression (Fig 2F). Finally, temperature shifts antagonistically regulate GFP expression within ~ 4 h (Fig 2G) thus highlighting how temperature‐controlled non‐productive splicing (i.e., AS‐decay) can generate rhythms in gene and protein expression.
Figure 2. Temperature‐dependent poison events globally regulate GE.

-
A.Temperature‐dependent poison events in Hnrnpdl (left, see also Fig 1C), Hnrnph3 (middle), and Cirbp (right). For each target, on top a simplified exon–intron structure is given and below Sashimi plots show the distribution of raw sequencing reads. Exon–exon junction reads are indicated by the numbers connecting the exons. Below, sequence conservation across placental species is indicated. Hnrnpdl exhibits a heat‐included exon that leads to a PTC, Hnrnph3 a cold‐skipped exon that leads to a frameshift, and in Cirbp, heat leads to inclusion of an alternative transcript end, which leads to formation of a PTC.
-
B.For each gene from A, the triplicate normalized read counts in DMSO (left y‐axis, green) and the percentage of the poison isoform in CHX (right y‐axis, orange) are plotted at the two temperatures. Line represents mean PSI.
-
C.Correlation of poison isoform inclusion and GE levels for all RBPs with temperature‐dependent AS‐decay skipped exon events. Shown are the log2 fold change (FC) in GE vs the ΔPSI of the poison isoform between the CHX samples. Top shows the GE change between the DMSO samples and bottom between the CHX samples. R 2 and P (deviation from zero slope) are indicated. N = 24.
-
D.GFP splicing reporter. The GFP encoding sequence is interrupted by an intron and contains destabilizing mRNA (AU‐rich) and protein (PEST) elements (based on a luciferase reporter by (Younis et al., 2010)). Insertion of an alternative exon makes GFP expression splicing‐dependent, as exon inclusion (with or without a STOP, red asterisks) abolishes the GFP signal (see Fig EV2B).
-
E.Splicing reporters, containing only an intron (GFP ctrl, #1) or additionally temperature‐dependent exons with ~ 1 kB surrounding introns. Representative radioactive RT–PCRs (bottom) confirm cold‐induced inclusion of Ythdf3 exon 7 (#2) and human Synerg exon 21 (#3) after 24 h at the indicated temperatures. Representative images are derived from three independent gels. See SourceDataForFigure2E for uncropped versions of these gels.
-
F–G.GFP expression was quantified using FACS and is shown relative to GFP ctrl. In (F) transfected cells were incubated at the indicated temperatures for 24 h (n = 3, mean ± SD). In (G) cells were incubated directly after transfection for 12 h at 39°C (blue) or 33°C (red) and then shifted for 2, 4, 8 and 12 h (n = 3, mean ± SD).
Data information: See also Fig EV2.
Source data are available online for this figure.
Figure EV2. Temperature‐dependent poison events globally regulate GE.

- Global correlation of poison isoform inclusion and GE levels for all genes with temperature‐dependent AS‐decay skipped exon events. Shown are the log2 fold change (FC) in GE vs the ΔPSI of the poison isoform between the CHX samples. Left shows the GE change between the DMSO samples and right the between the CHX samples at the two temperatures. R 2 and P (deviation from zero slope) are indicated, N = 254.
- Exon inclusion abolishes the GFP signal. Replacing the central 27 nucleotides of exon 7 in the Ythdf3 reporter (GFP Ythdf3 b‐GLO, #4) results in almost complete exon inclusion (RT–PCR, bottom right). A cytometric analysis comparing this mutant (#4, > 95% exon inclusion) and the GFP ctrl (#1, almost complete skipping) confirms that exon inclusion abolishes the GFP signal.
Temperature‐controlled AS‐decay is evolutionarily conserved across endotherms
We next elucidated the evolutionary conservation of temperature‐regulated AS‐decay. For SR proteins, it was previously described that these have NMD‐inducing exons with ultraconserved regions (Lareau et al., 2007) but their function was only linked to producing homeostatic GE levels. We hypothesized that these ultraconserved regions likely have additional functionalities and could be involved in controlling temperature‐dependent expression of SR proteins via AS‐decay. We indeed detect poison variants for most SR proteins, ranging from temperature‐insensitive to strongly temperature‐dependent (Fig EV3A). Two extreme examples for heat‐induced and cold‐induced non‐productive splicing events are ultraconserved exons in Srsf2 and Srsf10. For Srsf2, we observed and validated strong heat‐induced inclusion of exon 3 (46% ΔPSI in CHX, Figs 3A, left and EV3B). Inclusion of exon 3 generates a second exon junction complex downstream of the canonical stop, which therefore becomes a PTC (Sureau et al., 2001; Lejeune & Maquat, 2005). In contrast, we observed almost cold‐restricted generation of a non‐productive isoform for Srsf10, (19% ΔPSI in CHX, Figs 3A, right and EV3C). The splicing event in Srsf10 represents an exciting example for autoregulatory control of two competing 5′ splice sites (Meinke et al, 2020). The upstream 5′ splice site in exon 2 is recognized by the minor spliceosome and splices exclusively to exon 4, allowing the formation of productive Srsf10 mRNA. In contrast, the downstream 5′ splice site is recognized by the major spliceosome and coupled to exon 3 inclusion thereby introducing a PTC encoded by the additional sequence from exon 2. The strength of this PTC depends on the usage of an alternative polyadenylation signal in exon 3. In hepatocytes, the exon 3 isoform is strongly stabilized by CHX, correlating with splicing from exon 3 to exon 4. In Hek293 cells, the polyadenylation site in exon 3 is used resulting in only mild stabilization of the respective isoform via CHX (see Fig 3B). The CHX‐stabilized isoforms in Srsf2 and Srsf10 are also strongly stabilized upon SMG6/7 knock‐down (Fig EV3D), arguing for degradation via NMD.
Figure EV3. Temperature‐dependent AS‐decay is evolutionary‐conserved.

- Comparing temperature‐dependent AS‐decay (ΔPSI of 38–34°C in CHX) of SR proteins in primary mouse hepatocytes. The poison event in Srsf1 (*) escaped the computational analysis; data are derived from radioactive RT–PCR.
- Validation of Srsf2 temperature‐dependent AS‐decay by radioactive RT–PCR as described in Fig 1C. Box plots show median PSI of validation PCRs of four different samples from at least two different mice; whiskers show min to max. Individual data points are shown as circles. Statistical significance was determined by two‐way ANOVA and is indicated by asterisks: adjusted P values: not significant (ns), ***P < 0.001, ****P < 0.0001.
- Validation of Srsf10 temperature‐dependent AS‐decay by radioactive RT–PCR as described in A. Box plots show median PSI of validation PCRs of four different samples from at least two different mice; whiskers show min to max. Individual data points are shown as circles. Statistical significance was determined by two‐way ANOVA and is indicated by asterisks: adjusted P values: not significant (ns), *P < 0.05, **P < 0.01, ****P < 0.0001.
- Sashimi plots for SRSF2 (top) and SRSF10 (bottom) confirming increased poison exon inclusion upon SMG6/SMG7 double KD (light gray) compared to control (black).
- Heatmap for SR protein expression in D. melanogaster. Normalized mean GE in ovaries kept at 18 or 29°C (n = 2, (Fast et al, 2017)) is shown for each SR protein. Note cold‐induced expression of the SRSF2 homolog SC35.
- Sashimi plots indicating heat‐induced (red, top) or cold‐induced (blue, bottom) generation of a low‐abundance splicing isoform in cold‐induced Rbp1‐like (top) or heat‐induced Rbp1 (bottom).
Figure 3. Temperature‐dependent AS‐decay is evolutionarily conserved.

- SR protein specific temperature‐dependent AS‐decay. Sashimi plots are shown for heat‐induced poison exon inclusion for Srsf2 (left) and cold‐induced poison isoform formation for Srsf10 (right). Quantification of GE and AS are presented in the center (as in Fig 2B), lines represent mean.
- Poison exon inclusion for Srsf2 (left) and Srsf10 (right) in a 24 h temperature rhythm in human cells. Hek293 cells were pre‐entrained with square‐wave temperature cycles (12 h 34°C/12 h 38°C) for 48 h. For the last 24 h, cells were treated with DMSO or CHX every 4 h and harvested after 4 h and analyzed by splicing sensitive RT–PCR (n = 3, mean ± SD). White area: 34°C; Red area: 38°C. In Hek293 cells, inclusion of Srsf10 exon 3 is coupled to polyadenylation making it a weak decay target.
- AS‐decay of human targets at 34°C (blue) and 38°C (red) was investigated after a square‐wave temperature regime (see B). RNAs were harvested after 56 and 68 h, respectively (n = 3, mean ± SD). Statistical significance was determined by two‐way ANOVA and is indicated by asterisks: adjusted P values: not significant (ns), **P < 0.01, ***P < 0.001, ****P < 0.0001.
- Intron conservation of alternative cassette exons. Shown are average placental conservation scores of introns surrounding all alternative exons (black, N = 46,901), AS‐decay exons (yellow, N = 453) and temperature‐regulated AS‐decay exons (green, N = 139).
- Quantification of Srsf2 AS‐decay in hamster, rabbit, and chicken cells (n = 2, mean ± SD). Statistical significance was determined by two‐way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Data information: In (C) and (E) Box plots show median PSI of validation PCRs, whiskers show min to max. Individual data points are shown as circles. See also Fig EV3.
The combined data indicate antagonistic effects of temperature on different SR proteins. Again, the heat‐induced NMD exon inclusion of Srsf2 correlates with decreased GE in heat and vice versa for Srsf10 (Fig 3A, center). To investigate whether temperature cycles can lead to rhythmic splicing of NMD‐inducing exons, we applied a modified simulated body temperature regime (Brown et al., 2002; Preussner et al., 2017) to human Hek293 cells. Consistent with our data from mouse hepatocytes, the corresponding AS events in Srsf2 and Srsf10 respond to 24‐h square‐wave 34/38°C temperature rhythms in human Hek293 (Fig 3B), showing a cell‐type‐independent and evolutionarily conserved regulation of SR proteins through (circadian) temperature changes. Furthermore, we have validated additional temperature‐dependent poison isoforms for Hnrnpdl, Cirbp and Hnrnph3 in Hek293 cells (Fig 3C), consistent with high evolutionary conservation (see conservation track in Fig 2A). Importantly, we find that all temperature‐controlled AS‐decay exons as well as several 100 nucleotides in the surrounding introns are extremely conserved across placental organisms (Fig 3D and conservation tracks in Sashimi plots in Figs 2A and 3A), arguing for an evolutionarily vital function. Consistent with Thomas et al, 2020, this conservation is present in all AS‐decay exons and is not specific for temperature‐dependent events. Finally, we exemplarily investigated temperature‐controlled Srsf2 AS in hamster, rabbit, and chicken cell lines (chicken as a non‐placental endothermic organism), and strikingly find exon 3 inclusion promoted by heat and CHX in all cases (Fig 3E), thus demonstrating conserved regulation in all investigated endothermic organisms. The percentage of heat‐induced NMD exon inclusion varies between 8% in rabbit cells or human Hek293 and up to 80% in primary mouse hepatocytes, indicating species‐ or tissue‐specific differences in SRSF2 expression or activity. Interestingly, we also find temperature‐dependent GE for most SR proteins in the invertebrate ectotherm Drosophila melanogaster (Fig EV3E). While we do not detect a poison isoform in SRSF2 (SC35) without a decay inhibitor, increased expression in cold is consistent with our data from endotherms. Additionally, we detect temperature‐regulated low‐abundance splicing isoform in the strongest temperature‐regulated genes RBP1 and RBP1‐like, indicating splicing‐controlled RNA stability (Fig EV3F). In summary, we find that temperature‐dependent poison exon inclusion in SR proteins is evolutionarily conserved and anticorrelates with SR protein expression.
Temperature rhythms are sufficient to generate GE rhythms depending on AS‐NMD
To investigate whether external 24‐h rhythms in temperature are sufficient to generate rhythmic GE, we analyzed Srsf2 and Srsf10 GE in response to square‐wave temperature rhythms (Fig 4A). Both genes clearly show 24‐h rhythms in gene expression, with Srsf2 showing higher expression during the cold phase (heat‐induced NMD) and Srsf10 showing higher expression during the warm phase (cold‐induced NMD; wild‐type [WT] in Fig 4B and C). Next, we generated CRISPR/Cas9‐edited cell lines lacking either Srsf2 exon 3 or Srsf10 exon 3 (see also Meinke et al., 2020) and confirmed removal of the respective exons on RNA level (Fig EV4A and B). Upon removal of Srsf10 exon 3, only the upstream 5′ splice site, excluding the PTC, is used. Applying square‐wave temperature rhythms to these cell lines, we find that removing the temperature‐dependent exon is sufficient to abolish rhythms driven by external temperature cycles in both cases (ΔE3 in Fig 4B and C), showing that conserved NMD‐inducing alternative exons in SR proteins indeed are necessary for their temperature‐dependent GE. To investigate a potential function of AS‐NMD controlled SR protein expression on temperature entrainment of the circadian clock, we investigated temperature‐controlled expression of the core clock components Bmal1 and Per1 via RT–qPCR in control cells and in our cell line lacking SRSF10 exon 3. Temperature cycles induce 24‐h rhythmic gene expression of Bmal1 in both cell lines, but basal expression is strongly altered in the cell line lacking the SRSF10 NMD exon (Fig 4D). Consistent with temperature entrainment data from Liu et al., 2013, in WT cells Bmal1 expression is lowest at the beginning of the cold phase when Per1 expression is highest (Fig 4D and E). In contrast, in the edited cell line we find highest expression of both Bmal1 and Per1 in the middle of the cold phase. There is an additional reduction of Per1 expression in the middle of the warm phase in both cell lines (Fig 4E), which might be due to the short pre‐entrainment of only 48 h. Together, these data strongly argue for a direct function of SRSF10—and the NMD exon—in controlling temperature entrainment of the circadian clock.
Figure 4. Temperature‐dependent AS‐decay is necessary for rhythmic GE.

-
A.WT Hek293 and cell lines lacking either Srsf2 or Srsf10 exon 3 (Δpoison exon) through CRISPR/Cas9‐mediated genome editing (see also Fig EV4) were temperature entrained at three consecutive days of square‐wave temperature rhythm (12 h at 34 and 38°C, respectively). Time points of harvest are indicated by arrows.
-
B, C.Rhythmic Srsf2 (B) and Srsf10 (C) productive mRNA levels (relative to Gapdh) were analyzed by RT–qPCR. For each clone, expression is normalized to time point 52 h, (B) n = 3, (C) n = 4, mean ± SEM. For Srsf10 two independently generated clones were investigated (ΔE3 A and B). Statistical significance was determined by unpaired t‐test and is indicated by asterisks: P values: **P < 0.01, ****P < 0.0001.
-
D, E.Comparing temperature‐induced expression of the core clock components Bmal1 (D) and Per1 (E) in WT and Srsf10 ΔE3 cells (relative to Gapdh, normalized to time point 52 h). Statistical significance was determined by unpaired t‐test and is indicated by asterisks: P values: *P < 0.05, ***P < 0.001, ****P < 0.0001. n = 3, mean ± SEM.
Data information: White area: 34°C; Red area: 38°C. See also Fig EV4.
Figure EV4. Temperature‐dependent AS‐decay is necessary for rhythmic GE.

-
A, B.CRISPR/Cas9 strategy for cell lines lacking poison exons for Srsf2 (A) and Srsf10 (B). Lack of poison exons in clonal Srsf2 CRISPR/Cas9‐edited cell lines was confirmed by splicing sensitive RT–PCR (bottom). WT and mutant cell lines were treated with DMSO or CHX at 38°C and inclusion of the poison isoform was investigated by splicing sensitive RT–PCR (A). Investigation of Srsf10 dn‐E3 splicing in samples from Fig 4C. AS was analyzed by RT–PCR. A Gapdh PCR was performed simultaneously and served as a loading control. A representative gel image is shown. The asterisk indicates an unspecific product.
Temperature‐dependent AS‐decay regulates expression levels of SR proteins in Arabidopsis thaliana
The pervasive identification of temperature‐controlled AS‐decay in distinct SR proteins (and other RBPs) indicates an evolutionary advantage in homeotherms. To span a broader evolutionary spectrum, we next asked whether a similar mechanism is also in place for poikilotherms, which experience much greater differences in (body) temperature (10°C during a day–night cycle and often 50°C in seasonal changes compared to 1–2°C change in body temperature in mice). We therefore examined temperature‐dependent SR protein splicing and expression in A. thaliana, encoding 18 SR proteins (based on (Barta et al, 2010)) with evolutionary‐conserved regions triggering AS‐decay (Palusa & Reddy, 2010; Richardson et al, 2011; Rauch et al, 2014). We analyzed a publicly available RNA‐Seq time course dataset from plants kept at 20°C for 24 h and then shifted to 4°C for the next 24 h, while being under a 12‐h dark–light regime (Calixto et al, 2018), allowing us to distinguish between light‐ and temperature‐dependent effects. When assessing changes in GE, we strikingly find that all 18 SR proteins show a clear temperature‐dependent regulation (Fig 5A). Some genes additionally show a light‐dependent expression pattern, but temperature generally is the dominant signal. About half of the regulated SR proteins show higher GE at 20°C, while the other half is cold‐induced. Interestingly, we find at least one example for both warm‐ and cold‐expressed SR genes in all but one SR protein subfamilies (Fig 5B), which may suggest partially complementing functions. Within the RNA‐Seq data, we find clear evidence for decay‐inducing AS that is antagonistically regulated to GE in 50% of temperature‐regulated SR proteins (Fig EV5A and B), for example, in At‐SR34 and At‐SCL33 (Fig 5C), consistent with previous notions (Staiger & Brown, 2013; Calixto et al., 2018). In all tested examples, temperature‐dependent splicing and GE changes were confirmed by RT–PCR from independently generated plant samples (Figs 5D and EV5C). To systematically characterize temperature‐dependent AS‐decay events in plants, we first used RNA‐Seq data of CHX‐treated plants to identify poison events (Drechsel et al, 2013) and then quantified temperature regulation of these targets in samples without decay inhibition (data from (Calixto et al., 2018)). Although detection of poison isoforms is challenging without stabilization of the respective isoforms, we were able to identify 80 of these isoforms that were clearly affected by temperature (Table EV1). Consistent with our findings in mouse, there is a highly significant anti‐correlation between poison isoform inclusion and GE (Fig 5E). This includes a temperature‐controlled AS‐decay event in the snRNP assembly factor At‐GEMIN2 (Fig EV5D), which was previously identified as a key regulator of downstream temperature‐dependent clock‐related splicing events (Schlaen et al, 2015). In conclusion, these data expand our findings from the mammalian to the plant system, identifying temperature‐dependent AS‐decay as an evolutionary pervasive mechanism that adapts SR protein expression to changing temperatures. During mammalian evolution, AS‐decay in SR genes arose independently, evolutionary rapid and in a SR protein specific manner (Lareau & Brenner, 2015), thus showing that cis‐regulatory elements regulating temperature‐dependent AS‐decay independently arose in homeotherms and poikilotherms. This suggests a broad evolutionary advantage of temperature‐dependent SR protein expression and is indicative for an evolutionary‐conserved and evolutionary‐adapted temperature sensor controlling SR protein activity.
Figure 5. Temperature‐dependent AS‐decay of SR proteins is conserved in plants.

- Normalized GE values of plant SR proteins in a 48‐h time course. In the first day, plants were kept at 20°C (red) and shifted to 4°C (blue) for the second day. Plants were under a 12‐h dark/light regimen (gray/yellow bars). Warm‐ and cold‐expressed genes are indicated.
- Arabidopsis thaliana SR proteins divided in their subfamilies and classified as warm‐ or cold‐expressed.
- Sashimi plots as shown in Fig 2A (without conservation score) for decay‐inducing AS events upon cold (At‐SCL33, left) or warm temperature (At‐SR34, right). Data shown represent, from top to bottom, warm‐dark, warm‐light, cold‐dark, and cold‐light conditions as indicated on the right of the plots.
- For validation, A. thaliana were incubated for 3 days either at 20 or 4°C and RNA was investigated for gene expression (green, left y‐axis) and poison isoform formation (orange, right y‐axis). Gene expression of At‐SCL33 (left) or At‐SR34 (right) is shown relative to Ipp2. Lines represent mean, and individual data points are shown.
- Correlation of poison isoform inclusion and GE levels for temperature‐dependent poison skipped exon events. Shown are the log2 fold change (FC) in GE vs the ΔPSI of the poison isoform. Decay targets were identified by using data from CHX‐treated plants. R 2 and P (deviation from zero slope) are indicated. N = 80 (see main text and Materials and Methods for details).
Data information: See also Fig EV5A–D.
Figure EV5. Temperature‐dependent AS‐decay of SR proteins in plants and in mice.
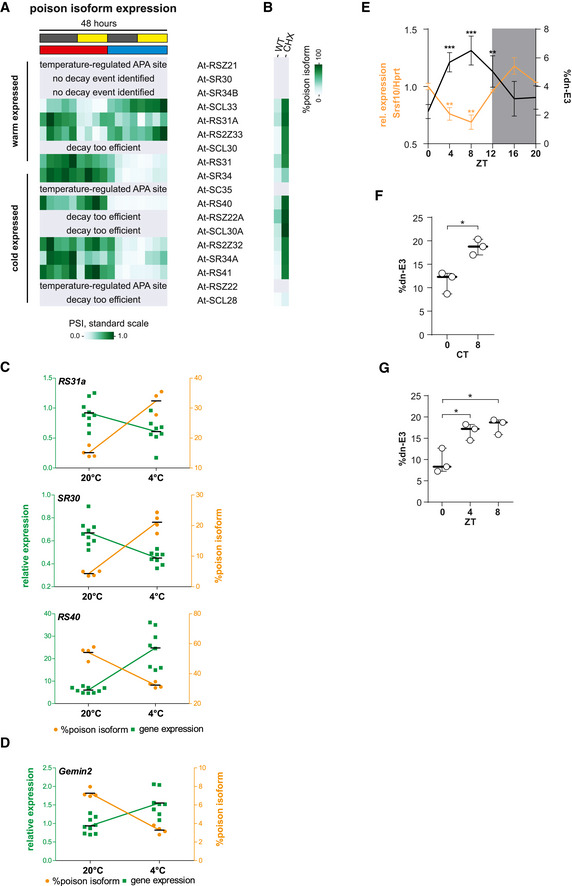
- Normalized poison isoform expression of plant SR proteins as in Fig 5A. In this dataset, without inhibition of decay pathways, the identification of poison isoforms is likely incomplete (compare to B). In three cases, a clear temperature‐regulated alternative polyadenylation (APA) event occurred but was not quantified (also indicating a post‐transcriptional origin of temperature‐dependent GE). In two cases, no poison event could be identified. Four times decay seemed to be too efficient, as no poison isoform could be identified without CHX treatment.
- Poison isoform inclusion in WT‐ or CHX‐treated plants for the same genes as in A. Here, the amount of the respective isoforms was compared in control and CHX‐treated samples (based on another RNA‐Seq dataset (Drechsel et al., 2013), see Table EV1). Note that most temperature‐dependent isoforms from A are stabilized upon CHX treatment.
- Validation RT–PCRs and qPCRs of poison events in further plant SR proteins as shown in Fig 5D. For each case, poison isoform inclusion correlates with lower GE. Lines represents mean.
- Validation of an AS‐decay event in GEMIN2, in which cold‐induced inclusion of the poison isoform correlates with lower GE. Lines represents mean.
- Correlation of rhythmic productive Srsf10 mRNA expression (orange, left y‐axis) and exon 3 inclusion (black, right y‐axis) in mouse cerebellum samples from the indicated ZTs. Splicing was analyzed using radioactive RT–PCR of at least three mice per time point. Expression was determined by RT–qPCR and is shown relative to Hprt and ZT0 (mean of at least three mice ± SEM). Student's unpaired t‐test‐derived P values **P < 0.01, ***P < 0.001 indicate difference from ZT0.
- Rhythmic Srsf10 AS persists in constant darkness. Mice were kept in constant darkness for 24 h and sacrificed at the indicated circadian times (CTs) of the following subjective day. RT–PCR analysis as in A (n = 3, median ± min to max). Student's unpaired t‐test‐derived P values *P < 0.05.
- Entrained Srsf10 AS. Mice were 8‐h phase delayed and sacrificed at the indicated ZTs on the 4th day. RT–PCR analysis as in A (n = 3, median ± min to max). Student's unpaired t‐test‐derived P values *P < 0.05.
Body temperature‐regulated AS‐decay generates 24‐h rhythms in GE
Finally, we address the question to which extent rhythmic GE in vivo is depending on naturally occurring body temperature cycles. We therefore compared the acrophase (time of maximal GE) of heat‐ and cold‐induced genes across a circadian cycle in mouse liver (Atger et al, 2015). Strikingly, we find that genes with higher expression at 38°C in hepatocytes predominantly peak during the night when mice are active and have a higher body temperature. In contrast, cold‐induced genes from hepatocytes rather peak during the colder day (Fig 6A, top), providing evidence for a direct function of body temperature cycles in generating 24‐h rhythmic GE. Consistent with a function of temperature‐dependent AS‐decay in this mechanism, we furthermore find that genes with a cold‐induced poison exon peak during the night, while genes with a heat‐induced poison exon tend to peak during the day (Fig 6A, bottom). Of note, of the 254 genes with temperature‐induced poison exons, 102 were found to exhibit cyclic expression.
Figure 6. AS‐decay generates GE rhythms in vivo .
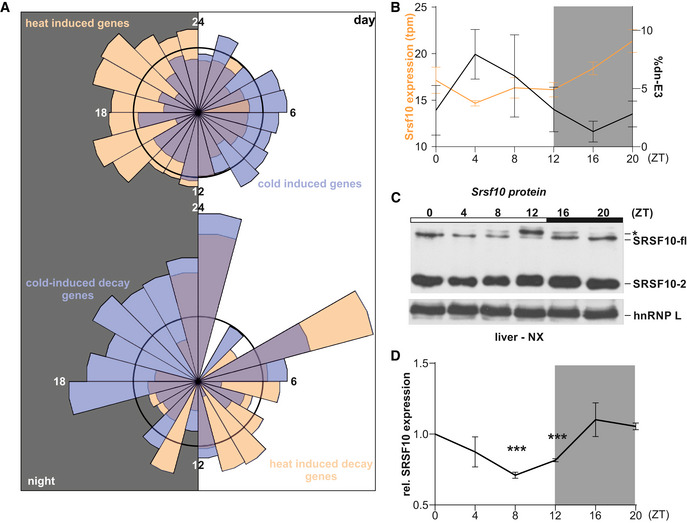
- Top: Rose plot visualizing the acrophases (based on rhythmic genes in the liver transcriptome from (Atger et al., 2015)) of genes exhibiting heat‐induced (orange, n = 797) and cold‐induced (blue, n = 828) upregulation of GE in mouse hepatocytes, respectively. Bottom: Acrophases of genes with cold‐induced (blue, n = 55) and heat‐induced (orange, n = 47) poison exon events, respectively. Numbers indicate respective ZT, with the left half representing night (at which mice are active and have an elevated body temperature) and the right half representing day (at which mice are asleep with lower body temperature). The black circles represent expected normal distribution if data would be randomly sampled.
- Correlation of rhythmic Srsf10 expression (orange, left y‐axis) and exon 3 inclusion (black, right y‐axis) in mouse liver samples from the indicated ZTs (n = 4, mean ± SEM). Quantifications were obtained by RNA‐Seq analysis of datasets from (Atger et al., 2015).
- Representative Western blot of SRSF10 protein levels from mouse liver nuclear extracts (NX) from different ZTs. The different SRSF10 variants are highlighted on the right. SRSF10‐fl and SRSF10‐2 are the result of differential last exon usage. hnRNP L was used as a loading control. The asterisks could represent hyperphosphorylated SRSF10‐fl through higher CLK activity during the day.
- Quantification of SRSF10‐fl + SRSF10‐2 relative to ZT0 and hnRNP L (mean of at least 3 mice ± SEM). Student's unpaired t‐test‐derived P values ***P < 0.001.
Data information: Gray areas indicate night time (dark). See also Fig EV5E–G.
As one particular example, we analyzed Srsf10 expression in vivo in detail and find a clear time of the day‐dependent formation of the non‐productive Srsf10 isoform (Fig 6B, right y‐axis). Consistent with our data from cultured hepatocytes, we observe antagonistic 24‐h rhythms in GE across a circadian day in vivo in liver (Fig 6B, left y‐axis). This is not a tissue‐specific but a general effect, as shown by validations in samples from mouse cerebellum confirming increased non‐productive splicing in combination with lower GE during the day (Fig EV5E). Similar to other temperature‐driven AS events (Preussner et al., 2014), we find that Srsf10 AS persists in constant darkness and is entrainable to a new light‐dark regime (Fig EV5F and G), highlighting common, e.g., body temperature‐dependent, regulatory principles. Finally, we found that AS‐decay‐driven rhythms in GE are also reflected at the protein level by confirming daytime‐dependent SRSF10 protein expression in mouse liver (Fig 6C and D). In summary, these data suggest a fundamental role of mammalian body temperature in generating rhythmic GE in vivo, which is independent from the classical, circadian clock‐mediated generation of 24‐h rhythms (Fig 7).
Figure 7. A model of two independent cycles generating rhythmic GE.

Inner ring (gray): Classical transcription–translation feedback loop or the central circadian clock, involving the transcription activator CLOCK and BMAL1 and the transcription targets/repressors Period (Per) and Cryptochrome (Cry). Outer ring (green/yellow): Model of rhythmic GE driven via AS‐decay. AS regulates the formation of productive stable or unproductive instable isoforms, thereby regulating the abundance of SR protein mRNA and protein. An additional regulatory layer is achieved through temperature‐mediated control of SR protein activity (active SR proteins are indicated by an asterisks), resulting in rhythmic GE of SR proteins themselves and output target genes. GE in response to continuous (top) or single (bottom) input signal (indicated by arrows) is shown in the center.
Discussion
Here we report that temperature‐regulated AS coupled to mRNA degradation occurs frequently in diverse genes from evolutionary distant species. We furthermore show that these splicing events are sufficient to generate temperature‐dependent GE changes and to induce 24‐h rhythms in GE. Together, these data are consistent with a model where body temperature‐controlled AS‐decay represents a second, core clock‐independent generator of 24‐h rhythmic GE. Classical circadian cycles in GE are driven by a transcription‐translation feedback loop (Fig 7, inner ring). Here, transcriptional activation trough CLOCK/BMAL1 results in accumulation of PERIOD (PER) and CRYPTOCHROME (CRY). When a critical protein level is reached, these factors dimerize, enter the nucleus, and repress their own GE. This results in low mRNA and protein expression, again allowing activation through CLOCK and BMAL1. Together with Per and Cry, many other Clock‐controlled genes oscillate through this feedback loop. Circadian oscillations are robust to environmental changes and continue to oscillate with only mildly affected amplitude in the absence of rhythmic stimuli (Fig 7, center). In a second and largely independent cycle, body temperature induces rhythmic GE through AS‐decay (Fig 7, outer ring). Here, body temperature cycles control the activity and AS‐decay‐mediated autoregulation of SR proteins. An SR protein that is inactive due to its phosphorylation level results in the accumulation of its mRNA and (inactive) protein. Upon a switch to the activating body temperature, phosphorylation of the SR protein changes through altered CLK activity (Haltenhof et al., 2020) and the activated SR proteins promotes poison exon inclusion within their own pre‐mRNA. Degradation of the unproductive mRNA isoform then leads to low but active protein levels. Only upon a second switch to the inactivating temperature, predominant formation of the productive isoforms becomes possible, again resulting in accumulation of the respective SR protein. As for the classic circadian feedback loop, autoregulation of SR proteins represents the core machinery, which then controls other genes containing poison exons to also cycle in response to rhythmic body temperature changes. As long as the body temperature oscillates, AS‐decay‐driven GE also cycles in a circadian‐like manner (Fig 7, center). This however requires continuous input and the rhythmicity is lost under constant conditions, which sets it apart from classic circadian GE.
In addition to SR proteins, we identified temperature‐controlled poison isoforms in many other RBPs. Mechanistically, it will be interesting to investigate, whether these splicing events are directly controlled by SR proteins or whether temperature also controls the activity of other RBPs (e.g., by phosphorylation or other post‐translational modifications). The presence of temperature‐regulated poison isoforms in many RBPs is indicative of an interlocked network of RBPs to post‐transcriptionally adapt GE to temperature, representing a potential layer of signal amplification. In mammals, this network requires sensors and modulators that are sensitive enough to respond to temperature differentials of around 1°C, to be able to respond to daily changes in body temperature. In recent work, we have characterized CLK kinases as the direct molecular sensor of such temperature changes (Haltenhof et al., 2020). Interestingly, we find in evolutionary distant species that groups of SR proteins are antagonistically affected by temperature, such that one group is activated by hyperphosphorylation, whereas this leads to inactivation of another group of SR proteins. While we observe this regulatory pattern in evolutionary distant species such as plants, fly, and humans, pointing to an important functionality, it remains unknown, how phosphorylation of SR proteins can have opposing effect on their activity.
Autoregulatory feedback loops were initially characterized as a mechanism allowing homeostatic regulation of SR proteins (Lareau et al., 2007; Ni et al., 2007). Here, we report that the same autoregulatory feedback loop can produce rhythms in GE if an input signal that controls SR protein activity or expression is present. This has intriguing consequences for SR protein target genes, as activation of a protein reduces total levels of the same protein. Target genes with high‐affinity binding sites might therefore be bound and stronger activated (including the own pre‐mRNA) while target genes with low affinity binding sites are not bound and therefore rather repressed.
At first sight, the existence of two independent 24‐h rhythms appears puzzling, but these two rhythms are fundamentally different: While classical circadian rhythms depend on transcription‐translation feedback loops and are robust to minor ambient changes (Gonze et al, 2002), temperature‐induced rhythms are generated post‐transcriptionally and can quickly adapt GE in response to changing temperatures. Therefore, temperature‐induced rhythms can stabilize circadian rhythms under circadian temperature cycles (Morf et al., 2012) but can additionally adapt GE in response to sudden temperature changes (especially for mRNAs with low half‐life time), i.e., under disease‐associated stress conditions, and are therefore able to integrate all temperature‐changing signals into complex GE programs. In mammals, this could involve body temperature changes associated with the female hormone cycle (Nagashima, 2015) or with aging (Keil et al, 2015). Furthermore, the interplay of these two mechanisms (e.g., through SR protein mediated control of clock gene expression, Fig 4D and E) could be involved in seasonal control of GE by integrating light and temperature signals. For example, At‐RS40 exhibits a fascinating upregulation of GE in cold‐light conditions (Fig 5A) and could therefore realize specific effects during winter days.
Homeothermic and poikilothermic organisms experience very different amplitudes in body temperature, and it is therefore a striking finding that in mouse/human (1–2°C) and plants (> 20°C) an almost identical mechanism controls SR protein levels. The evolution of AS‐NMD in SR proteins (Lareau & Brenner, 2015) indicates an evolutionary independent origin of the temperature‐controlled exons/introns. Mechanistically, it will therefore be interesting to investigate how cis‐regulatory elements, SR proteins, and kinases are evolutionary adapted to allow control of SR protein expression in the body temperature range of diverse organisms. The evolutionarily pervasive identification of temperature‐controlled AS‐NMD in SR proteins is indicative for an important function in temperature adaptation. Additionally, the fact that ultraconserved elements within SR proteins have evolved rapidly and are effectively “frozen” in mammals and birds (Bejerano et al, 2004), which are all endothermic homeotherms, represents a strong connection to endothermy. Because of the high sensitivity of temperature‐controlled AS‐decay, this mechanism could be involved in setting or maintaining a constant body temperature. The clinical importance of ultraconserved exons triggering degradation is furthermore highlighted by their potential tumor suppressive function (Thomas et al., 2020) and the association of NMD isoforms with neuronal diseases (Jaffrey & Wilkinson, 2018). Therefore, the de novo identification of temperature‐restricted poison isoforms might reveal novel candidates for antisense oligonucleotide‐based therapies in human (Bennett et al, 2019). Overall, we describe a widespread mechanism that integrates temperature changes to induce AS‐coupled mRNA decay cycles and generate GE rhythms from plants to human.
Materials and Methods
Mouse maintenance
All animal experiments were performed with C57BL/6 mice in accordance with institutional and governmental recommendations and laws. Mice were kept under constant 12‐h light–dark conditions. RNA samples across a circadian day, from constant darkness or after jet‐lag were previously generated (Preussner et al., 2014). Mice of both genders were used. For preparation of RNA, tissues were quickly removed, frozen in liquid nitrogen, and homogenized in RNATri (Bio&Sell). For preparation of nuclear extracts, we first prepared single‐cell suspensions of freshly isolated liver samples.
Tissue culture cells
The HEK293T cell line has been present in the laboratory for over 5 years and is maintained in liquid nitrogen. Early passage aliquots are thawed periodically. Rabbit RK‐13, Chinese hamster CHO, and chicken enterocyte 8E11 cell lines were a kind gift from Dusan Kunec (FU Berlin). For all cell lines, cell morphology and growth is routinely assessed and corresponds to the expected phenotype. Cell cultures are tested for mycoplasma contamination monthly using a PCR‐based assay. HEK293T and 8E11 cell lines were maintained in DMEM medium containing 10% FBS and Pen/Strep (Invitrogen). RK13 cells were maintained in EMEM medium containing 10% FBS and Pen/Strep (Invitrogen). CHO cells in Ham's F12 medium containing 10% FBS and Pen/Strep. All cell lines were usually maintained at 37°C and 5% CO2, except for 8E11 cells (39°C). For square‐wave temperature cycles, we used two incubators set to 34 and 38°C and shifted the cells every 12 h. See (Preussner et al., 2017) for an image of the square‐wave temperature cycles with temperatures and time points. For chicken cells, we used 34°C and 40°C (as 37°C degrees would already be a reduced temperature). Transfections of Hek293T using Rotifect (Roth) were performed according to the manufacturer's instructions. Cycloheximide (Sigma) was used at 40 µg/ml final concentration or DMSO as solvent control.
To isolate primary mouse hepatocytes, liver was perfused with PBS and digested using Collagen digestion solution. Liver was transferred into a Petri dish, and cells were liberated by mechanical force. Cells were washed three time with Williams Medium E and 10% FCS and plated.
Arabidopsis thaliana
To confirm temperature‐dependent alternative splicing and gene expression in A. thaliana, we isolated RNA from 28‐day‐old Col‐0 plants kept only at 20°C or 31‐day‐old Col‐0 plants kept at 4°C for the last 3 days. For RNA isolation using TRIzol (see below), we used ~ 100 mg material from leaves.
RT–PCR and RT–qPCR
RT–PCRs were done as previously described (Preussner et al., 2014). Briefly, RNA was extracted using RNATri (Bio&Sell) and 1µg RNA was used in a gene‐specific RT reaction. For analysis of minigene splicing, the RNA was additionally digested with DNase I and re‐purified. Low‐cycle PCR with a 32P‐labeled forward primer was performed; products were separated by denaturing PAGE and quantified using a Phosphoimager and ImageQuantTL software. For qRT–PCR, up to four gene‐specific primers were combined in one RT reaction. qPCR was then performed in a 96‐well format using the ABsolute QPCR SYBR Green Mix (Thermo Fisher) on Stratagene Mx3000P instruments. qPCRs were performed in duplicates, mean values were used to normalize expression to a housekeeping gene (mouse: Hprt, human: Gapdh, plants: Ipp2; ΔCT), and Δ(ΔCT)s were calculated for different conditions. See Table EV2 for primer sequences.
RNA‐Seq analysis and bioinformatics
Mapping of reads to reference genomes was performed using STAR version 2.5.3a (Dobin et al, 2012). Reference genomes mm10 (mouse), hg38 (human), BDGP6 (D. melanogaster), and TAIR10 (A. thaliana) were applied. Gene expression analysis was performed using Salmon version 0.11.3 transcript quantification (Patro et al, 2017) followed by DESeq2 version 1.22.2 quantification (Love et al, 2014) both used according to their documentation.
Whippet version 0.11 (Sterne‐Weiler et al., 2018) was used to obtain splicing ratios and transcript per million GE quantifications. To obtain splicing quantifications of AS‐decay events, index creation was supplemented with mapped reads of CHX‐treated sequencing samples (see Table EV1 for plant accession numbers) and the low TSL flag was not set. A splice event was considered significantly different between two conditions with a |ΔPSI| > 15%, probability > 85% and on average more than 10 junction reads. For AS‐decay events of SR proteins that could not be properly quantified by Whippet, PSI values were calculated based on junction read counts (e.g., Srsf7). Downstream analyses were performed using Python3, bash, and R scripts. Most relevant Python packages used were pandas (general secondary data analysis, (McKinney, 2010)), numpy (numerical operations, (Walt et al, 2011)), Matplotlib (data visualization, (Hunter, 2007)), and scikit‐learn (principle component analysis, (Pedregosa et al, 2011)). Molecular function analysis was performed using PANTHER version 14.1 (Mi et al, 2013) with all genes expressed above 1 TpM as background. Sashimi plots were generated using a customized version of ggsashimi (Garrido‐Martin et al, 2018), which additionally displays conservation scores. Junction reads with low count numbers were removed for clarity.
Comparison of CHX‐dependent events in mouse hepatocytes and HeLa cells upon SMG6/SMG7 double knock‐down were performed as described in the following: Coordinates of CHX‐dependent events were converted from mm10 to hg38 using the LiftOver tool. AS was investigated for knock‐down and control HeLa samples as described above. The Whippet results were then queried for events that matched on coordinate level with the lifted over AS‐decay events. In these overlapped list, 168 of 603 events were significantly differential upon SMG6/SMG7 knock‐down (28%). See also Table EV1 for the events and quantifications.
Phylogenetic P value conservation score data of placental organisms were downloaded from UCSC (mm10 phyloP60way placental, (Siepel et al, 2005; Pollard et al, 2010)) and manipulated with BEDOPS version 2.4.26 and bedtools version 2.26.0 (Quinlan & Hall, 2010; Neph et al, 2012).
For mouse data, splicing events were classified into the following groups:
Events that show differential splicing upon temperature (34 vs 38°C between DMSO or CHX conditions).
Events that show differential splicing upon CHX treatment (DMSO vs CHX in 34 or 38°C conditions) and no temperature‐dependent differential splicing (34 vs 38°C in CHX).
Events that show differential splicing upon CHX treatment (DMSO vs CHX in 34 or 38°C conditions) and additional temperature‐dependent differential splicing (34 vs 38°C in CHX).
Events that do not fall into any of the above categories.
Splicing events in groups 2) and 3) were identified as AS‐decay events. The temperature‐regulated events were further divided into cold‐induced and heat‐induced AS‐decay events (considering directionality of the CHX‐induction). AS‐decay events that showed different directionality upon CHX treatment at the two temperatures were omitted from the analysis (these made up < 2% of CHX‐induced events).
For A. thaliana, AS‐decay events were identified by using independent RNA‐Seq data from CHX‐treated plants. These data did not include a temperature shift and therefore the classification between temperature‐dependent and temperature‐independent AS‐decay events has been done using temperature shifted but not CHX‐treated data. Otherwise, the classification was performed as for mouse.
For preparation of rose plots, gene acrophases (time of highest expression) from (Atger et al., 2015) were used and combined with information about temperature regulation on expression (based on DESeq2 analysis, Padj < 0.05) and AS‐decay level (based on Whippet and post‐analysis, see above). Genes were sorted into 24 bins that represent 1 h each. As the data are highly skewed, normalization was needed. Therefore, the percentage of genes falling into each bin was calculated for all genes obtained from (Atger et al., 2015), and the subsets of heat‐expressed and cold‐expressed genes as well as those of genes with cold‐induced and heat‐induced decay events. The percentages for each bin in the subsets were then divided by this bin's percentage of overall genes. The black circles shown in Fig 6A represent the normalization line (expected proportion of genes if no effect was present).
GFP splicing reporter
To generate a temperature‐dependent splicing reporter, we adapted a luciferase reporter from (Younis et al, 2010) to the GFPC1 vector. We first transferred the destabilizing PEST and AU‐rich sequences to the GFPC1 vector using XhoI and BamHI, thereby destroying the XhoI site. In a next step, we inserted the intron from the luciferase reporter into the GFP open reading frame at position 1,344, thereby generating strong 5′ and 3′ splice sites, via PCR. During this step, we introduced a XmaI and a novel XhoI site into the intron, allowing subsequent cloning of temperature‐dependent exons with surrounding introns. Mouse Ythdf3 exon 7 with an artificial stop (360 nt upstream intron..AAATAAAGGAGAGCAAAACAGTTGTTTCATTGT.. 350 nt downstream intron) or human Synerg exon 21 (167 nt upstream intron..AAAGAAGAG AAGCCTGCAGAAGAACATCCTAAAAAA.. 984 nt downstream intron) were inserted into the intron using XmaI and XhoI. Interestingly, replacing 27nt of Ythdf3 exon 4 via beta‐globin sequence results in abolished temperature‐dependent AS, complete exon inclusion, and absence of GFP signal (Fig EV2B). Plasmids were transfected into Hek293 cells and investigated either for AS using splicing sensitive RT–PCRs with primers binding to GFP upstream (5′‐CACATGAAGCAGCACGACTT) and downstream (5′‐TCCTTGAAGTCGATGCCCTT) of the inserted intron or for protein expression using FACS. To normalize for general effects of temperature on splicing efficiency and GFP expression/folding, each time point/temperature is shown normalized to the GFP intron‐only reporter.
Generation of CRISPR/Cas9 modified cell lines
For genome‐engineering in Hek293T cells, sequences flanking the conserved exons of Srsf2 or Srsf10 (human) sgRNA candidates in silico using the Benchling tool (the Srsf10 cell line was originally introduced in (Meinke et al., 2020)). Upstream and downstream of each exon at least one pair of oligos for the highest ranked candidate sgRNA (Ran et al, 2013) was synthesized and subcloned into the PX459 vector (kindly provided by Stefan Mundlos). sgRNA sequences are available on request. Cells were transfected in 6‐well plates using Rotifect following the manufacturer's protocol. Forty‐eight hours after transfection, the transfected cells were selected with 1 μg/ml puromycin and clonal cell lines were isolated by dilution (Ran et al., 2013). Genomic DNA was extracted using DNA extraction buffer (200 mM Tris pH 8.3, 500 mM KCl, 5 mM MgCl2, 0.1% gelatin in H2O), and a PCR was performed using gene‐specific primers to confirm the exon knockout on DNA level. In promising clones, the exon knockout was additionally confirmed after RNA isolation by splicing sensitive PCR.
Western Blot
Nuclear fractionations (NX) were performed as previously described (Heyd & Lynch, 2010). SDS–PAGE and Western blotting followed standard procedures. Western blots were quantified using the ImageQuant TL software. The following antibodies were used for Western blotting: hnRNP L (4D11, Santa Cruz), SRSF10 (T‐18, Santa Cruz).
Quantification and statistical analysis
Figure legends contain information on repetitions and statistical tests used.
Data and software availability
RNA sequencing data are available under GSE158882 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158882).
Contact for reagent and resource sharing
Please contact M.P. (mpreussner@zedat.fu-berlin.de) for reagents and resources generated in this study. Accession numbers to all RNA‐Seq datasets used in this study are noted in Table EV1.
Author contributions
SM, GG, MS, and MP performed experiments. AN and MP performed the bioinformatics analysis. DS provided plant material. BT performed RNA sequencing. AN, MP, and FH designed the study, planned experiments, analyzed data, and wrote the manuscript with help from SM. MP and FH conceived and supervised the work.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Source Data for Figure 2
Acknowledgements
The authors would like to thank Jessica Stock, Alena Lohnert, Kevin Huolt, and Hoonsung Cho for their contribution to this work as rotation students. Furthermore, we would like to thank Dirk Hincha for discussing AS‐decay in plants, Tom Haltenhof for the isolation of primary hepatocytes, the SeqCore facility for generation of RNA Sequencing data and the HPC Service of ZEDAT, Freie Universität Berlin, for computing time. This work was funded through DFG grants HE5398/4‐2 and 278001972 ‐ TRR 186 to FH. MP is funded by a stipend from the Peter Traudl Engelhorn Foundation. Open access funding enabled and organized by ProjektDEAL.
EMBO Reports (2020) 21: e51369.
Contributor Information
Florian Heyd, Email: florian.heyd@fu-berlin.de.
Marco Preußner, Email: mpreussner@zedat.fu-berlin.de.
References
- Atger F, Gobet C, Marquis J, Martin E, Wang J, Weger B, Lefebvre G, Descombes P, Naef F, Gachon F (2015) Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci USA 112: E6579–E6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta A, Kalyna M, Reddy AS (2010) Implementing a rational and consistent nomenclature for serine/arginine‐rich protein splicing factors (SR proteins) in plants. Plant Cell 22: 2926–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D (2004) Ultraconserved elements in the human genome. Science (New York, NY) 304: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Bennett CF, Krainer AR, Cleveland DW (2019) Antisense oligonucleotide therapies for neurodegenerative diseases. Annu Rev Neurosci 42: 385–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley Brenton R, Blencowe Benjamin J (2013) Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Zumbrunn G, Fleury‐Olela F, Preitner N, Schibler U (2002) Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 12: 1574–1583 [DOI] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS (2010) Temperature as a universal resetting cue for mammalian circadian oscillators. Science (New York, NY) 330: 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto CPG, Guo W, James AB, Tzioutziou NA, Entizne JC, Panter PE, Knight H, Nimmo HG, Zhang R, Brown JWS (2018) Rapid and dynamic alternative splicing impacts the arabidopsis cold response transcriptome. Plant Cell 30: 1424–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Bonneau F, Schüssler S, Eppinger E, Conti E (2014) Phospho‐dependent and phospho‐independent interactions of the helicase UPF1 with the NMD factors SMG5‐SMG7 and SMG6. Nucleic Acids Res 42: 9447–9460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Karousis ED, Bourquin J, Bruggmann R, Mühlemann O (2017) Transcriptome‐wide identification of NMD‐targeted human mRNAs reveals extensive redundancy between SMG6‐ and SMG7‐mediated degradation pathways. RNA (New York, NY) 23: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2012) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G, Kahles A, Kesarwani AK, Stauffer E, Behr J, Drewe P, Ratsch G, Wachter A (2013) Nonsense‐mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25: 3726–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast I, Hewel C, Wester L, Schumacher J, Gebert D, Zischler H, Berger C, Rosenkranz D (2017) Temperature‐responsive miRNAs in Drosophila orchestrate adaptation to different ambient temperatures. RNA (New York, NY) 23: 1352–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido‐Martin D, Palumbo E, Guigo R, Breschi A (2018) ggsashimi: Sashimi plot revised for browser‐ and annotation‐independent splicing visualization. PLoS Comput Biol 14: e1006360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A, Saini C, Curie T, Emmenegger Y, Rando G, Gosselin P, Gotic I, Gos P, Franken P, Schibler U (2015) The systemic control of circadian gene expression. Diabetes Obes Metab 17(Suppl 1): 23–32 [DOI] [PubMed] [Google Scholar]
- Goldammer G, Neumann A, Strauch M, Muller‐McNicoll M, Heyd F, Preussner M (2018) Characterization of cis‐acting elements that control oscillating alternative splicing. RNA Biol 15: 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V, Jordan P (2015) Posttranscriptional regulation of splicing factor SRSF1 and its role in cancer cell biology. Biomed Res Int 2015: 287048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonze D, Halloy J, Goldbeter A (2002) Robustness of circadian rhythms with respect to molecular noise. Proc Natl Acad Sci USA 99: 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotic I, Omidi S, Fleury‐Olela F, Molina N, Naef F, Schibler U (2016) Temperature regulates splicing efficiency of the cold‐inducible RNA‐binding protein gene Cirbp. Genes Dev 30: 2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltenhof T, Kotte A, De Bortoli F, Schiefer S, Meinke S, Emmerichs AK, Petermann KK, Timmermann B, Imhof P, Franz A et al (2020) A conserved kinase‐based body temperature sensor globally controls alternative splicing and gene expression. Mol Cell [DOI] [PubMed] [Google Scholar]
- Heyd F, Lynch KW (2010) Phosphorylation‐dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol Cell 40: 126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N, Longman D, Cáceres JF (2016) Mechanism and regulation of the nonsense‐mediated decay pathway. Nucleic Acids Res 44: 1483–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD (2007) Matplotlib: a 2D graphics environment. Comput Sci Eng 9: 90–95 [Google Scholar]
- Hurt JA, Robertson AD, Burge CB (2013) Global analyses of UPF1 binding and function reveal expanded scope of nonsense‐mediated mRNA decay. Genome Res 23: 1636–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Wilkinson MF (2018) Nonsense‐mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat Rev Neurosci 19: 715–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil G, Cummings E, de Magalhães JP (2015) Being cool: how body temperature influences ageing and longevity. Biogerontology 16: 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15 (Spec No 2): R271–R277 [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science (New York, NY) 338: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U (2007) System‐driven and oscillator‐dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE (2007) Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Lareau LF, Brenner SE (2015) Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol Biol Evol 32: 1072–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE (2005) Mechanistic links between nonsense‐mediated mRNA decay and pre‐mRNA splicing in mammalian cells. Curr Opin Cell Biol 17: 309–315 [DOI] [PubMed] [Google Scholar]
- Lindeboom RGH, Supek F, Lehner B (2016) The rules and impact of nonsense‐mediated mRNA decay in human cancers. Nat Genet 48: 1112–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu W, Murakawa Y, Yin J, Wang G, Landthaler M, Yan J (2013) Cold‐induced RNA‐binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Rep 3: 2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Qian M, Tang X, Hu W, Sun S, Li G, Zhang S, Meng F, Cao X, Sun J et al (2019) SIRT7 couples light‐driven body temperature cues to hepatic circadian phase coherence and gluconeogenesis. Nature Metabolism 1: 1141–1156 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke‐Andersen S, Jensen TH (2015) Nonsense‐mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 16: 665–677 [DOI] [PubMed] [Google Scholar]
- Manley JL, Krainer AR (2010) A rational nomenclature for serine/arginine‐rich protein splicing factors (SR proteins). Genes Dev 24: 1073–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, de Leone MJ, Torchio J, Reichel M, Staiger D (2018) Beyond transcription: fine‐tuning of circadian timekeeping by post‐transcriptional regulation. Genes (Basel) 9: 616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney WT (2010) Data structures for statistical computing in Python Proceedings of the 9th Python in Science Conference, pp 51–56 [Google Scholar]
- Meinke S, Goldammer G, Weber AI, Tarabykin V, Neumann A, Preussner M, Heyd F (2020) Srsf10 and the minor spliceosome control tissue‐specific and dynamic SR protein expression. Elife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M (2012) Nascent‐Seq reveals novel features of mouse circadian transcriptional regulation. Elife 1: e00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41: D377–D386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morf J, Rey G, Schneider K, Stratmann M, Fujita J, Naef F, Schibler U (2012) Cold‐inducible RNA‐binding protein modulates circadian gene expression posttranscriptionally. Science (New York, NY) 338: 379–383 [DOI] [PubMed] [Google Scholar]
- Nagashima K (2015) Thermoregulation and menstrual cycle. Temperature (Austin) 2: 320–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S et al (2012) BEDOPS: high‐performance genomic feature operations. Bioinformatics 28: 1919–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr (2007) Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense‐mediated decay. Genes Dev 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa SG, Reddy AS (2010) Extensive coupling of alternative splicing of pre‐mRNAs of serine/arginine (SR) genes with nonsense‐mediated decay. New Phytol 185: 83–89 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias‐aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V et al (2011) Scikit‐learn: machine learning in Python. J Mach Learn Res 12: 2825–2830 [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preussner M, Wilhelmi I, Schultz AS, Finkernagel F, Michel M, Moroy T, Heyd F (2014) Rhythmic U2af26 alternative splicing controls PERIOD1 stability and the circadian clock in mice. Mol Cell 54: 651–662 [DOI] [PubMed] [Google Scholar]
- Preussner M, Heyd F (2016) Post‐transcriptional control of the mammalian circadian clock: implications for health and disease. Pflugers Arch 468: 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preussner M, Goldammer G, Neumann A, Haltenhof T, Rautenstrauch P, Muller‐McNicoll M, Heyd F (2017) Body temperature cycles control rhythmic alternative splicing in mammals. Mol Cell 67: 433–446.e434 [DOI] [PubMed] [Google Scholar]
- Preussner M, Heyd F (2018) Temperature‐controlled rhythmic gene expression in endothermic mammals: all diurnal rhythms are equal, but some are circadian. BioEssays 40: e1700216 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR‐Cas9 system. Nat Protoc 8: 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch HB, Patrick TL, Klusman KM, Battistuzzi FU, Mei W, Brendel VP, Lal SK (2014) Discovery and expression analysis of alternative splicing events conserved among plant SR proteins. Mol Biol Evol 31: 605–613 [DOI] [PubMed] [Google Scholar]
- Refinetti R, Menaker M (1992) The circadian rhythm of body temperature. Physiol Behav 51: 613–637 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Raes J, Izaurralde E (2006) Nonsense‐mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem Sci 31: 639–646 [DOI] [PubMed] [Google Scholar]
- Ri H, Lee J, Sonn JY, Yoo E, Lim C, Choe J (2019) Drosophila CrebB is a substrate of the nonsense‐mediated mRNA decay pathway that sustains circadian behaviors. Mol Cells 42: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DN, Rogers MF, Labadorf A, Ben‐Hur A, Guo H, Paterson AH, Reddy AS (2011) Comparative analysis of serine/arginine‐rich proteins across 27 eukaryotes: insights into sub‐family classification and extent of alternative splicing. PLoS One 6: e24542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Morf J, Stratmann M, Gos P, Schibler U (2012) Simulated body temperature rhythms reveal the phase‐shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev 26: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaen RG, Mancini E, Sanchez SE, Perez‐Santangelo S, Rugnone ML, Simpson CG, Brown JW, Zhang X, Chernomoretz A, Yanovsky MJ (2015) The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc Natl Acad Sci USA 112: 9382–9387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhmantsir I, Sehgal A (2019) Splicing the clock to maintain and entrain circadian rhythms. J Biol Rhythms 34: 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R (2012) Translation drives mRNA quality control. Nat Struct Mol Biol 19: 594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S et al (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Brown JW (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne‐Weiler T, Weatheritt RJ, Best AJ, Ha KCH, Blencowe BJ (2018) Efficient and accurate quantitative profiling of alternative splicing patterns of any complexity on a laptop. Mol Cell 72: 187–200.e186 [DOI] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stévenin J, Soret J (2001) SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J 20: 1785–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrez SS, Sharma RD, Jain V, Siddiqui AA, Mukhopadhyay A (2017) Differential alternative splicing coupled to nonsense‐mediated decay of mRNA ensures dietary restriction‐induced longevity. Nat Commun 8: 306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Polaski JT, Feng Q, De Neef EJ, Hoppe ER, McSharry MV, Pangallo J, Gabel AM, Belleville AE, Watson J et al (2020) RNA isoform screens uncover the essentiality and tumor‐suppressor activity of ultraconserved poison exons. Nat Genet 52: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt S, Colbert SC, Varoquaux G (2011) The NumPy Array: a structure for efficient numerical computation. Comput Sci Eng 13: 22–30 [Google Scholar]
- Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K et al (2013) Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154: 583–595 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang Y, Sun Y, Yu J, Wang P, Ma H, Chen S, Ma L, Zhang D, He Q et al (2017) Up‐Frameshift protein UPF1 regulates Neurospora crassa circadian and diurnal growth rhythms. Genetics 206: 1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis I, Berg M, Kaida D, Dittmar K, Wang C, Dreyfuss G (2010) Rapid‐response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol Cell Biol 30: 1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Source Data for Figure 2
Data Availability Statement
RNA sequencing data are available under GSE158882 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158882).


