Abstract
The thyroid gland regulates growth and metabolism via production of thyroid hormone in follicles composed of thyrocytes. So far, thyrocytes have been assumed to be a homogenous population. To uncover heterogeneity in the thyrocyte population and molecularly characterize the non‐thyrocyte cells surrounding the follicle, we developed a single‐cell transcriptome atlas of the region containing the zebrafish thyroid gland. The 6249‐cell atlas includes profiles of thyrocytes, blood vessels, lymphatic vessels, immune cells, and fibroblasts. Further, the thyrocytes show expression heterogeneity, including bimodal expression of the transcription factor pax2a. To validate thyrocyte heterogeneity, we generated a CRISPR/Cas9‐based pax2a knock‐in line that monitors pax2a expression in the thyrocytes. A population of pax2a‐low mature thyrocytes interspersed in individual follicles can be distinguished. We corroborate heterogeneity within the thyrocyte population using RNA sequencing of pax2a‐high and pax2a‐low thyrocytes, which demonstrates 20% differential expression in transcriptome between the two subpopulations. Our results identify and validate transcriptional differences within the presumed homogenous thyrocyte population.
Keywords: CRISPR/Cas9, heterogeneity, single‐cell, thyroid gland, zebrafish
Subject Categories: Development & Differentiation, RNA Biology
This study molecularly characterizes the zebrafish thyroid gland and surrounding structures at single‐cell resolution. It identifies the major cell‐types and potential cross‐talk and uncovers genetic heterogeneity in thyroid follicular cells.
Introduction
The thyroid gland produces hormones thyroxine (T4) and triiodothyronine (T3) that regulate body metabolism, growth, and development. Thyroid dysfunction afflicts almost 100 million people worldwide (Taylor et al, 2018). Hypothyroidism can be efficiently managed by lifelong hormone replacement therapy, while hyperthyroidism is treated with antithyroid medication, surgery, or ablation, depending on the underlying disorder. Congenital hypothyroidism if left untreated may result in profound adverse effects on development, including mental retardation, goiter, or dwarfism.
The thyroid gland is an endocrine organ with an intricate structure enabling production, storage, and release of the thyroid hormones. It contains numerous variable‐sized spherical follicles composed of thyroid follicular epithelial cells or thyrocytes. The thyrocytes generate the thyroid hormones in a multi‐step process. They secrete and store thyroglobulin (TG) in the lumen of the follicles. Additionally, they intake iodide from the blood via sodium‐iodide symporter (NIS/Slc5a5). At the interface between thyrocytes and the lumen, thyroid peroxidase (TPO) expressed by the cells catalyzes the coupling of iodide to tyrosyl residues of TG. Iodinated TG is absorbed back into the thyrocyte and cleaved by cysteine proteases in lysosomes to form T4 and T3 (Brix et al, 1996). Though the machinery responsible for the production of thyroid hormones by thyrocytes is well established, it remains unknown whether all the thyrocytes resident in a follicle are equally capable of generating thyroid hormones. In other words, the extent of molecular homogeneity between individual thyrocytes has not yet been investigated.
Additionally, the thyroid gland contains many cell types with potential roles in modulating thyrocyte functionality. The gland contains an extensive distribution of blood vessels, which carry iodide to the thyrocytes and carry thyroid hormones away from them. The thyroid follicles are separated by a mesenchymal cell population, called connective tissue septa, which also divides the gland into lobules. The mammalian thyroid gland also contains parafollicular epithelial cells, or C cells, that synthesize and secrete the hormone calcitonin. These parafollicular epithelial cells are, however, located outside the thyroid gland in fish and amphibians (Alt et al, 2006). Further, the presence of immune cells and innervation has been demonstrated within the thyroid gland (Nonidez, 1931; Linehan et al, 2001). Though we have a considerable understanding of these cell types on a histological level, we still lack the molecular characterization of the thyroid gland cell ensemble. This extends to an incomplete appreciation of the impact of the diverse cell populations on thyroid follicular cell physiology.
To uncover the diversity within the thyrocyte population and further characterize the surrounding tissue at cellular resolution, we develop the first atlas of the thyroid gland at single‐cell resolution. For this, we build on the progress in single‐cell transcriptomics (Svensson et al, 2018) to transcriptionally profile thousands of individual cells isolated from the thyroid gland of juvenile and adult zebrafish. We demonstrate that these profiles comprehensively represent the cells present in the zebrafish thyroid gland. Utilizing the expression profiles of discrete cell populations, we build an intercellular signaling network to uncover communication between thyrocytes and the surrounding tissue. Further, we demonstrate the segregation of thyrocytes into two transcriptionally distinct subpopulations based on the expression level of a transcription factor. Finally, to enable easy access to the data, we have made the zebrafish thyroid gland atlas available for online browsing.
Results
Single‐cell transcriptomics of the zebrafish thyroid gland
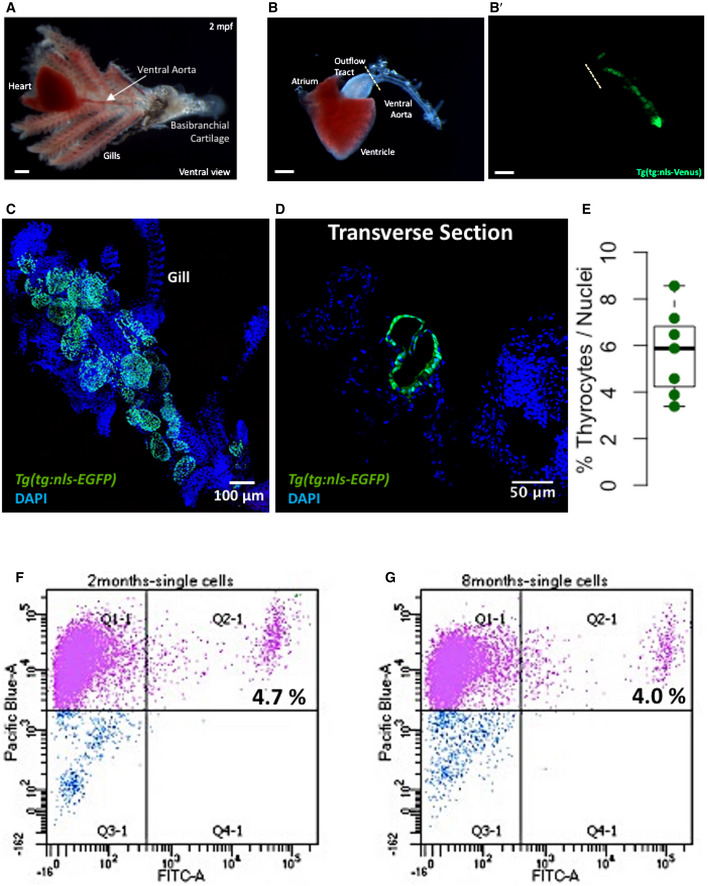
The zebrafish thyroid gland is composed of follicles scattered in the soft tissue surrounding the ventral aorta (Fig 1A and B). Ventral aorta extends from the outflow tract of the zebrafish heart and carries blood from the ventricle to the gills. Dissection of the ventral aorta associated region (detailed in Methods section) provided us with tissue that included the thyroid follicles and parts of zebrafish gills (Fig 1C). Using Tg(tg:nls‐EGFP) transgenic line, which labels thyrocytes with nuclear green fluorescence (Fig 1D), we estimated presence of 5.9 ± 1.9% thyrocytes within the dissociated region (Fig 1E).
Figure 1. Isolation of zebrafish thyroid gland.
-
A, BA brightfield image showing the zebrafish thyroid gland along with surrounding tissue. The thyroid follicles reside in the soft tissue surrounding the ventral aorta, which extends from the outflow tract of the heart into the gills toward the basibranchial cartilage in the lower jaw. The thyroid follicular cells, or thyrocytes, are labeled in green in the Tg(tg:nls‐mVenus‐NTR) transgenic line (B’). Scale bars: 500 µm.
-
CMaximum intensity projection of 3D confocal stack obtained from the dissected thyroid gland of Tg(tg:nls‐EGFP) animal and labeled with DAPI.
-
DConfocal scan of a transverse section across the dissected thyroid gland from Tg(tg:nls‐EGFP) animal at 3 mpf. Sections were stained with DAPI to visualize cells surrounding thyroid follicles.
-
EBoxplot depicting the proportion of thyrocytes present in transverse sections obtained from three Tg(tg:nls‐EGFP) animals at 3 mpf. Each dot represents a transverse section. The Tukey boxplot marks the 25th percentile, median, and 75th percentile with whiskers extending from smallest to largest values.
-
F, GRepresentative FACS plot of single cells from Tg(tg:nls‐mVenus‐T2A‐NTR) animals at 2 mpf (F) and 8 mpf (G). Calcein (Pacific Blue) labels live cells, while green fluorescence (FITC) labels thyrocytes. Percentage values represent proportion of calcein+ thyrocytes within total calcein+ cells.
Source data are available online for this figure.
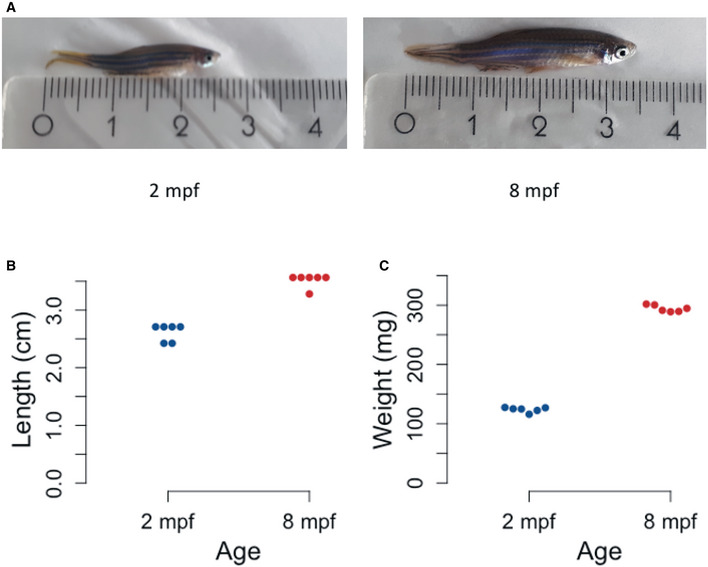
To generate the molecular catalogue of the thyroid gland at cellular resolution, we sampled the organ from two ages of zebrafish: 2 months post‐fertilization (mpf) and 8 mpf (Fig EV1A). The time points span juvenile to adult transition in zebrafish, with animals containing fully differentiated functional organs at both stages. By 2 mpf, the juvenile animals have completed morphogenesis, but are yet to reach sexual maturity. The animals sampled at 2 mpf were on average 2.6 cm in length and 123.8 mg in weight. In contrast, fish at 8 mpf are sexually mature adults, with an average length of 3.5 cm and an average weight of 294.4 mg (Fig EV1B and C). To characterize the organ cell types in an unbiased manner, we dissected out the entire thyroid gland (Fig 1B and C) from six animals at each stage and prepared the single‐cell suspension for cDNA library preparation. To guide thyroid gland dissection, we utilized the Tg(tg:nls‐mVenus‐T2A‐NTR) zebrafish reporter line (McMenamin et al, 2014) that labels thyrocytes with bright yellow fluorescent protein (Fig 1B’). The micro‐dissected tissue was dissociated using enzymatic digestion. The single‐cell suspension was stained with calcein, which specifically labels live cells with blue fluorescence. The live cells were then enriched using FACS (Fig 1F–G) to limit false‐positive signals from dead and/or ruptured cells (AlJanahi et al, 2018). Thyrocytes consisted of around 4% of the alive cells at both stages, comparable to the percentage obtained by immunofluorescence analysis (Fig 1E). Twelve thousand live cells, pooled from six animals, were collected in separate tubes according to age and profiled using droplet‐based high‐throughput single‐cell RNA sequencing provided by 10× Genomics (Macosko et al, 2015; Zheng et al, 2017). Droplet‐based methods encapsulate cells with single‐Poisson distribution (Zheng et al, 2017). This leads to approximately 50% cell capture rate, which is the ratio of the number of cells detected by sequencing and the number of cells loaded. The 10× Genomics pipeline uses molecule and cell‐specific barcoding allowing transcript quantification without amplification bias (Kivioja et al, 2011; Islam et al, 2014). Using the Cell Ranger bioinformatics pipelines, the resulting Next‐Generation Sequencing libraries were mapped to the zebrafish genome, de‐multiplexed according to their cellular barcodes and quantified to generate gene/cell UMI (unique molecular identifier) count tables. The Cell Ranger pipeline provided us with 13,106 sequenced cells from 24,000 input cells (54.6 % cell capture rate). Quality‐based exclusion of single‐cell transcriptomes was implemented based on mean library size, percentage of mitochondrial reads, and number of genes detected per cell. On average, we detected 6,012 UMIs and 1,303 genes per cell (Fig EV2). The process recovered in total 6,249 cells out of 13,106 sequenced cells (47.7% retention rate), providing single‐cell transcriptomic profiles for 2,986 and 3,263 individual cells for 2 mpf and 8 mpf, respectively.
Figure EV1. Physical characteristics of zebrafish at 2 mpf and 8 mpf.
-
ARepresentative images of zebrafish at 2 mpf (left) and 8 mpf (right). The ruler depicts length in cm.
-
BDotplot representing the length of individual animals at each stage. Y‐axis represents the length of six zebrafish from mouth to end of fin in cm.
-
CDotplot representing the weight of individual animals at each stage. Y‐axis represents the weight of six zebrafish in mg.
Figure EV2. Quality control parameters for cells present in the zebrafish thyroid gland atlas.
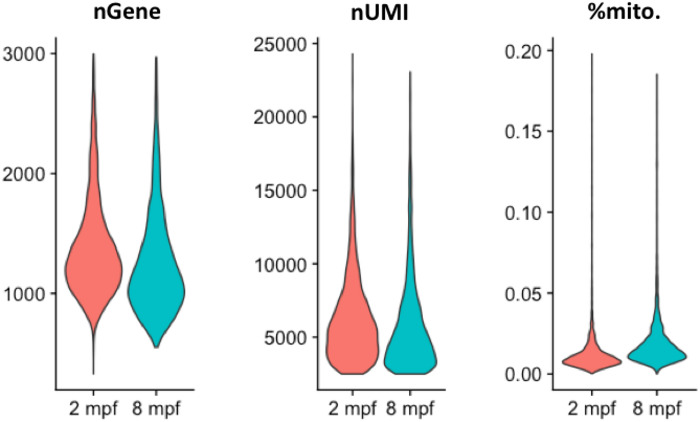
Violin plots depicting the number of genes (nGene), number of unique RNA molecules detected (nUMI—number of Unique Molecule Identifiers) and the percentage of reads mapped onto the mitochondrial genome for the cells profiled in the zebrafish thyroid gland atlas.
Identification of cell types present in the zebrafish thyroid gland
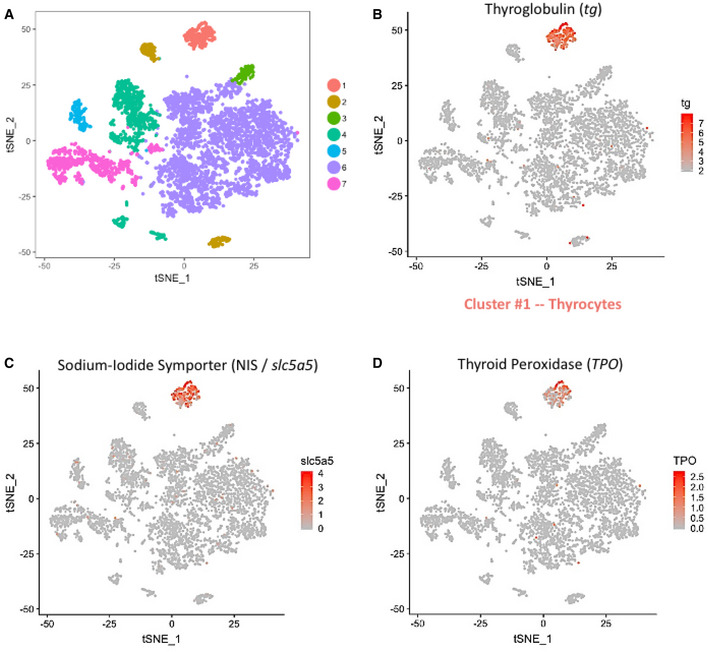
To aid with visualization of the zebrafish thyroid gland single‐cell RNA‐Seq (scRNA‐Seq) data, we projected the cellular profiles onto t‐distributed stochastic neighbor embedding (t‐SNE) plots, a non‐linear dimensionality reduction technique (Maaten & Hinton, 2008) (Fig 2A). Using unsupervised graph‐based clustering, we identified seven clusters for the thyroid gland. Using the expression of genes involved in thyroid hormone production, we could identify one of the clusters as thyroid follicular cells (Fig 2B–D). Specifically, the cluster displayed high relative expression of tg gene, which was further enriched by background correction (Appendix Figure S1), thereby demonstrating that the cells represented differentiated thyroid follicular cells. The cluster, labeled as thyrocytes, contains 267 cells. This represents 4.2% of the total cells recovered after quality control, similar to the proportion of thyrocytes quantified in the dissociated tissue by imaging and FACS (Fig 1E–G), suggesting lack of thyrocyte loss during the sequencing procedure.
Figure 2. Single‐cell RNA‐Seq of the zebrafish thyroid gland.
-
At‐SNE plot displaying the 6,249 single cells profiled in the zebrafish thyroid gland atlas. The colors represent cell clusters denoting a specific cell type.
-
B–DCluster #1 represents the thyrocytes that express tg, slc5a5 (NIS) and tpo. The color scale represents the normalized expression counts for each gene ranging from lowest (gray) to highest (red).
To define the identity of the remaining cell clusters, we generated cluster‐specific marker genes by performing differential gene expression analysis (Fig 3A) (Table EV1). For four clusters, the marker genes included one or more known cell type–specific identifiers. This included gpr182 for endothelial cells; acta2 for musculature; fcer1gl for immune cells; and ponzr3 for cells from zebrafish gills (Fig 3B–E). Based on these cell identifiers, the atlas includes 233 endothelial cells, 135 muscle lineage cells, 914 immune cells, and 199 cells from zebrafish gills. Notably, the endothelial cell cluster includes blood vessels (flt1 and kdrl) and lymphatic vessels (mrc1a, prox1a, flt4, and lyve1b) (Fig EV3); while the immune cell cluster includes macrophages (mpeg1.1 and mfap4), neutrophils (lyz), and lymphocytes (il4, il13, and il11b) (Fig EV4).
Figure 3. Gene expression signature of the different cell types in the zebrafish thyroid gland.
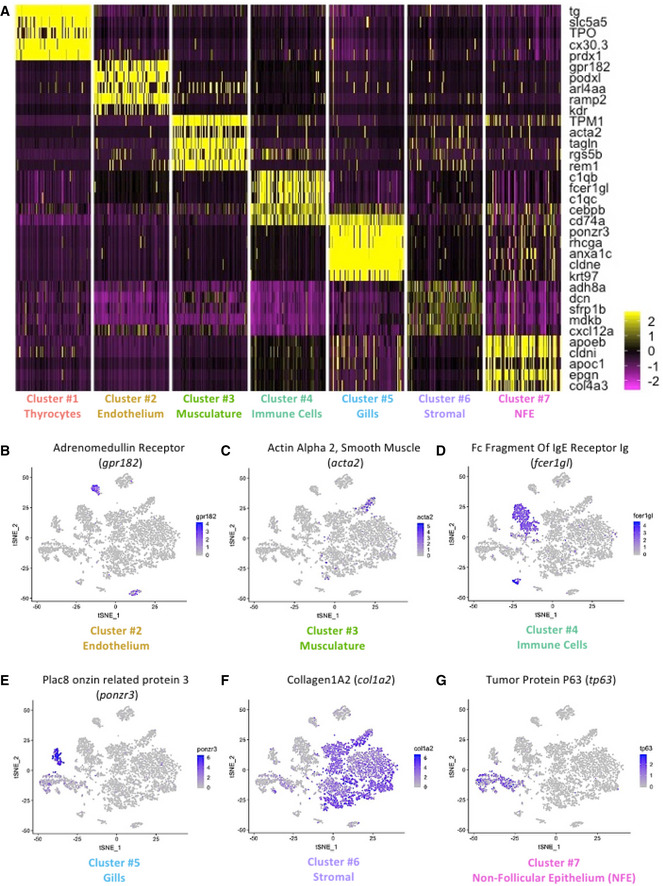
-
AHeatmap depicting five genes specifically expressed in each one of the seven clusters of the zebrafish thyroid gland atlas. The color scale represents expression as a z‐score ranging from −2 (purple) to 2 (yellow). NFE: Non‐Follicular Epithelium.
-
B–Gt‐SNE plots overlaid with the expression of a gene specific to each of the cluster. The endothelium cluster (cluster #2) is a mix of blood vessels and lymphatic vessels (see Appendix Fig S4), while the immune cell cluster (cluster #4) is a mix of macrophages, neutrophils, and lymphocytes (see Appendix Fig S5). The color scale represents the normalized expression counts for each gene ranging from lowest (gray) to highest (blue).
Figure EV3. Cells belonging to blood vessels and lymphatic vessels are present in the zebrafish thyroid gland atlas.
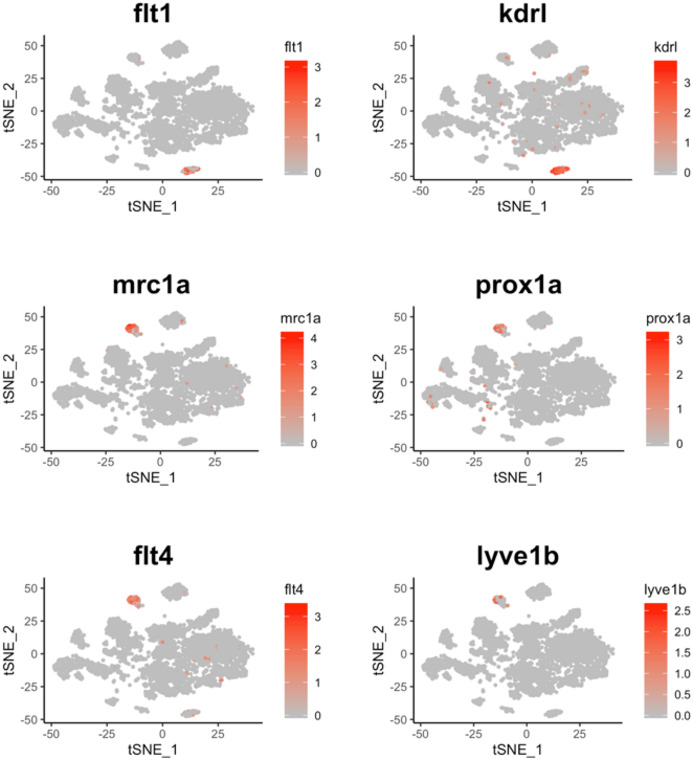
t‐SNE plot overlaid with gene expression for genes specific to blood vessels (flt1 and kdrl) and lymphatic vessels (mrc1a, prox1a, flt4, and lyve1b). The color scale represents the normalized expression counts for each gene ranging from lowest (gray) to highest (red).
Figure EV4. Cells belonging to diverse hematopoietic lineage are present in the zebrafish thyroid gland atlas.
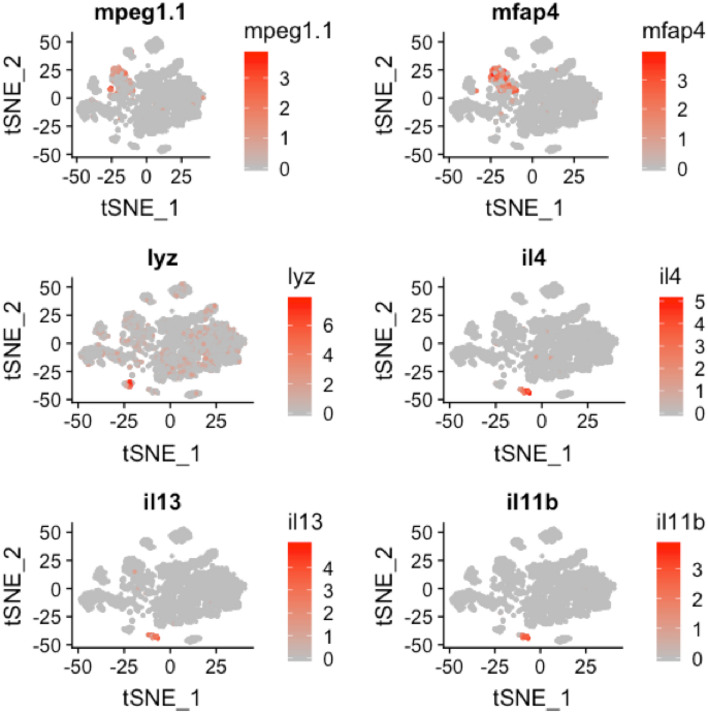
t‐SNE plot overlaid with gene expression for genes specific to macrophages (mpeg1.1 and mfap4), neutrophils (lyz), and lymphocytes (il4, il13, and il11b). The color scale represents the normalized expression counts for each gene ranging from lowest (gray) to highest (red).
For the remaining two clusters (number six and seven), we identified marker genes that hinted toward identity of the cell type. Specifically, col1a2 and tp63 enriched in cluster number six and seven, respectively (Fig 3F–G), are known markers of fibroblasts (Denton et al, 2001; Sánchez‐Iranzo et al, 2018) and epithelial tissue (Barbieri & Pietenpol, 2006; Reischauer et al, 2009; Lisse et al, 2016). We performed gene ontology (GO) enrichment analysis of the marker genes to aid with classification (Appendix Fig S2). Cluster six demonstrated an enrichment of "extracellular matrix structural constituent", "connective tissue development", and "extracellular space", confirming the presence of tissue fibroblasts in this cluster. Thus, we labeled cluster six as "Stromal" cells. Cluster seven displayed an enrichment of "cell motility", "cell migration", and "epithelium development", suggestive of epithelial cells. Notably, cluster seven displayed expression of cytokeratins (krt5, krt14) and E‐cadherin (cdh1) (Table EV1). Hence, we labeled cluster seven as "Non‐Follicular Epithelium (NFE)", to distinguish them from the thyroid follicular epithelial cells. Our data contains 3670 stromal cells and 831 non‐follicular epithelial cells.
We validated the presence of blood vessels, macrophages, and stromal cells in the thyroid gland using tissue‐specific transgenic lines (Fig 4A–C). Immunofluorescence (IF) analysis demonstrated physical proximity between thyrocytes and blood vessels (Fig 4A). Notably, we observed a subset of macrophages in direct contact with thyroid follicles (Fig 4B). In addition, we visualized NFE by immunostaining against TP63 antibody (Fig 4D), which revealed NFE scattered throughout the gills and in the region adjacent to the follicles. Thus, the IF analysis successfully confirmed the presence of different cell types identified in the single‐cell atlas.
Figure 4. Immunofluorescence‐based visualization of cell types surrounding zebrafish thyroid follicles.
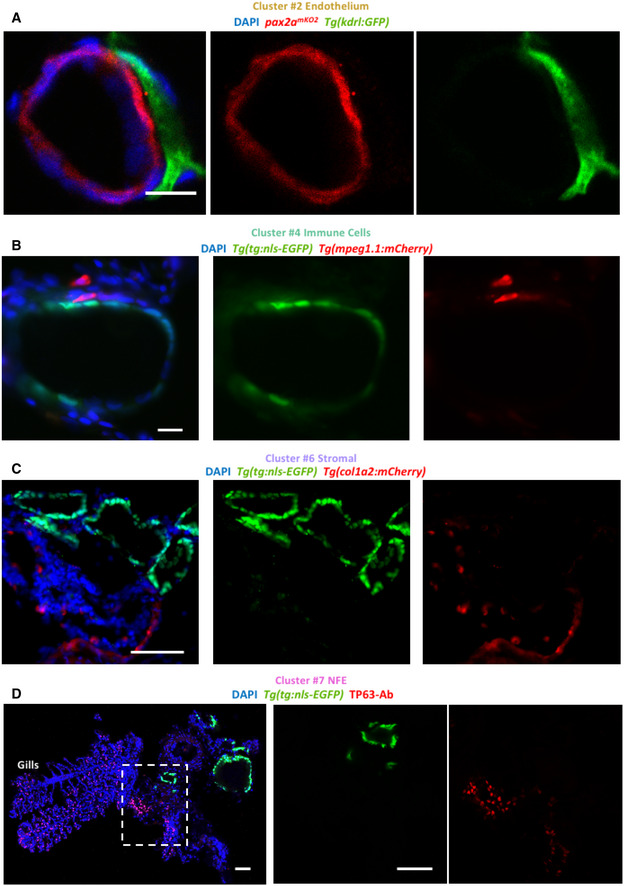
-
A–DImages show immunofluorescence labeling of thyroid gland from adult zebrafish. Transverse sections were utilized for imaging. The organ was isolated from tissue‐specific transgenic lines to allow marking of a particular cell type adjacent to the thyroid follicle. Blood vessels were marked using Tg(kdrl:EGFP) (A), macrophages using Tg(mpeg1.1:mCherry) (B), and stroma using Tg(col1a2:mCherry) (C). Thyrocytes were labeled with pax2amKO2 expression in (A) (described in Fig 7) and Tg(tg:nls‐EGFP) expression in (B, C). (D) NFE was labeled using antibody against TP63 in sections of the thyroid gland isolated from Tg(tg:nls‐EGFP) animals. Gills are marked based on their morphological appearance. DAPI labels nuclei. Scale bars: 10 µm (A, B), 50 µm (C, D).
Our marker gene identification further established additional genes enriched in a single cell type in the thyroid gland (Fig 3A) (Table EV1). For instance, we identified cx30.3, a connexin gene, and prdx1, a gene involved in the antioxidant response, to be specifically expressed in the thyrocytes. To enable further investigation of the clusters and gene expression profiles, we have developed an interactive webtool for online browsing (https://sumeet.shinyapps.io/zfthyroid/).
Development of autocrine and paracrine signaling networks in the thyroid gland using known ligand–receptor interactions
Having defined the cell types of the thyroid gland, we quantified potential cell–cell interactions between thyrocytes and all cell types present in the organ (Fig 5A) based on a reference list of approximately 3,100 literature‐supported interactions containing receptors and ligands from receptor tyrosine kinase (RTK), extracellular matrix (ECM)‐integrin, chemokine, and cytokine families (Ramilowski et al, 2015). Although anatomical barriers between cell types are not modeled in this analysis, we restricted the analysis to secreted ligands for NFE, stroma, and gills—cell types that are physically separated from thyrocytes (Fig 4C–D). For the remaining cell types, secreted and cell membrane‐tethered ligands were considered. The expression patterns of ligand–receptor pairs revealed a dense intercellular communication network (Fig 5B). The network consisted of 272 ligands expressed on different cell types with a corresponding receptor expressed on the thyrocytes (Table EV2). For instance, the stromal cells express the ligand lpl (Lipoprotein Lipase) that signals through the lrp2a (zebrafish homologue of Megalin) receptor (Fig 5C). Stromal and smooth muscle cells express dcn (Decorin) whose receptor met is expressed by thyrocytes. Further, the ligand cyr61 is broadly expressed in the thyroid gland and adjoining gills, with one of its receptors, itgb5, an integrin isoform, expressed specifically by the thyrocytes. The identified interactions also include autocrine signaling. For example, the ligand sema3b and its receptor nrp2a are both present on thyrocytes. GO analysis for identified ligand–receptor pairs revealed genes involved in "PI3K‐Akt signaling pathway", "MET signaling", and "integrin binding" (Appendix Fig S3).
Figure 5. Connectome of the zebrafish thyroid gland identifies a dense intercellular signaling network.
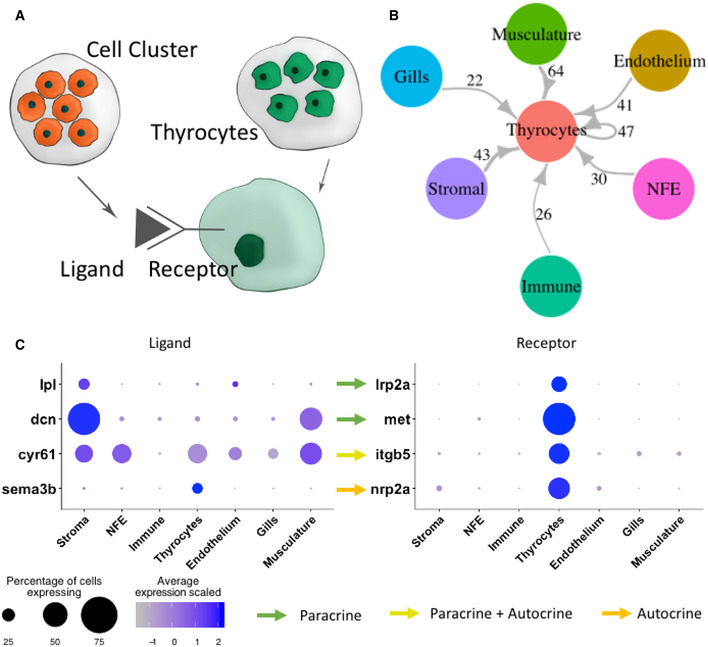
-
ATo build a connectome for the atlas, the ligands expressed specifically in each cell type were matched with their corresponding receptors in the thyrocytes.
-
BA highly connected intercellular interaction network is identified by the connectome. The number of ligand–receptor pairs identified between two cell types is denoted alongside the arrows. For NFE, Gills, and Stromal cells, the connectome was restricted to secreted ligands.
-
CA dotplot depicting examples of paracrine and autocrine signaling in the thyroid gland. The size of the dots represent percentage of cells expressing the gene in a particular cluster, while the color scale represents average expression of the gene in the cluster after scaling.
Thyrocytes display transcriptional heterogeneity
Next, we characterized the transcriptional differences within the thyrocyte population. For this, we bioinformatically isolated the thyrocytes and performed analysis of genetic entropy, a measure of the degree of transcriptional uncertainty. This revealed transcriptional heterogeneity in 231 genes in the thyrocyte population (Fig 6A) (Table EV3). Genes displaying statistically significant entropy (P‐value < 0.05) included pax2a and ctsba. Notably, for the thyrocyte population, the expression of tg displays a unimodal distribution, while the expression of pax2a and ctsba displays bimodal distribution (Fig 6B, Appendix Fig S4).
Figure 6. Thyrocytes display transcriptional heterogeneity.
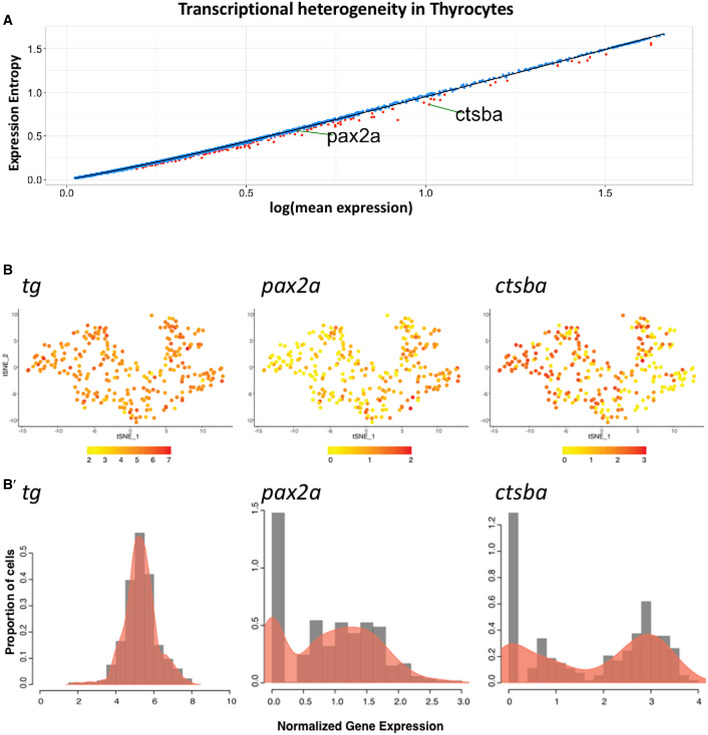
-
ADot plot depicting expression entropy on y‐axis against average gene expression on x‐axis for the thyrocyte population. Each dot depicts a gene, with red dots depicting genes that show statistically significant (P < 0.05) difference in entropy from expected value. Expected value is represented by black regression line. pax2a and ctsba are marked on the graph.
-
Bt‐SNE plot of the thyrocyte cluster with expression of tg,pax2a, and ctsba. The color scale represents the normalized expression counts for each gene ranging from lowest (yellow) to highest (red). B’ Histogram overlaid with density plots depicting the distribution of normalized gene expression for tg, pax2a, and ctsba.
Cathepsin B (mammalian homologue of ctsba) is a cysteine protease that is involved in the processing of iodinated thyroglobulin to T4 and T3 in the thyrocyte lysosomes (Brix et al, 1996, 2001). Moreover, fusion of Cathepsin B and EGFP has been previously used to track thyroid hormone processing lysosomes in rat thyroid epithelial cell lines (Linke, 2002).
The gene pax2a belongs to the PAX (paired box DNA binding) domain containing family of transcription factors. The heterogeneity in pax2a expression is notable, as pax2a is an important regulator of thyrocyte development (Wendl et al, 2002). Zebrafish thyroid primordium expresses pax2a at 24 hpf (Wendl et al, 2002), which is required for specification of the thyroid follicles (Pfeffer et al, 1998; Porazzi et al, 2009). Consequently, zebrafish lacking pax2a fail to develop thyroid follicles (Wendl et al, 2002), which is similar to the Pax8 knockout phenotype in mouse (Mansouri et al, 1998). The low expression of pax2a in a subset of thyrocytes suggests the presence of a thyrocyte subpopulation with a distinct gene expression signature.
Generation of pax2a knock‐in reporter line
To validate the heterogeneity among the zebrafish thyrocytes, we focused on the expression of pax2a transcription factor. We generated a knock‐in line by inserting monomeric Kusabira‐Orange 2 (mKO2) fluorescent protein to the 3’ end of the endogenous pax2a genomic location (Fig 7A). The mKO2 coding sequence was inserted in‐frame with the C terminus of pax2a, separated by a T2A‐cleavable linker. This leads to generation of two proteins, PAX2A and mKO2, from the pax2a regulatory sequence. The two proteins could display differential protein turnover characteristics. The pax2apax2a‐T2A‐mKO2 (abbreviated as pax2amKO2) reporter expression overlapped with PAX2A antibody staining in a majority of regions at 9.5 h post‐fertilization (Fig 7B). Moreover, the knock‐in line displayed mKO2 fluorescence in the otic vesicle, mid‐hindbrain boundary, optic stalk, pronephros, and the thyroid gland (Fig 7C–F, Movie EV1, Appendix Fig S5), mimicking known expression of pax2a during zebrafish development (Kesavan et al, 2017). The difference in levels of PAX2A antibody staining and mKO2 fluorescence could be attributed to different half‐lives of the two proteins. In order to assess whether the dynamics of mKO2 expression would follow modifications in the expression of endogenous pax2a, we used CRISPR/Cas9 technology to generate F0 knockouts (also known as Crispant (Trubiroha et al, 2018)) of pax2a gene in our pax2a mKO2 line. The crispants displayed defects in thyroid morphogenesis (Fig 7G and H), mimicking the phenotype of pax2a loss‐of‐function mutation (Wendl et al, 2002). Live imaging of crispants at 55 hpf revealed strong decrease of mKO2 expression (Fig 7G and H), thereby corroborating the faithful recapitulation of pax2a expression by the newly generated reporter line.
Figure 7. pax2amKO2 knock‐in line faithfully reports pax2a expression and knock‐down.
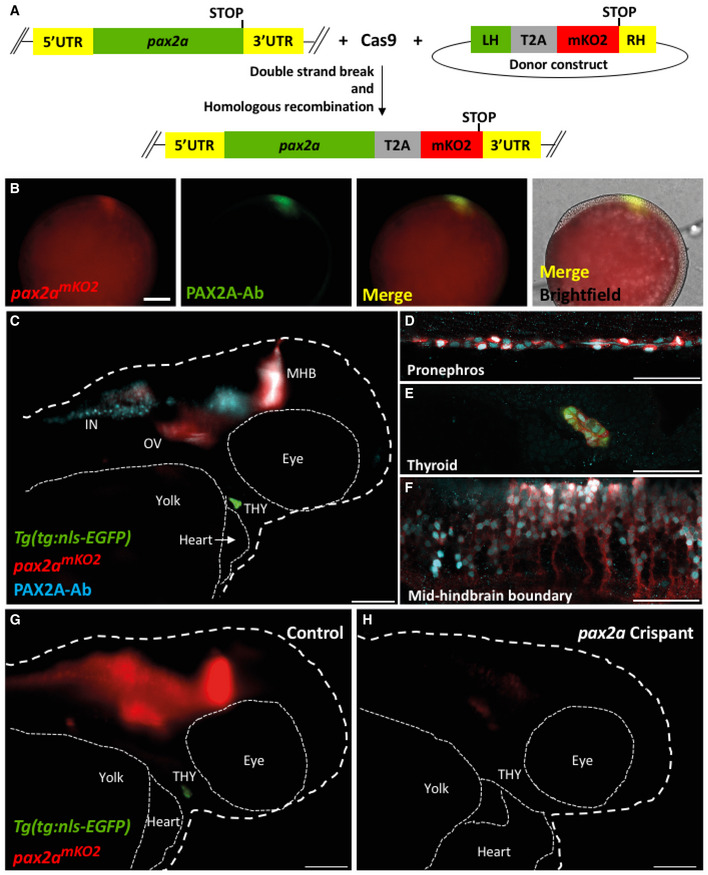
-
ASchematic of the knock‐in strategy used to generate the pax2amKO2 line. Double strand break was induced between the penultimate codon and the STOP codon of pax2a gene using CRISPR/Cas9. DNA repair integrates the donor construct at the site of double strand break, resulting in a pax2a reporter line. The donor construct contains T2A‐mKO2 reporter cassette flanked by left homology (LH) and right homology (RH) arms.
-
BWhole mount immunofluorescence of 9.5hpf pax2a mKO2 embryos stained with anti‐mKO2 antibody (red) and anti‐PAX2A antibody (green). Anterior is to the left, and dorsal side is to the top. Scale bar: 0.1 mm.
-
CWhole mount immunofluorescence of 55 hpf pax2amKO2; Tg(tg:nls‐EGFP) stained with PAX2A antibody (PAX2A‐Ab) displays an overlap of mKO2 and PAX2A‐Ab signal. The otic vesicle (OV), mid‐hindbrain barrier (MHB), interneurons (IN), and thyroid gland (THY) is labeled. White dashed line represents the outline of the zebrafish larvae. Scale bar: 100 µm. Anterior to the right.
-
D–FConfocal microscopy imaging of a sagittal section of a 55 hpf pax2amKO2 (red); Tg(tg:nls‐EGFP) (green) embryos immunostained with PAX2A antibody (cyan) showing co‐localization of mKO2 and pax2a in the pronephros (D), thyroid gland (E), and mid‐hindbrain barrier (F). In the thyroid gland, mKO2 (red), PAX2A‐Ab (cyan), and thyrocyte‐specific GFP (green) show co‐localization. Scale bar: 50 µm.
-
G, HSnapshots from live imaging of 55 hpf pax2a mKO2; Tg(tg:nls‐EGFP) embryos injected with sgRNA targeting pax2a coding sequence. The anterior part of a representative control embryo (G) is shown alongside a representative crispant (H). Crispants display a strong reduction of mKO2 fluorescence, as well as an absence of GFP signal suggesting absence of thyroid (THY) tissue. White dashed line represents the outline of the zebrafish larvae. Scale bar: 100 µm. Anterior to the right.
Segregation of thyrocyte subpopulations based on pax2a reporter expression
Upon investigating the fluorescence expression of the pax2a reporter in the thyroid gland of adult zebrafish, we found strong and specific expression of pax2a reporter in the thyrocytes lining the thyroid follicles (Fig 8A–D). Although a majority of thyrocytes displayed uniform expression of pax2a reporter, we could identify a small population of pax2amKO2‐Low thyrocytes (Fig 8B–D). The pax2amKO2‐Low thyrocytes were not segregated, but scattered throughout the gland, thereby suggesting a mixing of the two thyrocyte subpopulations.
Figure 8. Immunofluorescence‐based validation of thyrocyte heterogeneity.
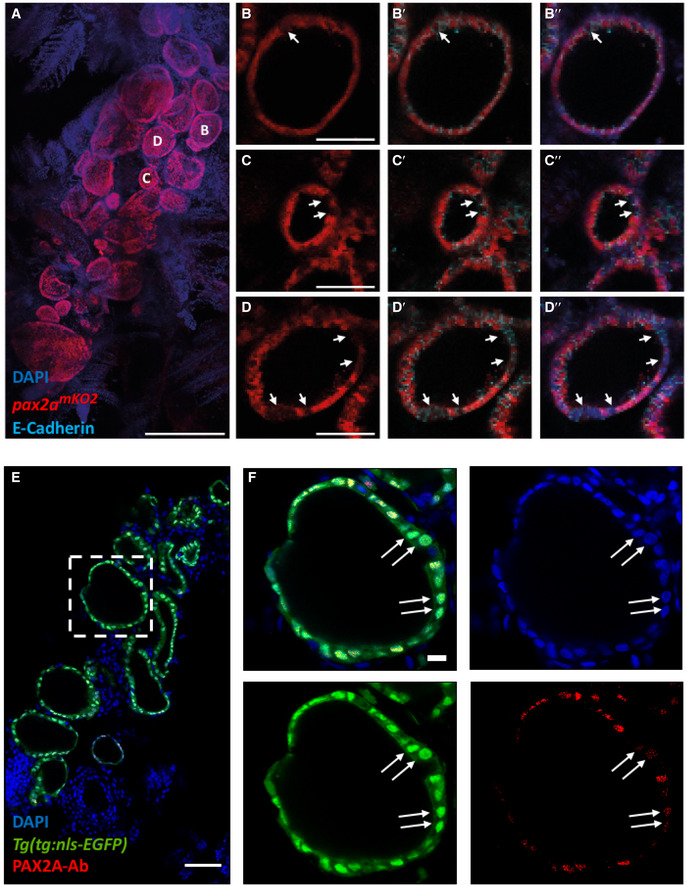
-
A–DAnalysis of 3 mpf thyroid gland from pax2a mKO2 zebrafish shows heterogeneity in pax2a reporter expression. (A) Whole mount confocal imaging of a 3 mpf pax2a mKO2 thyroid labeled with mKO2 (red), E‐cadherin (cyan, not shown in "A" for clarity reasons) and DAPI (dark blue) for nuclear localization. (B–D) Optical sections of three follicles, with mKO2‐Low cells labeled with arrows. E‐cadherin (B’–D’) and DAPI (B’’–D’’) staining shows that absence of mKO2 signal does not correspond to an absence of cells. Anterior to the bottom of the pictures.
-
E, F(E) Confocal image of thyroid gland section from Tg(tg:nls‐EGFP) at 4 mpf stained with PAX2A antibody and DAPI. The dotted region is displayed at high magnification in (F). Arrows marks thyrocytes displaying low PAX2A staining. Notably, PAX2A‐Low thyrocytes display tg‐driven EGFP expression, demonstrating their differentiated status. Data information: Scale bars: 250 µm (A), 50 µm (B–E), 10 µm (F).
To validate pax2a expression heterogeneity at a protein level, we performed immunostaining against PAX2A in thyroid glands obtained from Tg(tg:nls‐EGFP) animals (Fig 8E). For antibody staining, we utilized 8 µm thin sections of the thyroid gland to ensure uniform antibody penetration to all cells. Confocal imaging of the stained sections demonstrated the presence of PAX2A‐Low and PAX2A‐High thyrocytes (Fig 8F). Notably, both PAX2A‐Low and PAX2A‐High cells display tg promoter‐driven EGFP expression, thereby confirming their differentiated status.
Further, to quantify the proportions of pax2amKO2‐Low and pax2amKO2‐High thyrocytes, we performed FACS analysis on pax2amKO2; Tg(tg:nls‐EGFP) double transgenic line (Fig 9A–C). The Tg(tg:nls‐EGFP) zebrafish line labels the thyrocyte population in green fluorescence (Trubiroha et al, 2018). We restricted our analysis to the thyrocyte population by gating for GFP+ cells in the thyroid gland (Fig 9A). Within the thyrocyte population, the cells displayed a normal distribution of GFP fluorescence; however, thyrocytes could be split into two subpopulations based on the levels of pax2a reporter expression (Fig 9B and C). Specifically, 75% of thyrocytes (202 out of 268 cells) displayed pax2amKO2‐High fluorescence, while 25% of thyrocytes (66 out of 268 cells) displayed pax2amKO2‐Low fluorescence levels.
Figure 9. Transcriptome profiling of pax2amKO2‐High and pax2amKO2‐Low thyrocytes.
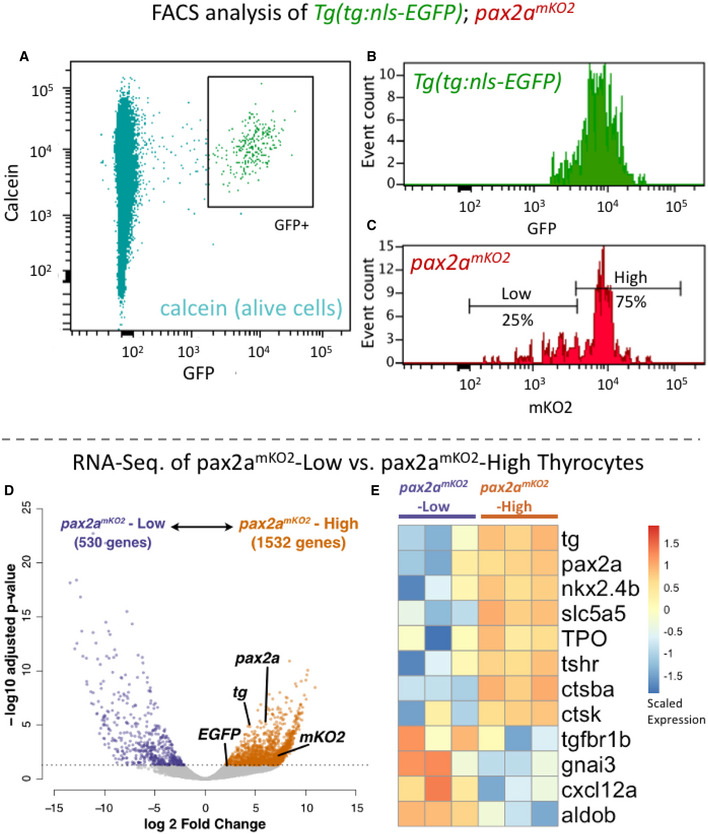
-
A–CCells from the thyroid gland of 5 mpf Tg(tg:nls‐GFP); pax2amKO2 animals were labeled with calcein (live cell marker) and analyzed using FACS. (A) A FACS plot showing calcein on y‐axis and GFP on x‐axis. The box encompassing the GFP+ cells represents the thyrocyte population, which was gated for further analysis. (B) Histogram showing the distribution of GFP intensity in thyrocytes. (C) Histogram showing the distribution of mKO2 intensity in thyrocytes. Thyrocytes were selected by gating for GFP+ population. Horizontal lines indicate the mKO2‐Low and mKO2‐High expression level, with percentage values representing proportion of thyrocytes with mKO2‐Low and mKO2‐High expression.
-
D, ERNA‐Seq analysis of transcriptome isolated from pax2a mKO2‐High and pax2amKO2‐Low subpopulations. (D) Volcano plot representing the differential expression of genes between the two thyrocyte subpopulations. Dots for pax2a, tg, mKO2, and EGFP are marked. Dots for genes that display significant differential expression (DESeq2, adjusted P‐value < 0.05) are colored. (E) Heatmap displaying the expression of selected genes in the two subpopulations. Biological replicates are represented by individual columns.
Next‐generation sequencing of pax2amKO2‐High and pax2amKO2‐Low thyrocytes
To investigate molecular factors that differ between pax2a mKO2‐High and pax2a mKO2‐Low thyrocytes, we performed bulk RNA sequencing on FACS‐enriched subpopulations (Fig 9C). Three biological replicates were sequenced at a depth of 10 million reads for each subpopulation. During analysis, quality control was performed by filtering genes that displayed expression lower than 0.5 counts per million (CPM) in all six samples. This provided us with expression levels of 10,062 genes for differential gene expression analysis, which revealed differential expression of 2062 genes (Fig 9D) (Table EV4). This corresponds to changes in 20.5 % of detected transcriptome between the two subpopulations.
Specifically, pax2a mKO2‐High population displayed enrichment in the expression of pax2a, mKO2 (driven from pax2a locus), tg and EGFP (driven from tg locus) (Fig 9D). Differential expression of tg was observed in‐spite of the fact that tg was detected as one of the top 15 highly expressed gene in both pax2a mKO2‐High and pax2a mKO2‐Low populations (Fig EV5). The high expression of tg in both populations confirms the identity of the cells as thyrocytes.
Figure EV5. Gene expression in pax2a mKO2‐Low and pax2a mKO2‐High thyrocyte subpopulations.
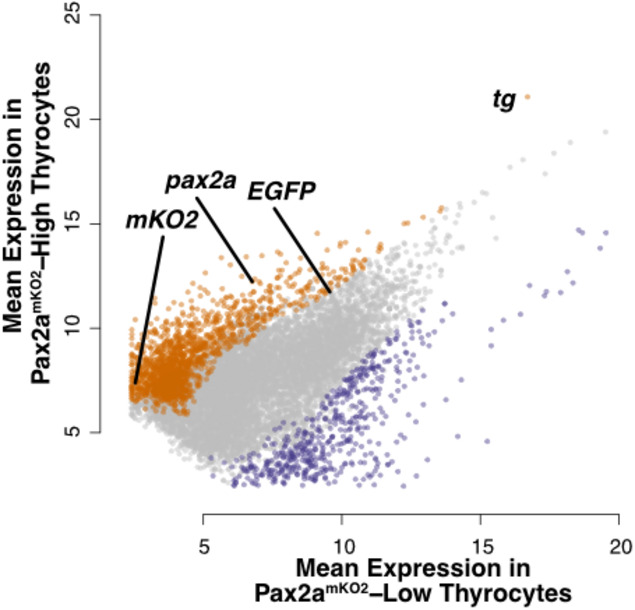
Dotplot depicting the mean expression of genes in pax2a mKO2‐High (y‐axis) and pax2amKO2‐Low (x‐axis) subpopulations. Genes enriched in pax2a mKO2‐High and pax2a mKO2‐Low subpopulations are labeled in orange and violet, respectively. Dots representing pax2a, tg, mKO2, and EGFP are marked.
In addition, pax2a mKO2‐High population displayed enrichment in various genes involved in thyrocyte function, including nkx2.4b, slc5a5 (NIS), TPO, tshr, ctsba, and ctsk (Cathepsin K) (Fig 9E). Gene ontology (GO) analysis revealed enrichment in genes related to "translation", "macromolecule biosynthetic process", "focal adhesion", and "MAPK signaling" (Appendix Fig S6). The pax2a mKO2‐Low population, in contrast, displayed enrichment in genes related to "Oxidative phosphorylation", "Inositol phosphate metabolism", and "ATP metabolism". Notably, pax2a mKO2‐Low population had higher expression of tgfbr1b (receptor for TGF‐ß pathway), gnai3 (an inhibitor of cAMP activity), aldob (enzyme involved in glycolytic process), and cxcl12a (a cytokine) (Fig 9E).
In summary, RNA‐Seq of thyrocyte population segregated by pax2a expression levels, along with the analysis of pax2a knock‐in line and immunofluorescence imaging of PAX2A protein, validates the identification of thyrocyte heterogeneity within our single‐cell RNA‐Seq data and clearly demonstrates, for the first time, the presence of transcriptionally diverse subpopulations of thyrocytes present in the thyroid gland.
Discussion
Previous studies on mammalian thyroid gland have identified functional and morphological heterogeneity between the thyroid follicles (Smeds et al, 1987; Struder et al, 1989; Baptist et al, 1995). Using histological analysis, functionally active follicles were identified as being outlined with tall columnar thyrocytes, while inactive follicles were marked by an outline of low cuboidal to almost squamous thyrocytes (Studer et al, 1978; Gerber et al, 1987). However, two open questions remain unanswered in the field: (a) What are the genes responsible for the functional heterogeneity; and (b) Do all thyrocytes in a follicle display uniform functional capacity. Here, by applying unbiased single‐cell gene expression analysis to the thyroid gland for the first time, we have identified transcriptional heterogeneity within the thyrocyte population. Further, we demonstrate that transcriptionally diverse thyrocytes are present in the same follicle. Thyrocytes differ in the expression levels of a transcription factor pax2a and a cysteine protease Cathepsin B (ctsba) (Fig 6), among other genes (Table EV3). Cathepsin B is particularly notable as it enables the liberation of thyroid hormone from thyrocytes by proteolytic processing of thyroglobulin (Brix et al, 1996, 2001).
We validate the heterogeneity among the thyrocytes using a newly generated knock‐in reporter line for pax2a gene (Fig 7). The knock‐in reporter line was generated using CRISPR/Cas9‐based insertion of mKO2 fluorescent protein in the endogenous pax2a genomic location. The pax2a knock‐in line faithfully recapitulates the embryonic expression of pax2a gene (Movie EV1, Fig 7B–F). Using the pax2a reporter line to characterize the adult thyroid gland, we demonstrate the presence of pax2amKO2‐Low thyrocytes in the follicles (Fig 8). Notably, pax2amKO2‐Low and pax2amKO2‐High thyrocytes are present in the same follicle (Fig 8C, D and F), raising the possibility of contact‐mediated interactions between the two subpopulations.
Further, the knock‐in line allowed us to segregate the thyrocyte population based on pax2a expression levels (Fig 9A–C). RNA‐Seq of the two subpopulations revealed enrichment of thyrocyte functional machinery in the pax2a mKO2‐High population (Fig 9E). Our result suggests that the gland may be divided into functional and resting thyrocytes. It would be of interest to build on this study and investigate the functional and replicative differences among the two subpopulations of thyrocytes. Moreover, it would be important to elucidate the dynamics and plasticity between the two subpopulations as thyrocytes might shuffle between the two states.
Our single‐cell transcriptomics atlas provides a comprehensive genomics resource to study the zebrafish thyroid gland in unprecedented detail. We performed unbiased profiling of the thyroid gland, without enrichment for a specific cell type. This allowed us to capture yet poorly characterized cell populations within the thyroid gland. Specifically, we provide the molecular characteristics of the stromal tissue present in the zebrafish thyroid gland. The stromal cells (Fig 4C) display enrichment of extracellular matrix (ECM)‐related genes (Table EV1) and are possibly homologous to the mesenchymal connective septa found in the mammalian thyroid gland. The connective septa help cluster the thyroid follicles into lobules. Notably, the expression of fgf ligands from the mesenchymal septa cells has been implicated in lobe formation during mouse thyroid gland development (Liang et al, 2018). It would be of interest to test whether similar morphological clustering of the thyroid follicles exists in zebrafish, and the role the stromal cells play during development and growth of the thyrocytes.
Our atlas further identifies a non‐follicular epithelial (NFE) cell population present near the zebrafish thyroid follicles. A subset of NFE are present in the gills (Fig 4D) and may potentially represent a progenitor population for the gills, similar to the TP63+ basal layer in the zebrafish (Guzman et al, 2013) and mammalian (Yang et al, 1999) epithelium. We also observe NFE outside the gills (Fig 4D), which may play a different role. It is interesting to note that epithelial cells apart from follicular and parafollicular cells have been observed in the mammalian thyroid gland. In a report from Dr. E. Baber published in 1876 (Baber, 1876), histological examination of the dog thyroid gland displayed the presence of cells “beside the stroma, lymphatics, blood vessels, & cells between the vesicles”. Dr. Baber labeled the cells as "parenchyma" and noted the existence of “numerous cells differing markedly in size and shape from the epithelial cells amongst which they lie” (Baber, 1876). In 1907, Dr. Sophia Getzowa described an epithelial cells containing structure called the Solid Cell Nests (SCN) of the thyroid (Getzowa, 1907). SCN are lumen containing irregular structures located within the thyroid in mammals (Harach, 1988). SCN contain two types of epithelial cells: main cells and C‐cells, expressing TP63 and calcitonin respectively (Ríos Moreno et al, 2011). Notably, the NFE cells we identified in the zebrafish thyroid gland are marked with TP63 expression (Fig 3G, 4D), raising the possibility of their homology with the main cells of the SCN. C‐cells, however, exist in the ultimobranchial bodies, which lies outside the thyroid gland in zebrafish. The ultimobranchial bodies are the zebrafish homologues of parafollicular cells and are located as a pair of follicles on top the sinus venous, adjacent to the atrium and esophagus (Alt et al, 2006). Cells adjacent to the atrium were removed during our dissections (Fig 1B). Additionally, NFE cells identified in our atlas do not express the zebrafish homologue of calcitonin (calca) (Table EV1). Thus, it is unlikely that the NFE cells we have identified would be related to cells of parafollicular origin. Currently, the developmental origin of NFE cells and their role in thyroid gland is unclear. To study the cell population, transgenic zebrafish reagents driving expression using the tp63 regulatory region (Rasmussen et al, 2015) could be utilized in future.
To survey the communication between thyrocytes, the functional unit of the thyroid gland, and the other cell types present in the thyroid gland, we constructed a cellular interaction network (Fig 5B). The network was built by matching the expression of ligands in the diverse cell types with the expression of receptor in the thyrocytes (Table EV2) (Ramilowski et al, 2015). Based on literature survey, we manually identified multiple interacting genes that have been implicated in thyroid diseases. For instance, the ligand Decorin (dcn) is expressed by the stromal cells and musculature, with its receptor, MET, present on thyrocytes (Fig 5C). Decorin, a secreted proteoglycan, is considered a “guardian from the matrix” (Neill et al, 2012), as it is an antagonist of growth factor signaling. Importantly, Decorin expression has been reported to be downregulated in thyroid cancer samples (Martínez‐Aguilar et al, 2016). Thus, stromal cells could modulate Decorin to control thyrocyte growth. Further, interactions for CYR61 (associated with Graves’ Disease (Planck et al, 2014)), LRP2/ Megalin (involved in thyroglobulin processing (Marinò & McCluskey, 2000)), and NRP2 (associated with thyroid cancer metastasis (Tu et al, 2016)) were identified (Fig 3C). The hypothesis generated by the theoretical ligand–receptor interaction network can be tested in vivo in zebrafish or in vitro by manipulation of thyrocytes in thyroid organoid models (Antonica et al, 2012) to gain valuable insight into thyroid gland homeostasis.
The current atlas is restricted to healthy juvenile and adult thyroid gland. The two stages represent a period of slow growth in zebrafish. Thus, genes driving cellular proliferation might be repressed at these stages. Additionally, the low number of cells per cluster obtained at each stage restricts an in‐depth analysis of the transcriptional difference with age. In future, it would be of interest to extend the atlas by increasing cell numbers and by including single‐cell transcriptomics from embryonic and old fish, providing a comprehensive resource for development, homeostasis and aging of the thyroid gland. It would be of further interest to profile zebrafish models of thyroid disorder (Anelli et al, 2017; Chopra et al, 2019) to understand the cellular and molecular changes underlying organ dysfunction. Combined with the power of CRISPR/Cas9 based screen that we have established for the thyroid gland (Trubiroha et al, 2018), this resource will provide a roadmap for the functional elucidation of cell type–specific programs during thyroid gland growth and homeostasis.
In summary, our work provides the first molecular map of the zebrafish thyroid gland at cellular resolution. The atlas contains the molecular characterization of the thyroid gland stromal population and identification of non‐follicular epithelial cells and demonstrate the transcriptional heterogeneity among zebrafish thyrocytes. Further, by constructing cell–cell communication network, the atlas provides clues into tissue dynamics present within the organ. Finally, the dataset has been made available for browsing via an interactive webtool (https://sumeet.shinyapps.io/zfthyroid/). We hope that our efforts will expand the understanding of thyrocytes as a collection of heterogenous endocrine cell population, providing a complex picture of the diversity in thyrocyte identity and function.
Materials and Methods
Zebrafish strains and husbandry
Wild‐type or transgenic zebrafish of the outbred AB, WIK, or a hybrid WIK/AB strain were used in all experiments. Zebrafish were raised under standard conditions at 28 °C. Animals were chosen at random for all experiments. Published transgenic strains used in this study were Tg(tg:nls‐mVenus‐T2A‐NTR) (McMenamin et al, 2014), Tg(tg:nls‐EGFP) (Trubiroha et al, 2018), Tg(kdrl:GFP) la116 (Choi et al, 2007), Tg(mpeg1.1:mCherry) gl23 (Ellett et al, 2011), and Tg(col1a2:LOXP‐mCherry‐NTR)cn11 (referred as Tg(Col1a2:mCherry)) (Sánchez‐Iranzo et al, 2018). Experiments with Tg(tg:nls‐mVenus‐T2A‐NTR) were conducted in accordance with the Animal Welfare Act and with permission of the Landesdirektion Sachsen, Germany (DD24‐5131/346/11, DD24‐5131/346/12, DD24.1‐5131/476/2, TVV21/2018, and all corresponding amendments). Zebrafish husbandry and experiments with all transgenic lines will be performed under standard conditions as per the Federation of European Laboratory Animal Science Associations (FELASA) guidelines (Aleström et al, 2020), and in accordance with institutional (Université Libre de Bruxelles (ULB)) and national ethical and animal welfare guidelines and regulation, which were approved by the ethical committee for animal welfare (CEBEA) from the Université Libre de Bruxelles (protocols 578N‐579N).
Dissection of the zebrafish thyroid gland
The dissection of thyroid gland in zebrafish was performed by using the ventral aorta as a reference (Fig 1A and B). In zebrafish, the thyroid follicles sit loosely in soft tissue around the ventral aorta. Ventral aorta connects to the outflow tract that further joins with the heart ventricle. During dissociation, cells connected to the ventral aorta, including parts of zebrafish gills (Fig 1C) were kept intact to avoid injuring the organ during dissociation.
In detail, zebrafish were euthanized in 0.2% Tricaine (MS‐222, Sigma E10521) solution. Using fine forceps, the lower jaw was separated from the upper jaw and disconnected from the gut by pinching near the gills. The dissected tissue was carefully cleaned by removing muscle, skin, pectoral fin, and lateral cartilages of the lower jaw. The cleaned tissue when observed from the ventral side under brightfield clearly shows the ventral aorta as a thick pink blood vessel extending from the heart toward the basibranchial cartilage (Fig 1A). Next, the surrounding gills are pinched off using fine forceps, taking care to keep the ventral aorta intact (Fig 1B). This leaves small parts of gills connected to the ventral aorta (Fig 1C). Lastly, the ventral aorta is disconnected from the outflow tract by pinching with fine forceps (dashed line in Fig 1B).
Single‐cell suspension of zebrafish thyroid gland
Single‐cell suspension of zebrafish thyroid gland was performed by adapting the cell dissociation protocol outlined in Singh et al., Scientific Reports, 2018 (Singh et al, 2018). In brief, the thyroid gland was collected and dissociated into single cells by incubation in TrypLE (Thermo Fisher, 12563029) with 0.1% Pluronic F‐68 (Thermo Fisher, 24040032) at 37°C in a benchtop shaker (Biosan TS‐100) set at 450 rpm for 45 min. Following dissociation, TrypLE was inactivated with 10% FBS, and the cells pelleted by centrifugation at 500 g for 10 min at 4°C. The supernatant was carefully discarded and the pellet re‐suspended in 500 μl of HBSS (without Ca, Mg) + 0.1% Pluronic F‐68. To remove debris, the solution was passed over a 30 µm cell filter (Miltenyi Biotec, 130‐041‐407). To remove dead cells, calcein violet (Thermo Fisher, C34858) was added at a final concentration of 1 µM and the cell suspension incubated at room temperature for 20 min. The single‐cell preparation was sorted with the appropriate gates, including excitation with UV (405 nm) laser for identification of alive cells (calcein+) (Fig 1F and G). FACS was performed through 100 µm nozzle.
Single‐cell profiling of the zebrafish thyroid gland
For single‐cell RNA‐seq of the zebrafish thyroid gland using the 10× Genomics platform, cell suspension was prepared as mentioned above from the thyroid glands of six 2‐month post‐fertilization and six 8‐month post‐fertilization Tg(tg:nls‐mVenus‐T2A‐NTR) animals. The cell suspension was adjusted with Hanks’ Balanced Salt Solution (without calcium and magnesium) to a density of 800 cells/µl and diluted with nuclease‐free water according to the manufacturer’s instructions to yield 12,000 cells. Subsequently, the cells were carefully mixed with reverse transcription mix before loading the cells on the 10× Genomics Chromium system (Zheng et al, 2017). After the gel emulsion bead suspension underwent the reverse transcription reaction, emulsion was broken and DNA purified using Silane beads. The complementary DNA was amplified with 10 cycles, following the guidelines of the 10× Genomics user manual. The 10× Genomics single‐cell RNA‐seq library preparation—involving fragmentation, dA tailing, adapter ligation, and indexing PCR—was performed based on the manufacturer’s protocol. After quantification, the libraries were sequenced on an Illumina NextSeq 550 machine using a HighOutput flowcell in paired‐end mode (R1: 26 cycles; I1: 8 cycles; R2: 57 cycles), thus generating ~45 mio fragments. The raw sequencing data were then processed with the "count" command of the Cell Ranger software (v.2.1.0) provided by 10× Genomics with the option "–expect‐cells" set to 10,000 (all other options were used as per default). To build the reference for Cell Ranger, zebrafish genome (GRCz10) and gene annotation (Ensembl 87) were downloaded from Ensembl and the annotation was filtered with the "mkgtf " command of Cell Ranger (options: "–attribute = gene_biotype:protein_coding– attribute = gene_biotype:lincRNA –attribute = gene_biotype:antisense"). Genome sequence and filtered annotation were then used as input to the "mkref " command of Cell Ranger to build the appropriate Cell Ranger Reference.
Analysis of single‐cell RNA‐Seq of the zebrafish thyroid gland
The raw data generated from 10× Chromium pipeline was clustered using Seurat 2.3.4 (Butler et al, 2018) using the recommended analysis pipeline. Briefly, the raw data as UMI counts were log‐normalized, regressed to remove the effect of library size and mitochondrial counts, and scaled. Highly variable genes were identified for PCA analysis and graph‐based clustering using shared nearest neighbor (SNN). For clustering, the first five principal components (PCs) were utilized as they displayed significant deviation from uniform distribution as accessed by JackStraw analysis. Further, a resolution of 0.3 for SNN was used for clustering. Marker genes identified for each cluster were used to classify the cell type.
Development of intercellular signaling network
Development of intercellular signaling network for zebrafish was performed as described in Cosacak et al (2019). Briefly, ligands expressed in 20% of a cell population were identified. A connection between cell type and thyrocyte was made if the expression of the corresponding receptor was identified in 20% of thyrocytes. The connectome contains secreted and membrane‐tethered ligands. For cell types that do not physically contact the thyrocytes (gills, NFE and stroma), membrane‐tethered ligands were manually removed from the connectome.
Background correction for thyrocyte gene expression
Supervised background correction for the thyrocyte population was performed using DecontX (Yang et al, 2020). As input, normalized data and clustering information from Seurat were used. The method using Bayesian approach to model gene expression as a mixture of expression in the expected cell population plus background expression accessed from remaining cell types. Background noise is removed, which likely resembles free mRNA released from injured and ruptured cells. As background correction required clustering information, the corrected data were not utilized for re‐clustering to avoid circular use of the data.
Genetic entropy analysis for thyrocyte population
Quantification of genetic entropy was performed using ROGUE (Ratio of Global Unshifted Entropy; preprint: Liu et al, 2019). As input, raw counts of thyrocytes that passed quality control were used. Default parameters were used for analysis. The algorithm provided a measure of entropy (degree of uncertainty/ heterogeneity), along with P‐value, within the population.
Generation of knock‐in pax2apax2a‐T2A‐mKO2 zebrafish line
For generation of pax2a reporter line, we designed a single‐guide RNA (sgRNA) targeting the STOP codon of the pax2a coding sequence (GCTGCGATGGTAACTAGTGG). We then generated a donor construct in which the sequence encoding for the monomeric Kusabira‐Orange (mKO2) protein was fused to a viral 2A peptide linker. This reporter cassette was flanked by left (1,000 bp) and right (2000bp) homology arms of the pax2a genomic DNA region around the stop codon therefore preventing the sgRNA from cutting the donor construct. sgRNA design, production, and validation were done as previously described (Varshney et al, 2015; Trubiroha et al, 2018). Wild‐type embryos were injected with 3 nl of the injection mix containing the sgRNA (final concentration 80 ng/µl), the donor construct (final concentration 7.5 ng/µl), the protein Cas9 (recombinant cas protein from S. pyogenes PNA Bio CP01, final concentration 100 ng/µl), and KCL (final concentration 200 mM). Upon homologous recombination of this reporter construct in the endogenous locus, pax2a‐expressing cells were fluorescently labeled by mKO2. This pax2apax2a ‐T2A‐mKO2 line is referenced as pax2a mKO2 in the text.
Generation of pax2a crispants
Somatic mutagenesis of pax2a gene was carried out exactly as mentioned in Trubiroha et al (2018). Briefly, sgRNA targeting the exon 2 of pax2a was generated as described in the publication. Following the strategy described in the publication, Cas9 protein along with sgRNA was injected in one‐cell stage of zebrafish embryos for disruption of pax2a gene. Non‐injected animals were used as controls.
Tissue collection
To facilitate confocal imaging of the thyroid gland, the organ was manually dissected from fish as previously described and fixed. Fish were killed in Tricaine followed by dissection of the gland, which was fixed by immersion in 4% paraformaldehyde (PFA) + 1% Triton X overnight at 4°C. The gland was washed 2–3 times in PBS to remove PFA before proceeding.
Quantification of proportion of thyrocytes within the dissected tissue
To quantify the proportion of thyrocytes within the dissected tissue, the gland was dissected from Tg(tg:nls‐EGFP) animals and fixed as described above. The fixed tissue was permeabilized by three washes with 1% PBT (1× PBS + 1% Triton X‐100). Nuclei were stained by immersing the tissue in 1 µg/ml Hoechst prepared in 1× PBS for 2 h at room temperature. The tissue was immersed in 30% sucrose solution overnight at 4°C, embedded in Tissue Freezing Medium (Leica 14020108926) and frozen at −80°C. Thin sections (8 µm) were obtained using cryostat (Leica CM3050 S), collected on frosted glass slides (Thermo Scientific 12362098), and covered with glass coverslip of #1 thickness (Carl Roth GmbH NK79.1) using mounting media (Dako S3023). The sections were imaged on Zeiss LSM 780 confocal microscope. Confocal images were analyzed in Fiji using the following step: threshold using "IsoData" to distinguish signal from background, "watershed" transformation to separate joined nuclei and "measure" function to obtain nuclei count. With this, the green channel (number of thyrocyte nuclei) and blue channel (total number of nuclei) was measured for seven transverse sections obtained from three animals. Percentage was calculated by taking the ratio of thyrocyte nuclei to total nuclei.
Immunofluorescence and image acquisition
Whole mount immunofluorescence was performed on thyroid gland collected as described above. The collected samples were permeabilized in 1% PBT (Triton X‐100) and blocked in 4% PBTB (BSA). Primary and secondary antibody stainings were performed overnight at 4°C. Primary antibodies used in this study were anti‐PAX2A (rabbit, Genetex GTX128127) at 1:250, anti‐EGFP (chicken, Abcam ab13970) at 1:1,000, anti‐E‐Cadherin (mouse, BD bioscience cat 610181) at 1:200, anti‐monomeric Kusabira‐Orange 2 (mouse, MBL amalgam M‐168‐3M) at 1:200, anti‐monomeric Kusabira‐Orange 2 (rabbit, MBL amalgam PM051M) at 1:250, and anti‐p63 (mouse, Santa Cruz Biotechnology 4A4) at 1:200. Secondary antibodies at 1:250 dilutions used in this study were Alexa Fluor 488 anti‐chicken (Jackson ImmunoResearch laboratories 703‐545‐155), Alexa Fluor 647 anti‐rabbit (Jackson ImmunoResearch laboratories 711‐605‐152), Alexa Fluor 647 anti‐mouse (Jackson ImmunoResearch laboratories 715‐605‐150), CyTM3‐conjugated anti‐rabbit (Jackson ImmunoResearch laboratories 711‐165‐152), and CyTM3‐conjugated anti‐mouse (Jackson ImmunoResearch laboratories 715‐165‐150). When needed, nuclei were staining using DAPI at a 1:1,000 dilution. Samples were mounted in NuSieve™ GTG™ Agarose (Lonza cat50080) and imaged on a glass bottom FluoroDish™ (WPI FD3510‐100) using a Zeiss LSM 780 confocal microscope or Leica DMI 6000b microscope. ImageJ was used to add scale bars and PowerPoint was used for adding arrows and labels.
FACS‐based reporter analysis
For analyzing the levels of pax2amKO2 by FACS, single‐cell suspension from the thyroid gland of 5 mpf Tg(tg:nls‐GFP); pax2amKO2 animals was prepared as described earlier and stained with 1 µM calcein violet (Thermo Fisher, C34858). Cells were sorted and analyzed using FACS‐Aria II (BD Bioscience). Thyrocytes were selected by gating for calcein+GFP+ population, and mKO2 expression level recorded for analysis.
Gene ontology (GO) analysis
Gene ontology (GO) analysis was performed using DAVID (Huang et al, 2007). The list of genes was uploaded on the web browser of DAVID and statistically significant (P < 0.05) GO terms were identified using default parameters.
Transcription profiling and analysis of pax2amKO2‐High and pax2amKO2‐Low thyrocytes
For transcriptional profiling of pax2amKO2‐High and pax2amKO2‐Low thyrocytes, mRNA was isolated from cell populations using ReliaPrep™ RNA Miniprep Systems (Promega, Z6011). For this, cells were passed through FACS as outlined above and approximately 100 cells were collected directly in the lysis buffer provided in the kit. mRNA was isolated as outlined in the kit protocol. Three biological replicates for each population.
For sequencing, mRNA was reverse transcribed to cDNA and amplified SMART‐seq V4 Ultra Low Input RNA kit for Sequencing (Takara Bio USA, Inc) following the manufacturer’s protocol. Amplified cDNA was further processed according to TruSeq Sample Preparation v.2 Guide (Illumina) and paired end‐sequenced (2 × 75 bp) on the HiSeq 2500 (Illumina) at a depth of 10 reads per sample. Raw reads were trimmed using trim‐galore (Martin, 2011), mapped using HiSAT2 (Kim et al, 2015) against the GRCz11 zebrafish genome (in which mKO2 and EGFP sequences were added), and counted using FeatureCounts (Liao et al, 2014). For normalization, differential gene expression analysis, and GO analysis, iDEP version 0.92 was utilized (Ge et al, 2018).
Statistical analysis
Statistical analysis was performed using R. Power analysis was not performed to estimate sample sizes. Animals were randomly chosen for experimentation. No animals were excluded from analysis. Blinding was not performed during analysis. Analysis of normal distribution was performed using Shapiro–Wilk test.
Author contribution
SPS conceptualized the project. NN, GK, CL, and MB provided reagents and animals for single‐cell RNA sequencing. SPS, SR, AK, JB, and AP performed the single‐cell RNA sequencing. SPS, SEE, VD, and SC analyzed and interpreted the data. SPS developed the online browser. PG and BH generated the pax2a knock‐in line. PG and MS analyzed the pax2a reporter line and MP‐M and IG collected immunofluorescence images. AL and FL performed bulk RNA‐Seq experiment, with data analysis being performed by SPS. SPS wrote the first draft and SC, PG, and SEE edited the manuscript. SPS, NN and SC acquired funding for the project. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Movie EV1
Source Data for Expanded View/Appendix
Appendix
Source Data for Figure 1
Acknowledgements
We thank members of the Costagliola and Singh laboratory for comments on the manuscript, members of Center for Regenerative Therapies Dresden (CRTD) fish, FACS and sequencing facility, and members of IRIBHM fish facility for technical assistance. We thank J.‐M. Vanderwinden from the Light Microscopy Facility and Christine Dubois from the FACS facility for technical assistance at ULB. Bulk RNA sequencing was performed at the Brussels Interuniversity Genomics High Throughput core (BRIGHTcore: www.brightcore.be). We are grateful to Priyanka Oberoi for illustrations. P.G is Fund for Research in the Industry and the Agriculture (FRIA) Research fellow; M.S. is FNRS Research Fellow (34985615 ‐ THYSCEFA); S.C. is FNRS Senior Research Associate. V.D. acknowledges grants from the Fond Naets (J1813300), the Fondation Contre le Cancer (2016‐093) and FNRS (EQP/OL U.N019.19, J006120F). Work by M.B., C.L., and G.K. was supported by grants to M.B. from the Deutsche Forschungsgemeinschaft (BR 1746/11‐1) and European Union (European Research Council AdG Zf‐BrainReg). N.N. received funding from the DFG–Center for Regenerative Therapies Dresden at TU Dresden and the German Center for Diabetes Research (DZD), as well as research grants from the German Research Foundation (DFG) and the European Foundation for the Study of Diabetes. Work by S.P.S. was supported by MISU funding from the FNRS (34772792 – SCHISM). This work was supported by grants from the Belgian National Fund for Scientific Research (FNRS) (FRSM 3‐4598‐12; CDR‐J.0145.16, GEQ U.G030.19), the Fonds d’Encouragement à la Recherche de l’Université Libre de Bruxelles (FER‐ULB).
EMBO Reports (2020) 21: e50612.
Contributor Information
Sabine Costagliola, Email: scostag@ulb.ac.be.
Singh Sumeet Pal, Email: sumeet.pal.singh@ulb.ac.be.
Data availability
The datasets produced in this study are available in the following databases:
RNA‐Seq data: Gene Expression Omnibus GSE133466 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133466).
RNA‐Seq data: Gene Expression Omnibus GSE153197 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153197)
Interactive bowser: Shinyapps (https://sumeet.shinyapps.io/zfthyroid/)
References
- Aleström P, D’Angelo L, Midtlyng PJ, Schorderet DF, Schulte‐Merker S, Sohm F, Warner S (2020) Zebrafish: Housing and husbandry recommendations. Lab Anim 54: 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AlJanahi AA, Danielsen M, Dunbar CE (2018) An Introduction to the analysis of single‐cell RNA‐sequencing data. Mol Ther Methods Clin Dev 10: 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt B, Reibe S, Feitosa NM, Elsalini OA, Wendl T, Rohr KB (2006) Analysis of origin and growth of the thyroid gland in zebrafish. Dev Dyn 235: 1872–1883 [DOI] [PubMed] [Google Scholar]
- Anelli V, Villefranc JA, Chhangawala S, Martinez‐McFaline R, Riva E, Nguyen A, Verma A, Bareja R, Chen Z, Scognamiglio T et al (2017) Oncogenic BRAF disrupts thyroid morphogenesis and function via twist expression. Elife 6: e20728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonica F, Kasprzyk DF, Opitz R, Iacovino M, Liao X‐H, Dumitrescu AM, Refetoff S, Peremans K, Manto M, Kyba M et al (2012) Generation of functional thyroid from embryonic stem cells. Nature 491: 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baber EC (1876) XXI. Contributions to the minute anatomy of the thyroid gland of the dog. Philos Trans R Soc London 166: 557–568 [Google Scholar]
- Baptist M, Dumont JE, Roger PP (1995) Intercellular heterogeneity of early mitogenic events: cAMP generalizes the EGF effect on c‐Fos protein appearance but not on MAP kinase phosphorylation and nuclear translocation in dog thyroid epithelial cells. Exp Cell Res 221: 160–171 [DOI] [PubMed] [Google Scholar]
- Barbieri CE, Pietenpol JA (2006) p63 and epithelial biology. Exp Cell Res 312: 695–706 [DOI] [PubMed] [Google Scholar]
- Brix K, Lemansky P, Herzog V (1996) Evidence for extracellularly acting cathepsins mediating thyroid hormone liberation in thyroid epithelial cells. Endocrinology 137: 1963–1974 [DOI] [PubMed] [Google Scholar]
- Brix K, Linke M, Tepel C, Herzog V (2001) Cysteine proteinases mediate extracellular prohormone processing in the thyroid. Biol Chem 382: 717–725 [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 36: 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen J‐N (2007) FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol 304: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra K, Ishibashi S, Amaya E (2019) Zebrafish duox mutations provide a model for human congenital hypothyroidism. Biol Open 8: bio037655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosacak MI, Bhattarai P, Reinhardt S, Petzold A, Dahl A, Zhang Y, Kizil C (2019) Single‐Cell Transcriptomics Analyses of Neural Stem Cell Heterogeneity and Contextual Plasticity in a Zebrafish Brain Model of Amyloid Toxicity. Cell Rep 27: 1307–1318 [DOI] [PubMed] [Google Scholar]
- Denton CP, Zheng B, Shiwen X, Zhang Z, Bou‐Gharios G, Eberspaecher H, Black CM, de Crombrugghe B (2001) Activation of a fibroblast‐specific enhancer of the proalpha2(I) collagen gene in tight‐skin mice. Arthritis Rheum 44: 712–722 [DOI] [PubMed] [Google Scholar]
- Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ (2011) mpeg1 promoter transgenes direct macrophage‐lineage expression in zebrafish. Blood 117: e49–e56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge SX, Son EW, Yao R (2018) iDEP: an integrated web application for differential expression and pathway analysis of RNA‐Seq data. BMC Bioinformatics 19: 534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber H, Peter HJ, Studer H (1987) Age‐related failure of endocytosis may be the pathogenetic mechanism responsible for ‘cold’ follicle formation in the aging mouse thyroid. Endocrinology 120: 1758–1764 [DOI] [PubMed] [Google Scholar]
- Getzowa S (1907) Über die Glandula parathyreodeaa, intrathyreoideale Zellhaufen derselben und Reste des postbranchialen Körpers. Virchows Arch Pathol Anat Physiol Klin Med 188: 181–235 [Google Scholar]
- Guzman A, Ramos‐Balderas JL, Carrillo‐Rosas S, Maldonado E (2013) A stem cell proliferation burst forms new layers of P63 expressing suprabasal cells during zebrafish postembryonic epidermal development. Biol Open 2: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harach HR (1988) Solid cell nests of the thyroid. J Pathol 155: 191–200 [DOI] [PubMed] [Google Scholar]
- Huang D, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA (2007) The DAVID Gene Functional Classification Tool: a novel biological module‐centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S (2014) Quantitative single‐cell RNA‐seq with unique molecular identifiers. Nat Methods 11: 163–166 [DOI] [PubMed] [Google Scholar]
- Kesavan G, Chekuru A, Machate A, Brand M (2017) CRISPR/Cas9‐Mediated Zebrafish Knock‐in as a Novel Strategy to Study Midbrain‐Hindbrain Boundary Development. Front Neuroanat 11: 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Vähärautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J (2011) Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 9: 72–74 [DOI] [PubMed] [Google Scholar]
- Liang S, Johansson E, Barila G, Altschuler DL, Fagman H, Nilsson M (2018) A branching morphogenesis program governs embryonic growth of the thyroid gland. Development 145: dev146829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Linehan SA, Martínez‐Pomares L, da Silva RP, Gordon S (2001) Endogenous ligands of carbohydrate recognition domains of the mannose receptor in murine macrophages, endothelial cells and secretory cells; potential relevance to inflammation and immunity. Eur J Immunol 31: 1857–1866 [DOI] [PubMed] [Google Scholar]
- Linke M (2002) Trafficking of lysosomal cathepsin B–green fluorescent protein to the surface of thyroid epithelial cells involves the endosomal/lysosomal compartment. J Cell Sci 115: 4877–4889 [DOI] [PubMed] [Google Scholar]
- Lisse TS, Middleton LJ, Pellegrini AD, Martin PB, Spaulding EL, Lopes O, Brochu EA, Carter EV, Waldron A, Rieger S (2016) Paclitaxel‐induced epithelial damage and ectopic MMP‐13 expression promotes neurotoxicity in zebrafish. Proc Natl Acad Sci USA 113: E2189–E2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li C, Li Z, Wang D, Ren X, Zhang Z (2020) An entropy‐based metric for assessing the purity of single cell populations. Nat Commun 11: 3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G (2008) Visualizing data using t‐SNE. J Mach Learn Res 9: 2579–2605 [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM et al (2015) Highly Parallel Genome‐wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161: 1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Chowdhury K, Gruss P (1998) Follicular cells of the thyroid gland require Pax8 gene function. Nat Genet 19: 87–90 [DOI] [PubMed] [Google Scholar]
- Marinò M, McCluskey RT (2000) Megalin‐mediated transcytosis of thyroglobulin by thyroid cells is a calmodulin‐dependent process. Thyroid 10: 461–469 [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.journal 17: 10 [Google Scholar]
- Martínez‐Aguilar J, Clifton‐Bligh R, Molloy MP (2016) Proteomics of thyroid tumours provides new insights into their molecular composition and changes associated with malignancy. Sci Rep 6: 23660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMenamin SK, Bain EJ, McCann AE, Patterson LB, Eom DS, Waller ZP, Hamill JC, Kuhlman JA, Eisen JS, Parichy DM (2014) Thyroid hormone‐dependent adult pigment cell lineage and pattern in zebrafish. Science 345: 1358–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill T, Schaefer L, Iozzo RV (2012) Decorin: a guardian from the matrix. Am J Pathol 181: 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonidez JF (1931) Innervation of the thyroid gland. II. Origin and course of the thyroid nerves in the dog. Am J Anat 48: 299–329 [Google Scholar]
- Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M (1998) Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development 125: 3063–3074 [DOI] [PubMed] [Google Scholar]
- Planck T, Shahida B, Sjögren M, Groop L, Hallengren B, Lantz M (2014) Association of BTG2, CYR61, ZFP36, and SCD gene polymorphisms with Graves’ disease and ophthalmopathy. Thyroid 24: 1156–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porazzi P, Calebiro D, Benato F, Tiso N, Persani L (2009) Thyroid gland development and function in the zebrafish model. Mol Cell Endocrinol 312: 14–23 [DOI] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppman E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B et al (2015) A draft network of ligand‐receptor‐mediated multicellular signalling in human. Nat Commun 6: 7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JP, Sack GS, Martin SM, Sagasti A (2015) Vertebrate epidermal cells are broad‐specificity phagocytes that clear sensory axon debris. J Neurosci 35: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischauer S, Levesque MP, Nüsslein‐Volhard C, Sonawane M (2009) Lgl2 executes its function as a tumor suppressor by regulating ErbB signaling in the zebrafish epidermis. PLoS Genet 5: e1000720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos Moreno MJ, Galera‐Ruiz H, De Miguel M, López MIC, Illanes M, Galera‐Davidson H (2011) Inmunohistochemical profile of solid cell nest of thyroid gland. Endocr Pathol 22: 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Iranzo H, Galardi‐Castilla M, Sanz‐Morejón A, González‐Rosa JM, Costa R, Ernst A, Sainz de Aja J, Langa X, Mercader N (2018) Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc Natl Acad Sci USA 115: 4188–4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Janjuha S, Chaudhuri S, Reinhardt S, Kränkel A, Dietz S, Eugster A, Bilgin H, Korkmaz S, Zararsız G et al (2018) Machine learning based classification of cells into chronological stages using single‐cell transcriptomics. Sci Rep 8: 17156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds S, Peter HJ, Jörtsö E, Gerber H, Studer H (1987) Naturally occurring clones of cells with high intrinsic proliferation potential within the follicular epithelium of mouse thyroids. Cancer Res 47: 1646–1651 [PubMed] [Google Scholar]
- Struder H, Peter HJ, Gerber H (1989) Natural Heterogeneity of Thyroid Cells: The Basis for Understanding Thyroid Function and Nodular Goiter Growth. Endocr Rev 10: 125–135 [DOI] [PubMed] [Google Scholar]
- Studer H, Forster R, Conti A, Kohler H, Haeberli A, Engler H (1978) Transformation of normal follicles into thyrotropin‐refractory ‘cold’ follicles in the aging mouse thyroid gland. Endocrinology 102: 1576–1586 [DOI] [PubMed] [Google Scholar]
- Svensson V, Vento‐Tormo R, Teichmann SA (2018) Exponential scaling of single‐cell RNA‐seq in the past decade. Nat Protoc 13: 599–604 [DOI] [PubMed] [Google Scholar]
- Taylor PN, Albrecht D, Scholz A, Gutierrez‐Buey G, Lazarus JH, Dayan CM, Okosieme OE (2018) Global epidemiology of hyperthyroidism and hypothyroidism. Nat Rev Endocrinol 14: 301–316 [DOI] [PubMed] [Google Scholar]
- Trubiroha A, Gillotay P, Giusti N, Gacquer D, Libert F, Lefort A, Haerlingen B, De Deken X, Opitz R, Costagliola S (2018) A rapid CRISPR/Cas‐based mutagenesis assay in zebrafish for identification of genes involved in thyroid morphogenesis and function. Sci Rep 8: 5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu D‐G, Chang W‐W, Jan M‐S, Tu C‐W, Lu Y‐C, Tai C‐K (2016) Promotion of metastasis of thyroid cancer cells via NRP‐2‐mediated induction. Oncol Lett 12: 4224–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M et al (2015) High‐throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res 25: 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendl T, Lun K, Mione M, Favor J, Brand M, Wilson SW, Rohr KB (2002) Pax2.1 is required for the development of thyroid follicles in zebrafish. Development 129: 3751–3760 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C et al (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718 [DOI] [PubMed] [Google Scholar]
- Yang S, Corbett SE, Koga Y, Wang Z, Johnson WE, Yajima M, Campbell JD (2020) Decontamination of ambient RNA in single‐cell RNA‐seq with DecontX. Genome Biol 21: 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J et al (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8: 14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Movie EV1
Source Data for Expanded View/Appendix
Appendix
Source Data for Figure 1
Data Availability Statement
The datasets produced in this study are available in the following databases:
RNA‐Seq data: Gene Expression Omnibus GSE133466 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133466).
RNA‐Seq data: Gene Expression Omnibus GSE153197 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153197)
Interactive bowser: Shinyapps (https://sumeet.shinyapps.io/zfthyroid/)


