Abstract
Respiratory chains are crucial for cellular energy conversion and consist of multi‐subunit complexes that can assemble into supercomplexes. These structures have been intensively characterized in various organisms, but their physiological roles remain unclear. Here, we elucidate their function by leveraging a high‐resolution structural model of yeast respiratory supercomplexes that allowed us to inhibit supercomplex formation by mutation of key residues in the interaction interface. Analyses of a mutant defective in supercomplex formation, which still contains fully functional individual complexes, show that the lack of supercomplex assembly delays the diffusion of cytochrome c between the separated complexes, thus reducing electron transfer efficiency. Consequently, competitive cellular fitness is severely reduced in the absence of supercomplex formation and can be restored by overexpression of cytochrome c. In sum, our results establish how respiratory supercomplexes increase the efficiency of cellular energy conversion, thereby providing an evolutionary advantage for aerobic organisms.
Keywords: bioenergetics, competitive fitness, cryo‐EM , mitochondria, respiratory chain supercomplexes
Subject Categories: Metabolism, Structural Biology, Organelles
Mitochondrial respiratory supercomplexes are found in various organisms, however their physiological function remains elusive. This study reveals that supercomplexes enhance mitochondrial electron transfer, resulting in increased competitive fitness.
Introduction
The mitochondrial respiratory chain represents a sophisticated system of multi‐subunit complexes that mediate cellular energy conversion. In mammalian cells, the mitochondrial respiratory chain (MRC) is formed by four distinct complexes (CI–CIV) (Lobo‐Jarne & Ugalde, 2018). Together with the ATP synthase (CV), these complexes form the oxidative phosphorylation (OXPHOS) system, in which electron transfer to molecular oxygen is coupled with the formation of an electrochemical gradient over the inner mitochondrial membrane to drive ATP synthesis (Mitchell, 1966). Given its importance for cellular energy conversion, dysfunction of the OXPHOS system causes various human diseases including neuromuscular and neurodegenerative disorders (Kauppila et al, 2017; Kawamata & Manfredi, 2017).
Three major models have been proposed for the structural organization of the MRC: the solid‐state, the liquid‐state (fluid), and the plasticity model (Acín‐Pérez et al, 2008; Milenkovic et al, 2017; Lobo‐Jarne & Ugalde, 2018). The latter consolidates the previous two models and is based on findings from the past two decades, showing that MRC complexes are not randomly distributed within the inner mitochondrial membrane, but can be assembled into supramolecular structures termed supercomplexes (SCs) (Schägger & Pfeiffer, 2000). Hence, the plasticity model describes a dynamic equilibrium between free complexes and SCs to adapt energy conversion to cellular needs (Acin‐Perez & Enriquez, 2014). SCs exist in a wide range of species, including mammals, plants, yeast, and some bacteria (Milenkovic et al, 2017). But despite extensive research, the physiological significance of respiratory SCs remains controversial. Several hypotheses on their role have been suggested. An attractive but highly debated possibility involves SC‐mediated substrate channeling operating such that each SC sequesters and retains its own subpopulation of the mobile electron carriers cytochrome c (Cyt c) and, for mammalian SCs, ubiquinone (Blanchi et al, 2004; Lapuente‐Brun et al, 2013). However, no confining structure within SCs is reported to retain Cyt c or ubiquinone and accumulating biophysical evidence rather suggests free diffusion of these electron carriers (Blaza et al, 2014; Milenkovic et al, 2017; Fedor & Hirst, 2018; Hirst, 2018). Further, SCs were suggested to participate in the regulation of respiratory activity (Greggio et al, 2017), reduction of oxidative stress (Maranzana et al, 2013), as well as assembly and stabilization of individual MRC complexes (Schägger et al, 2004; Moreno‐Lastres et al, 2012). The latter is based on the observation that defects in CIII or CIV can cause CI deficiency, suggesting a structural interdependence between the affected complexes, which might be mediated by SCs (Acín‐Pérez et al, 2004; Schägger et al, 2004; Diaz et al, 2006). Although it was subsequently reported that cells lacking CIV were shown to still retain significant amounts of assembled CI (Balsa et al, 2012), it has been recently disclosed that multiple pathways exist for SC assembly and that the presence of COX1, but not necessarily holo‐CIV, is sufficient to stabilize CI (Lobo‐Jarne et al, 2020; Timón‐Gómez et al, 2020). CI ab initio assembly was described to be only finalized when its nascent form is incorporated into SCs (Moreno‐Lastres et al, 2012). However, by using complexome analysis of assembly intermediates in the steady‐state, others suggested that SCs are not required as scaffolding unit, as CI assembly is completed prior to SC formation (Guerrero‐Castillo et al, 2017). In addition, SC formation is proposed to prevent protein aggregation in the highly protein‐rich mitochondrial inner membrane (Blaza et al, 2014). Several studies have investigated the physiological effects of absence of the mammalian supercomplex assembly factor COX7A2L or SCAF1. Absence of this factor specifically impairs formation of the supercomplex CIII2/CIV, but, importantly, does not impact the respirasomes CI/CIII2/CIVn. In human cells, ablation of COX7A2L has no effect on bioenergetics (Perez‐Perez et al, 2016; Lobo‐Jarne et al, 2018), but provokes changes in nutrient sensing (Balsa et al, 2019). The controversy extents to mouse models, in which energy metabolism effects have been observed or not (Shiba et al, 2017; Mourier et al, 2014; Lapuente‐Brun et al, 2013), and a recent zebrafish model in which abnormal fat deposition was observed (García‐Poyatos et al, 2020). Because respirasomes are maintained in these model systems, they do not allow to analyse the physiological relevance to assemble the respiratory chain into higher ordered structures. Hence, several contested hypotheses on the role of SC formation exist, none of which has been sufficiently experimentally evidenced due to the lack of a model system entirely lacking SCs. Thus, the fundamental question remains: What is the purpose of supercomplexes and why do they exist?1
Results and Discussion
A detailed structure of the CIII–CIV interface allows to design supercomplex‐disrupting mutations
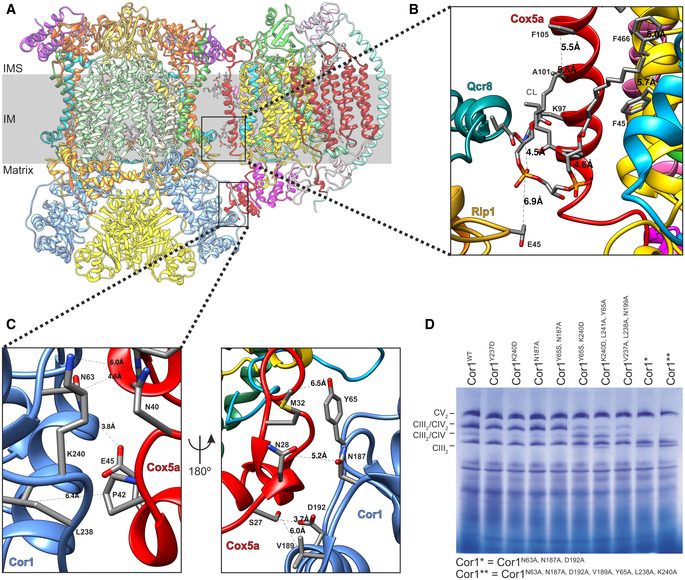
To address this enigma, we aimed to disrupt SC formation by mutation of key residues in the interaction interface of CIII2/CIV SCs of Saccharomyces cerevisiae. To this end, we improved the resolution of our previous SC structure (Rathore et al, 2019). This yielded a CIII2/CIV density map with an overall resolution of 3.17 Å, and, more importantly, after particle subtraction of CIII2, the CIV density was solved to an overall resolution of 3.41 Å (Fig EV1A–D, Table EV1). These improvements allowed us to create a more detailed model of several CIV subunits (Figs 1A and EV2A–D). Moreover, we could confirm that the protein–lipid–protein interactions between Cox5a and Rip1 contained cardiolipin (CL) (Fig 1B), as previously suggested (Hartley et al, 2019). The improved resolution of the matrix‐localized protein–protein interactions between the CIII‐subunit Cor1 and the CIV protein Cox5a also enabled unequivocal identification of the residues connecting the two complexes (Fig 1C). To gain insights into the physiological function of SCs and explore the consequences of their absence, we performed a variety of alanine‐substitutions in the CIII‐subunit Cor1 to disrupt the CIII–CIV interaction (Fig 1D). Two of these mutants, namely Cor1N63A, N187A, D192A (hereafter Cor1*), and Cor1N63A, N187A, D192A, Y65A, V189A, L238A, K240A (Cor1**), lacked higher molecular weight complexes containing both CIII and CIV, thus revealing complete SC disruption (Fig 1D).
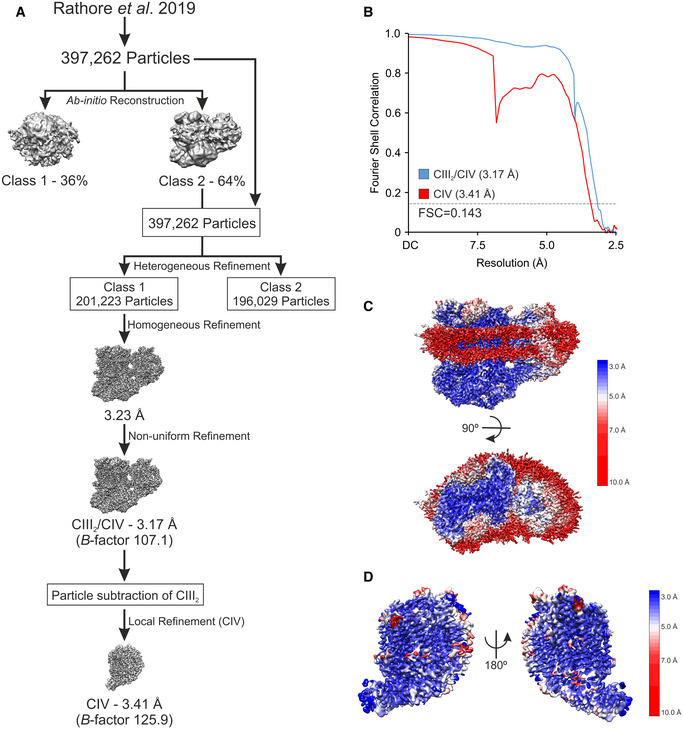
Figure EV1. Workflow of cryo‐EM processing.
-
ASchematic illustration of Workflow of cryo‐EM processing.
-
BGold‐standard Fourier shell correlation (FSC) of the CIII2/CIV supercomplex and the CIV density maps after final refinements. FSC = 0.143 is presented as dashed line.
-
C, DLocal resolution maps of the CIII2/CIV supercomplex viewed along the mitochondrial inner membrane (IM) and from the mitochondrial intermembrane space (IMS) side (C), and the CIV density after particle subtraction of CIII2 and the digitonin micelle viewed from two rotations along the mitochondrial inner membrane (D).
Figure 1. A detailed structure of the CIII‐CIV interface allows to design supercomplex‐disrupting mutations.
-
ASide view of the overall structure of the Saccharomyces cerevisiae CIII2/CIV supercomplex and its position in the mitochondrial inner membrane (IM). IMS: intermembrane space.
-
B, CZoom‐in of the CIII2/CIV interaction sites with residues and distances annotated. The inner membrane protein–lipid–protein interactions (Rip1‐CL-Cox5a), where Rip1 is marked in gold, cardiolipin (CL) in gray, and Cox5a in red (B), and the mitochondrial matrix, protein–protein interaction of Cor1 (blue), and Cox5a (red) (C) are shown.
-
DBlue‐native gel electrophoresis of S. cerevisiae strains with indicated mutations in Cor1.
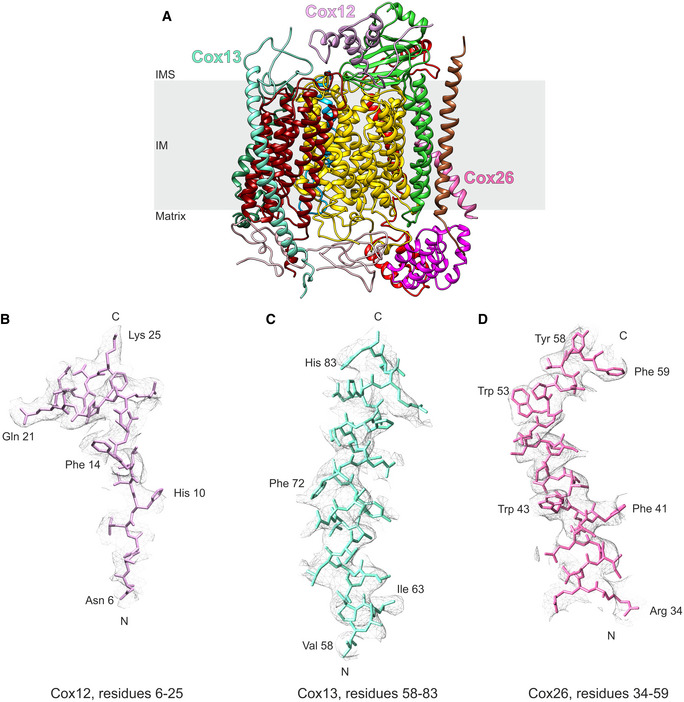
Figure EV2. Example densities of CIV subunits Cox12, Cox13, and Cox26.
-
ARibbon diagram of CIV with Cox12, Cox13, and Cox26 annotated. IM: inner membrane; IMS: intermembrane space.
-
B–DExample densities of Cox12 (B), Cox13 (C), and Cox26 (D) demonstrating parts of the subunits that improved due to the re‐processing of the data set.
Characterization of strains lacking respiratory supercomplexes
In addition to the Cor1‐Cox5a protein–protein interaction, CIII and CIV also interact with each other through a CL molecule situated between Cox5a and Rip1 (Fig 1B). As CL is thought to be crucial for the structural and functional integrity of SCs (Zhang et al, 2002), we generated strains devoid of CRD1, encoding cardiolipin synthase. Immunoblotting after BN‐PAGE confirmed that the Cor1 variants lacked SCs also in CRD1 deletion strains (Fig 2A). However, the absence of CL per se did not abolish SC formation in our experimental setup, revealing that the Cor1–Cox5a interaction is necessary and sufficient for forming SCs. Interestingly, CIII2 levels were reduced in strains expressing Cor1*, but not in Cor1** (Fig 2A). Likewise, optical spectra analyses revealed a major decrease of heme b in the Cor1* strain compared with Cor1 wild‐type (Cor1WT) and Cor1** (Fig 2B, Table EV2). Western blot analyses (Fig 2C–E) confirmed reduced levels of Cyt b and other CIII subunits (Rip1, Qcr7, and Cor1) in the Cor1* mutant, whereas CIV subunits (Cox1, Cox2, Cox5, Cox12, and Cox13) were not affected (Fig 2C and D). Of note, the Cor1** mutant, despite also being unable to form SCs, had a normal accumulation of CIII subunits. Hence, we decided to focus on the Cor1** strain for further analyses. As Cor1** was sufficient to disrupt SCs and absence of CL did not inhibit SC formation, we also excluded crd1Δ strains from further experiments to avoid potential secondary effects caused by the lack of this important mitochondrial phospholipid.
Figure 2. Characterization of strains lacking respiratory supercomplexes.
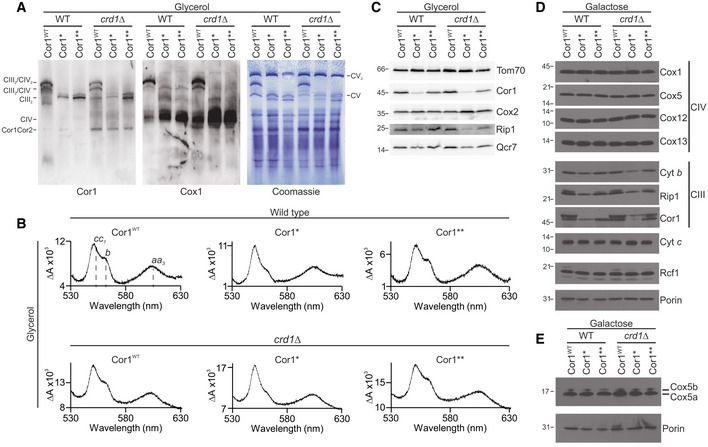
-
ABlue‐native gel electrophoresis and immunoblots of cells expressing the wild‐type form of Cor1 (Cor1WT), as well as the mutants Cor1N63A, N187A, D192A (Cor1*) and Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**) in wild‐type (WT) background as well as in cells lacking CRD1 (crd1Δ). Blots were probed with antibodies against Cor1 to visualize Complex III (CIII), as well as Cox1, to monitor Complex IV (CIV). CV: Complex V; Cor1Cor2: multimer consisting of Cor1 and Cor2.
-
BReduced‐minus‐oxidized difference spectra of strains described in (A). Heme peaks are indicated for Cor1WT in WT background.
-
C–ESteady‐state protein levels of strains as described in (A) cultivated on YP media containing glycerol (C) or CM media with galactose as carbon source (D, E).
Abrogation of supercomplexes decreases competitive fitness
Our engineered Cor1** mutant disrupts SCs formation without affecting individual complexes, thus presenting an advanced tool to study the physiological function of SCs. Hence, we next evaluated the growth rates and chronological lifespan under fermentable (glucose) or respiratory carbon sources (glycerol) in clonal cultures. Unexpectedly, we did not find any significant differences between the Cor1WT and mutant (Cor1**) strains (Fig 3A and B). In line, absence of SC formation had only modest effects on oxidative stress and cell death on glucose and no alterations were observed under respiratory conditions (Fig EV3A–F). Since SC evolved in many different organisms across life kingdoms, we hypothesized that SC formation confers an apparent, selectable advantage. To test this, we determined competitive fitness to mimic a more natural scenario. Hygromycin and clonNAT selection cassettes were chromosomally integrated into Cor1WT and Cor1** cells, respectively, and both strains were inoculated with identical optical density in the same culture. Colony‐forming units on antibiotic selection plates were determined to evaluate the prevalence of either strain in the culture over time (Fig 3C). While no disadvantage of SC disruption was detected during fermentation, growth competition on respiration massively reduced the prevalence of Cor1** in culture (Fig 3D). To avoid potential artefacts, we exchanged the selection markers in a second set of strains and obtained similar results (Fig 3E). We therefore conclude that SC formation results in increased competitive fitness, providing a selectable trait that favored the establishment of SCs during evolution.
Figure 3. Abrogation of supercomplexes decreases competitive fitness.
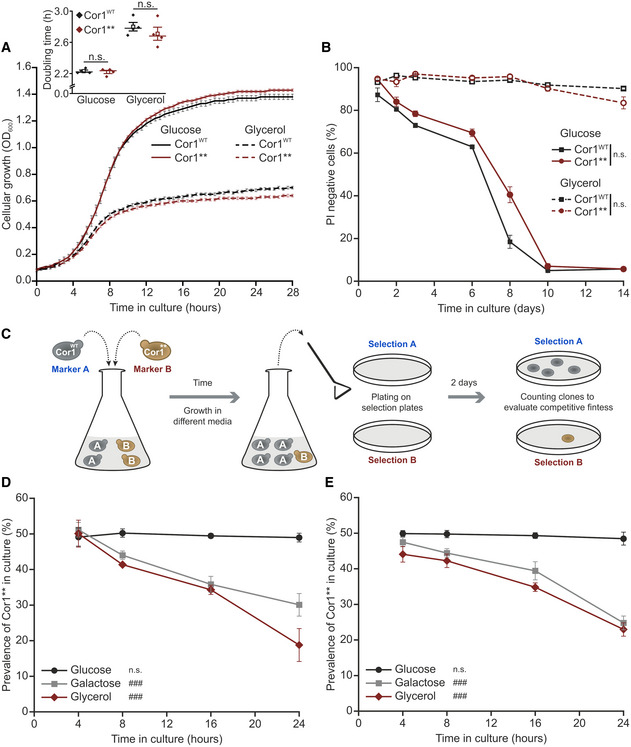
-
AGrowth analysis of strains expressing the wild‐type form of Cor1 (Cor1WT) or the mutant Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**). Growth curves and calculated doubling times from the exponential phase (insert) are presented. Cells were cultivated in media containing either glucose or glycerol as carbon source.
-
BDetermination of chronological lifespan via propidium iodide (PI) staining of cells described above.
-
CScheme of the experimental setup to evaluate competitive fitness.
-
D, EAnalysis of competitive fitness as described in (C). Cor1WT strains harbored a chromosomally integrated hygromycin selection cassette, whereas Cor1** contained a clonNAT selection cassette (D). Subsequently, selection markers were switched as a control (E).
Figure EV3. Disruption of supercomplexes does not modulate oxidative stress or cell death.
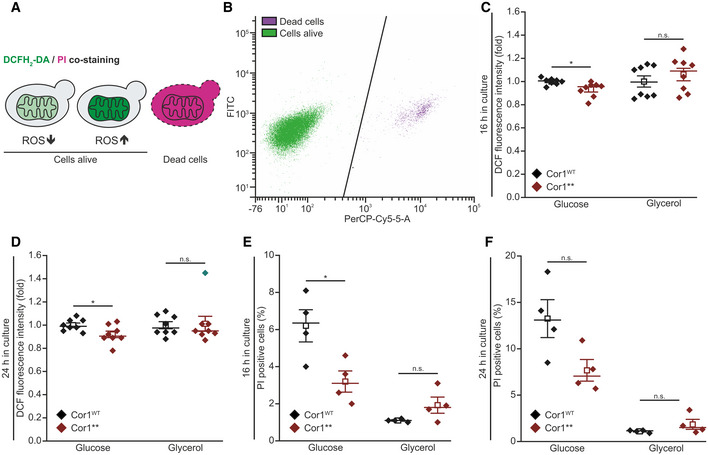
-
A, BScheme of 2′,7′‐dichlorodihydrofluorescein diacetate (DCFH2‐DA), and propidium iodide (PI) staining and gating strategy for flow cytometry. PI is taken up by cells upon loss of membrane integrity, thus indicating dead cells. Non‐fluorescent DCFH2‐DA is oxidized to fluorescent 2′,7′‐dichlorofluorescein (DCF) upon oxidative stress (A). Hence, the DCF mean fluorescence intensity of PI‐negative cells was applied as a measure for oxidative stress. The threshold used to separate PI‐negative and PI‐positive cells is shown as black line in (B).
-
C, DFlow cytometric quantification of oxidative stress indicated via DCFH2‐DA/PI counterstaining of cells expressing the wild‐type form of Cor1 (Cor1WT), as well as the mutant Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**). Cells were analyzed 16 h (C) and 24 h (D) after inoculation in media either containing glucose or galactose as indicated. To visualize fold values, normalization was performed to Cor1WT cells of the respective medium used.
-
E, FAnalysis of cell death via flow cytometric quantification of PI stained cells as described above. Experiments were performed 16 h (E) and 24 h (F) after inoculation, and the percentages of PI‐positive cells are presented.
Lack of supercomplexes impairs electron transport between CIII and CIV
To characterize the molecular mechanism by which SC formation mediates increased competitive fitness, we evaluated the impact of SC disruption on mitochondrial function. While living cells lacking SCs maintained normal mitochondrial transmembrane potential, oxygen consumption, and ATP levels (Fig EV4A–H), we observed an increased maximal activity of CIV and attenuated CIII activity for the Cor1** mutant in solubilized mitochondria (Fig 4A and B). These results prompted us to investigate electron transfer efficiency of isolated mitochondria (Fig EV5A) utilizing different substrates. When using NADH, which transfers electrons to coenzyme Q via Nde1 and Nde2, but bypasses CII, decreased rates of respiration were detected in both basal (state 2) and ADP‐driven (state 3) conditions (Fig 4C). Similar observations were obtained with a combination of succinate and glycerol‐3‐phosphate (G3P) as substrate, which feed electrons via CII and G3P‐dehydrogenase to coenzyme Q for subsequent delivery to CIII (Fig 4D). Of note, the respiratory control ratios (state 3/state 2), a measure for the coupling between substrate oxidation and phosphorylation, were similar between mitochondria from Cor1WT and Cor1** cells (Fig 4E). We hypothesized that this impairment of the respiratory chain might be caused by inefficient electron transfer from CIII to CIV via Cyt c. As neither heme c nor Cyt c content was reduced in Cor1** mutants compared with Cor1WT expressing cells (Fig 2B and D, and Table EV2), this deficiency in electron transfer is potentially due to increased diffusion distance between the separated MRC complexes. We tested this by analyzing substrate oxidation in mitoplasts (mitochondria with permeabilized outer membrane, thereby exposing the Cyt c binding sites to the solvent) with and without administration of exogenous oxidized Cyt c. As seen for intact mitochondria, NADH oxidation was severely decreased in mitoplasts derived from Cor1** strains (Fig 4F). Importantly, the addition of exogenous Cyt c completely corrected the defect (Fig 4F and G). Taken together, these results demonstrate a decreased efficiency by which Cyt c transfers electrons from CIII to CIV upon disruption of respiratory SCs.
Figure EV4. Absence of supercomplex formation does not impact mitochondrial function in living cells.
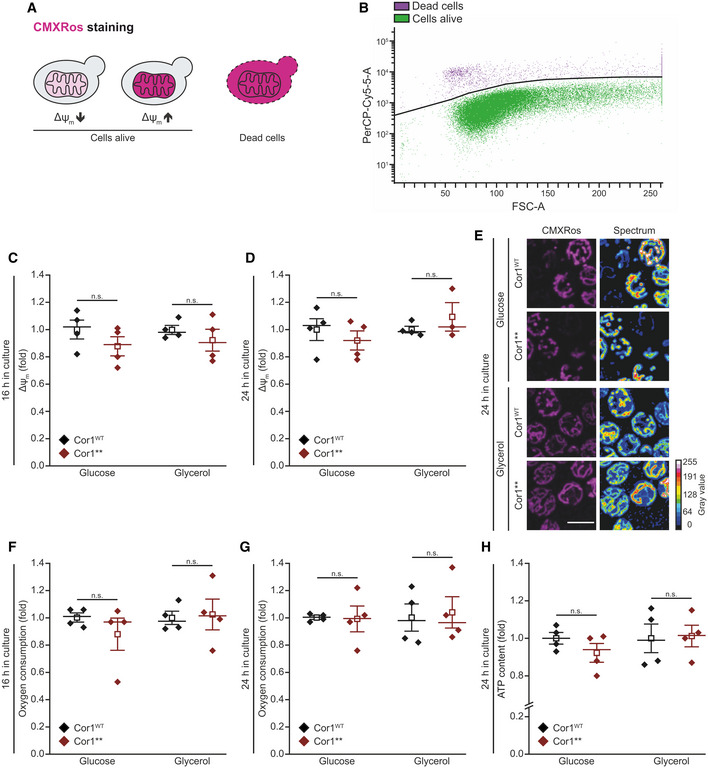
-
A, BScheme of Mitotracker CMXRos staining and gating strategy for flow cytometric analysis. The fluorescence probe is taken up by mitochondria depending on the transmembrane potential (Δψm,). Dead cells accumulate the dye due to a lack of membrane integrity (A) and are excluded from the analysis. The threshold used to separate dead and living cells is shown as black line in (B). The mean fluorescence intensity of cells alive is quantified as a readout for Δψm.
-
C–EAnalysis of Δψm via Mitotracker CMXRos staining of cells expressing the wild‐type form of Cor1 (Cor1WT), as well as the mutant Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**). Cells were cultivated in CM media containing indicated carbon sources. Flow cytometric quantification 16 h (C) and 24 h (D) after inoculation are shown, as well as representative confocal micrographs (Z‐projections) after 24 h (E). Normalization in (C) and (D) was performed to Cor1WT cells of the respective medium used.
-
F, GOxygen consumption quantified in intact cells. Strains described in (C) were analyzed 16 h (F) and 24 h (G) after inoculation. Normalization of data was performed as stated in (C) to present fold values.
-
HCellular ATP content measured from cells described in (C). The measurement was performed with cells cultivated for 24 h in CM media containing the indicated carbon sources. Normalization was performed as stated in (C).
Figure 4. Lack of supercomplexes impairs electron transport from CIII to CIV .
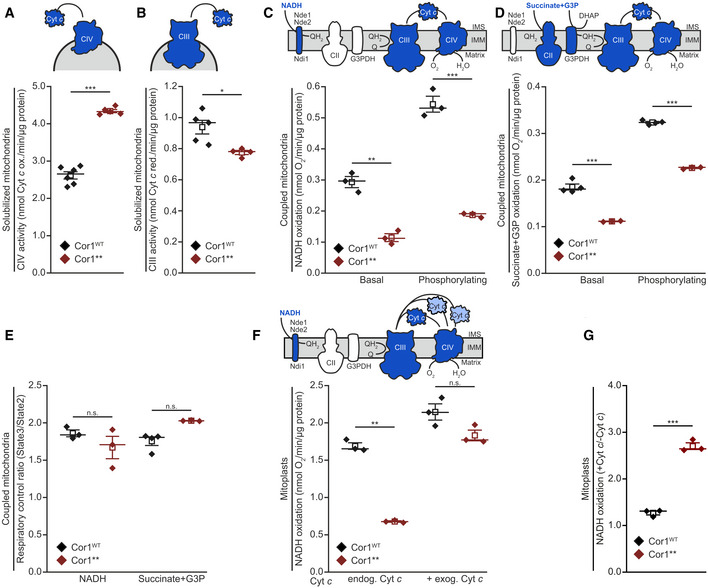
-
A, BSpectrophotometric measurement of Cyt c oxidase (CIV; A) and NADH Cyt c reductase (CIII; B) activities in solubilized mitochondrial extracts. Mitochondria were isolated from cells expressing the wild‐type form of Cor1 (Cor1WT) or the mutant Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**).
-
C, DPolarographic measurement of KCN‐sensitive oxygen consumption, driven by NADH (C) or succinate + glycerol‐3-phosphate (G3P; D) in isolated coupled mitochondria of indicated strains. Measurements were performed in the absence (basal respiration) or presence (phosphorylating condition) of ADP.
-
ERespiratory control ratio calculated from polarographic measurements of KCN‐sensitive oxygen consumption driven by NADH and succinate + glycerol‐3-phosphate (G3P) in isolated coupled mitochondria of indicated strains.
-
F, GPolarographic measurement of KCN‐sensitive oxygen consumption driven by NADH in mitoplasts in the absence (endog. Cyt c) or presence (+ exog. Cyt c) of exogenous oxidized Cyt c. NADH oxidation (E) and the calculated ratio +Cyt c/−Cyt c of substrate oxidation (F) are visualized.
Figure EV5. The lack of supercomplexes impairs electron transport from CIII to CIV .
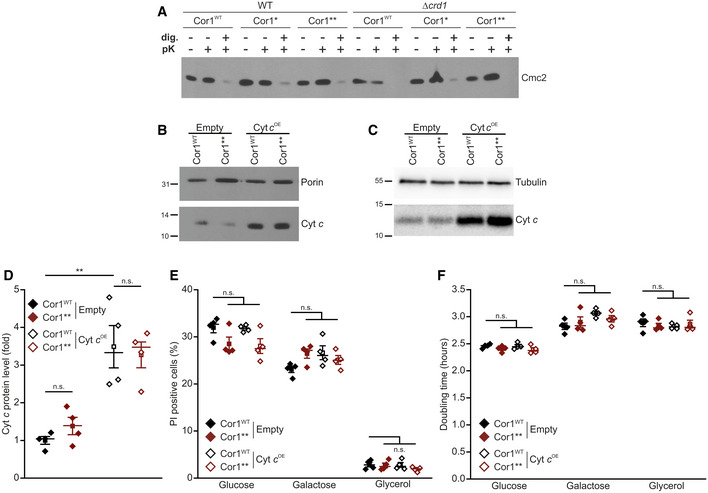
- Assays to verify the intactness of the outer mitochondrial membrane. Mitochondria were isolated from strains expressing the wild‐type form of Cor1 (Cor1WT), as well as the mutants Cor1N63A, N187A, D192A (Cor1*) and Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**) in wild‐type (WT) background as well as in cells lacking CRD1 (crd1Δ). Intactness of the outer mitochondrial membrane was tested by following the stability of the intermembrane protein Cmc2 1 h after treatment with 6 μg/ml proteinase K (pK). Negative control samples were created by membrane permeabilization with 0.5% digitonin (dig.).
- Immunoblot analysis of isolated mitochondria from strains expressing the wild‐type form of Cor1 (Cor1WT) or the mutant Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**). Cells either overexpressed cytochrome c (Cyt c OE) or contained the empty plasmid (Empty) as a control. Blots were probed with antibodies against Cyt c, as well as porin as loading control.
- Immunoblot analysis as described in (B) from intact cells. Blots were probed with antibodies against Cyt c, as well as tubulin as loading control.
- Densitometric quantification of immunoblots validating overexpression of cytochrome c (Cyt c OE) in strains expressing either the wild‐type form of Cor1 (Cor1WT) or the mutant Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**). The Cyt c signal was normalized to tubulin intensities as a loading control, followed by normalization to Cor1WT strains harboring the empty vector to present fold values.
- Flow cytometric quantification of loss of membrane integrity as visualized via propidium iodide (PI) staining of cells described in (D). Cells were cultivated in CM media either containing glucose, galactose or glycerol as carbon source.
- Doubling time of strains described in (E).
Next, we tested whether an increase in endogenous Cyt c levels can restore electron transfer in intact mitochondria in a similar way as observed upon exogenous administration of Cyt c to mitoplasts. We therefore isolated mitochondria from cells overexpressing Cyt c (Fig EV5B). Indeed, higher levels of Cyt c could restore NADH oxidation rates in strains lacking SCs at both basal and phosphorylating conditions (Fig 5A). This therefore showed that increased levels of Cyt c can compensate for the decreased electron transfer efficiency upon SC disruption. Inspired by these results, we next set out to test whether increased level of Cyt c can also restore competitive fitness of cells lacking SCs. In intact cells, overexpression of Cyt c led to a 3.5‐fold increase in protein level compared to control cells (Fig EV5C and D). Despite being a pro‐apoptotic factor upon release into the cytosol in both yeast and mammals (Guaragnella et al, 2012), Cyt c overexpression did not significantly affect cellular growth rates nor cell death within 24 h after inoculation (Fig EV5E and F). This allowed us to determine whether increased Cyt c protein levels can correct the reduced competitive fitness upon SC disruption. Indeed, elevated Cyt c levels restored competitive fitness defects in the Cor1** cells on non‐fermentable carbon sources (Fig 5B–D). In aggregate, our data demonstrate that SC formation mediating close proximity between individual complexes allows efficient electron transfer via Cyt c. Thereby, SC formation constitutes a fitness advantage that enforced the formation of these macromolecular structures during evolution.
Figure 5. Cytochrome c overexpression restores competitive fitness of strains lacking supercomplexes.
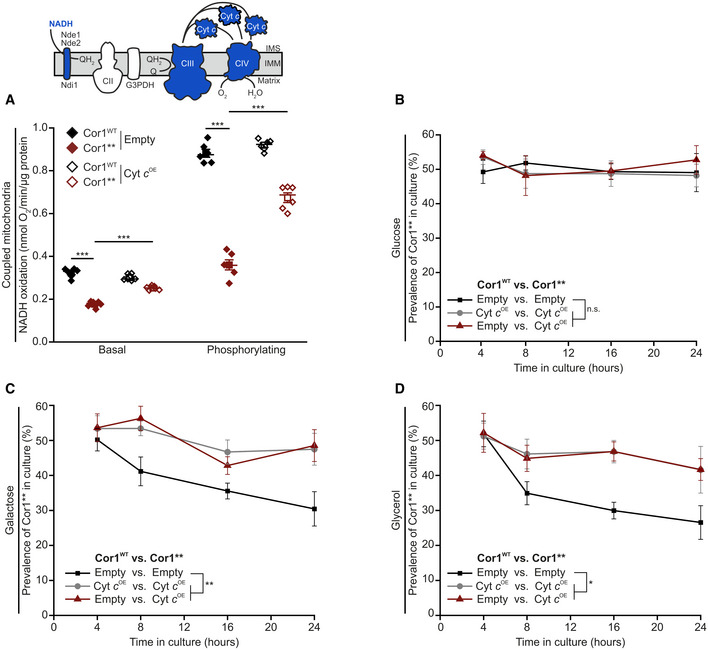
-
APolarographic measurement of KCN‐sensitive oxygen consumption, driven by NADH in isolated coupled mitochondria. Mitochondria were isolated from cells expressing the wild‐type form of Cor1 (Cor1WT) or the mutant Cor1N63A, N187A, D192A, V189A, Y65A, L238A, K240A (Cor1**). Thereby, strains either overexpressed cytochrome c (Cyt c OE) or harbored the empty plasmid as a control (Empty). Measurements were performed in the absence (basal respiration) or presence (phosphorylating condition) of ADP.
-
B–DAnalysis of competitive fitness of strains described in (A). Cor1WT strains harbored a chromosomally integrated clonNAT selection cassette, whereas Cor1** contained a hygromycin selection cassette. Strains were cultivated in CM media containing glucose (B), galactose (C), or glycerol (D) as carbon source.
Since the discovery of mitochondrial respiratory SCs two decades ago, several hypotheses on their functional and physiological relevance have been suggested, including increased electron transfer rates through substrate channeling (Blanchi et al, 2004; Lapuente‐Brun et al, 2013). However, biophysical evidence argues against strict substrate channeling (Blaza et al, 2014; Fedor & Hirst, 2018). Accordingly, the decreased coupling efficiency identified in our work cannot be caused by an impairment of a strictly defined substrate channeling operating by Cyt c trapped in a SC. The distance between the respective Cyt c binding sites between CIII and CIV in SC configuration (70 Å in yeast SC as compared to 100 Å in the mammalian respirasome) renders direct electron transfer via one single bound Cyt c molecule impossible (Rathore et al, 2019). Importantly, our data are in line with a scenario where electron transfer by Cyt c between CIII and CIV is more efficient in the SC configuration than between the separated complexes (Trouillard et al, 2011; Hirst, 2018; Stuchebrukhov et al, 2020). Hence, our data comply with a scenario where the main physiological function of SCs is to bring individual MRC complexes in close proximity, allowing a more efficient electron transfer via the mobile electron carriers between the complexes.
Our data further show that this rather simple function of SCs has extensive implications for the evolution of aerobic organisms. In clonal cultures, the absence of SC formation did not provoke a massive defect in several physiological parameters, including ATP levels, oxidative stress, or chronological life span, thereby disputing the suggestion that SCs play a role in reducing reactive oxygen species (Maranzana et al, 2013). Thus, the electron transfer deficiency caused by SC disruption does either not exceed a critical threshold that would result in an detectable OXPHOS impairment, or it might be counteracted by an upregulation of protective genetic programs to compensate cytotoxicity (Melber & Haynes, 2018; Suhm et al, 2018). In both cases the formation of SCs presents the favorable solution, explaining why these structures arose during evolution. The main driving force of evolution is the competition for resources. In the case of yeast in its natural habitat, this involves competition with other yeast strains and bacteria that populate the same environment. Cells that can use the actual conditions best and consequently contain the most efficient pathways to exploit certain substrates will prevail. Indeed, when applying competitive fitness analyses cells lacking SCs were outcompeted only under conditions requiring respiration, an effect that could be compensated by overexpression of Cyt c. This illustrates that the increased electron transport efficiency, which enhances OXPHOS, is an evolutionary advantage favouring the formation of SCs.
In summary, our results demonstrate that the formation of SCs is important for efficient electron transfer between CIII and CIV by positioning the Cyt c binding sites of both complexes in close vicinity, a conserved feature of SC architecture found in multiple organisms. Thereby, SC formation confers a competitive fitness advantage for aerobic organisms, which constituted a selectable trait during evolution.
Material and Methods
Yeast strains used in this study
All stains in this study were isogenic to the intronless W303 strain MRSI0 (Gruschke et al, 2011) and are listed in the Table EV3. COR1 and CRD1 were disrupted with homologous recombination using a Kanamycin resistance cassette and a TRP1 selection cassette, respectively. The primers used for this approach (COR1_fw; COR1_rev, CRD1_fw and CRD1_rev) are described in Table EV4. Cor1 mutations were ordered from GeneArt Gene Synthesis (Thermo Fisher) and cloned into a pRS305 integrative plasmid, where the variants of the COR1 gene were inserted together with 300 bp upstream and 200 bp downstream of the open reading frame using the restrictions sites BamHI and SalI. COR1‐containing plasmids were linearized with AflII and integrated into the LEU2 locus of the genome with lithium acetate transformation method (Gietz & Woods, 2002). For overexpression of yeast Cyt c in experiments evaluating competitive fitness, the CYC1 gene was amplified from yeast genomic DNA (primers: CYC1_fw and CYC1_rev) and cloned into a pCM190 plasmid using a Gibson Assembly Cloning Kit (New England BioLabs; E5510S) following the supplier's protocol. Thereby, pCM190 was amplified with the primers pCM190_fw and pCM190_rev. For the biochemical experiments, the CYC1 gene under the control of its endogenous promoter was sub‐cloned in the episomal plasmid YEplac112 as a XmaI‐HindIII fragment from the Yep351‐CYC1 (ST13) construct reported in (Barrientos et al, 2003).
Culture conditions
Cells were grown in full media (YP; 1% yeast extract, 2% peptone) or complete minimal (CM) media (0.17% yeast nitrogen base, 0.5% (NH4)2SO4, 30 mg/l of all amino acids—except 80 mg/l histidine and 200 mg/l leucine—30 mg/l adenine and 320 mg/l uracil), as indicated. Media were supplemented with respective carbon sources (2% glucose, 2% galactose or 2% glycerol), and 2% agar was added for solid media. Where applicable, 300 mg/l hygromycin B or 100 mg/l nourseothricin was added after autoclaving for selection.
For assessing cellular consequences of SC disruption, overnight cultures were grown for 16–20 h at 28°C, 145 rpm in CM media containing glucose in glass eprouvettes and used for inoculating the main culture in 10 ml CM medium in baffled 100 ml Erlenmeyer flasks to an OD600 of 0.1. Samples were taken at indicated time points and further processed as described below. Of note, at least four different clones per strain were applied for the analysis to consider clonogenic variation. To suppress Cyt c overexpression in respective strains, doxycycline (40 μg/ml) was added to agar plates and expression was induced by inoculation of cells in media without doxycycline.
Cryo‐EM image processing
Particles from a dataset previously acquired (Rathore et al, 2019) were re‐processed using an updated version of the software cryoSPARC (Punjani et al, 2017) (Fig EV1A; Table EV1). In brief, 397,262 particles with a calibrated pixel size of 1.06 Å per pixel were imported into cryoSPARC v2.2. An initial model of the CIII2/CIV respiratory SC was created using 253,478 particles in the ab initio function of the software before all the particles were 3D‐classified using the heterogeneous refinement tool with a low‐pass filtered (30 Å) version of that model as a reference. This resulted in a class of 201,223 particles that we continued to use. Non‐uniform refinement of these particles generated a 3.17 Å density map (GSFSC = 0.143; B‐factor: −107.1; Fig EV1B), which was used to model the CIII2/CIV respiratory SC. From the atomic coordinates of CIII2, and CIV, two masks with a resolution of 7 Å and 3 pixels extension (6 pixel soft edge) was created using RELION3 (Zivanov et al, 2018) and imported to cryoSPARC. Using the particle subtraction tool and local refinement tool to subtract the signal from CIII2 and refine CIV alone, we could generate a 3.41 Å density map (GSFSC = 0.143; B‐factor: −125.9; Fig EV1B), which was used to model the atoms of CIV. Using cryoSPARCs local resolution function (FSC = 0.5), we estimated the local resolution for both classes (Fig EV1C and D).
Model building, validation, and figure preparation
The 3.17 Å (FSC = 0.143) density map of the CIII2/CIV SC was used for model building the SC in Coot v0.8.8 (Emsley et al, 2010). The previous structure of the CIII2/CIV subunit [PDB: 6GIQ; (Rathore et al, 2019)] was used as a starting reference for all model building. The amino acid positions of both subunits were manually corrected to the new density in Coot after the reference had been rigid‐body fitted to the new map. Additional improvement of the CIV subunits was built using the 3.41 Å (FSC = 0.143) density map. The phospholipid in the interface previously described could here be identified and placed correctly. Restrained model refinement was done with PHENIX v1.13 phenix.real_space_refine (Liebschner et al, 2019) with additional restraints applied for all ligands. These restraints were made using the eLBOW function in PHENIX. First, the separate CIII2 and CIV subunits were refined individually before being combined to perform the final refinement. Settings applied for the rounds of global real‐space refinement were as follows: five macro cycles with secondary‐structure, rotamer, Ramachandran, and Cβ‐torsion restraints. The final model was inspected again in Coot and validated using MolProbity (Chen et al, 2010). Additional refinement statistics are provided in Table EV1.
All figures of the cryo‐EM maps and atomic models were made in UCSF Chimera v1.13.1 (Pettersen et al, 2004), as well as UCSF ChimeraX (Goddard et al, 2018) and processed with CorelDRAW X8.
Isolation of mitochondria
Mitochondria were isolated according to (Meisinger et al, 2006) with slight modifications. In brief, yeast cells were grown either in CM media supplemented with galactose or YP media, containing glycerol to early‐ or mid‐exponential phase (OD600 ≈ 0.8 or 2.0), respectively. Cells were harvested by centrifugation (3,000 g, 5 min) and washed once with distilled water before being resuspended (2 ml/g cell wet weight) in MP1 buffer (0.1 M Tris, 10 mM dithiothreitol, pH 9.4) and incubated for 10 min at 30°C. Cells were then washed with 1.2 M sorbitol, before being resuspended (6.7 ml/g wet cell weight) in MP2 buffer (20 mM potassium phosphate, 0.6 M sorbitol, pH 7.4), containing 3 mg/g of cell wet weight zymolyase 20T. Spheroplasts were created by incubation for 1 h at 30°C and harvested by centrifugation (3,000 g, 5 min, 4°C). After careful resuspension in 13.4 mg/g of cell wet weight in ice‐cold homogenization buffer (10 mM Tris, 0.6 M sorbitol, 1 mM ethylenediaminetetraacetic acid, 1 mM phenylmethylsulfonyl fluoride, pH 7.4), homogenization was performed by ten strokes with a Teflon plunger (Sartorius Stedim Biotech S.A.). Homogenates were centrifuged at 3,000 g for 5 min at 4°C, and the supernatants were subsequently centrifuged at 17,000 g for 12 min at 4°C. Pelleted mitochondria were resuspended in isotonic buffer (20 mM HEPES, 0.6 M sorbitol, pH 7.4) to a concentration of 10 mg/ml.
The intactness of the outer mitochondrial membrane was tested by following the stability of the intermembrane protein Cmc2 2 h after treatment with 6 μg/ml proteinase K. Negative control samples were created by membrane permeabilization with 0.5% digitonin.
Blue‐Native PAGE and immunoblot analysis
Isolated mitochondria (100 μg) were centrifuged at 16,000 g for 10 min at 4°C, and the resulting pellet was resuspended in 15 μl lysis buffer (50 mM Bis‐Tris, 100 mM KCl, 2 mM Aminohexanoic acid, 1 mM EDTA, 1× Complete Protease Inhibitor cocktail (Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF), 12% glycerol, 2% digitonin, pH 7.2) and incubated for 10 min on ice. After centrifugation (16,000 g, 10 min, 4°C), 1.5 μl of sample additive (5% G‐250) was added, and the samples were loaded on a 3–12% precast native gel (Invitrogen). For subsequent immunoblot analysis, a transfer on PVDF membranes was conducted for 2 h at 200 mA.
For the analysis of steady‐state protein levels, cells were harvested when reaching OD600 of 1.5, resuspended in 0.1 NaOH and incubated for 5 min at room temperature (RT). After centrifugation (16,000 g, 5 min, RT), the pellet was resuspended in 50 μl of SDS sample buffer (63 mM Tris, 2% SDS, 10% glycerol, 0.1% β‐mercaptoethanol and 0.0005% bromophenol blue, pH 6.8) and incubated for 3 min at 95°C. After centrifugation (14,000 g, 2 min, RT), 25 μl of the supernatants was applied for standard SDS–PAGE. All antibodies used in this study are listed in the Table EV5.
UV‐VIS spectroscopy
Optical spectra (400–650 nm) of isolated mitochondria were recorded using a Cary4000 UV‐Vis spectrophotometer (Agilent Technologies). In brief, 200 μg of isolated mitochondria grown in YP media containing glycerol were lysed in 20 μl of lysis buffer (50 mM KPi pH 7.4, 150 mM KCl, 1× Complete Protease Inhibitor cocktail (Roche), 1 mM PMSF, 1% n‐Dodecyl β‐D‐maltoside) for 20 min at 4°C, before being mixed with 130 μl of dilution buffer (50 mM KPi pH 7.4, 150 mM KCl) in a microcuvette and measured with a spectrophotometer. To get the reduced spectra, a small amount of sodium dithionite was added to the cuvette before the spectra were measured again. Heme concentrations were determined from the differential spectrum (reduced‐minus‐oxidized) from three separate measurements and preparations. The concentration of a‐type hemes (aa 3) was determined by applying the absorption coefficient ε = 23.2/mM/cm (Vanneste, 1966) at ΔA605, using a linear trend line between 577–630 nm to calculate the base of the peak. Similarly, the bases of heme b (ΔA562) and the c‐type hemes cc 1 (ΔA553) peaks were determined from a trend line between 540–577 nm. The concentrations of these hemes were determined as previously described (Guergova‐Kuras et al, 1999), using the formulas:
To separate heme c from the heme cc 1 peak, mitoplasts were created by swelling the mitochondria in hypotonic buffer (50 mM KPi pH 7.4) for 30 min, before adjusting its concentration of KCl to 500 mM. Mitoplasts were pelleted (25,000 g, 10 min, 4°C), and the supernatant was used for spectroscopic measurements as described above. Heme c concentrations were determined by applying the absorption coefficient ε = 21.8/mM/cm using a trend line between 540–577 nm to calculate its base peak.
Polarographic measurement of substrate oxidation
Using a Clark‐type oxygen electrode (Hansatech Instruments, Norfolk, UK), KCN‐sensitive oxidation of different substrates was analyzed in isolated mitochondria as described recently (Ocampo et al, 2010). Coupled mitochondria (30 μg) were resuspended in 750 μl isotonic respiration buffer (0.6 M mannitol, 20 mM HEPES pH 7.0, 10 mM H3PO4 pH 7.4, 2 mM MgCl2, 1 mM EGTA, and 0.1% BSA) and transferred into the polarographic chamber, which was subsequently supplemented with respiratory substrates (10 μl of 0.1 M NADH, 10 μl of 0.4 M succinate + 10 μl of 0.5 M glycerol‐3‐phosphate, or 9.4 μl of 0.4 M ascorbate + 6 μl of 12 mM N,N,N′,N′‐tetramethyl‐p‐phenylenediamine (TMPD)). Following the measurement of state 2 respiration, 10 μl of 20 mM ADP was admixed, and state 3 respiration was validated. Three μl of 80 mM KCN solution were finally added to assess the specificity of the assay. To analyze maximum uncoupled respiration, similar experiments were performed in hypotonic respiratory buffer containing 10 mM H3PO4 (pH 7.4) and 1 mM EDTA. In some experiments performed in these conditions, oxidized cytochrome c (3 μl of 1% solution (w/v)) was added to assess exogenous cytochrome c‐stimulated respiration. To control for the complete oxidation of our cytochrome c stock, we verified that no increase in respiration occurred when cytochrome c was added to uncoupled mitochondria in the absence of substrates.
Spectrophotometry of respiratory chain activity
Isolated mitochondria were also used for spectrophotometric assays performed at 24°C to measure KCN‐sensitive cytochrome c oxidase (CIV) activity and antimycin A‐sensitive cytochrome c reductase (CIII) activity, as described in (Barrientos et al, 2009). Briefly, CIII enzymatic activity was measured in 1 ml 20 mM phosphate buffer pH 7.4 containing 0.05% oxidized cytochrome c, 0.4 μM KCN and 50 μM decyl‐ubiquinol (coenzyme Q reduced with lithium borohydride). The reaction was started by addition of approximately 30 μg of mitochondrial proteins permeabilized with final 0.5% sodium deoxycholate. Addition of the CIII inhibitor antimycin A (80 μM final concentration) was used to test reaction specificity. Similarly, CIV enzymatic activity was measured in 1 ml 20 mM phosphate buffer pH 7.4 containing 0.08% sodium dithionite‐reduced cytochrome c. The reaction was started by addition of approximately 20 μg of mitochondrial proteins permeabilized with final 0.5% sodium deoxycholate. Addition of the CIV inhibitor KCN (0.24 μM final concentration) was used to test reaction specificity. For both reaction, absorption was recorded at 550 nm wavelength and specific activities were calculated using the cytochrome c extinction coefficient at 550 nm of 18.5/mM/cm.
Analysis of cellular growth
Cellular growth was analyzed with a Bioscreen CTM automated microbiology growth curve analysis system (Growth Curves, USA). Overnight cultures were grown as described above and used for inoculation in CM media containing indicated carbon sources to an OD600 of 0.1 in the suppliers “honeycomb microplates”. Thereby, a final volume of 250 μl per well was used and automatically measured every 30 min at 28°C and shaking at the maximum level. The respective medium without cells was applied as blank, and doubling time was calculated from growth curves during the logarithmic growth phase.
Analysis of oxidative stress and cell death
Oxidative stress was flow cytometrically quantified by simultaneous staining of cells with 2′,7′‐dichlorodihydrofluorescein diacetate (DCFH2‐DA; Sigma) and propidium iodide (PI; Sigma). Approximately 2 × 106 cells were collected by centrifugation in 96‐well plates and resuspended in 250 μl phosphate‐buffered saline (PBS, 25 mM potassium phosphate, 0.9% NaCl, pH 7.2) containing 100 μg/l PI and 100 mM DCFH2‐DA. PI is a fluorescence dye that is taken up in cells upon loss of membrane integrity, indicating cell death, and non‐fluorescent DCFH2‐DA is oxidized by reactive oxygen species and nitric oxide to fluorescent 2′,7′‐dichlorofluorescein (DCF), indicating overall oxidative stress. Cells were incubated for 10 min at RT in the dark, washed once in PBS, and 30,000 cells per sample were analyzed via a BD LSR Fortessa and FACSDivia software. Mean fluorescence intensity of the DCF from PI‐negative cells was evaluated as a readout for pre‐lethal oxidative stress and was normalized to control cells expressing the wild‐type form of Cor1 (Cor1WT) on respective media to present fold values. For quantification of cell death, the percentage of PI‐positive cells in the culture is visualized.
Analysis of mitochondrial transmembrane potential
Assessment of mitochondrial transmembrane potential (Δψm) of intact cells was performed according to (Aufschnaiter et al, 2018) with slight adaptations. In brief, approximately 2 × 106 cells were harvested in 96‐well plates and resuspended in 250 μl PBS containing 5% glucose and 200 nM Mitotracker CMXRos (Thermo Fisher Scientific). Cells were incubated for 10 min at RT in the dark, washed once in PBS, and analyzed by flow cytometry as described above. Dead cells that accumulated the dye due to a loss of membrane integrity were excluded from the analysis, and the mean fluorescence intensity of Mitotracker CMXRos of the remaining population was used as a readout for Δψm. Normalization to Cor1WT was done as described for DCFH2‐DA/PI staining.
Confocal microscopy
For monitoring Δψm via confocal microscopy, the specimens were prepared on agar slides to immobilize yeast cells after staining as described above. Samples were monitored with a Leica SP5 confocal laser scanning microscope, equipped with a Leica HCX PL Apo 63× NA 1.4 oil immersion objective. To efficiently monitor the 3‐dimensional mitochondrial network, Z‐stacks were recorded using 64 × 64 × 12.6 (x/y/z) nm sampling and Z‐projections were created with the open‐source software Fiji (Schindelin et al, 2012). Thereby, Gaussian filtering (xσ = yσ = zσ = 1), followed by background subtraction (rolling ball radius = 50 pixels) was conducted, and the pictures were visualized with the maximum‐intensity projection method. The dynamic range of presented figures was adapted by using the “Brightness/contrast” tool of Fiji. All pictures within an experiment were captured and processed with the same settings.
Measurement of oxygen consumption in intact cells
Oxygen consumption of intact yeast cells was investigated with a Fire‐Sting optical oxygen sensor system (Pyro Science) as described recently (Aufschnaiter et al, 2018). Oxygen sensor spots were mounted on 2 ml clear glass vials, filled with culture, hermetically sealed, and immediately used for analysis. Oxygen concentration in the vials was measured for 1 min, and the slope of the regression line was calculated. In parallel, the percentage of cells alive was determined via flow cytometric analysis of PI stained cells (as described above), and the total number of cells per sample was quantified with a CASY cell counting device (Schärfe Systems). These parameters were used to normalize the quantified oxygen consumption to the number of cells alive in the sample, which is expressed as fold change compared with Cor1WT cells cultivated in respective media.
Determination of cellular ATP content
Cellular ATP levels were quantified with a luminescent ATP detection assay kit (Abcam) as described recently (Aufschnaiter et al, 2018). Thereby, 1 ml of culture was harvested, and ATP was extracted with the hot ethanol method, where cells were flash‐frozen in liquid nitrogen, resuspended in 0.5 ml BES buffer (75% EtOH, 10 mM (NH4)2SO4) and incubated for 3 min at 90°C. After centrifugation for 20 min, 4°C with 16,000 g, samples were diluted 20‐fold in Tris buffer (20 mM, pH 8), and 150 μl were transferred into a low‐bind 96‐well plate and incubated for 5 min for adaption to RT. 50 μl of the supplied substrate solution were added in each well, and the plate was placed into a GloMax Multi detection system (Promega) for 10 min for incubation and dark adaption. The luminescence signal was then measured via integration over 10 s. Normalization to cells alive via PI staining and cell counting and calculation of fold values was done as described above for oxygen consumption assays.
Analysis of competitive fitness
For competitive fitness assays, a Hygromycin selection cassette was amplified from a pFA6a‐hphNT1 plasmid (Janke et al, 2004) with the primer HIS3_fw and HIS3_rev (listed in Table EV4) and integrated via homologous recombination into the HIS3 locus of Cor1WT and Cor1**. Similarly, the Nourseothricin (clonNAT) selection cassette was amplified from pFA6a‐natNT2 (Janke et al, 2004) with the same primer set and integrated into the HIS3 locus of Cor1WT and Cor1**. Separated overnight cultures of each strain (Cor1WT with Hygromycin resistance and Cor1* with clonNAT resistance) were used to inoculate a mixed culture with OD600 of 0.1 per strain. Thereby, CM media was used with indicated carbon sources. The mixed cultures were grown at 28°C, 145 rpm, and aliquots were taken at indicated time points and plated on both Hygromycin and clonNAT containing YP agar plates with glucose. After 2 days of incubation, colony‐forming units were counted and the prevalence of Cor1** was calculated as the percentage of colony‐forming units with resistance against clonNAT compared with colony‐forming units on Hygromycin and clonNAT containing agar plates. Of note, this experiment was conducted with a second set of strains, where selection markers were switched (Cor1WT resistant against clonNAT, Cor1** resistant against Hygromycin) to avoid artefacts mediated by the different selection cassettes.
Statistical analysis
Results are presented as line graphs, indicating mean ± standard error of the mean (s.e.m.), or dot plots with mean (square) ± s.e.m. and median (center line), as well as single data points. The number of n represents the amount of individual clones or mitochondrial preparations used for the analysis and is indicated in respective figure legends. Outliers were defined as data points outside the 2.2‐fold interquartile range and are highlighted in turquoise. Upon the presence of outliers, alternative non‐parametric tests were performed (for a detailed description of the statistical analysis of each experiment performed see Table EV6). The normal distribution of the data was assessed with a Shapiro–Wilk's test, and the homogeneity of variances was evaluated with a Levene's test (both analyzed with OriginPro 2020, OriginLab). A detailed description of the procedure upon violation of respective assumptions is presented in Table EV6. The means of three or more groups were compared upon the presence of one independent variable (genotype) with a one‐way Analysis of Variance (ANOVA) followed by Bonferroni post hoc test (analyzed with OriginPro 2020). Where applicable, a Kruskal–Wallis test with Bonferroni post hoc test was performed as non‐parametric alternative for a one‐way ANOVA (IBM SPSS statistics 26). Upon the presence of two independent variables (genotype and treatment), a two‐way ANOVA followed by a Bonferroni post hoc test was conducted with OriginPro 2020. To analyze the differences across two or more different groups in time‐dependent experiments, a two‐way ANOVA mixed design was conducted with genotype or media as between‐subject and time as within‐subject factor, followed by a Bonferroni post hoc test (Origin Pro 2020).
Significances for analyses with one independent variable are indicated with asterisks (***P < 0.001, **P < 0.01, *P < 0.05, n.s. P > 0.05), and for two independent variables main effects are displayed with diamonds (### P < 0.001, ## P < 0.01, # P < 0.05, n.s. P > 0.05) and simple main effects are depicted as asterisks (***P < 0.001, **P < 0.01, *P < 0.05, n.s. P > 0.05). Calculated P‐values are presented in Table EV6. Figures were created with OriginPro 2020 and further processed with CorelDRAW X8.
Author contributions
Conceptualization: MO, SB, and FF; Methodology: JB, AK, AB, FF, and MO; Validation: FF and MO; Formal Analysis: JB, AK, and SR; Investigation: JB, AK, SR, LM‐B, HD, JD, VK, and FF; Resources: AB, SB, FF, and MO; Data Curation: JB, AK, SR, FF, and MO; Writing—Original Draft: JB, AK, and MO; Writing—Review and Editing: JB, AK, HD, VK, AB, SB, FF, and MO; Visualization: JB, AK, and MO; Supervision: SB, FF, and MO; Project Administration: MO; Funding Acquisition: AK, VK, AB, SB, FF, and MO.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Acknowledgements
We would like to thank all members of our groups for stimulating discussions and the cryo‐EM facility at SciLifelab, Solna, Sweden, for support. The cryo‐EM data were collected at the Swedish national cryo‐EM facility funded by the Knut and Alice Wallenberg, Family Erling Persson, and Kempe Foundations. This work was supported by the Austrian Science Fund FWF (J4398‐B to AA; J4342‐B21 to VK; P27183‐B24 to SB), the Swedish Research Council (2014‐4116 and 2018‐03694 to MO; 2015‐05468 and 2019‐05249 to SB), the Knut and Alice Wallenberg foundation (2017.0091 to MO and SB; 2013.0006 to MO), Stiftelsen Olle Engkvist Byggmästare (194‐0681 to SB), NIH R35 Grant GM118141 (to A.B.), MDA Grant MDA‐381828 (to A.B.), and AHA Development Grant 14SDG20040003 (to F.F.).
EMBO Reports (2020) 21: e51015
See also: F den Brave & T Becker (December 2020)
Footnotes
Correction added on 8 December 2020, after first online publication: The last paragraph of the introduction and the references therein were added. This replaces the previous last two sentences of the introduction.
Contributor Information
Sabrina Büttner, Email: sabrina.buettner@su.se.
Flavia Fontanesi, Email: ffontanesi@med.miami.edu.
Martin Ott, Email: martin.ott@dbb.su.se.
Data availability
Cryo‐EM maps and atomic coordinates have been deposited at the Electron Microscopy Data Bank (EMD) and the Protein Data Bank (PDB) with the accession codes for the CIII2/CIV [EMD‐10847 (http://www.ebi.ac.uk/pdbe/entry/EMD-10847) and 6YMX (http://www.rcsb.org/pdb/explore/explore.do?structureId=6YMX)] and CIV model [EMD‐10848 (http://www.ebi.ac.uk/pdbe/entry/EMD-10848) and 6YMY (http://www.rcsb.org/pdb/explore/explore.do?structureId=6YMY)].
References
- Acin‐Perez R, Enriquez JA (2014) The function of the respiratory supercomplexes: the plasticity model. Biochim Biophys Acta 1834: 444–450 [DOI] [PubMed] [Google Scholar]
- Acín‐Pérez R, Bayona‐Bafaluy MP, Fernández‐Silva P, Moreno‐Loshuertos R, Pérez‐Martos A, Bruno C, Moraes CT, Enríquez JA (2004) Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell 13: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acín‐Pérez R, Fernández‐Silva P, Peleato ML, Pérez‐Martos A, Enriquez JA (2008) Respiratory active mitochondrial supercomplexes. Mol Cell, 32: 529–539 [DOI] [PubMed] [Google Scholar]
- Aufschnaiter A, Kohler V, Walter C, Tosal‐Castano S, Habernig L, Wolinski H, Keller W, Vögtle FN, Büttner S (2018) The enzymatic core of the parkinson's disease‐associated protein LRRK2 impairs mitochondrial biogenesis in aging yeast. Front Mol Neurosci 11: 205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsa E, Marco R, Perales‐Clemente E, Szklarczyk R, Calvo E, Landázuri MO, Enríquez JA (2012) NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab 16: 378–386 [DOI] [PubMed] [Google Scholar]
- Balsa E, Soustek MS, Thomas A, Cogliati S, García‐Poyatos C, Martín‐García E, Jedrychowski M, Gygi SP, Enriquez JA, Puigserver P (2019) ER and nutrient stress promote assembly of respiratory chain supercomplexes through the PERK‐eIF2α axis. Mol Cell 74: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Pierre D, Lee J, Tzagoloff A (2003) Cytochrome oxidase assembly does not require catalytically active cytochrome c . J Biol Chem 278: 8881–8887 [DOI] [PubMed] [Google Scholar]
- Barrientos A, Fontanesi F, Díaz F (2009) Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr Protoc Hum Genet Chapter 19: Unit19.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchi C, Genova ML, Castelli GP, Lenaz G (2004) The mitochondrial respiratory chain is partially organized in a supercomplex assembly: kinetic evidence using flux control analysis. J Biol Chem 279: 36562–36569 [DOI] [PubMed] [Google Scholar]
- Blaza JN, Serreli R, Jones AJY, Mohammed K, Hirst J (2014) Kinetic evidence against partitioning of the ubiquinone pool and the catalytic relevance of respiratory‐chain supercomplexes. Proc Natl Acad Sci USA 111: 15735–15740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Fukui H, Garcia S, Moraes CT (2006) Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol 26: 4872–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor JG, Hirst J (2018) Mitochondrial supercomplexes do not enhance catalysis by quinone channeling. Cell Metab 28: 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Poyatos C, Cogliati S, Calvo E, Hernansanz‐Agustín P, Lagarrigue S, Magni R, Botos M, Langa X, Amati F, Vázquez J et al (2020) Scaf1 promotes respiratory supercomplexes and metabolic efficiency in zebrafish. EMBO Rep 21: e50287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single‐stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE (2018) UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio C, Jha P, Kulkarni SS, Lagarrigue S, Broskey NT, Boutant M, Wang X, Conde Alonso S, Ofori E, Auwerx J et al (2017) Enhanced respiratory chain supercomplex formation in response to exercise in human skeletal muscle. Cell Metab 25: 301–311 [DOI] [PubMed] [Google Scholar]
- Gruschke S, Kehrein K, Römpler K, Gröne K, Israel L, Imhof A, Herrmann JM, Ott M (2011) Cbp3‐Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J Cell Biol 193: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S, Römpler K, Hildenbeutel M, Kehrein K, Kühl I, Bonnefoy N, Ott M (2012) The Cbp3‐Cbp6 complex coordinates cytochrome b synthesis with bc1 complex assembly in yeast mitochondria. J Cell Biol 199: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guaragnella N, Ždralevic M, Antonacci L, Passarella S, Marra E, Giannattasio S (2012) The role of mitochondria in yeast programmed cell death. Front Oncol 2: 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guergova‐Kuras M, Salcedo‐Hernandez R, Bechmann G, Kuras R, Gennis RB, Crofts AR (1999) Expression and one‐step purification of a fully active polyhistidine‐tagged cytochrome bc1 complex from Rhodobacter sphaeroides . Protein Expr Purif 15: 370–380 [DOI] [PubMed] [Google Scholar]
- Guerrero‐Castillo S, Baertling F, Kownatzki D, Wessels HJ, Arnold S, Brandt U, Nijtmans L (2017) The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab 25: 128–139 [DOI] [PubMed] [Google Scholar]
- Hartley AM, Lukoyanova N, Zhang Y, Cabrera‐Orefice A, Arnold S, Meunier B, Pinotsis N, Maréchal A (2019) Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc 1. Nat Struct Mol Biol 26: 78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildenbeutel M, Hegg EL, Stephan K, Gruschke S, Meunier B, Ott M (2014) Assembly factors monitor sequential hemylation of cytochrome b to regulate mitochondrial translation. J Cell Biol 205: 511–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J (2018) Open questions: respiratory chain supercomplexes‐why are they there and what do they do? BMC Biol 16: 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D, Fontanesi F, Barrientos A (2008) Exploring protein‐protein interactions involving newly synthesized mitochondrial DNA‐encoded proteins. Methods Mol Biol 457: 125–139 [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno‐Borchart A, Doenges G, Schwob E, Schiebel E et al (2004) A versatile toolbox for PCR‐based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962 [DOI] [PubMed] [Google Scholar]
- Kauppila TES, Kauppila JHK, Larsson NG (2017) Mammalian mitochondria and aging: an update. Cell Metab 25: 57–71 [DOI] [PubMed] [Google Scholar]
- Kawamata H, Manfredi G (2017) Proteinopathies and OXP HOS dysfunction in neurodegenerative diseases. J Cell Biol 216: 3917–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuente‐Brun E, Moreno‐Loshuertos R, Aciń‐Pérez R, Latorre‐Pellicer A, Colaś C, Balsa E, Perales‐Clemente E, Quirós PM, Calvo E, Rodríguez‐Hernández MA et al (2013) Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340: 1567–1570 [DOI] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ et al (2019) Macromolecular structure determination using X‐rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75: 861–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Barrientos A (2013) Transcriptional regulation of yeast oxidative phosphorylation hypoxic genes by oxidative stress. Antioxid Redox Signal 19: 1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo‐Jarne T, Nývltová E, Pérez‐Pérez R, Timón‐Gómez A, Molinié T, Choi A, Mourier A, Fontanesi F, Ugalde C, Barrientos A (2018) Human COX7A2L regulates Complex III biogenesis and promotes supercomplex organization remodeling without affecting mitochondrial bioenergetics. Cell Rep 25: 1786–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo‐Jarne T, Ugalde C (2018) Respiratory chain supercomplexes: structures, function and biogenesis. Semin Cell Dev Biol 76: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo‐Jarne T, Pérez‐Pérez R, Fontanesi F, Timón‐Gómez A, Wittig I, Peñas A, Serrano‐Lorenzo P, García‐Consuegra I, Arenas J, Martín MA et al (2020) Multiple pathways coordinate assembly of human mitochondrial complex IV and stabilization of respiratory supercomplexes. EMBO J 39: e103912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranzana E, Barbero G, Falasca AI, Lenaz G, Genova ML (2013) Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid Redox Signal 19: 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN (2006) Isolation of yeast mitochondria. Methods Mol Biol 313: 33–39 [DOI] [PubMed] [Google Scholar]
- Melber A, Haynes CM (2018) UPR mt regulation and output: a stress response mediated by mitochondrial‐nuclear communication. Cell Res 28: 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic D, Blaza JN, Larsson NG, Hirst J (2017) The enigma of the respiratory chain supercomplex. Cell Metab 25: 765–776 [DOI] [PubMed] [Google Scholar]
- Mitchell P (1966) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev 41: 445–502 [DOI] [PubMed] [Google Scholar]
- Moreno‐Lastres D, Fontanesi F, García‐Consuegra I, Martín MA, Arenas J, Barrientos A, Ugalde C (2012) Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab 15: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier A, Matic S, Ruzzenente B, Larsson NG, Milenkovic D (2014) The respiratory chain supercomplex organization is independent of COX7a2l isoforms. Cell Metab 20: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A, Zambrano A, Barrientos A (2010) Suppression of polyglutamine‐induced cytotoxicity in Saccharomyces cerevisiae by enhancement of mitochondrial biogenesis. FASEB J 24: 1431–1441 [DOI] [PubMed] [Google Scholar]
- Perez‐Perez R, Lobo‐Jarne T, Milenkovic D, Mourier A, Bratic A, García‐Bartolomé A, Fernández‐Vizarra E, Cadenas S, Delmiro A, García‐Consuegra I et al (2016) COX7A2L is a mitochondrial Complex III binding protein that stabilizes the III2+IV supercomplex without affecting respirasome formation. Cell Rep 16: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera – a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA (2017) CryoSPARC: algorithms for rapid unsupervised cryo‐EM structure determination. Nat Methods 14: 290–296 [DOI] [PubMed] [Google Scholar]
- Rathore S, Berndtsson J, Marin‐Buera L, Conrad J, Carroni M, Brzezinski P, Ott M (2019) Cryo‐EM structure of the yeast respiratory supercomplex. Nat Struct Mol Biol 26: 50–57 [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, De Coo R, Bauer MF, Hofmann S, Godino C, Brandt U (2004) Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem 279: 36349–36353 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba S, Ikeda K, Horie‐Inoue K, Nakayama A, Tanaka T, Inoue S (2017) Deficiency of COX7RP, a mitochondrial supercomplex assembly promoting factor, lowers blood glucose level in mice. Sci Rep 7: 7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchebrukhov A, Schäfer J, Berg J, Brzezinski P (2020) Kinetic advantage of forming respiratory supercomplexes. Biochim Biophys Acta 1861: 148193 [DOI] [PubMed] [Google Scholar]
- Suhm T, Kaimal JM, Dawitz H, Peselj C, Masser AE, Hanzén S, Ambrožič M, Smialowska A, Björck ML, Brzezinski P et al (2018) Mitochondrial translation efficiency controls cytoplasmic protein homeostasis. Cell Metab 27: 1309–1322 [DOI] [PubMed] [Google Scholar]
- Timón‐Gómez A, Garlich J, Stuart RA, Ugalde C, Barrientos A (2020) Distinct roles of mitochondrial HIGD1A and HIGD2A in respiratory complex and supercomplex biogenesis. Cell Rep 31: 107607 [DOI] [PubMed] [Google Scholar]
- Trouillard M, Meunier B, Rappaport F (2011) Questioning the functional relevance of mitochondrial supercomplexes by time‐resolved analysis of the respiratory chain. Proc Natl Acad Sci USA, 108: E1027–E1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste WH (1966) The stoichiometry and absorption spectra of components a and a3 in cytochrome c oxidase. Biochemistry 5: 838–848 [DOI] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W (2002) Gluing the respiratory chain together: cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem 277: 43553–43556 [DOI] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW (2018) New tools for automated high‐resolution cryo‐EM structure determination in RELION‐3. Elife 7: e42166 [DOI] [PMC free article] [PubMed] [Google Scholar]


