Abstract
Background
Infections caused by Enterobacteriaceae are mainly treated with the β-lactam antibiotics, nevertheless, the emergence of species with plasmid-borne β-lactamases has decreased the efficacy of these antibiotics. Therefore, continuing studies on the resistance pattern of different regions is important for assessment of proper antimicrobial therapy protocols. The study aimed to characterize extended-spectrum β-lactamase (ESBL) and AmpC β –lactamase (AmpC) producing Enterobacteriaceae isolated from community-acquired UTIs in Egypt.
Methods
Out of 705 urine samples, 440 Enterobacteriaceae isolates were investigated to detect ESBL and AmpC β -lactamases producers by phenotypic and molecular methods.
Results
Out of 440 Enterobacteriaceae isolates, 311 were identified as ESBL producers by phenotypic testing. ESBL genes were detected in 308 isolates. BlaCTX-M-type was the most prevalent 254 (81.6%), out of them blaCTXM-15 was the commonest (152, 48.8%) followed by blaCTX-M-1 (140, 45%), blaCTX-M-8 (72, 23.1%) and lastly blaCTX-M-2 (4, 1.3%).
blaTEM gene also was detected in a high rate (189, 60.7%). Two hundred and thirty-five (75.5%) of ESBL producers harbored blaCTX-M in combination with blaTEM and/or blaSHV genes. Multiple drug resistance in the ESBL-producers was significantly (P < 0.05) higher than in non–ESBL producers. Imipenem was the most effective drug against ESBL producers. Among 35 cefoxitin resistant isolates, 18 (51.4%) identified as carrying AmpC genes by multiplex PCR. Within AmpC β -lactamase genes, DHA gene was the predominant gene (15, 42.3%). CIT and MOX genes were also present, but in a low rate (5, 14.2% and 4, 11.4%) respectively. Co-existence of multiple AmpC genes was detected exclusively in K. pneumoniae isolates. E. coli isolates harbored DHA gene only. However, FOX gene was not detected in the study isolates. Seventeen of isolates carrying AmpC genes were also positive for ESBL genes.
Conclusion
The study shows that the prevalence of ESBL producing Enterobacteriaceae spread in south Egypt is alarming, however AmpC β -lactamase production is not so high.
Keywords: Enterobacteriaceae, AmpC β -lactamase, Extended-spectrum β-lactamase (ESBLs)
Background
Enterobacteriaceae are the most common pathogens causing urinary tract infections (UTIs) [1]. Increasing rates of antimicrobial resistance among Enterobacteriaceae strains decrease the options for empiric treatment of these infections [2]. These pathogens are the main bacteria found to be associated with extended-spectrum β-lactamase (ESBL) production [2]. Infections caused by ESBL-producing strains are considered a serious global health concern [3, 4] as these infections are associated with higher morbidity and mortality rates [5]. ESBL production is a mechanism of resistance in which the beta-lactam ring of antimicrobials such as penicillins and cephalosporins is hydrolyzed [6]. Until 2000s, blaSHV and blaTEM types of ESBLs used to be the commonest ESBL genotypes found in Enterobacteriaceae strains [7]. The corresponding genes were often found on plasmids that facilitate their rapid spread between different bacterial species [8, 9]. After that, blaCTX-M types were recorded as the commonest genotypes among Enterobacteriaceae strains causing human infections worldwide (particularly blaCTX-M-15) [10]. There are other variants of β-lactamases such as AmpC β -lactamase, that can mediate resistance to several antibiotics as penicillins, cephamycins (e.g., cefoxitin and cefotetan), and oxyimino-cephalosporins [11]. Resistance to broad-spectrum β-lactams mediated by ESBLs and AmpC β -lactamase enzymes has posed a great health burden [12], particularly in developing countries where the resistance rates are high. Additionally, drug use guidelines and studies on this issue are not enough in these countries [13]. Due to a lack of solid data regarding the emergence of ESBLs and AmpC β -lactamase enzymes from Egypt, particularly south Egypt, this study aimed to determine the prevalence of ESBLs and AmpC β -lactamase production in Enterobacteriaceae isolated from patients suffering from community- acquired UTIs and characterize these strains using phenotypic and genotypic assays.
Methods
Study design
This prospective study was conducted in the Department of Medical Microbiology and Immunology, Faculty of Medicine, Minia University, Egypt from June 2018 to December 2018. Urine samples were obtained by simple random sampling method from patients with suggested community-acquired UTI in 3 teaching hospitals in Minia, Egypt; Minia university hospital, Suzan Mubarak University hospital and Renal university hospital. The study included 705 patients of both sexes and different ages attending the outpatient’s clinics or admitted to the inpatient’s wards (who developed symptoms within 48 h of admission), who had no history of antibiotics use in the last 2 weeks. Demographic and clinical history of the patients were recorded. The samples were collected using the clean-catch midstream urine sampling technique.
Bacterial isolates
Calibrated 0.01 mL urine plastic loops were used to inoculate Urine samples on 5% blood agar and MacConkey agar plates. The plates were incubated for 24 h at 37 °C. Samples with suspected contamination and that had multiple organisms were excluded from the study. Urine samples with positive cultures with a colony count ≥ 105 colony-forming units per milliliter (CFU/mL) were only included. Out of 705 non repetitive samples included in the study, 440 isolates of Enterobacteriaceae were identified. Enterobacteriaceae isolates were identified by the standard biochemical tests including IMViC (indole, methyl red, Voges-Proskauer, citrate utilization), sugar fermentation, urease, and motility tests. The identified isolates were confirmed by chromogenic media (CHROMagar™ Orientation, Paris, France) and kept in trypticase soy broth with sterilized 15% glycerol at − 20 °C for further examination. The sample size was calculated using the formula advanced by Kish, 1965 [14], Basing on results of a previous study on the prevalence of ESBL and AmpC β -lactamase production in Egypt by Wassef et al., 2014 [15].
Antibiotic susceptibility testing
Disk diffusion method was used for identification of antibiotic susceptibility of the Enterobacteriaceae isolates to different antibiotics according to CLSI guidelines [16]. The used discs were; amoxicillin/clavulanic acid (AMC) 20 μg/10 μg, ceftazidime (CAZ) 30 μg, ceftriaxone (CRO) 30 μg, imipenem (IPM) 10 μg, amikacin (AK) 30 μg, gentamicin (CN)10 μg, nitrofurantoin (F) 300 μg, ciprofloxacin (CIP) 5 μg and cefoxitin (FOX) 30 μg (for detection of AmpC production) (Thermo Scientific™ Oxoid, UK). Resistance to three or more classes of antimicrobial agents is defined as Multiple drug resistance (MDR) [17].
Screening for ESBLs -producing strains
According to the CLSI guidelines, isolates with inhibition zone size ≤22 mm with ceftazidime (CAZ) 30 μg and ≤ 25 mm with ceftriaxone (CRO) 30 μg were suggested to be ESBL-producers and subjected to further phenotypic and genotypic examination. Double-Disc Synergy Test (DDST) was used for confirmation of ESBL production. Standard (0.5 McFarland) inoculum of the study isolates were inoculated on Mueller Hinton agar plates. Ceftazidime (CAZ) (30 μg) and ceftriaxone (CRO) 30 μg discs were applied on agar 1.5 cm away from the center of amoxicillin-clavulanic acid (AMC) (20 μg/10 μg) disc and incubated at 35 °C for 18 h. Positive result is identified when the zone of inhibition is extended towards AMC (20 μg/10 μg) disc > 5 mm [18].
Screening for AmpC β-lactamase-producing strains
Strains were screened using disk diffusion method in which cefoxitin (FOX) 30 μg disc was used. Isolates showing an inhibitory zone diameter ≤ 18 mm were suspected to be AmpC β-lactamase producers [19]. Disc Approximation Assay (D Test) was also performed; a blunting in the inhibitory zone (D shaped) around the CAZ (30 μg) towards the side of one of the inducers (IPM (10 μg), FOX (30 μg), and AMC (30 μg)) is considered as positive for inducible AmpC β-lactamase production [20].
Molecular characterization of ESBLs and plasmid mediated AmpC β-lactamase genes
DNA extraction was done using QIAamp Mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. All isolates that were phenotypically resistant to β-lactams were screened for ESBL genes by the polymerase chain reaction (PCR), Including blaTEM, blaSHV, blaCTX-M (1, 2, 8, 9, 15) genes. Presence of other resistance genes previously associated with plasmids encoding blaCTX-M-15 as aac(6′)-Ib-cr was screened by PCR. A multiplex PCR was used to examine the presence of plasmid-mediated AmpC genes, including; MOX, CIT, DHA, and FOX genes. Amplified products were resolved on 2% agarose gel electrophoresis and visualized under a UV transilluminator (Biometra, Germany). The primer sequences and amplification conditions are shown in Table 1. Amplified products (one sample for each gene) sequences were analyzed (Applied Biosystems, USA), according to the BLAST software of the National Library of Medicine (http://www.ncbi.nlm.nih.gov/blast).
Table 1.
PCR primers of the current study
| Gene name | Primer sequence | fragment Size (bp) | Annealing Temperature | Reference |
|---|---|---|---|---|
| blaTEM |
AAACGCTGGTGAAAGTA AGCGATCTGTCTAT |
822 | 58 | [21] |
| blaSHV |
ATGCGTTATATTCGCCTGTG TGCTTTGTTATTCGGGCCAA |
753 | 60 | [21] |
| blaCTX-M-1 |
GGT TAA AAA ATC ACT GCG TC TTG GTG ACG ATT TTA GCC GC |
850 | 55 | [22] |
| blaCTX-M - 9 |
ATG GTG ACA AAG AGA GTG CA CCC TTC GGC GAT GAT TCT C |
850 | 55 | [22] |
| blaCTX-M- 2 |
F CGACGCTACCCCTGCTATT R CCAGCGTCAGATTTTTCAGG |
552 | 52 | [23] |
| blaCTX-M − 8 |
TCGCGTTAAGCGGATGATGC AACCCACGATGTGGGTAG |
666 | 52 | [23] |
| blaCTX-M-15 |
CACACGTGGAATTTAGGGACT GCCGTCTAAGGCCATAAACA |
996 | 55 | [24] |
| MOX |
GCTGCTCAAGGAGCACAGGAT CAC ATT GAC ATA GGT GTG GTG C |
520 | 64 | [25] |
| FOX |
AAC ATG GGG TAT CAG GGA GAT G CAA AGC GCG TAA CCG GAT TGG |
190 | ||
| DHA |
AAC TTT CAC AGG TGT GCT GGG T CCG TAC GCA TAC TGG CTT TGC |
405 | ||
| CIT |
TGG CCA GAA CTG ACA GGC AAA TTT CTC CTG AAC GTG GCT GGC |
462 | ||
| aac(6_)-Ib |
F: TTGCGATGCTCTATGAGTGGCTA R: CTCGAATGCCTGGCGTGTTT |
482 | 55 | [26] |
Statistical analysis
Statistical analysis of demographic, clinical and laboratory data of study subjects was performed using SPSS for windows version 19.0 (IBM, USA). The chi -square test was used for analyzing categorical variables. P value < 0.05 was considered statistically significant (two-tailed).
Results
Demographic data and distribution of Enterobacteriaceae strains
A total of 440 Enterobacteriaceae strains were isolated from urine specimens of 440 patients suffering from UTI. The mean age of the patients was 38.8 ± 12.5 years (range, 5–60 years). A total of 299 (68%) were females and 141 (32%) were males. The majority of isolates were E. coli (303/440 (68.9%), followed by Klebsiella pneumoniae (K. pneumoniae) (71/440, 16.1%), Citrobacter spp. (40/440, 9.1%), Proteus spp. (15/440, 3.4%) and Enterobacter spp. (11/440, 2.5%).
Antimicrobial susceptibility and phenotypic identification
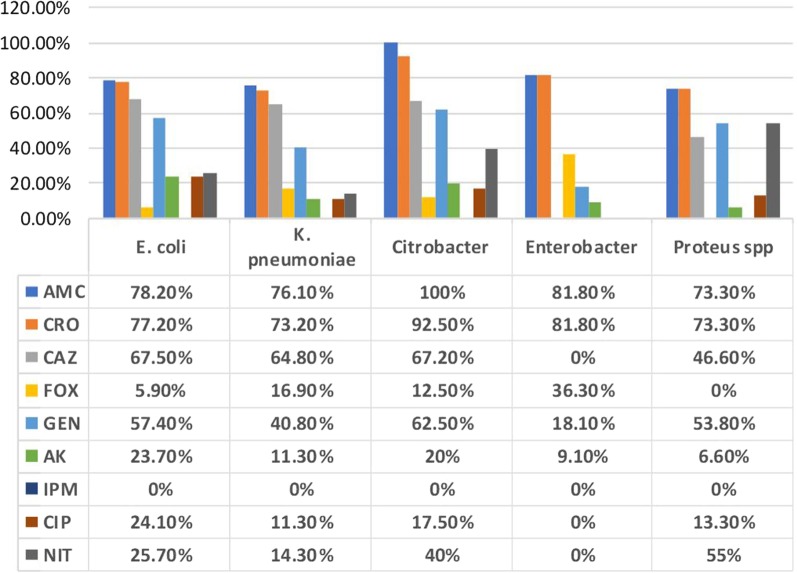
Among 440 Enterobacteriaceae isolates tested for antimicrobial susceptibility, the resistance rates were; AMC (351/440, 79.7%), CRO (343/440, 77.9%), CAZ (289/440, 67.8%), GEN (238/440, 54.3), AK (90/440, 20.4%), CIP (90/440, 20. 4%), NIT (110/440, 25%), and FOX (35/440, 7.9%). All isolates were sensitive to IPM (Fig. 1). Antimicrobial susceptibility and phenotypic tests identified 311 (70.6%) isolates as ESBL producers and 35 (7.9%) isolates as AmpC β-lactamase producers (cefoxitin resistant). Induction test gave no positive results at all. Regarding distribution among different species; the frequency of ESBL production was 211/311 (69.6%) in E. coli, 53/71 (74.6%) in K. pneumoniae, 40/40 (100%) in Citrobacter spp. and 7/15 (46.6%) in Proteus spp. isolates. However, the frequency of suggested AmpC β-lactamase production (cefoxitin resistant) was 18/311(5.8%) in E. coli, 12/71 (16.9) in K. pneumoniae, and 5/40 (12.5%) in Citrobacter spp. isolates.
Fig. 1.
Antimicrobials resistance patterns of 440 Enterobacteriaceae isolates from UTIs. AMC; Amoxicillin Clavulanic acid, CRO, Ceftriaxone, CTZ; Ceftazidime, FOX; Cefoxitin, CN; Gentamicin, AK; Amikacin, IMP; Imipenem, CIP; Ciprofloxacin, F; Nitrofurantoin
Genotypic characterization of ESBL producers
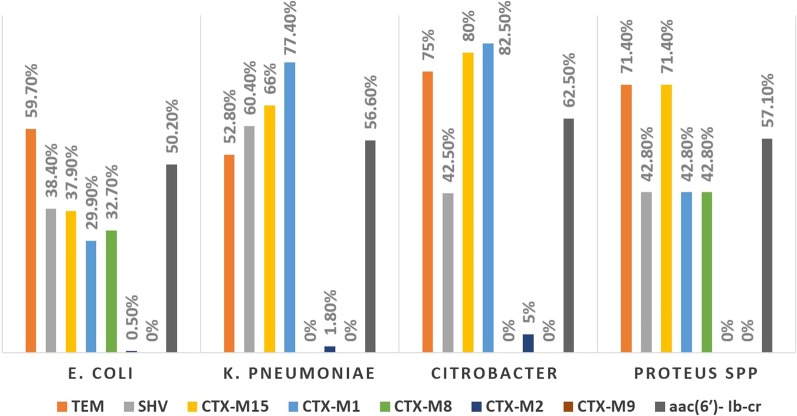
Out of 311 ESBL positive isolates, 308 (99%) isolates were positive for ESBL genes indicating high sensitivity of the phenotypic tests. blaCTX-M genes were detected in 254 (81.6%) isolates, out of them 19 (6.1%) harbored blaCTX-M alone, while the remaining 235 (75.5%) isolates harbored blaCTX-M in combination with blaTEM and/or SHV genes. However, 54 (17.3%) isolates were positive for blaTEM and/or blaSHV ESBL genes but negative for all blaCTX-M genes. The most prevalent gene among ESBL positive isolates was blaTEM gene (189, 60.7%), while within blaCTX-M genes, blaCTXM-15 was the most prevalent (152, 48.8%), followed by blaCTX-M-1 (140, 45%), blaCTX-M-8 (72, 23.1%) and lastly blaCTX-M-2 (4, 1.3%). The distribution of ESBL genes among different species is summarized in Fig. 2, Table 2 and (Additional file 1: Fig S1, Additional file 2: Fig S2, Additional file 3: Fig S3, Additional file 4: Fig S4). Frequency of aac(6′)-Ib-cr gene (responsible for resistance to AK and CIP) among ESBL producers was examined by PCR. A total of 165 (53%) isolates were positive aac(6′)-Ib-cr gene. The association between aac(6′)-Ib-cr gene and blaCTX-M genes was significant (p value < 0.01) (Table 3).
Fig. 2.
Distribution of resistance genes among 311 ESBL producing Enterobacteriaceae isolates
Table 2.
Frequency and combinations of ESBL genes among phenotypically identified ESBL- producing Enterobacteriaceae
| Genes | E. coli (n = 211) | K. pneumoniae (n = 53) | Citrobacter spp. (n = 40) | Proteus spp. (n = 7) | Total (n = 311) |
|---|---|---|---|---|---|
| blaCTX-M group | |||||
| CTX-M-15 alone | 38 (18%) | 0 (0%) | 0 (0%) | 0 (0%) | 38 (12.2%) |
| CTX-M-1 alone | 31(14.7%) | 6 (11.3%) | 3 (7.5%) | 1 (14.3%) | 41 (13.2%) |
| CTX-M-1 + 15 | 32 (15.2%) | 35 (66%) | 30 (75%) | 2 (28.5%) | 99 (31.8%) |
| CTX-M-8 alone | 59 (27.9%) | 0 (0%) | 0 (0%) | 0 (0%) | 59 (19%) |
| CTX-M-8 + 15 | 10 (4.7%) | 0 (0%) | 0 (0%) | 3 (42.8%) | 13 (4.2%) |
| CTX-M-2 alone | 1 (.5%) | 1 (1.8%) | 0 (0%) | 0 (0%) | 2 (0.6%) |
| CTX-M-2 + 15 | 0 (0%) | 0 (0%) | 2 (5%) | 0 (0%) | 2 (0.6%) |
| Total | 171 (81%) | 42 (79.2%) | 35 (87.5%) | 6 (85.7%) | 254 (81.7%) |
| Other β-lactamase genes | |||||
| blaSHV only | 15(7.1%) | 0(0%) | 1(2.5%) | 1(14.2%) | 17 (5.4%) |
| blaTEM only | 13(6.1%) | 3(5.6%) | 0 (0%) | 0(0%) | 16(5.1%) |
| blaTEM + SHV | 10(4.7%) | 7(13.2%) | 4(10%) | 0(0%) | 21(6.7%) |
| Combinations | |||||
| blaSHV+ CTX-M | 55(26.1%) | 20(37.7%) | 7(17.5%) | 1(14.2%) | 83(26.7%) |
| blaTEM + CTX-M | 102(48.3%) | 13(24.5%) | 21(52.5%) | 4(57.1%) | 140(45%) |
| TEM + SHV + CTX-M | 1(.4%) | 5(9.4%) | 5(12.5%) | 1(14.2%) | 12(2.2%) |
| CTX-M genes only | 13(6.1%) | 4(7.5%) | 2(5%) | 0(0%) | 19 (6.1%) |
Table 3.
Co-carriage of ESBLs genes and aac(6′)- Ib-cr gene in Enterobacteriaceae isolates
| aac(6′)-Ib-cr (n = 165) | Species | ESBL genes | Numbers of isolates |
|---|---|---|---|
| aac(6′)- Ib-cr associated with CTX-M group genes | E. coli | CTX-M-15 | 22 |
| E. coli | CTX-M-15 + 1 | 25 | |
| K. pneumoniae | CTX-M-15 + 1 | 24 | |
| Citrobacter spp | CTX-M-15 + 1 | 22 | |
| Proteus spp. | CTX-M-15 + 1 | 2 | |
| E. coli | CTX-M-1 | 19 | |
| K. pneumoniae | CTX-M-1 | 6 | |
| Citrobacter spp. | CTX-M-1 | 3 | |
| E. coli | CTX-M-8 | 2 | |
| Total | 125 (75.5%) | ||
|
aac(6′)-Ib-cr not associated with CTX-M group genes Total |
E. coli | SHV + TEM | 21 |
| E. coli | SHV | 15 | |
| E. coli | TEM | 2 | |
| Proteus spp. | SHV | 2 | |
| 40 (24.2%) | |||
| P value | < 0.01 |
Resistance pattern in ESBL genes carrying isolates and non-ESBL genes carrying isolates
The resistance rates to most of the antimicrobial agents were significantly higher in isolates carrying ESBLs genes than in isolates that don’t carry ESBL genes (p value< .05). However, the rate of resistance to cefoxitin and nitrofurantoin in the two groups did not differ significantly (p value > 0.05). (Table 4).
Table 4.
Resistance patterns in ESBL genes carrying isolates and non-ESBL genes carrying isolates
| Antibiotic | ESBL (N = 308) | non- ESBL (N = 132) | P value | ||
|---|---|---|---|---|---|
| AMC | 308 | 100% | 43 | 32.6% | < 0.001 |
| CRO | 308 | 100% | 35 | 26.5% | < 0.001 |
| CAZ | 308 | 100% | 2 | 1.5% | < 0.0001 |
| FOX | 30 | 9.7% | 5 | 3.7% | 0.06 |
| GEN | 225 | 73% | 13 | 9.8% | < 0.001 |
| AK | 90 | 29.2% | 0 | 0% | 0.02 |
| IPM | 0 | 0% | 0 | 0% | – |
| CIP | 90 | 29.2% | 0 | 0% | 0.02 |
| NIT | 88 | 28.5% | 22 | 16.6% | 0.08 |
| MDR | 88 | 28.5% | 2 | 1.5% | 0.04 |
AMC amoxicillin clavulanic acid, CRO ceftriaxone, CAZ ceftazidime, FOX cefoxitin, CN gentamicin, AK amikacin, IPM imipenem, CIP ciprofloxacin, F nitrofurantoin
Detection of AmpC β-lactamase genes
Among 35 isolates identified as AmpC -producers by phenotypic method, 18 (51.4%) were identified as carrying AmpC genes by multiplex PCR. Among AmpC genes, DHA gene was the commonest (15, 42.3%), while FOX gene was not detected in the isolates. ESBL genes were detected in 17/18 (94.4%) of AmpC genes-carrying isolates. (Table 5).
Table 5.
Frequency of AmpC genes among cefoxitin-resistant isolates and its combinations with ESBL genes
| AmpC genes | E. coli (n = 18) | K. pneumoniae (n = 12) | Citrobacter spp. (n = 5) | AmpC positive (n = 35) | Associated ESBL genes |
|---|---|---|---|---|---|
| MOX | 0(0%) | 1(8.3%) | 0(0%) | 1(2.8%) | CTX-M-1 + 15 |
| FOX | 0(0%) | 0(0%) | 0(0%) | 0(0%) | |
| DHA | 9 (50%) | 3(25%) | 0(0%) | 12(34.3%) |
CTX-M-15 (6) CTX-M-1 (3) TEM (2) No ESBL genes (1) |
| CIT | 0(0%) | 1(8.3%) | 0(0%) | 1(2.8%) | CTX-M-1 + 15+ TEM |
| DHA+ CIT | 0(0%) | 1(8.3%) | 0(0%) | 1(2.8%) | CTX-M-1 + 15 |
| MOX + CIT | 0(0%) | 1(8.3%) | 0(0%) | 1(2.8%) | CTX-M-1 + 15 + TEM |
| MOX + CIT+ DHA | 0(0%) | 2(16.6%) | 0(0%) | 2(5.6%) | CTX-M-1 + 15 |
| Total | 9(50%) | 9(75%) | 0(0%) | 18(51.4%) | (17/18, 94.4%) |
Discussion
Resistance of Enterobacteriaceae to third generation cephalosporins is a worldwide problem [27], which is mainly caused by ESBLs production. Production of additional β-lactamases (AmpC) also contributes to this problem, moreover, the presence of AmpC genes is often associated with multidrug resistance [10]. Previously, AmpC -β-lactamase has received less attention, but is now identified as an important cause of resistance in Enterobacteriaceae species [10]. Global spread of β-lactamases-producing strains gives a great importance to the study of these strains in community and hospitals for reassessment of the existing treatment protocols. In Egypt, multiple studies have investigated the prevalence of ESBLs among Enterobacteriaceae isolated from hospital and community acquired-UTIs [28–30]. However, little data exist on the frequency of co-existence of ESBLs and AmpC β-lactamase in different Enterobacteriaceae species isolated from community acquired-UTIs. The current study showed that 311/ 440 (70.6%) Enterobacteriaceae strains isolated from community acquired-UTIs are ESBL producers. This high frequency is comparable to a recent data reported by Hassuna et al., 2020 in our region, where 57.9% of E. coli isolated from community-acquired UTIs were ESBL producers [30]. On the other hand, our prevalence of ESBL-producing isolates is quite higher than that reported in several previous Egyptian studies; 17% by Fam et al., 2011 [28] and 38.8%, by Shash et al., 2019 [31], suggesting an increasing rate of ESBLs-producing Enterobacteriaceae spread in Egypt, that may be caused by extensive use of 3rd generation cephalosporines as empiric treatment in Egypt. The prevalence of ESBL production varies according to species, geographical areas, variations in infection control programs, different patterns of empiric antibiotic regimens and even over time. Moreover, selective pressure caused by the overuse of cephalosporins in some countries leads to the emergence of increasing rates of ESBLs production [32]. The prevalence of ESBL-production among species of our study was as follows; 100, 74.6, 69.6 and 46.6% of Citrobacter spp., K. pneumoniae, E. coli, and proteus spp. respectively. These findings disagree with some previous studies in Egypt, where ESBL-production was more frequent in E. coli isolates (17% E. coli and 1.2% of non-E. coli isolates) [28] and (97% E. coli, 82.6% K. pneumoniae and 82% Proteus) [33]. However, our finding was comparable with several studies from other African countries, that analyzed ESBL producing- Enterobacteriaceae isolated from different clinical samples. The prevalence in Uganda was 64.9% (72.7% K. pneumoniae and 58.1% E. coli) [34], in Burkina Faso was 58% (62.7% K. pneumoniae and 58.7% E. coli [35], and in Ethiopia 50.7% (52.2% E. coli and78.6% K. pneumoniae) [36]. However, our prevalence was higher than those found in USA, Europe [37], Australia [38], and also some Asian countries [39, 40]. ESBL producing Enterobacteriaceae isolates showed higher rates of resistance to all studied antimicrobials compared to the non-ESBL-producing isolates except for imipenem, where all tested isolates were imipenem-sensitive, that agrees with other Egyptian studies [30, 33]. On the other context, a recent study from our region reported that, (31%) of K. pneumoniae isolated from hospital infections were resistant to imipenem [41]. Although MDR rate among ESBL producers in the current study (28.5%) was lower than that reported in previous studies; (96.3%) [36] and (77.6%) [40], there was statistically significant increase in MDR rate reported in the ESBL-producers (28.5%) than that reported in the non-ESBL-producers (1.5%) (p value = 0.04). Out of 311 ESBL- producing isolates in the current study, 308 (99%) isolates were positive for ESBL genes, with blaCTX-M type as the most predominant. The frequency of community-acquired infections caused by blaCTX-M-producing strains have markedly increased in the last decade [42], that agrees with our findings, where blaCTX-M genes were detected in 254 (81.6%) of Enterobacteriaceae isolates. Within different blaCTX-M genes, blaCTXM-15 was the commonest, (152, 48.8%), followed by blaCTX-M-1 (140, 45%), then blaCTX-M-8 (72, 23.1%). Our results concur with several studies on hospital and community-acquired infections, those reported high prevalence of blaCTX-M genes, particularly blaCTX-M-15 among Enterobacteriaceae species in Egypt [28, 30, 33], Burkina Faso [35], Iran [38], Qatar [40] and Japan [43]. blaTEM and blaSHV-producing strains were reported previously as hospital pathogens until the late 1990s [42], however blaTEM and blaSHV gene were highly frequent among our isolates (189, 60.7%) and (133, 42.8%) respectively, this may be caused by previous contact with health care workers. This higher frequency of blaTEM gene in our report and also in a recent report from our region may indicate that blaTEM gene may be endemic in our locality [30]. Co-carriage of multiple ESBL genes in the same isolate was detected previously in Egypt [29, 30] and other countries; Burkina Faso [35], Qatar [40] and Iran [44], that concurs with our study, where 235 isolates (75.5%) harbored blaCTX-M in combination with blaTEM and/or blaSHV genes. AmpC β-lactamase production was identified phenotypically in 35 (7.9%) of the study isolates that was comparable with previous studies in Egypt [15, 45] and neighboring countries [46, 47]. However, another previous study in Egypt reported a higher rate (76.9%) [48]. AmpC genes were detected by multiplex PCR in 18/35 (51.4%) of cefoxitin resistant isolates, that disagrees with a previous study in Egypt that reported (88.46%) of cefoxitin resistant isolates were AmpC genes positive by PCR assay [48]. Among AmpC genes, DHA gene was the commonest (15/35, 42.3%), that disagrees with previous studies in Egypt, where CIT gene was the commonest [45, 48]. Co-carriage of AmpC genes was found exclusively in K. pneumoniae isolates that agrees with previous reports from Egypt and North Africa [15, 49]. Although FOX gene was commonly detected in previous Egyptian studies [15, 48], it is not detected at all in the current study. ESBL genes were detected in 17/18 (94.4%) of AmpC genes-carrying isolates, that was also reported previously [50]. The spread of ESBL genes is related to different mobile genetic elements, such as plasmid, transposons, and integrons. The co-carriage of ESBL and other-resistant genes in the same transposable genetic elements explain the co-resistance of ESBL producers to variable antibiotics. Our study investigated the frequency of aac(6′)-Ib gene among ESBL-producing Enterobacteriaceae, that was high rate (53%), particularly among blaCTX-M-carrying strains (75.5%). The association between aac(6′)-Ib-cr gene and blaCTX-M genes was statistically significant (p-value < 0.01). This finding may explain why resistance to CIP, CN and AK was significantly higher in ESBL producers than in the non-ESBL-producers, that findings are compatible with several previous studies [39, 51, 52].
Conclusion
Our study detected high prevalence of ESBL- production among isolated from community- acquired UTIs in south Egypt, however the prevalence AmpC β -lactamase production is low. Imipenem can be the drug of choice for community -acquired UTIs caused by these organisms. The blaCTX-M type was the predominant among ESBL-producing Enterobacteriaceae, especially in combination with blaTEM enzymes. β -lactamases production is an important cause of multiple drug resistance.
Supplementary Information
Additional file 1. Figure S1: Agarose gel electrophoresis (2%). lane 1; molecular size marker (100 bp), lanes: 2, 4,5,8,9 are positive for blaCTXM15 (996 bp).
Additional file 2. Figure S2: Agarose gel electrophoresis (2%). lane 1; molecular size marker (100 bp), lanes: 2, 4, 5, 6 are positive for blaCTX-M2 (552bp).
Additional file 3: Figure S3. Agarose gel electrophoresis (2%). lane 1; molecular size marker (100 bp), lanes: 5, 6 are positive for blaCTX-M8 (666bp).
Additional file 4: Figure S4. Agarose gel electrophoresis (2%). lane 1; molecular size marker (100 bp), lanes: 9, 10 are positive for blaCTX-M1 (850bp), lanes: 4, 5, 6 are positive for aac(6′)-Ib-cr gene (482 bp).
Acknowledgements
We thank the health care workers in Minia university hospitals for their cooperation.
Abbreviations
- AmpC
AmpC beta lactamase
- CFU
Colony-forming units
- DDST
Double-disc synergy test
- ESBLs
Extended-spectrum β-lactamases
- MDR
Multiple drug resistance
- UTIs
Urinary tract infections
Authors’ contributions
RMK designed the study and drafted the manuscript. ESM, RMK and SSA performed the experimental work and analyzed the data. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
All data generated or analyzed during this study are included in this article [and its supplementary information files].
Ethics approval and consent to participate
The study protocol was approved by the local ethics committee, Faculty of Medicine, Minia University. Written informed consents were obtained from all patients for the use of their samples.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13756-020-00856-w.
References
- 1.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brolund A. Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infect Ecol Epidemiol. 2014;4:24555. doi: 10.3402/iee.v4.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Centre for Disease Prevention and Control . Antimicrobial resistance surveillance in Europe 2014. Annual report of the European antimicrobial resistance surveillance network (EARSNet) Stockholm: ECDC; 2014. p. 2015. [Google Scholar]
- 4.Abayneh M, Tesfaw G, Abdissa A. Isolation of Extended-Spectrum β-lactamase- (ESBL-) Producing Escherichia coli and Klebsiella pneumoniae from Patients with Community-Onset Urinary Tract Infections in Jimma University Specialized Hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol. 2018;2018(13):4846159. doi: 10.1155/2018/4846159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker KJ, Lee YR, Klar AR. Clinical Outcomes of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae Infections with Susceptibilities among Levofloxacin, Cefepime, and Carbapenems. Can J Infect Dis Med Microbiol. 2018;2018:6. doi: 10.1155/2018/3747521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford PA. Extended-spectrum beta-lactamases in the 21 st century: characterization, epidemiology, and detection of this important resistance threat. J Clin Microbiol Rev. 2001;14(4):933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thenmozhi S, Moorthy K, Sureshkumar B, Suresh M. Antibiotic resistance mechanism of ESBL producing Enterobacteriaceae in clinical field: a review. Int J Pure Appl Biosci. 2014;2(3):207–226. [Google Scholar]
- 8.Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr Issues Mol Biol. 2015;17:11–21.7. [PubMed] [Google Scholar]
- 9.Zhao WH, Hu ZQ. Epidemiology and genetics of CTX-M extended-spectrumβ-lactamases in gram-negative bacteria. Crit Rev Microbiol. 2013;39(1):79–10. doi: 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby G. AmpC-lactamases. Clin Microbiol Rev. 2009;22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan T, Ng S, Teo L, Koh Y, Teok C. Evaluation of screening methods to detect plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis. Antimicrob Agents Chemother. 2009;53:146–149. doi: 10.1128/AAC.00862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grover N, Sahni AK, Bhattacharya S. Therapeutic challenges of ESBLS and AmpC beta-lactamase producers in a tertiary care center. Med J Armed Forces India. 2013;69:4–10. doi: 10.1016/j.mjafi.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byarugaba DK. Antimicrobial resistance in developing countries. New York: Springer; 2009. [DOI] [PubMed] [Google Scholar]
- 14.Kish L. Sampling organizations and groups of unequal sizes. Am Sociol Rev. 1965;30:564–572. doi: 10.2307/2091346. [DOI] [PubMed] [Google Scholar]
- 15.Wassef M, Behiry I, Younan M, El Guindy N, Mostafa S, Abada E. Genotypic identification of AmpC β-lactamases production in gram-negative bacilli isolates. Jundishapur J Microbiol. 2014;7(1):e8556. doi: 10.5812/jjm.8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI. Clinical and laboratory standards institute Performance standards for antimicrobial susceptibility testing. M100S. 2016;26:1–129. [Google Scholar]
- 17.Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–278. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 18.Jacoby GA, Medeiros AA. More extended Spectrum beta lactamases. Antimicrob Agents Chemother. 1991;35(9):1697–1704. doi: 10.1128/AAC.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polsfuss S, Bloemberg GV, Giger J, Meyer V, Bottger EC, Hombach M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2011;49(8):2798–2803. doi: 10.1128/JCM.00404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunne WM, Jr, Hardin DJ. Use of several inducer and substrate antibiotic combinations in a disk approximation assay format to screen for AmpC induction in patient isolates of Pseudomonas aeruginosa, Enterobacter spp., Citrobacter spp., and Serratia spp. J Clin Microbiol. 2005;43(12):5945–5949. doi: 10.1128/JCM.43.12.5945-5949.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, Bonomo RA. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob Agents Chemother. 2003;47:3554–3560. doi: 10.1128/AAC.47.11.3554-3560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckert C, Gautier V, Saladin-Allard M, et al. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob Agents Chemother. 2004;48:1249–1255. doi: 10.1128/AAC.48.4.1249-1255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum b-lactamases. J Antimicrob Chemother. 2006;57:154–155. doi: 10.1093/jac/dki412. [DOI] [PubMed] [Google Scholar]
- 24.Muzaheed DY, Adams-Haduch JM, Endimiani A, Sidjabat HE, Gaddad SM, Paterson DL. High prevalence of CTX-M-15–producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in southern India. J Antimicrob Chemother. 2008;61(6):1393–1394. doi: 10.1093/jac/dkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pe’rez-Pe’rez FJ, and Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 2002; 40:2153–2162. [DOI] [PMC free article] [PubMed]
- 26.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6_)-Ib-cr encoding a ciprofloxacin modifying enzyme. Antimicrob Agents Chemother. 2006;50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. Antimicrobial resistance: global report on surveillance: WHO; 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/. Accessed 17 May 2018.
- 28.Fam N, Leflon-Guibout V, Fouad S, Aboul-Fadl L, Marcon E, Desouky D, et al. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb Drug Resist. 2011;17:67–73. doi: 10.1089/mdr.2010.0063. [DOI] [PubMed] [Google Scholar]
- 29.Abdel-Moaty MM, Mohamed WS, Abdel-All SM, El-Hendawy HH. Prevalence and molecular epidemiology of extended spectrum Î2-lactamase producing Escherichia coli from hospital and community settings in Egypt. J App Pharm Sci. 2016;6(1):042–047. doi: 10.7324/JAPS.2016.600107. [DOI] [Google Scholar]
- 30.Hassuna NA, Khairalla AS, Farahat EM, Hammad AM, Abdel-Fattah M. Molecular characterization of Extended-spectrum β lactamase- producing E. coli recovered from community-acquired urinary tract infections in Upper Egypt. Sci Rep. 2020;10(1):2772. doi: 10.1038/s41598-020-59772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shash RY, Elshimy AA, Soliman MY, Mosharafa AA. Molecular characterization of extended-Spectrum β-lactamase Enterobacteriaceae isolated from Egyptian patients with community- and hospital-acquired urinary tract infection. Am J Trop Med Hyg. 2019;100(3):522–528. doi: 10.4269/ajtmh.18-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canton R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, et al. Prevalence and spread of extended-spectrum b-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14(1):144–153. doi: 10.1111/j.1469-0691.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- 33.Salah M, Azab M, Halaby H, Hanora A. Mutations in β lactamases detected in multidrug resistant gram-negative bacteria isolated from community acquired urinary tract infections in Assiut, Egypt. Afr J Microbiol Res. 2016;10:1938–1943. doi: 10.5897/AJMR2016.8150. [DOI] [Google Scholar]
- 34.Kateregga JN, Kantume R, Atuhaire C, Lubowa MN, Ndukui JG. Phenotypic expression and prevalence of ESBL-producing Enterobacteriaceae in samples collected from patients in various wards of Mulago hospital. Uganda BMC Pharmacol Toxicol. 2015;16(14):1–6. doi: 10.1186/s40360-015-0013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouedraogo AS, Sanou M, Kissou A, Sanou S, Solaré H, Kaboré F, Poda A, Aberkane S, Bouzinbi N, Sano I, Nacro B, Sangaré L, Carrière C, Decré D, Ouégraogo R, Jean-Pierre H, Godreuil S. High prevalence of extended-spectrum ß-lactamase producing enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infect Dis. 2016;16(11):326. doi: 10.1186/s12879-016-1655-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2019;8(15):39. doi: 10.1186/s13756-019-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoban DJ, Lascols C, Nicolle LE, Badal R, Bouchillon S, Hackel M, Hawser S. Antimicrobial susceptibility of Enterobacteriaceae, including molecular characterization of extended-spectrum beta-lactamase–producing species, in urinary tract isolates from hospitalized patients in North America and Europe: results from the SMART study 2009–2010. Diagn Microbiol Infect Dis. 2012;74(1):62–67. doi: 10.1016/j.diagmicrobio.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 38.Chua KYL, Stewardson AJ. Individual and community predictors of urinary ceftriaxone-resistant Escherichia coli isolates, Victoria, Australia. Antimicrob Resist Infect Control. 2019;8:36. doi: 10.1186/s13756-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azargun R, Sadeghi MR, Soroush Barhaghi MH, Samadi Kafil H, Yeganeh F, Ahangar Oskouee M, Ghotaslou R. The prevalence of plasmid-mediated quinolone resistance and ESBL-production in Enterobacteriaceae isolated from urinary tract infections. Infect Drug Resist. 2018;11(23):1007–1014. doi: 10.2147/IDR.S160720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eltai NO, Al Thani AA, Al-Ansari K, Deshmukh AS, Wehedy E, Al-Hadidi SH, Yassine HM. Molecular characterization of extended spectrum β -lactamases enterobacteriaceae causing lower urinary tract infection among pediatric population. Antimicrob Resist Infect Control. 2018;7:90. doi: 10.1186/s13756-018-0381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khairy RMM, Mahmoud MS, Shady RR, Esmail MAM. Multidrug-resistant Klebsiella pneumoniae in hospital-acquired infections: concomitant analysis of antimicrobial resistant strains. Int J Clin Pract. 2020;74(4):e13463. doi: 10.1111/ijcp.13463. [DOI] [PubMed] [Google Scholar]
- 42.Chong Y, Ito Y, Kamimura T. Genetic evolution and clinical impact in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Gen Evol. 2011;11:1499–1504. doi: 10.1016/j.meegid.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Chong Y, Shimoda S, Yakushiji H, Ito Y, Miyamoto T, Kamimura T, et al. Community spread of extended-spectrum β-lactamase-producing Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis: a long-term study in Japan. J Med Microbiol. 2013;62:1038–1043. doi: 10.1099/jmm.0.059279-0. [DOI] [PubMed] [Google Scholar]
- 44.Maleki N, Tahanasab Z, Mobasherizadeh S, Rezaei A, Faghri J. Prevalence of CTX-M and TEM β-lactamases in Klebsiella pneumoniae isolates from patients with urinary tract infection, Al-Zahra hospital, Isfahan, Iran. Adv Biomed Res. 2018;7(30):10. doi: 10.4103/abr.abr_17_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rensing KL, Abdallah HM, Koek A, Elmowalid GA, Vandenbroucke-Grauls CMJE, Al Naiemi N, van Dijk K. Prevalence of plasmid-mediated AmpC in Enterobacteriaceae isolated from humans and from retail meat in Zagazig, Egypt. Antimicrob Resist Infect Control. 2019;8(26):45. doi: 10.1186/s13756-019-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed SF, Ali MM, Mohamed ZK, Moussa TA, Klena JD. Fecal carriage of extended-spectrum beta-lactamases and AmpC-producing Escherichia coli in a Libyan community. Ann Clin Microbiol Antimicrob. 2014;13:22. doi: 10.1186/1476-0711-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdalhamid B, Albunayan S, Shaikh A, Elhadi N, Aljindan R. Prevalence study of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking inducible ampC from Saudi hospitals. J Med Microbiol. 2017;66(9):1286–1290. doi: 10.1099/jmm.0.000504. [DOI] [PubMed] [Google Scholar]
- 48.Helmy MM, Wasfi R. Phenotypic and molecular characterization of plasmid mediated AmpC β-lactamases among Escherichia coli, Klebsiella spp., and Proteus mirabilis isolated from urinary tract infections in Egyptian hospitals. Biomed Res Int. 2014;2014:171548. doi: 10.1155/2014/171548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chérif T, Saidani M, Decré D, Boutiba-Ben Boubaker I, Arlet G. Cooccurrence of Multiple AmpC β-Lactamases in Escherichia coli,Klebsiella pneumoniaee, and Proteus mirabilis in Tunisia. Antimicrob Agents Chemother. 2015;60(1):44–51. doi: 10.1128/AAC.00828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamma PD, Shahara SL, Pana ZD, Amoah J, Fisher SL, Tekle T Doi Y, Simner PJ. Molecular Epidemiology of Ceftriaxone Non-Susceptible Enterobacterales Isolates in an Academic Medical Center in the United States. Open Forum Infect Dis 2019 (11); 6(8). 2019 (11); 6(8). [DOI] [PMC free article] [PubMed]
- 51.Harajly M, Khairallah M-T, Corkill JE, Araj GF, Matar GM. Frequency of conjugative transfer of plasmid-encoded ISEcp1 - blaCTX-M-15 and aac(6′)-lb-cr genes in Enterobacteriaceae at a tertiary care center in Lebanon—role of transferases. Ann Clin Microbiol Antimicrob. 2010;9:19. doi: 10.1186/1476-0711-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peerayeh SN, Rostami E, Siadat SD, Derakhshan S. High rate of aminoglycoside resistance in CTX-M-15 producing Klebsiella pneumoniae isolates in Tehran. Iran Lab Med. 2014;45(3):231–237. doi: 10.1309/LMDQQW246NYAHHAD. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Figure S1: Agarose gel electrophoresis (2%). lane 1; molecular size marker (100 bp), lanes: 2, 4,5,8,9 are positive for blaCTXM15 (996 bp).
Additional file 2. Figure S2: Agarose gel electrophoresis (2%). lane 1; molecular size marker (100 bp), lanes: 2, 4, 5, 6 are positive for blaCTX-M2 (552bp).
Additional file 3: Figure S3. Agarose gel electrophoresis (2%). lane 1; molecular size marker (100 bp), lanes: 5, 6 are positive for blaCTX-M8 (666bp).
Additional file 4: Figure S4. Agarose gel electrophoresis (2%). lane 1; molecular size marker (100 bp), lanes: 9, 10 are positive for blaCTX-M1 (850bp), lanes: 4, 5, 6 are positive for aac(6′)-Ib-cr gene (482 bp).
Data Availability Statement
All data generated or analyzed during this study are included in this article [and its supplementary information files].