Abstract
X chromosome inactivation and genomic imprinting are two classic epigenetic regulatory processes that cause mono-allelic gene expression. In female mammals, mono-allelic expression of the long non-coding RNA gene X-inactive specific transcript (XIST) is essential for initiation of X chromosome inactivation upon differentiation. We have previously demonstrated that the central factor of super elongation complex-like 3 (SEC-L3), AFF3, is enriched at gamete differentially methylated regions (DMRs) of the imprinted loci and regulates the imprinted gene expression. Here, we found that AFF3 can also bind to the DMR downstream of the XIST promoter. Knockdown of AFF3 leads to de-repression of the inactive allele of XIST in terminally differentiated cells. In addition, the binding of AFF3 to the XIST DMR relies on DNA methylation and also regulates DNA methylation level at DMR region. However, the KAP1-H3K9 methylation machineries, which regulate the imprinted loci, might not play major roles in maintaining the mono-allelic expression pattern of XIST in these cells. Thus, our results suggest that the differential mechanisms involved in the XIST DMR and gDMR regulation, which both require AFF3 and DNA methylation.
Keywords: AFF3, XIST, X chromosome inactivation, DNA methylation
Introduction
In mammals, females (XX) have two copies of gene-rich X chromosomes, while males (XY) have only one copy of X chromosome and one copy of gene-poor Y chromosome. X chromosome inactivation (XCI) is a regulated developmental process by which mammalian females can silence one of the two X chromosomes, thus offsetting X-linked gene dosage imbalance between males and females (Lyon, 1961). The long non-coding RNA (lncRNA) XIST/Xist is expressed exclusively from the inactive X chromosome (Xi) and essential for initiation of XCI (Borsani et al., 1991; Brockdorff et al., 1991; Brown et al., 1991). Cis accumulation of XIST/Xist RNA on the X chromosome leads to chromosome wide heterochromatinization and gene silencing (Augui et al., 2011; Lee and Bartolomei, 2013).
One of the main tasks for dissecting the XCI process is to identify factors that regulate the expression of XIST/Xist gene. In murine pluripotent stem cells, Xist expression in repressed; while upon differentiation, Xist is randomly activated from either of the two X chromosomes in female, triggering inactivation of the Xist expressed X chromosome. The Xist gene on the active X chromosome remains ‘off’ status (Navarro et al., 2005; Sun et al., 2006). Several factors have been identified in regulating the Xist gene expression, and most of these factors are encoded within the X-Inactivation Center (XIC), which is located on X chromosome, encompassing the Xist gene, and controlling X chromosome inactivation. For example, the non-coding gene TSIX/Tsix within XIC is antisense to XIST/Xist. Although it has been demonstrated that human TSIX is not functional in XIST repression, murine Tsix has been shown to negatively regulate the expression and accumulation of Xist in cis (Lee and Lu, 1999; Migeon et al., 2002; Navarro et al., 2005; Sun et al., 2006). In contrast, the non-coding transcripts Jpx and Ftx within XIC are involved in Xist activation (Tian et al., 2010; Chureau et al., 2011). Upon differentiation, the E3 ubiquitin ligase RNF12 encoded by an X-linked gene within XIC is upregulated and targets the Xist repressor REX1 to degradation, therefore favoring Xist gene expression (Jonkers et al., 2009; Gontan et al., 2012). The zinc finger protein YY1 is a recently identified autosomal factor that binds to the unmethylated XIST/Xist promoter CpG island and directly activates the XIST/Xist promoter in both human and mouse (Makhlouf et al., 2014). Most of these studies focus on transcriptional regulation of the XIST/Xist gene at the onset of XCI following differentiation, while less is known about XIST/Xist regulation in terminally differentiated cells. In addition, a plethora of recent studies suggest that the XIST gene is dysregulated in various human cancers. It has been reported that XIST RNA can act as a tumor suppressor or activator in many cancers (Yildirim et al., 2013; Chen et al., 2017). Thus, the XIST gene is involved in important physiological and pathological processes. Therefore, it is essential to further explore the underlying mechanisms by which the mono-allelic expression pattern of XIST is maintained.
XCI and genomic imprinting are two classic epigenetic regulatory processes to ensure the mono-allelic expression of their target genes. These two processes are united by allelic control of gene expression in large chromatin domain by cis-acting master regulatory region (Lee and Bartolomei, 2013). We recently demonstrated that the central factor of super elongation complex-like 3 (SEC-L3), AFF3, is recruited to the gamete differentially methylated regions (gDMRs) of imprinted loci by the scaffold protein TRIM28 and its related heterochromatin H3K9 methylation machinery, and to the unmethylated enhancer of imprinted locus by Kruppel like zinc finger protein ZFP281, to regulate the expression of imprinted genes (Luo et al., 2016; Wang et al., 2017). In this study we found that AFF3 can also bind to the 5′ differentially methylated region (DMR) downstream of the XIST promoter and repress its expression in terminally differentiated cells. In contrast, the H3K9 methylation machineries, which are involved in genomic imprinting regulation, seem not to play major roles in XIST gene repression in these cells. In addition, the recruitment of AFF3 to the XIST 5′ promoter DMR is not dependent on H3K9 methylation machineries, but rely on the methylation status of the XIST DMR.
Results
AFF3 is required for the silencing of XIST in terminally differentiated cells
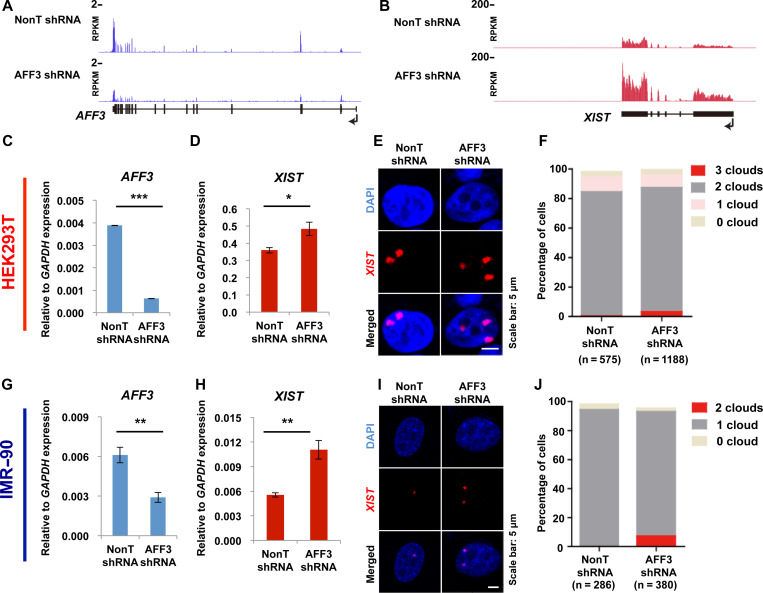
We have previously reported that the central factor of SEC-L3, AFF3, can bind to the master control regions of genomic imprinted loci and regulate the expression of imprinted genes (Luo et al., 2016). Since many similarities have been identified between the regulation of imprinted genes and the XCI master gene, XIST, we investigated the potential involvement of AFF3 in XIST gene expression. We first analyzed our previously published RNA-Seq datasets from HEK293T cells bearing control or AFF3 specific shRNA (Luo et al., 2012). The expression of XIST is upregulated following shRNA-mediated AFF3 knockdown (Figure 1A and B). To further validate this phenomenon, we conducted quantitative real-time PCR analysis after knockdown of AFF3 using an independent shRNA. Data confirmed that AFF3 knockdown increases XIST transcripts (Figure 1C and D).
Figure 1.
AFF3 is required for the silencing of XIST in terminally differentiated cells. (A) RNA-Seq showing the efficiency of shRNA-mediated AFF3 knockdown in HEK293T cells. (B) XIST RNA level increased by knockdown of AFF3 in HEK293T cells. (A and B) The y-axes represent RPKM values. RNA-Seq data in control and AFF3 knockdown HEK293T cells were downloaded from GSE34097. (C) RT-qPCR showing the efficiency of AFF3 knockdown by an independent shRNA in HEK293T cells. (D) RT-qPCR confirming the increase of XIST RNA level by an independent AFF3 shRNA in HEK293T cells. (E) XIST RNA FISH in control and AFF3 knockdown HEK293T cells. Scale bar, 5 μm. (F) Quantification of nuclei with XIST clouds in control and AFF3 knockdown HEK293T cells. The Chi-square test shows that the difference of cells containing three XIST clouds between control and AFF3 knockdown groups is significant. (G) RT-qPCR showing the efficiency of AFF3 knockdown in IMR-90 cells. (H) RT-qPCR showing an increase of XIST RNA level by AFF3 shRNA in IMR-90 cells. (I) XIST RNA FISH in control and AFF3 knockdown IMR-90 cells. Scale bar, 5 μm. (J) Quantification of nuclei with XIST clouds in control and AFF3 knockdown IMR-90 cells. The Chi-square test shows that the difference of cells containing two XIST clouds between control and AFF3 knockdown groups is significant. (C, D, G, and H) The expression of AFF3 and XIST was normalized to the expression of GAPDH. Results shown are technical replicates from representative biological replicates. Error bars represent standard deviations. Significant differences are marked with asterisks (t-test, *P < 0.05; **P < 0.01; ***P < 0.001).
We next asked whether increased XIST expression in AFF3-depleted HEK293T cells affects the formation of XIST RNA cloud on the inactive X chromosome (Xi). Control and AFF3-depleted cells were hybridized with fluorescent XIST RNA probes. HEK293T is a hypotriploid human female cell line, containing three copies of X chromosomes, two of which are inactive and the rest one is active. Our XIST RNA FISH also showed that the most majority of HEK293T cells contain two XIST RNA clouds within a single nucleus (Figure 1E and Supplementary Figure S1A). Consistent with the qPCR data, AFF3 knockdown leads to an increase in the number of cells with three XIST RNA clouds (Figure 1E and F; Supplementary Figure S1A).
The effects on XIST silencing after AFF3 knockdown were repeated in IMR-90 cells, which were derived from the lung of a female fetus. IMR-90 cells have a normal diploid karyotype with 46 chromosomes (46, XX), which could provide a closer condition to mimic a real function of AFF3 in XIST regulation in vivo. In line with the observation in HEK293T cells, knockdown of AFF3 in IMR-90 using two independent shRNAs against AFF3 also leads to an increase of XIST transcripts (Figure 1G and H; Supplementary Figure S1B and C). By FISH for XIST RNA, we observed that more than 98% of the diploid female fibroblasts IMR-90 displayed a single XIST RNA cloud (Figure 1I). AFF3 knockdown in IMR-90 leads to an increase in the number of cells with two XIST RNA clouds (Figure 1I and J). Therefore, from both cases we concluded that AFF3 inhibits XIST transcription and that AFF3 is involved in the maintenance of a proper X chromosome inactive state in female cells.
AFF3 binds to the methylated allele of XIST in female cells
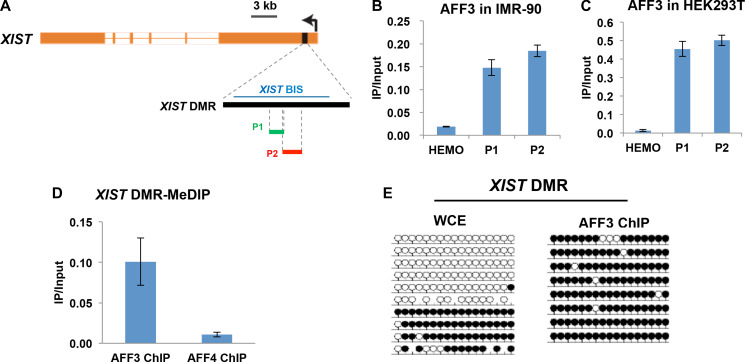
It has been shown that AFF3 can bind to the methylated gDMRs within the CpG islands of imprinted loci to regulate the expression of imprinted genes (Luo et al., 2016). Similar to imprinted genes, the XIST gene promoter CpG island is methylated on the active X chromosome (Xa), and unmethylated on Xi (Heard et al., 1993). We thus speculated that AFF3 regulates the expression of XIST gene through directly binding to the differentially methylated XIST promoter CpG island, hereinafter referred to as the XIST DMR. Quantitative PCR after chromatin immunoprecipitation (ChIP-qPCR) demonstrates that AFF3 is associated with the XIST DMR in both IMR-90 and HEK293T (Figure 2A–C). Pol II ChIP-qPCR in HEK293T at the XIST DMR and the GAPDH gene promoter region was performed as positive controls (Supplementary Figure S2A and B). To address whether AFF3 binds to the methylated allele, we performed methylated DNA immunoprecipitation (MeDIP) assay after AFF3 ChIP in HEK293T. The result demonstrated that the AFF3 bound XIST DMR is methylated (Figure 2D), suggesting that AFF3 has a binding preference for the methylated allele of XIST in female cells. To confirm the binding of AFF3 to the methylated allele of the XIST DMR, we further performed bisulfite-sequencing analysis of the XIST DMR from AFF3 ChIP DNA. AFF3 was found exclusively bound to the methylated allele of the XIST DMR (Figure 2A and E). This finding is also consistent with our previous finding that AFF3 is exclusively recruited to the methylated alleles of the DMR in the imprinted loci (Luo et al., 2016).
Figure 2.
AFF3 binds to the methylated allele of XIST in female cells. (A) Schematic illustration of the location of the XIST CpG island/XIST DMR, the amplified genomic regions of the XIST DMR, and the amplified genomic region of the XIST DMR after bisulfite treatment. (B and C) AFF3 is recruited to the XIST DMR in HEK293T (B) and IMR-90 (C) cells used in ChIP assay. The nonexpressed β-globin gene (HEMO) served as a negative control. Error bars represent standard deviations. (D) AFF3 binds to the methylated allele of the XIST DMR. MeDIP assays were performed using AFF3 or AFF4 ChIP DNA in HEK293T cells. AFF4 ChIP DNA was used as a negative control for the MeDIP assay. Primer pair P2 was used to amplify the XIST DMR. The HEMO gene served as a negative control for ChIP-qPCR. Error bars represent standard deviations. (E) Bisulfite-sequencing analysis of whole-cell extract (WCE; input for AFF3 ChIP) and AFF3 ChIP in HEK293T at the XIST DMR. Methylated and unmethylated cytosines are designated by filled and unfilled circles, respectively. Each line indicates a unique DNA clone. In contrast to WCE, the AFF3 ChIP DNA is predominantly methylated at the XIST DMR.
ENL and AF9, components of AFF3-containing complex SEC-L3, are also required for XIST silencing
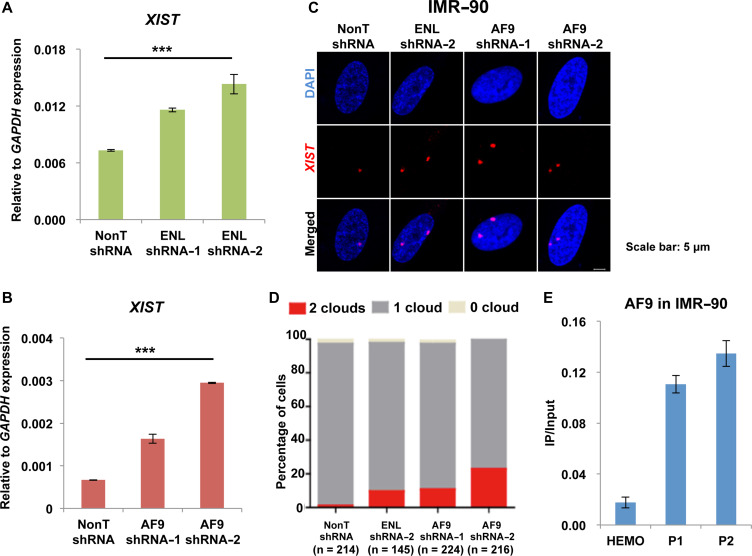
The YEATS domain proteins ENL and AF9 are subunits of the AFF3-containing complex SEC-L3 (Luo et al., 2012). It has also been reported that ENL and AF9 can interact with the components within PRC1 and mediate gene repression (Ui and Yasui, 2016). Since we found AFF3 can suppress XIST transcription, we speculated that its interacting proteins ENL and AF9 might also play similar roles. To test this hypothesis, we first examined the requirement of ENL or AF9 in XIST gene repression by performing ENL or AF9 knockdown experiments in IMR-90 cells (Supplementary Figure S3A and B). Knockdown of either ENL or AF9 leads to an increase in XIST RNA level (Figure 3A and B). ENL or AF9 knockdown in IMR-90 cells also results in an increase of the number of cells displaying two XIST RNA clouds, as shown by RNA FISH (Figure 3C and D). Our result further indicated that AF9 occupies the XIST DMR in IMR-90 cells by ChIP experiments (Figure 3E). Thus, it is highly possible that AF9 regulates XIST gene expression via its direct binding to the XIST DMR.
Figure 3.
ENL and AF9, components of AFF3-containing complex SEC-L3, are also required for XIST silencing. (A and B) RT-qPCR showing an increase of XIST RNA level by ENL (A) or AF9 (B) shRNA in IMR-90 cells. The expression of ENL and AF9 was normalized to the expression of GAPDH. Results shown are technical replicates from representative biological replicates. Error bars represent standard deviations. Significant differences are marked with asterisks (t-test, *P < 0.05; **P < 0.01; ***P < 0.001). (C) XIST RNA FISH in control, ENL knockdown, or AF9 knockdown IMR-90 cells. Scale bar, 5 μm. (D) Quantification of nuclei with XIST clouds in control, ENL knockdown, or AF9 knockdown IMR-90 cells. The Chi-square test shows that the differences of cells containing two XIST clouds between control and ENL knockdown or AF9 knockdown groups are significant. (E) ChIP-qPCR showing that AF9 binds to the XIST DMR in IMR-90 cells. The HEMO gene served as a negative control for ChIP-qPCR. Error bars represent standard deviations.
The recruitment of AFF3 to the XIST DMR is KAP1 independent
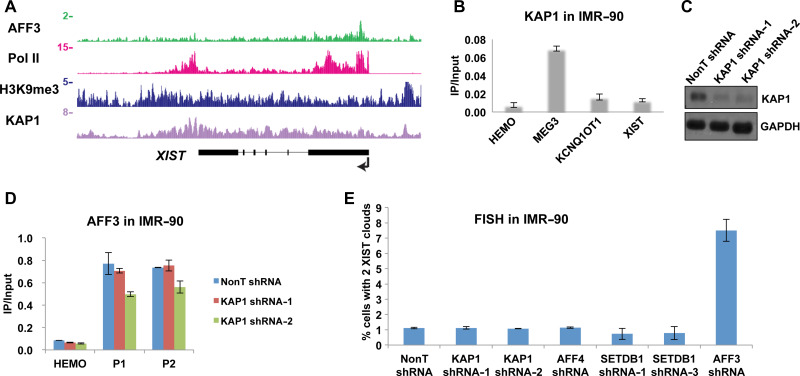
In the imprinted loci, AFF3 binds to the gDMRs that are both DNA and H3K9 methylated. KAP1 colocalizes with AFF3 and the H3K9 methyltransferase SETDB1 and recruits the two factors to the imprinted gDMRs (Quenneville et al., 2011; Luo et al., 2016). Here we performed AFF3 ChIP-Seq in HEK293T cells and compared the binding profile of AFF3, KAP1, and H3K9me3 across the XIST locus. Consistent with AFF3 ChIP-qPCR data, AFF3 is specifically located to the DMR downstream of the XIST promoter (Figure 4A). AFF3 and KAP1 co-occupy the gDMRs of the imprinted loci with H3K9me3. However, unlike AFF3, we found that KAP1 and H3K9me3 spread over the entire XIST gene, with no obvious peaks overlapping with the XIST DMR in both HEK293T and IMR-90 cells (Figure 4A and Supplementary Figure S4A). We also performed KAP1 ChIP in IMR-90 cells. In consistent with a previous study showing that KAP1 is enriched at paternal gDMRs in human cells, we found that KAP1 occupies the paternal MEG3 gDMR in IMR-90 cells (Tao et al., 2018). However, KAP1 is not detectable at the AFF3 occupied the XIST DMR in IMR-90 cells (Figure 4B). This suggests that the recruitment of AFF3 to the XIST DMR may not be mediated by KAP1. To test this hypothesis, we knockdowned KAP1 in IMR-90 cells. The efficiency of knockdown was confirmed by RT-qPCR and western blot (Figure 4C and Supplementary Figure S4B). We then performed AFF3 ChIP in IMR-90 cells depleted of KAP1. The occupancy of AFF3 at the XIST DMR is equally detected in both control and KAP1 knockdown cell line by ChIP-qPCR (Figure 4D). We also observed similar results in HEK293T cells (Supplementary Figure S4C). Thus, the data suggested that the recruitment of AFF3 to the XIST DMR is KAP1 independent in both IMR-90 and HEK293T cells.
Figure 4.
The recruitment of AFF3 to the XIST DMR is KAP1 and H3K9 methylation machineries independent. (A) ChIP-Seq showing AFF3, Pol II, H3K9me3, and KAP1 occupancies at the XIST locus in HEK293T cells. Pol II ChIP-Seq data were downloaded from GSE34097. H3K9me3 and KAP1 ChIP-Seq data were obtained from Encode project. (B) ChIP-qPCR showing that KAP1 is enriched in the paternal MEG3 gDMR, but not the AFF3 bound XIST DMR in IMR-90 cells. The HEMO gene served as a negative control for ChIP-qPCR. Error bars represent standard deviations. (C) Knockdown of KAP1 by lentiviral-mediated shRNA in IMR-90 cells. KAP1 protein levels were measured by western blotting. GAPDH was used as a loading control. (D) ChIP-qPCR showing that the occupancy of AFF3 at the XIST DMR remains unchanged after KAP1 knockdown in IMR-90 cells. The HEMO gene served as a negative control for ChIP-qPCR. Error bars represent standard deviations. (E) Quantification of nuclei with XIST clouds in control, KAP1 knockdown, AFF4 knockdown, SETDB1 knockdown, or AFF3 knockdown IMR-90 cells.
Since KAP1 and H3K9me3 occupy the XIST locus, we further examined the formation of XIST clouds after the depletion of KAP1 and SETDB1. The efficiency of SETDB1 knockdown in IMR-90 cells was confirmed by RT-qPCR and western blot (Supplementary Figure S5A and B). FISH experiments demonstrate that the percentage of IMR-90 cells with two XIST clouds remains the same after KAP1 or SETDB1 knockdown, which is different from what has been observed after AFF3 knockdown (Figure 4E). This further indicates that regulation of XIST gene expression by AFF3 is independent of KAP1 in IMR-90 cells.
In addition, we performed the knockdown of other H3K9 methyltransferase, such as SUV39H1, SUV39H2, and EHMT2 in IMR-90 cells. RT-qPCR analyses demonstrated that knockdown of these H3K9 methyltransferase does not lead to an obvious change of the XIST gene expression (Supplementary Figure S5C–F), suggesting that the H3K9 methylation machineries examined above might not be a major factor in regulating the mono-allelic expression of the XIST gene in the cells we tested.
The recruitment of AFF3 to the XIST DMR is DNA methylation dependent
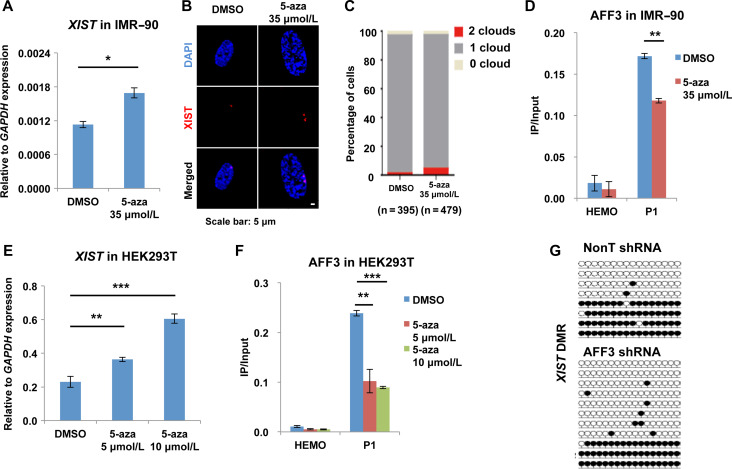
It has been well established that DNA hypomethylation can activate XIST gene expression. The DNA demethylating agent 5-Aza-2′-deoxycytidine (5-aza) can lead to the demethylation of the XIST DMR and XIST reactivation in somatic cells (Hansen et al., 1998; Tinker and Brown, 1998; de Araujo et al., 2014). Since AFF3 can bind to the methylated XIST DMR and repress XIST gene expression, we speculated that the binding of AFF3 might rely on the methylation status of the XIST DMR. To test this hypothesis, we treated IMR-90 cells with 5-aza and then performed AFF3 ChIP. 5-aza application leads to an ~50% increase of XIST gene expression and percentage of cells with two XIST clouds in IMR-90 cells (Figure 5A–C). The occupancy of AFF3 at the XIST DMR is reduced ~40% in IMR-90 cells after 5-aza treatment (Figure 5D). We then examined the requirement of DNA methylation for the binding of AFF3 to the XIST DMR in HEK293T cells. Consistently, 5-aza treatment in HEK293T results in an increase in XIST gene expression and loss of AFF3 binding to the XIST DMR (Figure 5E and F). We also analyzed the effects of AFF3 on DNA methylation status of the XIST DMR by performing bisulfite-sequencing analyses in control and AFF3 knockdown IMR-90 cells. The results indicated that AFF3 knockdown leads to a partial reduction of DNA methylation at the XIST DMR, but not the tested AFF3 unbound region (Figure 5G and Supplementary Figure S6A–C). Therefore, our results suggest a role for the AFF3-DNA methylation pathway in regulating the XIST DMR in terminally differentiated cells.
Figure 5.
The recruitment of AFF3 to the XIST DMR is DNA methylation dependent. (A) RT-qPCR showing that XIST RNA level is increased after treating IMR-90 cells with the demethylating agent 5-aza. The expression of XIST was normalized to the expression of GAPDH. Results shown are technical replicates from representative biological replicates. (B) XIST RNA FISH in DMSO and 5-aza-treated IMR-90 cells. Scale bar, 5 μm. (C) Quantification of nuclei with XIST clouds in DMSO and 5-aza-treated IMR-90 cells. The Chi-square test shows that the difference of cells containing two XIST clouds between DMSO and 5-aza groups is significant. (D) ChIP-qPCR showing that 5-aza treatment in IMR-90 cells leads to a reduction of AFF3 occupancy at the XIST DMR. The HEMO gene served as a negative control for ChIP-qPCR. (E) RT-qPCR showing that XIST RNA level is increased after treating HEK293T cells with the demethylating agent 5-aza. The expression of XIST was normalized to the expression of GAPDH. Results shown are technical replicates from representative biological replicates. (F) ChIP-qPCR showing that 5-aza treatment in HEK293T cells leads to a reduction of AFF3 occupancy at the XIST DMR. The HEMO gene served as a negative control for ChIP-qPCR. (A, D, E, and F) Error bars represent standard deviations. Significant differences are marked with asterisks (t-test, *P < 0.05; **P < 0.01; ***P < 0.001). (G) Bisulfite-sequencing analysis of DNA extracted from control and AFF3 knockdown IMR-90 cells at the XIST DMR. Methylated and unmethylated cytosines are designated by filled and unfilled circles, respectively. Each line indicates a unique DNA clone. The XIST DMR DNA methylation is slightly lost after AFF3 knockdown.
Discussion
Silencing one of the two X chromosomes in mammalian females is initiated by mono-allelic activation of the lncRNA XIST/Xist. Once the process is triggered off in early embryogenesis, mono-allelic expression of XIST/Xist is maintained in almost all of cells in female (Lyon, 1961; Augui et al., 2011; Lee and Bartolomei, 2013). In this study, we have found a role of AFF3 in maintaining the mono-allelic expression of XIST gene. AFF3 can directly bind to the XIST DMR, located at the CpG island downstream of the XIST promoter in terminally differentiated cells. The binding of AFF3 to the XIST DMR depends on DNA methylation. Knockdown of AFF3 leads to upregulation of XIST transcript level and an increase in cells containing one more extra RNA cloud. Our results also suggest that ENL and AF9, which can interact with AFF3, might play similar roles in XIST gene silencing.
Our results show that AFF3 knockdown leads to upregulation of XIST RNA level. However, after AFF3 knockdown only a small percentage of cells display one more extra RNA clouds, as detected by RNA FISH. A possible explanation for this observation is that, in addition to upregulation of XIST RNA level, formation of XIST clouds requires various chromatin modify enzymes to create epigenetic landscapes, which favor XIST accumulation in cis on X chromosome (Simon et al., 2013; da Rocha et al., 2014).
We have previously reported that the recruitment of AFF3 to the imprinted gDMRs requires both DNA methylation and KAP1, a scaffold factor assembling H3K9me3 machinery at gDMRs (Quenneville et al., 2011; Luo et al., 2016). At the imprinted gDMRs, DNA methylation and H3K9me3 are interdependent. Deleting ZFP57, the Krüppel associated box (KRAB) domain containing zinc finger protein recruiting KAP1 and its associated H3K9me3 machineries to gDMRs, leads to both loss of H3K9me3 and DNA demethylation at gDMRs; H3K9me3 hallmarks at gDMRs are not maintained in DNA methylases DNMT1/3a/3b triple knockout cells (Quenneville et al., 2011). Therefore, the recruitment of AFF3 to the imprinted gDMRs requires both DNA methylation and KAP1 in mouse embryonic stem cells (Luo et al., 2016). In this study, we found that the binding of AFF3 to the XIST DMR mainly relies on DNA methylation, but not KAP1. It needs to be further investigated whether non-requirement of KAP1 in recruiting AFF3 to the XIST DMR is due to lack of interdependency between DNA methylation and H3K9me3 at the XIST DMR, or due to KAP1 is not functioning as a scaffold for H3K9me3 machineries at the XIST locus.
Materials and methods
Cell culture
HEK293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (ExCell Bio). IMR-90 cells were cultured in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% FBS (Gibco). The cells were maintained under 5% CO2 at 37°C. 5-aza (Sigma) was dissolved in dimethyl sulfoxide (DMSO) for a stock solution and added to the culture medium fresh on the day of the experiment.
Lentivirus-mediated RNAi
Human AFF3, ENL, AF9, KAP1, SETDB1, EHMT2, SUV39H1, and SUV39H2 shRNA constructs were cloned into the pLKO.1 vector (Addgene #10878). The non-targeting shRNA construct (SHC002) was purchased from Sigma. Packaging to get lentivirus particles and infection were performed as described previously (Luo et al., 2016). shRNA target sequences used in this study are listed in Supplementary Table S1. Briefly, HEK293T cells were plated in 150-mm culture plate and co-transfected with 8 μg of the shRNA construct or non-targeting shRNA construct, 6 μg of psPAX2 packaging plasmids, and 2 μg of pMD2.G envelope plasmids using Lipofectamine 2000 (Thermo Fisher Scientific) according to manufacturer’s protocol. Lentiviral supernatants were collected 48 and 72 h after the transfection, filtered through 0.45-μm filters. IMR-90 and HEK293T cells were infected with filtered lentiviral supernatants together with polybrene (Sigma) at the concentration of 8 μg/ml. Twenty-four hours after infection, the cells were subjected to selection with 1.5 μg/ml (IMR-90) and 2 μg/ml (HEK293T) of puromycin for additional 72 h, respectively.
Antibodies and western blot
The antibodies to 5-methyl-cytosine (5mC) (Eurogentec, BI-MECY-0100), KAP1 (Abcam, ab10484), and SETDB1 (Proteintech, 11231-1-AP) were purchased. Antibodies to AFF3 and AFF4 were described previously (Luo et al., 2012). A fragment of human AF9 (amino acids 406–498) was expressed as a His-tag fusion protein in pET-16b, purified on NTA-agarose according to Qiagen’s protocol and by HPLC, and then sent to Genscript for immunization into rabbits. Whole-cell lysates for western blots were prepared by lysing cells in SDS-PAGE loading buffer.
qRT-PCR analysis
Total RNA was isolated with RNeasy kit (Qiagen), treated with RNase-free DNase I (New England Biolabs), and repurified with RNeasy columon. cDNAs were synthesized with the PrimeScript™ RT Master Mix (TaKaRa). The expression levels were measured with iTaq™ Universal SYBR® Green Supermix (Bio-Rad) on CFX96 (Bio-Rad). The relative expression levels of genes of interest were normalized to the expression of the housekeeping gene GAPDH. qRT-PCR primers used in this study are listed in the Supplementary Table S1.
RNA-Seq analysis
Reads from two biological replicates for each sample were aligned to the human genome UCSC hg19 and to gene annotations from Ensembl 67 using TopHat v2.0.9 (Trapnell et al., 2009). Cuffdiff v1.3.0 was used to quantify reads per kilobase million (RPKM) values, to perform differential expression analysis at FDR < 0.05, and to assess statistically sufficient read coverage for each gene (Trapnell et al., 2010). RNA-Seq reads were not extended for track figures and are shown at single base resolution.
ChIP and ChIP-Seq library preparation
ChIP assays were performed according to the previously described protocol (Lee et al., 2006). Briefly, a total of 5 × 107 HEK293T or IMR-90 cells were used per ChIP that cross-linked with 1% paraformaldehyde for 10 min at room temperature, and cross-linking was quenched by the addition of glycine. The fixed chromatin was sonicated into >200-bp fragments. Then 1% starting chromatin was saved as input sample. Immunoprecipitation with the specific antibodies was performed at 4°C overnight. The immune complex was collected by the protein A agarose beads and washed with RIPA buffer for five times. The chromatin was eluted, purified, and used as a template for qPCR or for ChIP-Seq libaray preparation. ChIP-qPCR primers used in this study are listed in the Supplementary Table S1. Libraries were prepared with NEBNext sample prep kit for the further next-generation sequencing.
ChIP-Seq analysis
ChIP-Seq reads were aligned to the human genome UCSC hg19 using the Bowtie aligner v0.12.9 allowing uniquely mapping reads only and allowing up to two mismatches (Langmead et al., 2009). Reads were extended to 150 bases toward the interior of the sequenced fragment and normalized to total reads aligned (reads per million; RPM). External sequencing data were acquired from GEO and Encode project as raw reads and aligned in the same way as internally sequenced samples. Peak detection was performed with MACS v1.4.2 (Zhang et al., 2008). Genome-wide sequencing datasets used in this study are listed in the Supplementary Table S2.
Bisulfite-sequencing analysis
Cell pellets were lysed in proteinase K digestion buffer (1 M Tris buffer, pH 8.0, 0.5 M EDTA, pH 8.0, 10% SDS) containing proteinase K at 50°C overnight. Genomic DNA was then isolated by phenol-chloroform method. The DNA samples were subjected to bisulfite treatment and purification using the EpiTect bisulfite kit (Qiagen). The purified DNA samples were then amplified using the primers corresponding to the XIST DMR and the AFF3 unbound region on Chromosome 21. Primers used for bisulfite-sequencing analysis are listed in the Supplementary Table S1. The PCR products were cloned into the pGEM-T easy vector (Promega). The clones were subjected to Sanger sequencing.
MeDIP
MeDIP assays were performed according to a previously described protocol (Ito et al., 2010). Briefly, The DNA fragments obtained with the specific antibody in ChIP were immunoprecipitated with the antibody against 5mC overnight at 4°C. MeDIP-qPCR primers used in this study are listed in the Supplementary Table S1.
RNA FISH
RNA FISH was performed according to Stellaris’ RNA FISH protocol (Biosearch Technologies). Briefly, cells were grown on 18-mm round coverglass in a 12-well plate, fixed by 3.7% formaldehyde for 10 min, and permeabilized by 70% ethanol for at least 1 h. Cells were then washed with Wash Buffer A (Biosearch Technologies), and hybridized with 100 μl Hybridization Buffer (Biosearch Technologies) containing Stellaris XIST FISH probes for at least 4 h at 37°C in a humidified chamber. Coverglasses were mounted on slides in Vectashield containing DAPI and visualized under fluorescent microscope. RNA FISH combined with immunofluorescence was performed according to Stellaris’ Sequential IF + RNA FISH protocol (Biosearch Technologies). Briefly, cells were grown on 18-mm round coverglass in a 12-well plate, fixed by 3.7% formaldehyde for 10 min, and permeabilized by 0.1% Triton X-100 in 1× PBS for 5 min. The cells were incubated with primary antibody for 1 h and then secondary antibody for another 1 h at room temperature, washed with PBS and fixed by 3.7% formaldehyde for 10 min. Cells were then washed with Wash Buffer A (Biosearch Technologies), and hybridized with 100 μl Hybridization Buffer (Biosearch Technologies) containing Stellaris XIST FISH probes for at least 4 h at 37°C in a humidified chamber. Coverglasses were mounted on slides in Vectashield containing DAPI and visualized under fluorescent microscope.
Supplementary Material
Funding
This work was supported by Thousand Young Talents Plan of China (5631006003 to C.L., 6231000011 to Z.L.), Natural Science Foundation of Jiangsu Province of China (BK20160026 to C.L., BK20160666 and BK20170020 to Z.L.), and Fundamental Research Funds for the Central Universities (3231007201 to C.L., 3231008201 to Z.L.).
Conflict of interest
none declared.
References
- Augui S., Nora E.P., and Heard E. (2011). Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12, 429–442. [DOI] [PubMed] [Google Scholar]
- Borsani G., Tonlorenzi R., Simmler M.C., et al. (1991). Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G.F., et al. (1991). Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351, 329–331. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Ballabio A., Rupert J.L., et al. (1991). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. [DOI] [PubMed] [Google Scholar]
- Chen D.L., Chen L.Z., Lu Y.X., et al. (2017). Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 8, e3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chureau C., Chantalat S., Romito A., et al. (2011). Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 20, 705–718. [DOI] [PubMed] [Google Scholar]
- da Rocha S.T., Boeva V., Escamilla-Del-Arenal M., et al. (2014). Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell 53, 301–316. [DOI] [PubMed] [Google Scholar]
- de Araujo E.S., Vasques L.R., Stabellini R., et al. (2014). Stability of XIST repression in relation to genomic imprinting following global genome demethylation in a human cell line. Braz. J. Med. Biol. Res. 47, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C., Achame E.M., Demmers J., et al. (2012). RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature 485, 386–390. [DOI] [PubMed] [Google Scholar]
- Hansen R.S., Canfield T.K., Stanek A.M., et al. (1998). Reactivation of XIST in normal fibroblasts and a somatic cell hybrid: abnormal localization of XIST RNA in hybrid cells. Proc. Natl Acad. Sci. USA 95, 5133–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Simmler M.C., Larin Z., et al. (1993). Physical mapping and YAC contig analysis of the region surrounding Xist on the mouse X chromosome. Genomics 15, 559–569. [DOI] [PubMed] [Google Scholar]
- Ito S., D’Alessio A.C., Taranova O.V., et al. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I., Barakat T.S., Achame E.M., et al. (2009). RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell 139, 999–1011. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., et al. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.T., and Bartolomei M.S. (2013). X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308–1323. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Johnstone S.E., and Young R.A. (2006). Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.T., and Lu N. (1999). Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99, 47–57. [DOI] [PubMed] [Google Scholar]
- Luo Z., Lin C., Guest E., et al. (2012). The SEC family of RNA Polymerase II elongation factors: gene target specificity and transcriptional output. Mol. Cell. Biol. 32, 2608–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Lin C., Woodfin A.R., et al. (2016). Regulation of the imprinted Dlk1-Dio3 locus by allele-specific enhancer activity. Genes Dev. 30, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373. [DOI] [PubMed] [Google Scholar]
- Makhlouf M., Ouimette J.F., Oldfield A., et al. (2014). A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 5, 4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon B.R., Lee C.H., Chowdhury A.K., et al. (2002). Species differences in TSIX/Tsix reveal the roles of these genes in X-chromosome inactivation. Am. J. Hum. Genet. 71, 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P., Pichard S., Ciaudo C., et al. (2005). Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 19, 1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S., Verde G., Corsinotti A., et al. (2011). In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.D., Pinter S.F., Fang R., et al. (2013). High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B.K., Deaton A.M., and Lee J.T. (2006). A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21, 617–628. [DOI] [PubMed] [Google Scholar]
- Tao Y., Yen M.R., Chitiashvili T., et al. (2018). TRIM28-regulated transposon repression is required for human germline competency and not primed or naive human pluripotency. Stem Cell Reports 10, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Sun S., and Lee J.T. (2010). The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143, 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker A.V., and Brown C.J. (1998). Induction of XIST expression from the human active X chromosome in mouse/human somatic cell hybrids by DNA demethylation. Nucleic Acids Res. 26, 2935–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., and Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui A., and Yasui A. (2016). Collaboration of MLLT1/ENL, Polycomb and ATM for transcription and genome integrity. Nucleus 7, 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shen Y., Dai Q., et al. (2017). A permissive chromatin state regulated by ZFP281-AFF3 in controlling the imprinted Meg3 polycistron. Nucleic Acids Res. 45, 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E., Kirby J.E., Brown D.E., et al. (2013). Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.