Abstract
The muscle RING-finger protein-1 (MuRF1) is an E3 ubiquitin ligase expressed in skeletal and cardiac muscle tissues and it plays important roles in muscle remodeling. Upregulation of MuRF1 gene transcription participates in skeletal muscle atrophy, on contrary downregulation of protein expression leads to cardiac hypertrophy. MuRF1 gene point mutations have been found to generate protein aggregate myopathies defined as muscle disorder characterized by protein accumulation in muscle fibers. We have discovered that MuRF1 turned out to be also a target for a new post-translational modification arbitrated by conjugation of SUMO1 and it is mediated by the SUMO ligases E2 UBC9 and the E3 PIASγ/4. SUMOylation takes place at lysine 238 localized at the second coiled-coil protein domain that is required for efficient substrate interaction for polyubiquitination. We provided evidence that SUMOylation is essential for MuRF1 nuclear translocation and its mitochondria accumulation is enhanced in hyperglycemic conditions delivering a stabilization of the overall SUMOylated proteins in cultured myocytes. Thus, our findings add this SUMO1 post-translational modification as a new concept to understand muscle disorders related to the defect in MuRF1 activity.
Keywords: TRIM63/MuRF1, muscle remodeling, SUMO, protein degradation, hyperglycemia
Introduction
SUMOylation is an enzymatic cascade of events where SUMO, the small ubiquitin-like modifier, a 101-amino acid protein, is covalently attached to lysine residues on target proteins. The reaction of SUMOylation requires E1 (hetero-dimer SUMO-activating, SAE1/2), E2 (SUMO-specific conjugating, Ubc9), and, in most cases, E3 (SUMO ligase, PIASs) enzymes (Geiss-Friedlander and Melchior, 2007). There are reported four SUMO mammalian paralogues, SUMO1 to SUMO4, each encoded by distinct genes. SUMO1 and SUMO2/3 are transiently and reversibly conjugated to substrate proteins by the same enzymatic machinery (Johnson, 2004; Gareau and Lima, 2010). However, it remains unclear whether SUMO4 is processed or conjugated to cellular proteins (Guo et al., 2004, 2005; Owerbach et al., 2005; Wang and She, 2008; Baczyk et al., 2017). These moieties can modify distinct targets although some proteins can be SUMOylated by either SUMO1 or SUMO2/3 (Tatham et al., 2001). The modifiers are expressed as immature peptides that must be cleaved before conjugation. This process is carried out by isopeptidases, known as SENPs or SUMO-specific proteases. The cleavage generates a C-terminal carboxyl group, SUMO-Gly-Gly-COOH that will be attached via an isopeptide bond, to a lysine (K) residue in the consensus sequence, ΨKxD/E, along with the targeted protein. Specifically, Ψ is a large hydrophobic residue, K is the target lysine, and D/E are acidic residues (Rodriguez et al., 2001). Nevertheless, proteomics studies have shown that a large amount of SUMOylated proteins do not contain the consensus sequence (Blomster et al., 2009; Golebiowski et al., 2009; Hsiao et al., 2009).
Protein SUMOylation is a dynamic route that can modulate various arrays of cellular processes. It includes genome stability (Gill, 2005), gene transcription, chromosome organization and function, DNA repair, nuclear transport (Johnson, 2004), cell cycle (Chung et al., 2004; Eifler and Vertegaal, 2015), proteins turnover (Weger et al., 2004), and enzyme activity (Dohmen, 2004; Meulmeester et al., 2008; Yang and Sharrocks, 2010; Masoumi et al., 2016; Chakraborty et al., 2017).
Members of the family of the tripartite motif (TRIM)-containing proteins could be defined as E3 ubiquitin ligases as they contain a RING-finger domain, one or two zinc-binding motifs named B-boxes and associated coiled-coil (CC) regions (Reymond et al., 2001). TRIM family proteins are involved in a wide range of biological processes and their alterations, concerning abundance and activity, are associated with different pathological conditions, such as neurodegenerative diseases, developmental disorders, cachexia, and cancer (Hatakeyama, 2011). TRIM63 also known as muscle-specific RING finger protein-1 (MuRF1) is an E3 ubiquitin ligase that is expressed selectively in cardiac and skeletal muscles (Bodine et al., 2001). This protein is upregulated during skeletal muscle atrophy and is capable of polyubiquitination and ubiquitin proteasome system-mediated degradation of sarcomeric muscle proteins like troponin I, and myosin heavy chain (Freemont, 2000; Kedar et al., 2004; Glass and Roubenoff, 2010). MuRF1 has been reported to localize at the Z-disk and M-line of the sarcomere (Centner et al., 2001) but also translocate to the nucleus of muscle fibers. In nuclei, the primary function is to influence gene expression through polyubiquitination and degradation of transcription factors. This nuclear translocation event was observed during muscle inactivity or unloading (Lange et al., 2005; Ochala et al., 2011). MuRF1 was also found in cardiomyocytes mitochondria to regulate the oxygen consumption (Mattox et al., 2014).
A delicate balance between protein synthesis and degradation is necessary for muscle remodeling and homeostasis. MuRF1 is transcriptionally upregulated under most atrophy-inducing conditions by FOXO transcription factors that can directly bind to the MuRF1 promoter and regulate MuRF1 protein expression to enhance muscle proteins degradation (Labeit et al., 2010; Files et al., 2012). Mutations in the gene encoding MuRF1 impart loss-of-function effects on E3 ligase activity. The consequential effects were reported to cause genetic muscle disorders like protein aggregate myopathies (PAMs), characterized by protein accumulation in muscle fibers (Olive et al., 2015a) and hypertrophic cardiomyopathy (HCM), defined by the presence of left ventricular hypertrophy (Gersh et al., 2011; Chen et al., 2012; Su et al., 2014).
MuRF1 not only associates with different cytoskeleton compartments such as microtubules, nuclear proteins, but also interacts with a network of diverse proteins implicated in sarcomeric assembly (troponin I and T, titin, telethonin, myosin light chain 2, and nebulin) (Witt et al., 2005), energy metabolisms (creatine kinase, mitochondria oxidative phosphorylation, and ATP regeneration) (Witt et al., 2005; Koyama et al., 2008), protein degradation (MuRF2 and MuRF1) (Bodine and Baehr, 2014), transcriptional regulation (McElhinny et al., 2002), and SUMO-mediated post-translational modification. Indeed, the RING domain of endogenous MuRF1 has been discovered to interact with SUMO3 (Dai and Liew, 2001) and with the E2 SUMO ligase, UBC9, (McElhinny et al., 2002). Recently, in our previous publication, we have shown MuRF1 as a protein that was found in a pull-down assay from diaphragm muscle tissue by using anti-SUMO1 antibodies (Namuduri et al., 2017), suggesting a possible involvement of MuRF1 in the SUMO pathway, but this role is still unknown. In the present study, for the first time, we used an innovative SUMO1 variant to identify MuRF1 a target for SUMO1 conjugation within the second CC domain necessary for the nuclear localization. We observed only a small fraction of the total amount of the MuRF1 to become SUMOylated, and its SUMOylation is enhanced by overexpression of UBC9 and PIASγ/4 enzymes. The MuRF1 protein conjugated by SUMO1 had mitochondria localization and increases when C2C12 cells were exposed to high glucose conditions. Finally, we detected a reduction of reactive oxidative species (ROS) in high glucose medium from cultured myocytes of a mouse expressing MuRF1 compared to myocytes expressing the SUMO conjugation mutation variant. We also described that MuRF1 has a positive effect to protect from a general deconjugation of SUMOylated protein in C2C12 cells exposed to high glucose as well as we observed in skeletal muscle tissue from diabetic mouse and MuRF1 KO.
Results
Identification of MuRF1 as a potential SUMO conjugation target
In our previous study, we identified the E3 ubiquitin ligase TRIM63/MuRF1 specifically bound to monoclonal SUMO1 antibodies, (Namuduri et al., 2017). To validate that MuRF1 is a SUMO target we performed a bacteria assay: BL21 bacteria were co-transformed with the prokaryotic pGEX-5X-1 plasmid where MuRF1 ORF was cloned in frame with the GST protein, together with the pSUMO plasmids containing the SUMO conjugation machinery (Figure 1A). A couple of slow migrating bands between 75 and 100 kDa, above the unmodified GFP-MuRF1 protein (66 kDa) were detected only with pSUMO1, pSUMO2, and pSUMO3 plasmids but not in mock transformation. This result suggested a potential conjugation of SUMOs to GFP-MuRF1 (Figure 2B, upper blot). Same lysates were probed with anti-6×-HIS antibodies to validate the efficiency of the SUMO conjugation machinery produced by the pSUMOs plasmids in the selected bacteria colonies. Corresponding immunoblot showed a correct pattern of bacteria SUMOylated proteins (Figure 1B, lower blot).
Figure 1.
SUMO conjugation assay performed with recombinant GST-MuRF1. (A). Scheme of the experimental SUMO conjugation assay setup in E. coli cells transformed with pGST-MuRF1 plasmid containing ampicillin resistance together with the pSUMO1, pSUMO2, or pSUMO3 conjugation system plasmid containing streptomycin resistance. (B) WB analyses of GFP-MuRF1 SUMOylation in E.coli BL21 cells. GFP-MuRF1 plasmid was cotransfected together with mock vector (M) and three pSUMO plasmids respectively for SUMO1 (lane 1), SUMO2 (lane 2), and SUMO3 (lane 3). Bacteria lysates were separated in a SDS-PAGE gel and membranes probed with anti-GST and anti-histidine antibodies. Unmodified GST-MuRF1 is indicated with an arrow and slow migrating bands related to potential SUMOylated MuRF1 species are shown with a square bracket. Total SUMOylated proteins were detected with anti-histidine antibodies in the pSUMOs plasmids-transformed bacteria lysates. (C) SUMO1 amino acid sequences. SUMO1 WT indicates the native protein sequence where the amino acid T (labeled in green) was mutated in K and R (marked in red) to generated SUMO1 mutants T95K and T95R, respectively. The black arrows indicate the closest trypsin cleavage side before the Gly-Gly residue. (D) Coomassie blue stained preparative gel of purified GST-MuRF1 from bacteria lysates cotransfected with GST-MuRF1 and mutated SUMO1 T95K and T95R. SUMO-GFP-MuRF1 bands indicated with the square brackets were isolated and sent to the MS/MS analysis.
Figure 2.
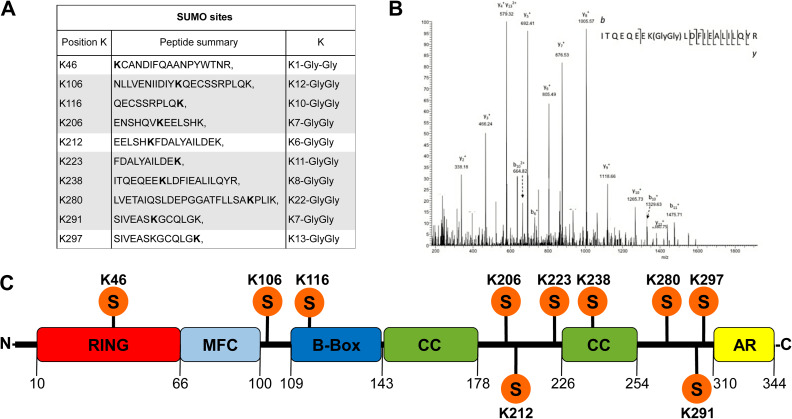
Identification of potential SUMO sites in the MuRF1 amino acid sequence. (A) Panel of identified SUMO sites using bioinformatics approaches (http://www.abgent.com/sumoplot combined with http://www.jassa.fr) and by MS/MS. Highlighted peptides had a reliably manual verification of the SUMO site position in MS/MS spectra. (B) MS/MS spectrum of the peptide with identified 238 K-GlyGly modification. (C) The structural domains of the MuRF1 protein are indicated with different colors with the positions of newly identified SUMO sites.
To associate the lysine residues involved in the conjugation of SUMO along MuRF1 protein, we adopted the strategy described in Impens et al. (2014). We generated SUMO1 variants in the SUMO1 ORF cloned in the pSUMO1 conjugation plasmid, where the wild-type amino acid T95 was replaced with a K or an R before the Gly-Gly residues (Figure 1C). Trypsin digestion generated SUMO-modified peptides with a GG tag easily identifiable by mass spectrometry on the lysine involved in the SUMO1 moieties conjugation. These mutations did not interfere with the SUMO1 conjugation efficiency as shown in a comparative western blot (WB) between lysates from bacteria transformed with pSUMO1-WT, -T95K, -T95R, and the two correspondent, not conjugable SUMO1 variants, -T95K ΔG and -T95R ΔG. A similar SUMOylation protein profile was observed in bacteria transfected with pSUMO1-WT, -T95K, and T95R but no SUMO conjugates were detected where SUMO1 were depleted of one Gly at the conjugation site (Supplementary Figure S1). To achieve our aim to identify SUMO sites in the MuRF1 sequence, we performed a massive purification of GST-MuRF1 protein with glutathione Sepharose beads, from lysates of bacteria previously co-transformed with pGEX-5X-1 MuRF1 plasmid together with the new pSUMO1 T95K or pSUMO1 T95R plasmids variant. Glutathione elutes containing the solubilized GST-tagged proteins were separated on a preparative SDS-PAGE gel and the bands between 75 and 100 kDa, corresponding to the putative SUMO1 modified MuRF1, were excised (Figure 1D) and analyzed with mass spectrometry.
Characterization of the SUMO1–MuRF1 conjugation site
From bioinformatics prediction of SUMO sites along MuRF1 protein, combined with the mass spectrometry results we identified 10 lysine residues involved in the SUMO conjugation (Figure 2A). The best MS/MS spectrum of modified peptides belonged to the SUMO site K238 (Figure 2B) localized in the second CC domain (Figure 2C), and the other designated lysines with the related spectra are shown in the Supplementary Figure S2A–K. The observed SUMOylation sites were identified in the GST-MuRF1 protein produced in bacteria where the folding could be different in case the same protein is expressed in a eukaryotic system. To confirm the potential SUMOylation in eukaryotic cells, GFP-MuRF1 was expressed in C2C12 and HeLa cells. In both eukaryotic systems, GFP-MuRF1 was detected predominantly with a single band migrating at 66 kDa together with three additional bands between 75 and 100 kDa. These various post-translational modifications of the GFP-MuRF1 are represented only within 5%–10%, of the total expressed protein (Figure 3A).
Figure 3.
Expression of GFP-MuRF1 in eukaryotic cells and identification of SUMO sites by site-direct mutagenesis kit. (A) Lysates from C2C12 and HeLa cells transfected with GFP plasmid alone (GFP+) or increasing amount of GFP-MuRF1 plasmids were probed with anti-GFP antibodies. (B) WB of C2C12 cells transfected with recombinant GFP-MuRF1 WT or mutant plasmids. K238R GFP-MuRF1 mutant lost the slow migrating bands as potential SUMO post-translational modifications. Asterisks show the unmodified GFP-MuRF1 protein and a squared bracket indicates the likely SUMOylated bands. Anti-GAPDH antibodies were used as protein loading control. Representative blots from three independently performed experiments are shown. (C–F) WB of HeLa cell transfected with mock, GFP, GFP-MuRF1, and GFP-MuRF1-K238R plasmids. Total cell lysates (10 μg, input, In) were loaded close to the correspondent immunoprecipitated samples using anti-GFP antibodies conjugated to Sepharose beads (IP). Membranes were incubated with anti-GFP (C), anti-SUMO1 (D), anti-SUMO2 (E), and anti-ubiquitin (F) antibodies. A single band is detected in the GFP-MuRF1 In and IP samples probed with anti-GFP antibodies, and a unique band is observed in the correspondent IP sample probed with anti-SUMO1 antibodies. Representative blots from two independently performed experiments are shown. (G–J) WB of C2C12 cells stably expressing HIS-tagged SUMO1 or SUMO2 transfected with mock, GFP-MuRF1 alone or in combination with Flag-PIAS1, Flag-PIASγ/4, c-myc-UBC9, and GFP-MuRF1-K238R in combination with Flag-PIASγ/4 plasmids. Cell lysates were immunoprecipitated with anti-GFP antibodies, separated by SDS-PAGE, and probed with anti-GFP (G–I) and anti-6×-HIS (H–J) antibodies. Arrows indicate the fraction of MuRF1 conjugated with SUMO1-HIS (G and H). Asterisks indicate the immunoprecipitated GFP-MuRF1 (G) and GFP-MuRF1-K238R (I). Hashtags did not show specific bands (H–J). Representative blots from two independently performed experiments are shown. (K) In vitro SUMOylation assay of GST-MuRF1 and GST-MuRF1-K238R. Purified GST-tagged proteins (200 ng) were used as template together with the indicated SUMO components for the SUMOylation reactions A–H. Products were separated by SDS-PAGE and membranes probed with anti-GST antibodies. MuRF1 SUMOylation was only observed in reactions A and E in the presence of SUMO1 as a slow migrating band between 75 and 100 kDa. Representative blots from two independently performed experiments are shown.
To define which lysine is involved in the SUMO conjugation, we generated GFP-MuRF1 mutants contained one of the 10 identified lysines into Arginine along the GFP-MuRF1 amino acid sequence. After sequencing, the corrected 10 plasmids codifying GFP-MuRF1 lysines mutants were transfected into C2C12 cells and the expression of the proteins was analyzed by WB where the membranes were probed with anti-GFP antibodies. Exclusively, the GFP-MuRF1 containing Arginine in the position 238 showed an entire absence of the slow migrating bands above the native GFP-MuRF1 protein. This result suggests that lysine 238 is the unique amino acid along the MuRF1 protein to become a target of SUMO1 conjugation. It localized in the second CC domain and this mutation does not affect the stability because the protein was not degraded, (Figure 3B). When GFP-MuRF1 is expressed either in bacteria and eukaryotic cells, we always observed more than one band above the native protein.
To determine which band corresponded to the SUMO conjugation site, we performed immunopurification assay with anti-GFP antibodies conjugated to Sepharose beads of GFP, GFP-MuRF1 and GFP-MuRF1-K238R proteins expressed in HeLa cells. Elutes were probed with anti-GFP, anti-SUMO1, anti-SUMO2, and anti-ubiquitin antibodies (Figure 3C–F). Only a single band migrated between 75 and 100 kDa was observed in the GFP-MuRF1 samples, either probed with anti-GFP and anti-SUMO1 antibodies, and no bands were detected in the GFP-MuRF1-K238R as well in the GFP and no transfected cells. We confirmed this post-translational modification with another assay by using a different cell background: C2C12 cells stably expressing 6×-HIS-tagged SUMO1 and SUMO2 (Supplementary Figure S5A–C). GFP-MuRF1 and GFP-MuRF1-K238R proteins were expressed in the two cell lines and immunoprecipitated with anti-GFP antibodies. Again, in C2C12 cells expressing SUMO1-6×-HIS and transfected with GFP-MuRF1 plasmid, immunoprecipitate samples with anti-GFP antibodies and probed with both anti-GFP and anti-6×-HIS shown only one slow migrating band between 75 and 100 kDa (Figure 3G and H). On the contrary, no post-translational modifications mediated by SUMO2 were observed when GFP-MuRF1 was produced in cells stably expressing SUMO2-6×-HIS (Figure 3I and J). We also approached an in vitro SUMOylation essay with commercial kits on bacterially expressed and purified GST-MuRF1 and GST-MuRF1-K238R. Only the reactions where SUMO1 was added as a reagent and without EDTA, shown a fraction of GST-MuRF1 conjugated to SUMO1, migrating between 75 and 100 kDa (Figure 3K). These results indicated and confirmed that MuRF1 is involved in a mono SUMO1 conjugation on the K238 lysine.
Identification of the enzymes involved in the SUMO1 conjugation of MuRF1
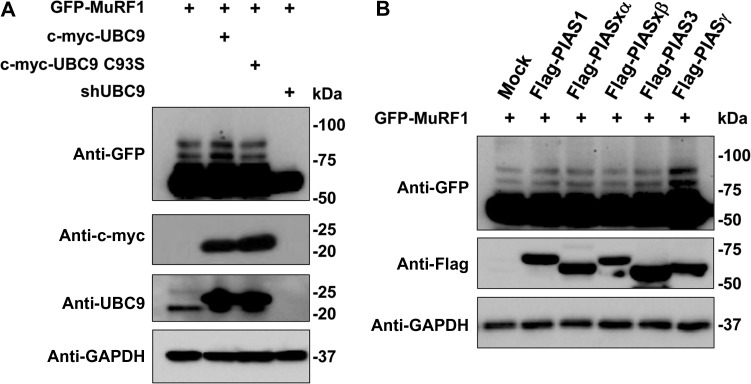
To identify which enzymes are engaged in the MuRF1 SUMO post-translational modification, GFP-MuRF1 plasmid was cotransfected in C2C12 cells together with c-Myc tagged UBC9 or with the correspondent catalytic mutant (C93S), or in C2C12 where the UBC9 transcript was abolished with specific shRNAs (Figure 4A). Only the overexpression of active UBC9 determined an increase of SUMOylated MuRF1 species, on the contrary, a complete reduction was shown in C2C12 cells where the UBC9 protein was knockdown by shRNA.
Figure 4.
Identification of enzymes involved in the conjugation of SUMO to GFP-MuRF1 protein. (A) C2C12 cells were transfected with GFP-MuRF1 plasmid alone or together with c-myc UBC9, c-myc UBC9 C93S, and shRNA UBC9. Lysates were probed with anti-GFP antibodies to detect the GFP-MuRF1 species, anti-c-myc to detect the UBC9 protein expression, and anti-UBC9 to show both endogenous and overexpressed c-myc-UBC9. (B) C2C12 cells were transfected with mock and different E3 SUMO ligases (PIAS1, PIASxα, PIASxβ, PIAS3, PIASγ/4). Total cell lysates were fractionated in a SDS-PAGE gel and membranes probed with anti-GFP to detect the MuRF1 GFP proteins with the potential SUMO-conjugated bands. Anti-Flag antibodies were used to identify the expression of Flag-tagged E3 SUMO ligases. Anti-GAPDH antibodies were used as protein loading control. Representative blots from three independently performed experiments are shown.
In parallel, GFP-MuRF1 was overexpressed in C2C12 cells, in the presence of the Flag-tagged E3 SUMO conjugating enzymes, PIAS1, PIASxα, PIASxβ, PIAS3, and PIASγ/4. Only the co-expression with PIASγ/4 showed an increase of GFP-MuRF1 SUMOylated proteins (Figure 4A). In addition, we performed an experiment where lysates from C2C12 cells stably expressing SUMO1- and SUMO2-6×-HIS previously transfected with GFP-MuRF1 together with Flag-PIAS1, Flag-PIASγ/4, and c-Myc-UBC9 were immunoprecipitated with anti-GFP antibodies and membranes were probed with anti-GFP and anti-6×-HIS antibodies. WB analysis shown an increase of the SUMO-MuRF1 fraction intensity only when UBC9 and PIASγ/4 were co-expressed in C2C12 cells expressing SUMO1 compared to mock or PIAS1 transfections (Supplementary Figure S5D and E). These results confirmed that the E2 SUMO UBC9 and the E3 PIASγ/4 are essential for the SUMO1 conjugation to MuRF1.
MuRF1 requires the lysine 238 for an efficient substrate degradation activity
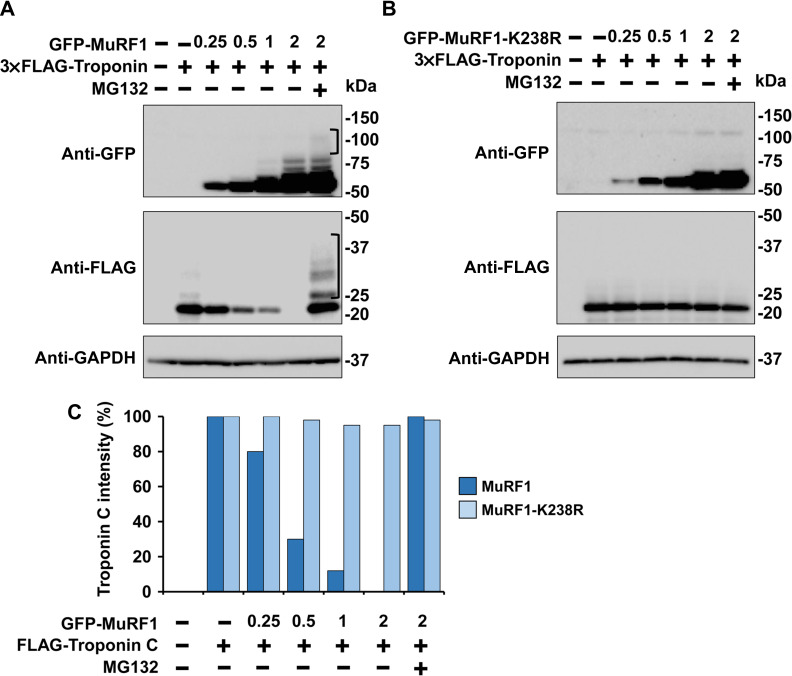
To address the question whether the lysine 238 influences the MuRF1 ubiquitination activity, we compared the enzymatic propriety of MuRF1 with the correspondent K238R mutant, in the degradation of Troponin C protein. For this purpose, cells were cotransfected with 4 μg of a plasmid codifying 3×Flag-Troponin C in the presence of 2 μg of mock vector and with different amounts (0.25, 0.5, 1, 2, 2 μg) of GFP-MuRF1 and GFP-MuRF1-K238R, respectively. One of the samples cotransfected with 2 μg of GFP-MuRF1 or GFP-MuRF1-K238R together with 4 μg of 3×Flag-Troponin C, were also treated for 8 h by 10 μM proteasome inhibitor, MG132. At 24 h post-transfection, C2C12 cells were harvested, and lysates were loaded in a SDS-PAGE gel. The fast activity of MuRF1 to promote Troponin C degradation was observed already when the enzyme was expressed in a low dose of 0.5 μg (Figure 5A) and Troponin C band intensity was reduced about of 70% compared to the signal without MuRF1 (Figure 5C). On the contrary, the presence of MG132 blocked the substrate degradation and bands corresponded to polyubiquitinated were detected above the Troponin C and weakly also over MuRF1 as a self-ubiquitination signal (Figure 5A). In contrast, the degradation of the substrate was impaired in MuRF1-K238R mutant protein also when it was expressed in high dose, 2 μg of plasmid, (Figure 5B). This result suggested that the mutation of lysine 238 to Arginine affects both the substrate and the MuRF1 self-ubiquitination probably due to protein conformational changes that generated a not functional E3 ubiquitin ligase or impairment of enzyme-substrate interaction.
Figure 5.
Assessment of GFP-MuRF1 and GFP-MuRF1-K238R activity on the Troponin C substrate performed in C2C12 cells. (A and B) C2C12 cells were transfected with 3×Flag-Troponin C plasmid alone (4 μg) or with different amounts of GFP-MuRF1 (A) and GFP-MuRF1-K238R plasmids (0.25, 0.5, 1, 2 μg) (B) in the absence (−) or presence (+) of the proteasome inhibitor MG132 (10 μM) for 8 h. Total cell lysates were fractionated by SDS-PAGE and membranes probed with anti-GFP to detect the GFP-MuRF1 protein expression. Anti-Flag antibodies were used to detect the presence of 3×FLAG-Troponin C. Anti-GAPDH antibodies were used as protein loading control. Brackets indicate the polyubiquitination smears on GFP-MuRF1 and 3×FLAG-Troponin C proteins during MG132 treatment. (C) Quantification of Flag-Troponin C protein from two independently performed experiments.
MuRF1 SUMOylation is required for the translocation to nuclei
To address the question about the effect of MuRF1 SUMOylation related to cellular localization, we performed confocal images of C2C12 cells transfected with GFP-MuRF1 and GFP-MuRF1-K238R. Following the GFP fluorescence, we classified the protein localization in three different groups: cytoplasm-nuclei, mitochondria, and cytoplasm aggregates, (Figure 6A). MuRF1 was homogenously distributed in the nuclei and the cytoplasm with a percentage of 78%, and in minor proportion, 10%, into mitochondria. Another small fraction of MuRF1, 12%, formed big aggregates in the cytoplasm, possibly due to its overexpression. MuRF1-K238R predominantly formed cytoplasm aggregates, 80%, but in the fraction of cytoplasm-nuclei group, 10%, more than 70%, of the cells shown the GFP fluorescence only in the cytoplasm with a complete absence in the nuclei (Figure 6B). Less than 10% of GFP fluorescence related to MuRF1-K238R was observed to localize into mitochondria. This result suggested that mutation of the unique SUMO conjugation site to MuRF1 impairs the nuclear translocation.
Figure 6.
Confocal microscopy analysis of GFP-MuRF1 and GFP-MuRF1-K238R cellular localization. C2C12 cells were transfected with GFP-MuRF1 and GFP-MuRF1-K238R. After fixation and DAPI staining, GFP fluorescence was observed with a confocal microscope to determine the protein localized in cytoplasm and nuclei, in mitochondria, or forming aggregates in the cytoplasm. (A) Cells were cultured in standard glucose medium (5.5 mM). (B) The statistical distribution of protein localization in transfected cells. About 10000 transfected cells were observed. The protein cellular localization is shown as a percentage on the total amount of transfected cells. (C) Mitochondria distribution of GFP-MuRF1 in C2C12 cells cultured in high glucose medium (25 mM). (D) Statistical cellular distribution of both proteins in transfected C2C12 cells cultured in high glucose medium. (E) WB of whole-cell lysates (TCL), nuclei (N), and mitochondria (M) fractions from C2C12 cells transfected with GFP-MuRF1 and GFP-MuRF1-K238R plasmids grown in normal (5.5 mM) or high (25 mM) glucose medium. Anti-GFP antibodies were used to determine the MuRF1 abundance along the cell fractions, while anti-GAPDH, anti-COX4, and anti-histone antibodies were used as cellular fraction markers. The experiment was performed in duplicate from two different sets of C2C12 transfections. Scale bar, 10 μm.
We then asked if it is possible to modulate the different MuRF1 cellular localization. To prove this, we transfected C2C12 cells with plasmids codifying GFP-MuRF1 and GFP-MuRF1-K238R and then grown in medium containing normal (1.0 g/L–5.5 mM) or high glucose (4.5 g/L–25 mM glucose) for 6 h. Surprisingly, the number of C2C12 cells transfected with MuRF1, with the GFP fluorescence localized in the mitochondria increased as much as 20% compared to the normal glucose medium culture, (Figure 6C and D). On contrary, no differences were detected with GFP-MuRF1-K238R compared with the normal glucose medium culture (Figure 6D). Interestingly, we noticed two distinct mitochondria shapes in the cells cultured in high glucose, where GFP-MuRF1 has the mitochondria localization. We counted the cells fraction with the GFP signal detected in mitochondria with a standard string shape distribution as 10%, (Figure 6C, left image). The remaining 20% of cells consisted mitochondria with a thick structure, and the intensity of GFP signal was 8 times more intense compared to the fluorescence from the classic mitochondria shape, (Figure 6C, right image). This result suggested a change in the mitochondria morphology due to an increased level of ROS as consequence of high glucose in the cell culture medium (Kamogashira et al., 2017). A co-localization of GFP fluorescence with MitoTrakers, a mitochondria marker, is shown in Supplementary Figure S3.
To further investigate which of the forms either the native or SUMOylated MuRF1 were present in the different cell compartments, we performed a cell fractionation of C2C12 cells transfected with GFP-MuRF1 and GFP-MuRF1-K238R grown in normal and high glucose medium for 6 h. With normal glucose medium, native form of GFP-MuRF1 was found distributed both in the nuclei and in mitochondria fractions. MuRF1 was only in native form in the nuclear fraction where as in mitochondria, we found it in both native and SUMOylated forms. In the case of high glucose medium treatment, the increasing number of myocytes with GFP-MuRF1 localized in the mitochondria observed with fluorescence analysis (Figure 6D) was confirmed with a presence of more intense band of SUMO GFP-MuRF1 identified in the mitochondrial fraction, (Figure 6E). We also validated the immunofluorescence results where in the nuclear fraction the abundance of GFP-MuRF1-K238R is considerably reduced since the band intensity is low compared to the total cell lysate fraction.
MuRF1 has a positive effect to protect the SUMO protein from deconjugation in myocytes exposed to high glucose
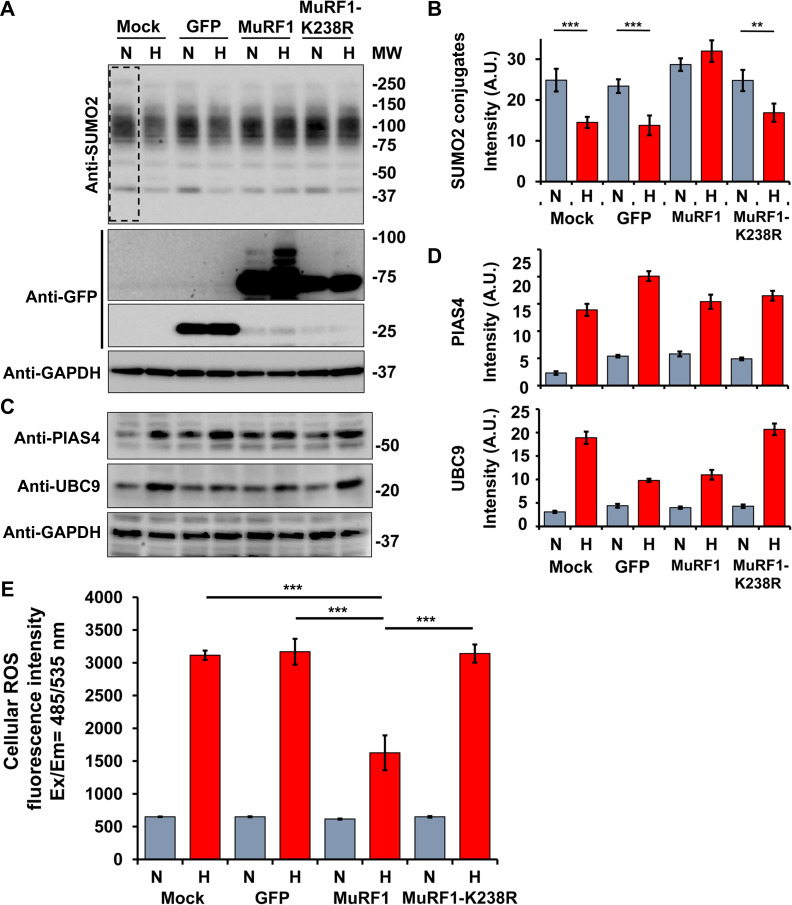
To determine the potential role and the biological significance of MuRF1 localization into mitochondria, we analyzed the level of total SUMO2-conjugated proteins in C2C12 cells transfected with mock and plasmids codifying GFP, GFP-MuRF1, and GFP-MuRF1-K238R. Twenty four hours after transfections cells were exposed to normal and high glucose medium for another 6 h. We adopted as a representative model to analyze the effects of ROS activity the quantification of the total endogenous SUMO2-conjugated proteins in the transfected cells. It has been reported that high level of ROS inhibited the conjugation of SUMO to substrates because the UBC9 activity was impaired (Bossis and Melchior, 2006).
High glucose culture medium treatment has instigated a significant decrease in overall SUMO2-conjugated proteins in C2C12 cells mock-transfected or expressing GFP alone or GFP-MuRF1-K238R. No differences were noticed in C2C12 overexpressing MuRF1 cultured in both normal and high glucose medium (Figure 7A and B). A concomitant increase of GFP-MuRF1 slow migrating bands was observed in cells grown in high glucose medium compared to normal glucose (Figure 7A). We also noted an increase in the endogenous protein level of the SUMO E2 ligase UBC9 and the SUMO E3 ligase PIASγ/4 in all C2C12 cells exposed for 6 h to high glucose medium (Figure 7C and D). To determine the effect of MuRF1 to preserve the deconjugation of endogenous SUMOylated protein against the increase of intracellular ROS, we performed ROS assay on C2C12 cells mock-transfected or expressing GFP, GFP-MuRF1, or GFP-MuRF1-K238R. A reduction of ~50% of cellular ROS was determined in C2C12 cells treated with high glucose medium expressing GFP-MuRF1 compared to mock, GFP, and GFP-MuRF1-K238R. Taken together, these findings demonstrated that MuRF1 has a positive effect to preserve the SUMO2 deconjugation of cellular proteins by reducing the level of cellular ROS.
Figure 7.
High glucose induces SUMOylation of MuRF1. (A) WB of empty vector transfected C2C12 cells (mock) or cells after transfection with GFP, GFP-MuRF1, and GFP-MuRF1-K238R plasmids cultured in normal (N) and high glucose (H) medium for 6 h. Cell lysates were fractionated by SDS-PAGE and membranes probed with anti-SUMO2 antibodies to determine the conjugation level of SUMOylated protein. SUMOylated proteins decreased in cells cultured in high glucose medium and transfected with GFP-MuRF1-K238R. Anti-GFP antibodies were used to identify the GFP and GFP-MuRF1 protein levels and anti-GAPDH antibodies were used as protein loading control. (B) Quantification of SUMO2 conjugates from three independent experiments and normalized with the loading control GAPDH. ***P < 0.001, **P < 0.01. (C) Membranes in a were also probed with anti-PIAS4 and anti-UBC9 antibodies to detect the endogenous level of two SUMO-conjugating enzymes. (D) Quantification of three independent experiments normalized with the correspondent GAPDH signal as loading control. PIAS4 and UBC9 protein levels increased in cells cultured in high glucose medium. Gray bars indicate normal glucose (5.5 mM) and red bars indicate high glucose (25 mM). (E) Cellular ROS detection in C2C12 cells transfected with mock, GFP, GFP-MuRF1, and GFP-MuRF1-K238R plasmids and incubated for 6 h with fresh normal glucose (N) and high glucose (H) medium. ROS measurement was determined with fluorescence intensity as the ratio between excitation and emission values from 485/535 nm wavelength. The experiment was performed in triplicated from of three independent transfection experiments. The fluorescence detected in the non-transfected cells was removed from all the values. Cellular ROS activity was reduced in C2C12 cells transfected with GFP-MuRF1 and incubated in high glucose for 6 h before the assay. ***P < 0.001.
The endogenous level of MuRF1 protein is not sufficient to contribute and protect the SUMO muscle protein from deconjugation in diabetic mice
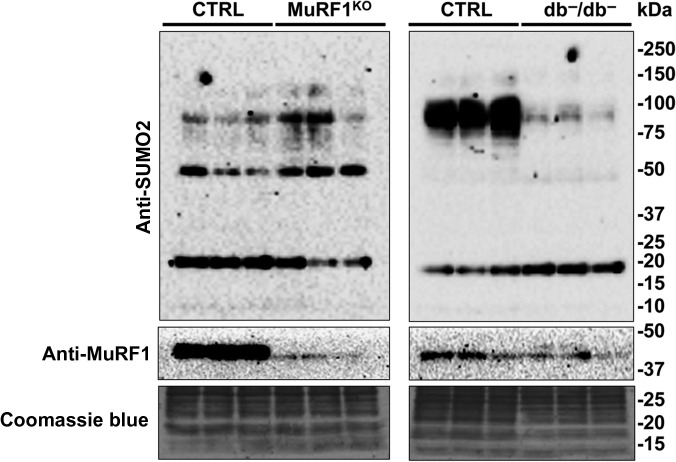
To investigate in a more physiological contest, another aspect of MuRF1's role in different mice models, we lysed diaphragm muscle from normal, MuRF1 KO and diabetic (db−/db−) young mice. We observed a significant reduction of SUMOylated muscle proteins only in diabetic mouse diaphragm but no differences were observed in the MuRF1 KO mice compared to the correspondent control mice (Figure 8). This result may suggest that, in vivo, the role of MuRF1 can be surmounted by MuRF2 or MuRF3 in MuRF1 KO mice (Witt et al., 2005), and in diabetic mice, the level of MuRF1 is not sufficiently high to protect the SUMO2-conjugated muscle proteins from the deconjugation as we have observed in cultured myocytes C2C12 cells.
Figure 8.
SUMO2 conjugates in diaphragm muscles from db−/db−, MuRF1 KO, and control mice. Diaphragm muscle protein lysates were fractionated by SDS-PAGE and membranes probed with anti-SUMO2 antibodies to determine the SUMO conjugation profile in the respiratory muscles from 8- to 10-week-old control and diabetic mice and from 32- to 36-week-old control and MuRF1 KO mice. Anti-MuRF1 antibodies were used to detect the endogenous protein level. A significant decrease in the total amount of SUMO2-conjugated proteins was observed in diabetic (db−/db−) diaphragm mice. Coomassie blue was used as protein loading control. Representative blots from two independently performed experiments with three mice for each group are shown.
Discussion
SUMOylation has been linked to the regulation of protein localization, activity, and stability. Previous studies reported that the muscle-specific E3 ubiquitin ligase MuRF1 localized in different cellular compartments without possessing any specific N-terminus peptide for subcellular sorting.
Our present study provided the first detailed molecular mechanism of SUMO1 conjugation to MuRF1 when it is overexpressed in murine myoblast C2C12 cells. This post-translational modification occurred to ~10% of the total native MuRF1 protein, and the SUMO enzymes UBC9 and PIASγ/4 catalyzed this reaction. The mass spectrometry analysis proved that SUMOylation occurred at the lysine 238 placed in the second Coil-coiled domain and it is essential for the protein translocation from cytoplasm to the nuclei. However, the MuRF1 protein located into the nuclei was found exclusively in the native form, suggesting that after nuclear translocation, SUMO1 was removed by a specific nuclear SUMO deconjugases. We validated this result with a MuRF1 mutated on the particular lysine residue, which is involved in the SUMO1 conjugation and this mutation had led to a total impairment of the nuclei localization of MuRF1.
Protein post-translational modifications are strategies routinely used by cells to expand their function but can also reflect the status quo of strived cellular activities under stressed conditions. It has been described that modulation of the SUMO pathway served as cellular stress antagonist in response to high glucose. For example, the effects of high glucose exposure in rat mesangial cells generated reactive oxygen species and activated the NF-κB inflammatory signaling through IκBα SUMOylation (Huang et al., 2013). In this study, in C2C12 cellular model grown in high glucose medium, the endogenous levels of UBC9 and PIASγ/4 proteins were upregulated. But despite this increase, we noticed a significant loss in the total SUMO2-conjugated proteins only in C2C12 cells not expressing GFP-MuRF1.
An explanation of this effect is related to the sensitivity of UBC9, the unique SUMO conjugating enzyme, to ROS. Indeed, high glucose levels generated an intensification of cellular ROS that reacted with the UBC9 catalytic domain and inhibited the enzymatic activity with a partial block in the SUMO conjugation pathway (Bossis and Melchior, 2006). Surprisingly, this impairment was not observed in C2C12 cells cultured in high glucose medium and transfected with GFP-MuRF1, where its over expression and mitochondria localization, contributed in decreasing the endogenous level of ROS (Mattox et al., 2014). As a result of this ROS modulation, the inactive fraction of UBC9 was significantly reduced and the status of the total SUMO2-conjugated cellular protein was preserved. This condition had also enhanced the endogenous level of PIASγ and contributed to the increase in the conjugation of MuRF1 by SUMO1 together with its mitochondria localization. This increase in the mitochondria localization was calculated to be at least 30% more compared to the C2C12 cell cultured in normal glucose medium. Here, we provided a novel and significant evidence suggesting a new role to MuRF1 as a protective part to maintaining the level of SUMOylated proteins in hyperglycemic conditions by modulating the ROS level in C2C12 cells. However, this protective role was not detected with the GFP-MuRF1-K238R SUMO mutant possibly because of improper protein folding and the massive protein cytoplasm aggregation. Further studies must be done to understand how MuRF1 with its SUMOylated fraction enter into mitochondria and interfere with the proteins and influence their functions. It is also important to dissect the different roles of the native and SUMO1 modified forms of MuRF1.
It has been reported that the two MuRF1 Coil-coiled domains are essential either for the interaction with substrates to be targeted by ubiquitin for protein degradation (Kedar et al., 2004) (Witt et al., 2005) and the formation of MuRF1 rod-shaped dimers (Franke et al., 2014). As consequence of those facts, we can wonder that the presence of the SUMO1 moiety attached to MuRF1 coil-coiled domain might change the structure and compelled the protein to lose the property to interact with its substrates or to form dimers but allows the nuclear translocation mediated by a not yet known receptor.
At the moment we were not able to demonstrate the effects promoted by SUMO1 attached to MuRF1 on the lysine 238 in terms of substrate interactions and its protein enzymatic activity with an in vitro experiment, due to the difficulty in purifying the sole GST-MuRF1-K238R SUMO1 from bacteria. On the other hand, the SUMO conjugation mutant MuRF1-K238R lost more than 95% of the Troponin C degradation activity compared to wild-type MuRF1. The impairment of the protein degradation activity can be explained as a consequence of change in the overall structure protein that can also correlate with the observation of intracellular aggregates that we detected in C2C12 cells expressing GFP-MuRF1-K238R.
This study also demonstrated a significant decrease in the total SUMO2-conjugated proteins in the respiratory muscle in young hyperglycemic (db−/db−) rodents compared to the control. The outcome can be linked to a consequence of high glucose level in the bloodstream combined with a substantial insulin resistance, which produced an increase of cellular ROS in this mice model (Perry et al., 2016). This alteration may affect the SUMO-conjugating machinery (Tempe et al., 2008; Rajan et al., 2012), particularly the activity of UBC9 (Bossis and Melchior, 2006), since no difference in the MuRF1 protein level was observed in these samples. This in vivo result confirmed our observation performed in myocytes C2C12 cells.
On the contrary, we found no alteration in the overall SUMO2-conjugated proteins in diaphragm muscle from MuRF1 KO mice compared to the correspondent control. One direct explanation is that MuRF2 and MuRF3 may cooperate in vivo with MuRF1 in the ubiquitin transfer to myofibrillar proteins and other well-known standard functions. Redundancy in MURF1 and MURF2 signaling might also explain why MURF1 KO mice have a similar phenotype, including a typical lifespan and fertility.
The Q247* (Olive et al., 2015b; Chen et al., 2012), T232M, and D254N (Su et al., 2014) MuRF1 gene mutations have been described to lead into severe muscle diseases as PAMs and human HCM. Those alterations are in closer proximity to the lysine 238 responsible for SUMOylation on the MuRF1 protein. We investigated the effects of those mutations in term of SUMO1 conjugation and speculated those muscle pathologies might be due to a lack of MuRF1 SUMOylation (data not shown). To achieve this result partially, we cloned the correspondent Q247* MuRF1 protein tagged with GFP, and we observed only the native form if compared to GFP-MuRF1 (Supplementary Figure S4). This result indicated that the MuRF1 fragment failed the property to become SUMOylated and consequently collapsed in functions related to the SUMO conjugation.
There are still open questions related to the new SUMO post-translational modification that we discovered. It is not clear why only a small portion of MuRF1 protein is SUMOylated, and the biological significance of the SUMOylated MuRF1 in the mitochondria remains unknown including the mitochondria complexes which are involved in its translocation (Kelley et al., 2002).
However, SUMOylation of MuRF1 adds a building block to crosstalk mechanisms between the ubiquitin and SUMO networks. Other reported examples include SUMOylation of the deubiquitinating enzyme USP25 which impairs binding to and hydrolysis of ubiquitin chains (Meulmeester et al., 2008), the E2 ubiquitin conjugation enzyme E2-25K, which prevents its interaction with the ubiquitin E1 enzyme (Pichler et al., 2005), and the E3 ubiquitin ligase RNF4 that target its substrates via SUMO/SIM interaction (Prudden et al., 2007; Uzunova et al., 2007).
In summary, our findings provide a novel paradigm for selective regulation of MuRF1 functions by SUMO1 mediated by UBC9-PIASγ/4 conjugating enzymes, which can be used as crucial SUMO regulators checkpoints with pharmacological relevance for treating MuRF1-mediated muscle atrophy of the myofibril.
Materials and methods
Chemicals
DL-dithiothreitol (DTT, D0632), N-ethylmaleimide (NEM, E1271), phenylmethanesulfonylfluoride (PMSF, P7626), iodoacetamide (I1149), IGEPAL CA-630 (NP40, I3021), isopropyl-β-D-thiogalaktosid (IPTG, I1000-10 from Saveen Werner), sodium deoxycholate monohydrate (DOC, D5670), Triton X-100 (T9284), sodium dodecyl sulfate (SDS, L3771), iodacetamide (I-1149), Tween-20 (P9416), ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA, E4884), Trizma base (T93349), dithiothreitol (DTT, D5545), ampicillin (ampicillin sodium salt, A0166), streptomycin (streptomycin sulfate salt, S9137), kanamycin (kanamycin solution, K-0254), poly-D-lysine hydrobromide (P-6407), paraformaldehyde (P-6148), and all oligonucleotides for cloning were from Sigma-Aldrich. Complete protease inhibitors cocktail tablets (protease inhibitors) were from Roche Diagnostic. Fluoroshield mounting medium with DAPI (ab104139) was from AbCam. Grade modified trypsin (V5073) was from Promega. Restriction enzymes BamHI (FD0054), XhoI (FD0694), NotI (FD0594), and KpnI (FD0524) and ligase (EL0011) were from Thermo Fischer Scientific.
Antibodies
The antibodies used for WB were: rabbit anti-SUMO1 (Y299, ab32058, 1:2000), rabbit anti-SUMO2+3 (EPR4602, ab109005, 1:2000), goat anti-UBE2I/UBC9 (ab21193, 1:2000), rabbit polyclonal anti-E3-SUMO-protein ligase PIASγ/4 (ab137500, 1:1000), rabbit monoclonal anti-MuRF1 (ab172479, 1:1000), mouse monoclonal anti-6×-HIS tag (ab18184, 1:3000) from AbCam; mouse anti-GAPDH (6C5, CB1001, 1:30000) from Merck Millipore (Darmstadt, Germany); mouse anti-GST (B-14, sc-138, 1:3000), mouse anti-GFP (B-2, sc-6669, 1:3000), mouse anti-GFP AC (B-2 conjugated to agarose, sc-6669 AC), mouse anti-c-Myc (9E10, sc-40, 1:3000), from Santa Cruz Biotechnology; mouse anti-FLAG M2 (F-3165, 1:5000) from Sigma-Aldrich; polyclonal swine anti-rabbit IgG (P0399), polyclonal rabbit anti-goat IgG (P044901), polyclonal rabbit anti-mouse IgG (P0260) horseradish peroxidases (HRP)-conjugated antibodies (1:5000 WB) from DAKO.
ORFs cloning in expression vectors
To clone the ORF of human MuRF1 in both prokaryotic pGEX-5X-1 (ampicillin resistance, 27-4584-01, GE Heathcare) and eukaryotic pEGFP-N1 (kanamycin resistance, 6085-1, Takara) plasmids, the truncate ORF of human MuRF1 Q247* in pEGFP-N1, and ne human Troponin C in eukaryotic p3XFLAG-CMV-10 (ampicillin resistance, E7658, Sigma-Aldrich), we performed a PCR with cDNA template generated from purified total RNA of human skeletal muscle cells (HSkMC,150-05 A, Sigma-Aldrich) using the correspondent designed primers for cloning (Supplementary Table S1). The PCR reactions were performed on 40 ng of template, 20 μM of primers (forward and reverse), 20 mM of dNTP’s mix, 1× High Fidelity Buffer, and 2 U of Pfu taq enzyme (Phusion high-fidelity PCR kit, F553S, Thermo Fischer Scientific) in a total volume of 50 μl. The PCR cycling program was: denaturation 95°C for 5 min, followed by 25 cycles of 95°C for 30 sec, 68°C for 30 sec, 72°C for 1 min with a final step of 72°C for 10 min elongation. Amplicons were purified in agarose gel and extracted by using gel extraction kit (GeneJET Gel Extraction Kit, K0692, Thermo Scientific) following the manufacture instructions. Ligase reactions were performed combining digested plasmids with inserts (molar ratio 1:3) in the presence of ligase with the appropriate buffer for 16 h at room temperature. Ligase reactions were used to transform competent DH5α and plated into the bacteria agar medium supplied with the appropriated antibiotic selection.
Bacteria SUMOylation assay
The plasmids codifying the complete SUMO conjugating system (pSUMO1, pSUMO2, and pSUMO3, with streptomycin resistance) were purchased from Addgene (http://www.addgene.org), plasmid IDs: 52258, 52259, 52260 (Weber et al., 2014). Those vectors contained distinctly SUMO1, SUMO2, and SUMO3 ORFs, N-terminus tagged with 6×-HIS and the ORFs of SUMO E1 dimers (SAE1-SAE2) and the SUMO E2 conjugating enzyme UBC9. Escherichia coli BL21 cells were co-transformed with the pGEX-5X-1 codifying MuRF1 together with the SUMO conjugating system plasmids. Selected colonies and bacteria cultures were grown in the presence of 100 mg/L of ampicillin and 50 mg/L of streptomycin. Protein expression was induced by the addition of the isopropyl-β-D-thiogalaktosid (IPTG, I1000-10 from Saveen Werner) to the final concentrations of 250 μM for 3 h at 30°C then transferred at 37°C for 30 min. Finally, cells were harvested by centrifugation (2500 g for 10 min at 4°C) and soluble protein fractions were extracted by sonication in lysis buffer (50 mM Tris-Cl pH 7.5, 300 mM NaCl, 0.1% NP40, 0.05% SDS, 1 mM DTT, 20 mM NEM, protease inhibitors). Crude lysates were clarified by centrifugation (11000 g for 10 min at 4°C) and separate with SDS-PAGE gel after denaturation in the presence of loading buffer.
Eukaryotic cell transfection and cell lyses
Murine myocytes (C2C12) and HeLa cells were grown in low glucose medium (1 g/L glucose D-6046, Sigma Aldrich) supplied with FBS 10% (F-0804, Sigma Aldrich) and penicillin and streptomycin (P-4333, Sigma Aldrich) and when occurred, C2C12 cells were grown in high glucose medium (4.5 g/L glucose D-5671). For transfection, 40%–50% confluent cells were transfected with the according plasmids using lipofectamine (R-0531, TurboFect transfection Reagents, Thermo Fischer Scientific). Cells were harvested after 24 h by centrifugation (3000 RPM for 10 min at 4°C) and washed with cold phosphate buffered saline (PBS) containing 0.2 M iodoacetamide. The soluble protein fractions were extracted with RIPA buffer (25 mM Tris-Cl pH 7.5, 50 mM NaCl, 0.5% NP40, 1 mM EDTA pH 8.5, 1 mM DTT, 20 mM NEM, protease inhibitors) and passed through a syringe 2–3 times. Crude lysates were clarified by centrifugation (11000 g for 15 min at 4°C) and protein concentration were measured and separate with SDS-PAGE gel after denaturation in presence of loading buffer.
GST protein purification for mass spectrometry analysis
E. coli BL21 cells were cotransformed with pGEX-5X-1-MuRF1 plasmid together with the SUMO conjugating system plasmids expressing the two variant SUMO1 versions, T95K and T95R (Impens et al., 2014). Induced bacteria were lysed and the GST-MuRF1 species were purified using the GST-beads according to the manufacturer protocol (17-0756-01, GE Healthcare). The purified GST-MuRF1 was separated by SDS-PAGE gel and stained with Coomassie blue. The bands corresponding to the GST-MuRF1 and the potential modified by SUMO between 75 and 100 kDa were cut and analyzed by mass spectrometry (Supplementary material).
Side direct mutagenesis
The site direct mutagenesis was performed on the pEGFP-N1 MuRF1, pCMV-cMyc-UBC9 and pSUMO1 plasmids with QuikChange II Site-Directed Mutagenesis Kit (200524, Agilent Technologies AH Diagnostic, Solna Sweden). The designed primers to generate the mutations are listed in Supplementary Table S1. The reaction contained 100 ng of plasmid template, 20 μM of each primer (forward and reverse), 20 mM of dNTP’s mix, 1× High Fidelity Buffer, 2 U of Pfu Taq DNA polymerase in the presence of 8% DMSO, in a total volume of 50 μl. The cycling program was: denaturation 95°C 3 min, followed by 16 cycles of 95°C for 30 sec, 65°C for 1 min, 72°C for 5 min with a final step of 72°C for 10 min elongation. After PCR samples were treated with 5 μl of Tango buffer and 1 μl of DpnI enzyme (ER1701, Thermo Fischer Scientific) at 37°C for 1 h. Then 20 μl of digested sample were used to transform 50 μl of DH5α bacteria and plated into selected LB agar plates. Plasmid DNAs were extracted and send to the sequence to confirm the nucleotide mutations.
Immunoprecipitation with anti-GFP antibodies conjugated to Sepharose beads
HeLa cells were transfected with pEGFP, pEGFP-MuRF1, and pEGFP-MuRF1-K238R plasmids. After 24 h from transfection, cells were rescued and washed twice with cold PBS containing 0.2 M of iodacetamide. Pellets were lysed with IP Lysis buffer (150 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA pH 8.5, 1% NP40, 0.01% SDS, 0.5% DOC, 1 mM DTT, 20 mM NEM and fresh protease inhibitor). After centrifugation for 10 min at 4°C at 10000 g, supernatants were precleared with 10 μl of mouse Ig beads (prewashed twice with IP Lysis buffer) for 1 h at 4°C under rotation and then left for 6 h with 20 μl of GFP-conjugated beads (prewashed twice with IP Lysis buffer) at 4°C under rotation. GFP beads were rescued by centrifugation (1000 g for 5 min at 4°C) and washed four times with IP Lysis buffer. Washed beads were resuspended with 80 μl of 2× loading buffer and separate with SDS-PAGE gel after denaturation.
Confocal images for MuRF1 intracellular localization
Twenty thousand cells were seeded in a 12-well plate over coverslips precoated with poly-D-lysine (P6407, Sigma-Aldrich). After 24 h, cells were transfected with 2 μg of pEGFP-MuRF1 plasmids. After additional 24 h, coverslips were fixed with 4% Paraformaldehyde (P6148, Sigma-Aldrich) solution in PBS mounted with Fluoroshield mounting medium with DAPI (ab104139, AbCam). Before fixation, cells were incubated for 20 min with MitoTracker reagent 250 nM (MitoTracker Deep Red 633, M22426, Thermo Fischer Scientific). Confocal microscope (LSM 510 META, Zeiss, Sweden) was used to acquire the images.
Muscle lysates for protein expression analysis
Frozen muscle biopsies were incubated with lyses buffer (50 mM Tris-Cl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% SDS, 0.5% DOC, 0.5% NP40, 1 mM DTT, 10 mM NEM, 20 mM Iodoacetamide, protease inhibitors) for 15 min on ice, then homogenized in a 1.5-ml tube with pestle (431-0098, VWR international AB). Muscle homogenizes were centrifuged for 20 min at 13000 g at 4°C. Clear supernatants were collected and protein concentration was measured with a Protein Assay kit (Bio-Rad Laboratories).
Reactive oxygen species detection
We used transfected C2C12 cells in a 96-well plate and follow the protocol instructions described in the manufacture manual of DCFDA/H2DCFDA—Cellular Reactive Oxygen Species Detection Assay Kit (ab113851, AbCam).
Muscle isolation from mice
Diaphragm muscles were isolated from 8- to 10-week-old BKS.Cg-Dock7m+/+Leprdb/J male mice, (Leprdb)wt/wt and (Leprdb)mut/mut, as the model of chronic hyperglycemia. This strain is used to model phases I–III of diabetes type II and obesity. Mice homozygous for the diabetes spontaneous mutation (Leprdb) manifest morbid obesity, chronic hyperglycemia, pancreatic beta cell atrophy, and become hypoinsulinemic. Obesity starts at 3–4 weeks of age. Elevated plasma insulin begins at 10–14 days and elevated blood sugar at 4–8 weeks. Mice were sacrificed by cervical dislocation and the respiratory muscles were immediately isolated and deeply frozen in liquid nitrogen and storage at −140°C.
Statistical analysis
One-way analyses of variance and Student t-test were used to compare multiple groups. P < 0.05 was considered statistically significant. Data are presented as average ± standard deviations.
Supplementary Material
Acknowledgements
UBC9 plasmid was a gift kindly provided by Dr Andrea Pichler (Max Planck Institute of Immunobiology and Epigenetics). pSCAI80 (pEFIRES-P-6His SUMO1) and pSCAI82 (pEFIRES-P-6His SUMO2) plasmids were gifts kindly provided by Dr Ron T. Hay (College of Life Sciences, Dundee University). Diaphragms from KO MuRF1 were gifts provided by Dr Jens Fielitz at Charité–Universitätsmedizin Berlin Experimental and Clinical Research Center (ECRC) at the Max-Delbrueck Center for Molecular Medicine (MDC).
Funding
This work was supported by Swedish Research Council 2013–3074 (Vetenskapsrådet) and Erik och Edith Fernströms foundation 2012 to S.G., G.H., and A.V.N., COST Proteostasis (BM1307-040917–087135) to S.G. and A.V.N., Taishan Scholar’s program to G.T., the National Natural Science Foundation of China (31671139 to M.J. and 31771284 to G.T.), Natural Science Foundation of Shandong Province, China (ZR2016JL026) to G.T., and the Key Research and Development Plan of Shandong Province (2017GSF18103) to M.J. G.H. is partially supported by KID Karolinska Institutet (C333802033). Also financial support from Åke Wiberg Foundation (M14-0127), Magnus Bergvall Foundation (2015-01200, 2016-01675), and Carl Trygger Foundation (CST 15:57) to S.B.L. are acknowledged.
Conflict of interest
none declared.
Author contributions
The study was conceived and supervised by S.G. G.H. performed bacteria transformation, protein purification for mass spectrometry, site direct mutagenesis, and enzymatic assays. G.H. and A.V.N. performed immunofluorescence assays, confocal microscopy, mouse diaphragm muscle lysates, and immunoblotting. G.H., A.V.N., and L.T. performed eukaryotic cell transfection, cell fractionation, and immunoprecipitation. G.S., A.F., and S.B.L. performed mass spectrometry and data analysis. M.J. and G.T. provided the hyperglycemic and control mice. S.G. performed the remained experiments, analyzed data, prepared figures, and wrote the manuscript with contributions from all authors.
References
- Baczyk D., Audette M.C., Drewlo S., et al. (2017). SUMO-4: A novel functional candidate in the human placental protein SUMOylation machinery. PLoS One 12, e0178056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomster H.A., Hietakangas V., Wu J., et al. (2009). Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol. Cell. Proteomics 8, 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine S.C., and Baehr L.M. (2014). Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 307, E469–E484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine S.C., Latres E., Baumhueter S., et al. (2001). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708. [DOI] [PubMed] [Google Scholar]
- Bossis G., and Melchior F. (2006). Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 21, 349–357. [DOI] [PubMed] [Google Scholar]
- Centner T., Yano J., Kimura E., et al. (2001). Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J. Mol. Biol. 306, 717–726. [DOI] [PubMed] [Google Scholar]
- Chakraborty J., Basso V., and Ziviani E. (2017). Post translational modification of Parkin. Biol. Direct 12, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.N., Czernuszewicz G., Tan Y., et al. (2012). Human molecular genetic and functional studies identify TRIM63, encoding Muscle RING Finger Protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ. Res. 111, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T.L., Hsiao H.H., Yeh Y.Y., et al. (2004). In vitro modification of human centromere protein CENP-C fragments by small ubiquitin-like modifier (SUMO) protein: definitive identification of the modification sites by tandem mass spectrometry analysis of the isopeptides. J. Biol. Chem. 279, 39653–39662. [DOI] [PubMed] [Google Scholar]
- Dai K.S., and Liew C.C. (2001). A novel human striated muscle RING zinc finger protein, SMRZ, interacts with SMT3b via its RING domain. J. Biol. Chem. 276, 23992–23999. [DOI] [PubMed] [Google Scholar]
- Dohmen R.J. (2004). SUMO protein modification. Biochim. Biophys. Acta 1695, 113–131. [DOI] [PubMed] [Google Scholar]
- Eifler K., and Vertegaal A.C. (2015). SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem. Sci. 40, 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Files D.C., D’Alessio F.R., Johnston L.F., et al. (2012). A critical role for muscle ring finger-1 in acute lung injury-associated skeletal muscle wasting. Am. J. Respir. Crit. Care Med. 185, 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke B., Gasch A., Rodriguez D., et al. (2014). Molecular basis for the fold organization and sarcomeric targeting of the muscle atrogin MuRF1. Open Biol. 4, 130172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P.S. (2000). RING for destruction? Curr. Biol. 10, R84–R87. [DOI] [PubMed] [Google Scholar]
- Gareau J.R., and Lima C.D. (2010). The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., and Melchior F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956. [DOI] [PubMed] [Google Scholar]
- Gersh B.J., Maron B.J., Bonow R.O., et al. (2011). 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 124, 2761–2796. [DOI] [PubMed] [Google Scholar]
- Gill G. (2005). Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15, 536–541. [DOI] [PubMed] [Google Scholar]
- Glass D., and Roubenoff R. (2010). Recent advances in the biology and therapy of muscle wasting. Ann. NY Acad. Sci. 1211, 25–36. [DOI] [PubMed] [Google Scholar]
- Golebiowski F., Matic I., Tatham M.H., et al. (2009). System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2, ra24. [DOI] [PubMed] [Google Scholar]
- Guo D., Han J., Adam B.L., et al. (2005). Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem. Biophys. Res. Commun. 337, 1308–1318. [DOI] [PubMed] [Google Scholar]
- Guo D., Li M., Zhang Y., et al. (2004). A functional variant of SUMO4, a new IκBκ modifier, is associated with type 1 diabetes. Nat. Genet. 36, 837–841. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S. (2011). TRIM proteins and cancer. Nat. Rev. Cancer 11, 792–804. [DOI] [PubMed] [Google Scholar]
- Hsiao H.H., Meulmeester E., Frank B.T., et al. (2009). ‘ChopNSpice,’ a mass spectrometric approach that allows identification of endogenous small ubiquitin-like modifier-conjugated peptides. Mol. Cell. Proteomics 8, 2664–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Xu L., Zhou X., et al. (2013). High glucose induces activation of NF-κB inflammatory signaling through IκBα sumoylation in rat mesangial cells. Biochem. Biophys. Res. Commun. 438, 568–574. [DOI] [PubMed] [Google Scholar]
- Impens F., Radoshevich L., Cossart P., et al. (2014). Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl Acad. Sci. USA 111, 12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S. (2004). Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382. [DOI] [PubMed] [Google Scholar]
- Kamogashira T., Hayashi K., Fujimoto C., et al. (2017). Functionally and morphologically damaged mitochondria observed in auditory cells under senescence-inducing stress. NPJ Aging Mech. Dis. 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedar V., McDonough H., Arya R., et al. (2004). Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc. Natl Acad. Sci. USA 101, 18135–18140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.E., He J., Menshikova E.V., et al. (2002). Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51, 2944–2950. [DOI] [PubMed] [Google Scholar]
- Koyama S., Hata S., Witt C.C., et al. (2008). Muscle RING-finger protein-1 (MuRF1) as a connector of muscle energy metabolism and protein synthesis. J. Mol. Biol. 376, 1224–1236. [DOI] [PubMed] [Google Scholar]
- Labeit S., Kohl C.H., Witt C.C., et al. (2010). Modulation of muscle atrophy, fatigue and MLC phosphorylation by MuRF1 as indicated by hindlimb suspension studies on MuRF1-KO mice. J. Biomed. Biotechnol. 2010, 693741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Xiang F., Yakovenko A., et al. (2005). The kinase domain of titin controls muscle gene expression and protein turnover. Science 308, 1599–1603. [DOI] [PubMed] [Google Scholar]
- Masoumi K.C., Marfany G., Wu Y., et al. (2016). Putative role of SUMOylation in controlling the activity of deubiquitinating enzymes in cancer. Future Oncol. 12, 565–574. [DOI] [PubMed] [Google Scholar]
- Mattox T.A., Young M.E., Rubel C.E., et al. (2014). MuRF1 activity is present in cardiac mitochondria and regulates reactive oxygen species production in vivo. J. Bioenerg. Biomembr. 46, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinny A.S., Kakinuma K., Sorimachi H., et al. (2002). Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J. Cell Biol. 157, 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E., Kunze M., Hsiao H.H., et al. (2008). Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619. [DOI] [PubMed] [Google Scholar]
- Namuduri A.V., Heras G., Mi J., et al. (2017). A proteomic approach to identify alterations in the small ubiquitin-like modifier (SUMO) network during controlled mechanical ventilation in rat diaphragm muscle. Mol. Cell. Proteomics 16, 1081–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochala J., Gustafson A.M., Diez M.L., et al. (2011). Preferential skeletal muscle myosin loss in response to mechanical silencing in a novel rat intensive care unit model: underlying mechanisms. J. Physiol. 589, 2007–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M., Abdul-Hussein S., Oldfors A., et al. (2015. a). New cardiac and skeletal protein aggregate myopathy associated with combined MuRF1 and MuRF3 mutations. Hum. Mol. Genet. 24, 3638–3650. [DOI] [PubMed] [Google Scholar]
- Olive M., Abdul-Hussein S., Oldfors A., et al. (2015. b). New cardiac and skeletal protein aggregate myopathy associated with combined MuRF1 and MuRF3 mutations. Hum. Mol. Genet. 24, 6264. [DOI] [PubMed] [Google Scholar]
- Owerbach D., McKay E.M., Yeh E.T., et al. (2005). A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 337, 517–520. [DOI] [PubMed] [Google Scholar]
- Perry B.D., Caldow M.K., Brennan-Speranza T.C., et al. (2016). Muscle atrophy in patients with Type 2 Diabetes Mellitus: roles of inflammatory pathways, physical activity and exercise. Exerc. Immunol. Rev. 22, 94–109. [PMC free article] [PubMed] [Google Scholar]
- Pichler A., Knipscheer P., Oberhofer E., et al. (2005). SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat. Struct. Mol. Biol. 12, 264–269. [DOI] [PubMed] [Google Scholar]
- Prudden J., Pebernard S., Raffa G., et al. (2007). SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan S., Torres J., Thompson M.S., et al. (2012). SUMO downregulates GLP-1-stimulated cAMP generation and insulin secretion. Am. J. Physiol. Endocrinol. Metab. 302, E714–E723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A., Meroni G., Fantozzi A., et al. (2001). The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.S., Dargemont C., and Hay R.T. (2001). SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659. [DOI] [PubMed] [Google Scholar]
- Su M., Wang J., Kang L., et al. (2014). Rare variants in genes encoding MuRF1 and MuRF2 are modifiers of hypertrophic cardiomyopathy. Int. J. Mol. Sci. 15, 9302–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham M.H., Jaffray E., Vaughan O.A., et al. (2001). Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374. [DOI] [PubMed] [Google Scholar]
- Tempe D., Piechaczyk M., and Bossis G. (2008). SUMO under stress. Biochem. Soc. Trans. 36, 874–878. [DOI] [PubMed] [Google Scholar]
- Uzunova K., Gottsche K., Miteva M., et al. (2007). Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167–34175. [DOI] [PubMed] [Google Scholar]
- Wang C.Y., and She J.X. (2008). SUMO4 and its role in type 1 diabetes pathogenesis. Diabetes Metab. Res. Rev. 24, 93–102. [DOI] [PubMed] [Google Scholar]
- Weber A.R., Schuermann D., and Schar P. (2014). Versatile recombinant SUMOylation system for the production of SUMO-modified protein. PLoS One 9, e102157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger S., Hammer E., and Heilbronn R. (2004). SUMO-1 modification regulates the protein stability of the large regulatory protein Rep78 of adeno associated virus type 2 (AAV-2). Virology 330, 284–294. [DOI] [PubMed] [Google Scholar]
- Witt S.H., Granzier H., Witt C.C., et al. (2005). MURF-1 and MURF-2 target a specific subset of myofibrillar proteins redundantly: towards understanding MURF-dependent muscle ubiquitination. J. Mol. Biol. 350, 713–722. [DOI] [PubMed] [Google Scholar]
- Yang S.H., and Sharrocks A.D. (2010). The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol. Cell. Biol. 30, 2193–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.