Abstract
Acquired drug resistance is the major reason why patients fail to respond to cancer therapies. It is a challenging task to determine the tipping point of endocrine resistance and detect the associated molecules. Derived from new systems biology theory, the dynamic network biomarker (DNB) method is designed to quantitatively identify the tipping point of a drastic system transition and can theoretically identify DNB genes that play key roles in acquiring drug resistance. We analyzed time-course mRNA sequence data generated from the tamoxifen-treated estrogen receptor (ER)-positive MCF-7 cell line, and identified the tipping point of endocrine resistance with its leading molecules. The results show that there is interplay between gene mutations and DNB genes, in which the accumulated mutations eventually affect the DNB genes that subsequently cause the change of transcriptional landscape, enabling full-blown drug resistance. Survival analyses based on clinical datasets validated that the DNB genes were associated with the poor survival of breast cancer patients. The results provided the detection for the pre-resistance state or early signs of endocrine resistance. Our predictive method may greatly benefit the scheduling of treatments for complex diseases in which patients are exposed to considerably different drugs and may become drug resistant.
Keywords: drug resistance, breast cancer, tipping point, dynamic network biomarker (DNB), molecular network, mRNA-seq
Introduction
Breast cancer, one of the most common cancers, is a heterogeneous, complex, interrelated disease that involves multi-factorial etiologies. The tumorigenesis of breast cancer is typically characterized by a combination of interactions between environmental (external) factors and a genetically susceptible host (internal factors) (Ou et al., 2010). Seventy percent of breast cancers are categorized as hormone-sensitive estrogen receptor (ER)-dependent tumors and initially respond to endocrine therapy, as tamoxifen treatment. However, approximately 30%–40% of tamoxifen-responsive tumors eventually acquire endocrine resistance after long-term treatment with this drug (Riggins et al., 2007; Musgrove and Sutherland, 2009). Tamoxifen resistance has been intensively studied both in vivo and in vitro at the molecular level. As a result, a number of mechanisms, such as the upregulation of membrane receptor kinases or dysregulation of the ER or PI3K pathways, have been proposed as the basis of tamoxifen resistance (Campbell et al., 2001; Gee et al., 2001; Knowlden et al., 2003; Creighton et al., 2008; Massarweh et al., 2008; Musgrove and Sutherland, 2009). Those studies indicate that drug resistance in breast cancer is caused by the modification of multiple molecules in the molecular network rather than by the alteration of individual molecules. In other words, drug resistance is the result of a cellular transition in which a molecular network is rewired to adapt to the drug environment. The prevalence of breast cancer as well as the growing economic and societal burden of treatment is making it urgently necessary to prevent the relapse of breast cancer. From the nonlinear dynamics viewpoint, the process of acquiring drug resistance typically has three stages, i.e. from a non-resistance state, through a pre-resistance state (or a tipping point) to a resistance state (Figure 1 and Supplementary Figure S1). Generally, the pre-resistance state can be reversed to the non-resistance state by appropriate treatment, but it is very difficult to return to the non-resistance state from the resistance state. Thus, the pre-resistance state is also the tipping point, after which the system undergoes an irreversible transition to the resistance state. In other words, acquiring drug resistance is typically a nonlinear process, with gradual changes during the non-resistance state but with drastic changes after the pre-resistance state. However, detecting the pre-resistance state or early signs of endocrine resistance is still a challenge because there are no significant differences between the non-resistance and pre-resistance states in terms of molecular signatures and clinical phenotypes (Chen et al., 2012), while irreversible complications after the tipping point may develop rapidly before the implementation of other treatment strategies (Saini et al., 2012). Therefore, it is of great importance to predict the phase shift in the response to tamoxifen treatment and to identify the related network responsible for such a critical phase shift, which is also crucial for a better understanding of drug resistance mechanisms.
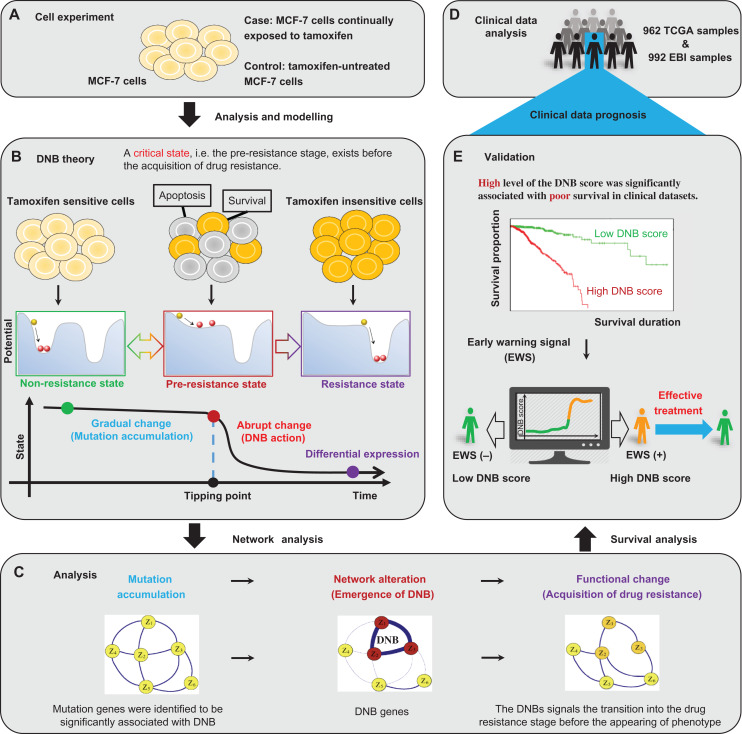
Figure 1.
The multi-states and tipping point of the tamoxifen resistance process. (A) The study is based on a set of time-course mRNA sequence data generated from the experiment of tamoxifen-treated ER-positive MCF-7 cell line. (B) According to the biological feature of the time-dependent progression of MCF-7 breast cancer cells exposed to tamoxifen, the process to acquire drug resistance is divided into three stages: i.e. a non-resistance state, a pre-resistance state (or the tipping point), and a resistance state. The non-resistance state is a steady or stable state with strong resilience or robust for small perturbations, representing a drug-sensitive stage. The pre-resistance state is defined as the limit of the non-resistance state but with a lower recovery rate or resilience from small perturbations. Such a pre-resistance state is the critical stage of drug resistance, which is crucial to the resistance process. The resistance state is another stable state usually also with strong resilience, where the system turns into a tamoxifen-insensitive stage and thus makes the drug ineffective. The DNB method can quantitatively identify the tipping point of this nonlinear process. (C) Through the DNB approach, the study reveals that the DNB network alteration is just prior to the observation of tamoxifen resistance, and follows the appearance of mutation genes. (D) Based on both TCGA and EBI samples, the clinical data analysis is processed and prepared for further validation. (E) The survival analysis shows that a higher level of the combined DNB score was significantly associated with poor survival in both TCGA and EBI datasets. The results suggest that the early-warning signal provided by DNB may benefit the implementation of appropriate treatment strategies.
The widespread utilization of high-throughput sequencing data in healthcare and clinical studies brings unprecedented opportunities to analyze disease progression at a system-wide or network level by exploiting high-dimensional dynamic data. However, the utility in leveraging these high-throughput data to integrate readily available biological datasets to detect the drug resistance transition has been relatively understudied. Preliminary studies have shown promising results in the biomedicine and bioinformatics fields (Chen et al., 2012; Liu et al., 2014a). In those works, the dynamic network biomarker (DNB) method was developed to detect the tipping point just prior to the drastic deterioration of complex diseases based on three statistical conditions derived from nonlinear dynamical theory (Li et al., 2015). The DNB method aims to identify the pre-disease state rather than the disease state, which is different from traditional biomarkers (Figure 1). Both theoretically and computationally, it has been shown that a group of highly correlated and strongly fluctuating molecules called DNB will appear when a biological system approaches a pre-disease state from a normal state (Chen et al., 2012; Liu et al., 2012). Such dynamic features can reliably quantify an imminent critical transition from observed data even if there is no significant difference in terms of the molecular concentrations or clinical phenotypes between the normal and pre-disease states (Figure 1). Indeed, a number of studies have revealed the role of such features before catastrophic shifts during the progression of many chronic diseases or biological processes (Litt et al., 2001; McSharry et al., 2003; Venegas et al., 2005; Hirata et al., 2010; He et al., 2012). In addition to the theoretical foundation, the DNB method has recently been successfully applied to real biological data to identify the early-warning signals of phase shifts during biological processes, such as the cell differentiation process (Richard et al., 2016), the process of cell fate decision (Mojtahedi et al., 2016), the critical transition in the immune checkpoint blockade-responsive tumor (Lesterhuis et al., 2017), the multi-stage deteriorations of T2D (Li et al., 2014), acute lung injury (Liu et al., 2014b), HCV-induced liver cancer (Liu et al., 2012), and many others (Zeng et al., 2014; Liu et al., 2014a; Tan et al., 2015; Chen et al., 2016). In this work, based on our mRNA sequence data of ER-positive MCF-7 breast cancer cells that were continually treated with tamoxifen up to 12 weeks (Figure 2A), we systematically analyzed the drug resistance process by the DNB approach from the aspects of both gene expression and mutation aspects and identified the critical state just before a tamoxifen-tolerance stage of the cancer cells. Specifically, by applying the DNB approach, a group of genes were identified to signal the critical change in the cellular state associated with the acquisition of drug resistance during 3–4 weeks of continual exposure to tamoxifen. Those DNB genes functioning at the tipping point are associated with the regulation of the cell cycle, DNA replication, and mismatch repair. Notably, by a gene variant analysis of the sequence data, we also found a number of mutations in the regulatory genes of the cell cycle, apoptotic signaling pathways and ER over 4–5 weeks, indicating that such a critical transition may be triggered by the interplay between mutant genes and DNB genes. The accumulated mutations eventually affect the DNB genes, which induce the drug resistance. Actually, further pathway and network analyses demonstrated that some mutant genes regulate the downstream DNBs that subsequently cause the change of transcriptional activities of other biomolecules, thereby enabling full-blown drug resistance.
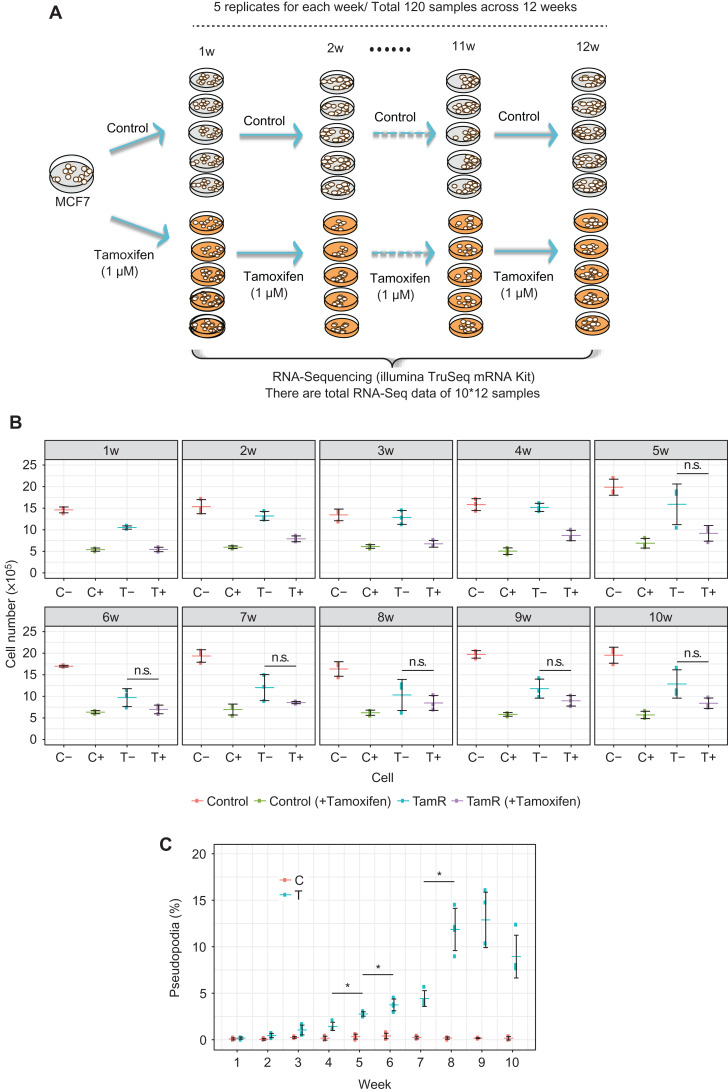
Figure 2.
Resistance acquisition and mRNA sequence data with long-term tamoxifen treatment of MCF-7 breast cancer cells. (A) MCF-7 cells were continually treated with tamoxifen for 12 weeks, with five biological replicates for each week. As controls, tamoxifen-untreated MCF-7 cells are cultured for 12 weeks, with five biological replicates for each week. RNA Sequencing was used to characterize gene expression patterns in 60 tamoxifen-treated samples and 60 control samples in this work. (B) Cell proliferation assay of parental control and TamR cells. Cells were grown in the media in the presence or absence of 1 μM tamoxifen. The significance of the growth inhibitory effect of tamoxifen was calculated with Welch’s test. Symbol * shows significant difference between the values with and without re-administration of tamoxifen at the same time points (P-value <0.05). (C) Cell transformation assay. The number of cells with elongated edges (pseudopodia) was counted. The percentage of the cells with pseudopodia in the total cell numbers was calculated. Symbol * indicates significant difference in the numbers of pseudopodia compared with the prior time point (P-value <0.05, Weltch’s test). Clearly, there are significant differences after the 5th week between TamR(−) and TamR(+), which indicates the phenotypic change or the tipping point around the 5th week for the tamoxifen resistance.
Actually, a number of studies on the molecular mechanisms of tamoxifen resistance have indicated that there are slow changes with a drastic phenotypic transition, during which the interplay of multiple altered molecules as a form of a molecular network, contributes to endocrine resistance (Campbell et al., 2001; Gee et al., 2001; Knowlden et al., 2003; Creighton et al., 2008; Musgrove and Sutherland, 2009), which also provide the evidence for this work. To summarize briefly, as the first nonlinear study on the critical point in the tamoxifen resistance process from a systems biology perspective, this work based on DNB not only identified the tipping point of the drug resistance process from the observed dynamical data but also revealed the crucial molecules that contribute to the resistance at the network level.
Results
We generated a series of mRNA-seq data from MCF-7 cells after tamoxifen administration to detect the tipping point during the acquisition of drug resistance. MCF-7 cells were continually treated with tamoxifen (TamR cells) for 12 weeks, with five biological replicates for each week. As controls, tamoxifen-untreated MCF-7 cells (control cells) were cultured for 12 weeks, with five biological replicates for each week. In total, the mRNA-seq data of 120 time-course samples were obtained using the HiSeq2500 (Illumina) sequencing platform (Figure 2A). An assay of the cellular phenotype associated with long-term TamR cells was performed with triplicates (Figure 2B and C).
Gene expression patterns and functions change significantly at approximately the 5th week based on traditional bioinformatics analyses
We first used the mRNA-seq data to characterize the gene expression patterns in 60 TamR samples and 60 control samples over the 12-week period. Our analysis included 4135 genes whose reads per kilobase per million mapped reads (RPKM) values were dynamically diverse over the time-course in the TamR samples, with a standard deviation >3. A hierarchical clustering algorithm was used to group the samples based on similarities in the patterns in which the expression varied over these genes (Figure 3A). The detailed clustering of 120 samples is shown in Supplementary Figure S1. As expected, the 60 control samples were found clustered together. Notably, two distinct clusters were observed in the 60 TamR samples. The first cluster included all the samples from weeks 1 to 4, the expression patterns of which were quite different from those of the second cluster, which included all the samples from weeks 5 to 12. However, the expression patterns at the 5th week exhibited a different pattern between the two clusters, immediately after the tipping point identified by DNB at the 4th week. Our analysis of the gene expression showed that in the tamoxifen-treated samples, the 5th week marked a special stage after the tipping point in the phenotypic traits in terms of the gene expression.
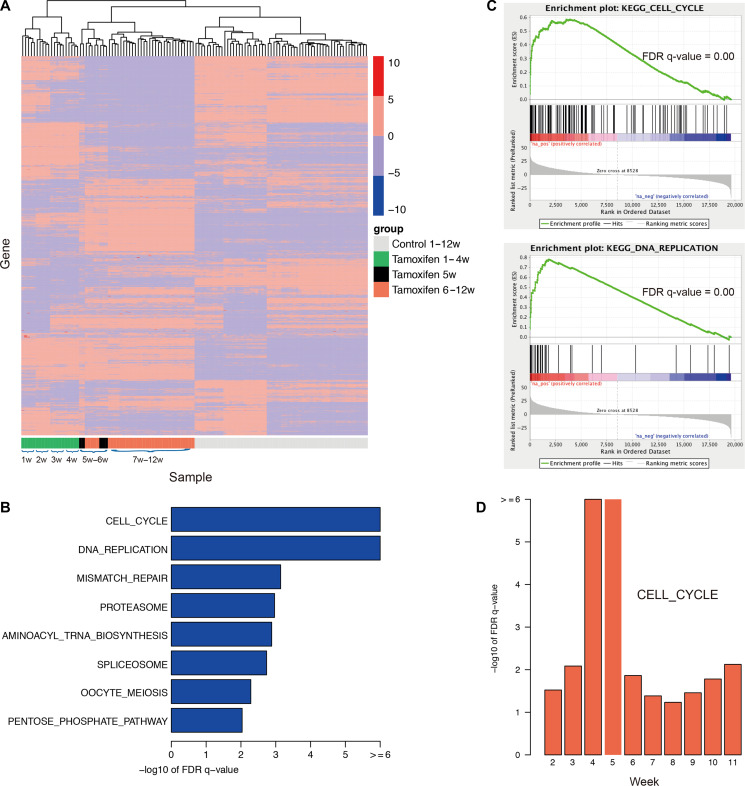
Figure 3.
mRNA expression patterns and drug resistance-associated pathways by traditional bioinformatics analyses. (A) Gene expression patterns over the time-course of both TamR and parental cells (120 samples with five replicates for each week over 12 weeks). Each row in the heatmap represents a specific gene with standard deviation (SD) > 3, whose expression is normalized across the column, with high expression shown in red and low expression shown in blue. Clearly, the gene expression pattern drastically changes at the 5th week, which is different from other weeks. (B) Eight enriched pathways with the FDR q-value <0.01 and the output of GSEA for the top two pathways. (C) GSEA graphs for two enriched pathways, cell cycle and DNA replication. (D) The cell cycle pathway is statistically associated with the occurrence of MCF-7 drug resistance, gaining the greatest statistical value at the 4th/5th week. The traditional bioinformatics analyses identified the change in expression patterns and functions over 12 weeks.
We then divided the TamR samples into two groups, with the first group consisting of all the samples from weeks 1–4 and the second group consisting of all the samples from weeks 6–12 and excluding the samples from the 5th week. Differentially expressed genes between the two groups were identified using DESeq2 (Love et al., 2014); the top 1000 differentially expressed genes are shown in Supplementary Table S1. We further applied the tool Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) to determine the functions enriched based on a pre-ranked list of all the genes, which were sorted according to the statistical significance of the differential expression defined by DESeq2. Eight KEGG pathways (Kanehisa and Goto, 2000) with the FDR q-value <0.01 (Figure 3B and C) were enriched, including the DNA replication pathway and cell cycle pathway. This result suggested that tamoxifen resistance in MCF-7 cells was triggered by the ectopic re-activation of the cell cycle and DNA replication together with other functions, resulting in rapid cell proliferation and growth beginning at the 5th week.
To assess whether these pathways were statistically associated with the occurrence of drug resistance, which began to occur at the 5th week, we generated a series of fake points by setting before-the-5th-week points b1, b2, and b3 (representing the 2nd, 3rd, and 4th weeks, respectively) or after-the-5th-week points a1, a2, a3, a4, a5 and a6 (representing 6th, 7th, 8th, 9th, 10th, and 11th weeks, respectively). The cells were reclassified into two groups based on the fake points, and the same analyses were performed. We then investigated the enrichment functions by comparing these fake points with the real point (the 5th week). Notably, we found that the cell cycle pathway was significantly associated with the occurrence of drug resistance, with a remarkably high FDR q-value of –log10 at the 4th–5th weeks (Figure 3D). Most of the genes functioning in the cell cycle pathway were highly expressed after the 5th week, including CCND1, MCM6, MCM4, CDC6, and CDC25A, which are shown in red (Supplementary Figure S2). Among these, CCND1 is a known oncogene in breast cancer. The mismatch repair pathway was shown to be specifically associated with the 4th–5th weeks (Supplementary Figure S3). These findings further demonstrated that the occurrence of tamoxifen resistance in MCF-7 cells was represented by the activation of functions such as the cell cycle pathway, DNA replication pathway, and mismatch repair pathways.
Cell proliferation assays indeed showed that the TamR cells grew slowly during the 1st–2nd weeks, but started to proliferate rapidly from the 3rd–4th weeks in the presence or absence of the re-administration of tamoxifen (Figure 2B). The TamR cells began to be significantly insensitive to newly added tamoxifen at the 5th week and then became stable from the 8th week (P < 0.05). The parental cells were sensitive to the drug treatment throughout the time period. The cell proliferation assays thus suggested that the acquisition of tamoxifen resistance may begin around the 4th–5th weeks. To gain more insight into the relationship between the cellular phenotype and tamoxifen resistance, we examined the cellular phenotype. Pseudopodia is a phenotypic marker associated with the invasiveness and migration of cells; therefore, we quantified the number of cells that had pseudopodia. The result indicated that the number of cells having pseudopodia started to increase significantly from the 5th week (Figure 2C).
The tipping point of tamoxifen resistance is the 4th week identified by DNB
In this study, we identified the early-warning signal for the critical transition of MCF-7 breast cancer cells to a drug-tolerance stage, during continual treatment with tamoxifen. The gene expression information from mRNA-seq was used to identify the pre-resistance state with the associated DNB prior to the irreversible drug resistance state. The DNB method was expected to detect the tipping point of the phase shift in the response to drugs as well as improve the sensitivity and specificity of the treatment strategy by delivering the longitudinal trends in the data. Specifically, as shown in Figure 4, from the observed mRNA-seq time-course data during the 12-week period, we identified the DNB (Supplementary Table S2) as well as the tipping point around the 3rd–4th week after continual exposure to tamoxifen. Clearly, the DNB score drastically increased when the system approached the tipping point, indicating the critical state; i.e. there were strongly amplified and highly correlated fluctuations of DNB molecules near the pre-resistance state, in contrast to the non-resistance and resistance states (Figure 4A). DNB molecules are considered as the functional genes for this critical state. In addition, by applying the landscape DNB algorithm (Supplementary Information) to the expression data of the tamoxifen-treated MCF-7 cells, we generated a local DNB score for each gene at each week. As shown in Figure 4B, strong early-warning signals of the tipping point were detected at the period of the 3rd–4th weeks, which validated the result of Figure 4A. It should be noted that, the drastic increase of the DNB score was detected at the period of the 3rd–4th weeks (Figure 4A and B), while cells from the 3rd and 4th weeks were cultured under the same condition (see section Materials and methods). In other words, the tipping point at the 4th week was identified by the collective dynamical behavior of DNB genes rather than the batch effects.
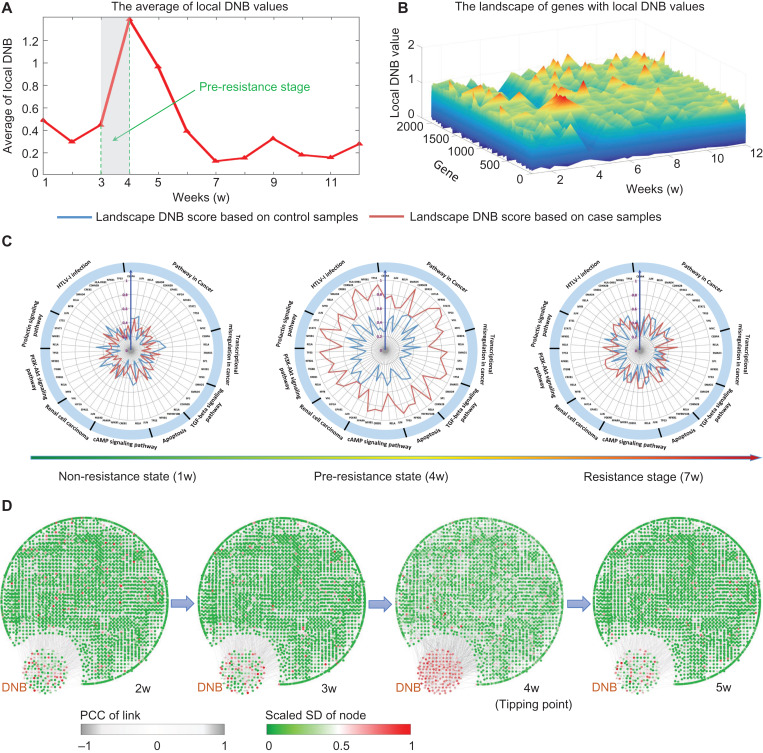
Figure 4.
The tipping point of drug resistance process revealed by DNB method. (A) The abrupt increase of the average DNB score clearly signals that there is a tipping point located around 3–4 weeks after the exposure of tamoxifen, after which the system undergoes the drastic transition to the resistance state. (B) The landscape curve of the local DNB method again indicates that 3–4 weeks after the exposure of tamoxifen are the critical transition period from the tamoxifen non-resistance state into a resistance state. (C) Some genes with top significant changes in local DNB scores around the identified transition period (3–4 weeks) are illustrated in three consecutive radar plots, in which the local DNBs generated from case and control samples are showed. The red curve represents the local DNB scores from the case samples while the green curve is that from the control data. At the pre-resistance stage, the DNB scores of case samples are significant. It can be seen that these top significant genes are enriched in pathways closely relevant to breast cancer. Besides, the sensitivity is demonstrated from the comparison between case and control local DNB curves. (D) Based on the mRNA-seq data of MCF-7 breast cancer cells, we illustrate dynamical evolution of the whole gene network (the co-expressed network). DNB genes are intentionally put together. It can be seen that by using DNB, an early-warning signal of the resistance from a network perspective can be detected at 3–4 weeks after the exposure of tamoxifen. The whole dynamics across the 12 weeks are given in Supplementary Figure S4.
In Figure 4C, the radar plots present the dynamical change in the local DNB scores of certain genes that were enriched in the pathways that are closely related to cancer progression. The local DNB scores of the TamR data increased considerably, while there were no significant changes in those of the control data. Furthermore, these identified genes showed that their collective behavior might lead to the dysfunction of some signaling pathways and thus affect the cellular function of drug resistance.
Figure 4D illustrates the dynamical evolution of the entire group of DNB molecules in terms of the network based on the observed mRNA-seq data. We found a group of genes that were strongly correlated with a high DNB score near the 4th week, while other molecules showed no significant signal during the entire time period (also see Supplementary Figure S4). Interestingly, members of the DNB group behaved similarly to other molecules after the system transitioned to the resistance state after the 4th week, which implied that these DNB genes mainly facilitated the biological functions only around the tipping point at the 4th week.
To further reveal the functions of the critical transition at the 4th week, we analyzed differentially expressed genes between the periods before and after the tipping point, i.e. the 1st–3rd weeks vs. the 5th–12th weeks. As shown in Supplementary Figure S8A, comparing with only 19.41% background turnover rate, there were over 66.67% turnover genes (i.e. significant changes of gene expressions from high to low or from low to high values, Supplementary Table S3) in the DNB-associated network (constructed from STRING) (Szklarczyk et al., 2015) just after passing the tipping point, which indicated the drastic transition of the biological system mediated by the DNB. Furthermore, in Supplementary Figure S8B, we present the statistical significance of the identified DNB genes. The DNB group arose with differential expression levels according to both Student’s t-test and the fold change when the system approached the tipping point.
The computational results agree well with the experiment and functional analysis. Immediately after the tipping point identified by DNB at the 4th week, the expression patterns at the 5th week were significantly different from those at other time points (Figure 3A). The first significant change in the numbers of pseudopodia was observed at the 5th week (Figure 2C). Moreover, the functional analysis suggested that the tamoxifen resistance in MCF-7 cells was triggered by the ectopic re-activation of the cell cycle and DNA replication together with other functions, resulting in rapid cell proliferation and growth beginning at the 5th week. Therefore, the DNB signaled the critical transition into drug resistance state.
Drug-driven mutations are identified during the 4th–5th weeks by mutation analyses
The evolving nature of human tumors enables the emergence of new mutations in patients after drug administration, leading to the development of drug resistance through the acquisition of a growth advantage. Therefore, mutations aligned with the tipping point of endocrine resistance can be considered as another type of early-warning sign before full-blown drug resistance.
To identify such mutations, we next employed GATK to analyze gene variants in the mRNA-seq data. In our case, there are three types of mutations: random mutation, MCF-7 mutation, and drug-driven mutation. Random mutations occur both in the control samples and the TamR samples. MCF-7 mutations originally occurred in the parental cell line, while drug-driven mutations specifically occur in the TamR cells (see section Materials and methods).
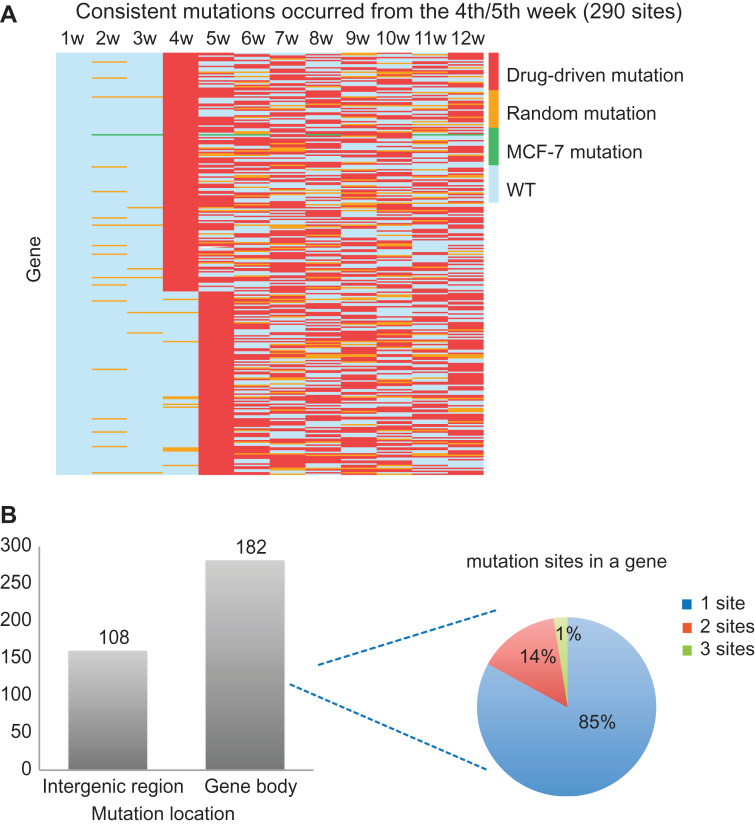
We further examined those drug-driven mutations that may confer drug resistance. The hypothesis was that these mutations would appear around the tipping point and persistently exist over the following weeks. Therefore, we focused on those drug-driven mutations that occurred from the 4th–5th weeks and existed persistently for at least 4 weeks thereafter (Supplementary Table S4). In total, 290 drug-driven mutation sites were identified, of which 197 were mutated from the 4th week and 93 were mutated from the 5th week (Figure 5A). Among the 290 mutation sites, 108 were in the intergenic region and 182 were in the gene body region covering 159 genes (Figure 5B, the left panel). An analysis of the genes harboring these mutation sites showed that 85% of the genes had a one-mutation site, 14% had two sites, and 1% had three sites (Figure 5B, the right panel). Two of the mutant genes were found to interact with the DNBs.
Figure 5.
Drug resistance-associated mutations. (A) Drug-driven mutations consistently existing from the 4th/5th week that may confer drug resistance, sorted by mutation occurrence time. Each row in the heatmap represents a specific mutation site, and each column represents a week. (B) The genome location of these mutations and their distribution in gene: 108 mutations are in the intergenic region, and 182 mutations are in the gene body region covering 157 genes; 85%, 14%, or 1% of genes contain one, two, or three mutations, respectively.
The regulations from drug-driven mutations to DNB genes
The interplay between the mutant genes and DNBs may promote the development of drug resistance. Since the functional consequences of mutations occurring in the region of exons are more predictable, we especially focused on 15 genes with drug-driven mutations that occurred in the gene exons (Supplementary Table S5). Among them, there were nine genes (red-colored genes in Supplementary Table S5) with functions associated with expression regulation, cell death, or cell growth and proliferation. Moreover, by investigating the KEGG pathways, the following five genes were found to interact with the downstream of DNBs, potentially influencing the progression of drug resistance with the DNB partners of these genes. Note that some of mutant and DNB genes are overlapped, such as PRICKLE2.
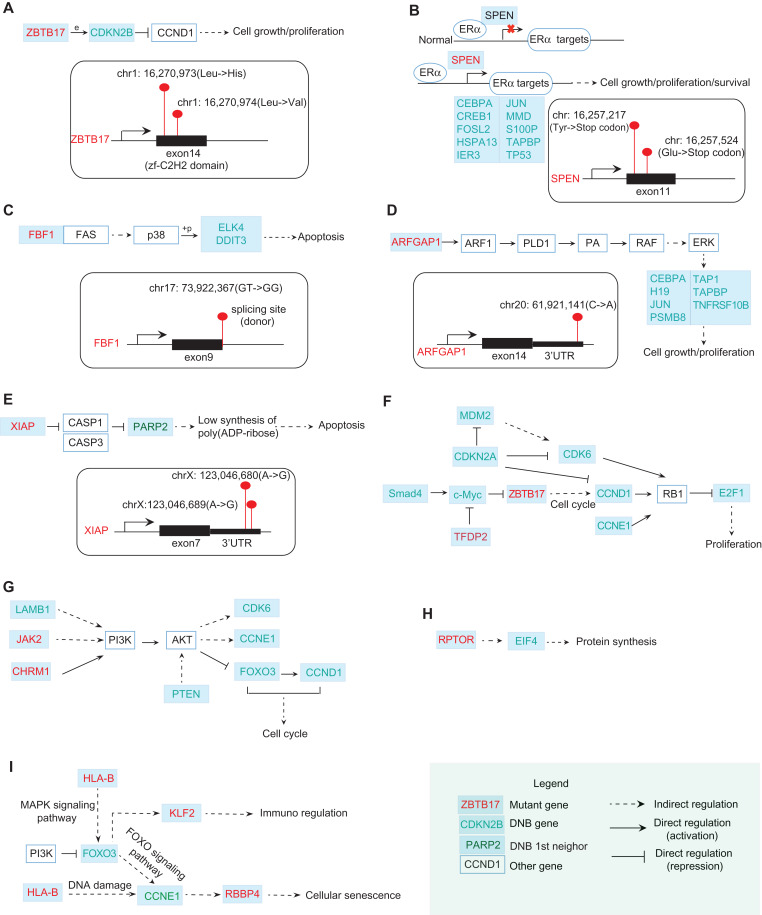
In Figure 6A, ZBTB17 encodes the zinc finger protein Miz1, a Myc-interacting protein. In the resistant cells, two sites in the region of the 14th exon, located in the zf-C2H2 domain, were detected to have missense mutations (chr1: 16,270,973A→T in the DNA resulting in Leu→His in the protein; chr1: 16,270,974G→C in the DNA resulting in Leu→Val in the protein). Miz1 directly regulates the expression of p15, which is encoded by the DNB gene CDKN2B. p15 is a direct repressor of cyclin D (CCND1), a member of the cyclin protein family, which is involved in regulating cell cycle progression. CCND1 has been identified as an oncogene in breast cancer. In our data, CCND1 was significantly upregulated in the resistant cells (P = 1.056e−282).
Figure 6.
The functional relations between mutation genes and DNB genes. (A) ZBTB17 with two missense mutations occurring in its zf-C2H2 domain can directly regulate the expression of a DNB gene CDKN2B. By directly repressing Cyclin D (CCND1), CDKN2B is involved in regulating cell cycle progression. (B) In normal breast cells, SPEN binds to ERα and negatively regulates the transcription of ERα targets. SPEN with nonsense mutations may re-activate ERα targets including DNB genes CEBPA, CREB1, FOSL2, etc. (C) A Fas-binding factor FBF1 has a mutation at the splicing site. Fas can induce the apoptosis by indirectly regulating some TFs. Among those TFs, two of DNB genes ELK4 and DDIT3 are involved. (D) ARFGAP1 with a mutation at 3′UTR may stimulate cell growth and proliferation by indirectly activating ERK for regulating some of DNB genes downstream, including CEBPA, H19, JUN, etc. (E) XIAP with two mutations at its 3′UTR encodes an X-linked inhibitor of apoptosis. One of the DNB neighbors at the first level, PARP2, is involved in this pathway. (F) From KEGG pathway in cancer, DNB genes SMAD4 and c-Myc regulate the mutant gene ZBTB17, which in turn affects DNB genes CCND1, E2F1 and thus induces the proliferation. (G) From PI3K–AKT signaling pathway, DNB genes LAMB1 and mutant genes JAK2, CHRM1 regulate the downstream DNB genes FOXO3, CCNE1, CDK6, etc. (H) The interplay between mutant gene RPTOR and DNB gene EIF4 takes part in the protein synthesis process. (I) The interplay between mutant genes HLA-B, RBBP4 and DNB genes FOXO3, CCNE1 is also involved in the cellular senescence pathway. The mutant genes above may interplay with DNBs to promote the development of drug resistance. In other words, the accumulated mutations eventually affect the DNB genes that subsequently cause the change of transcriptional landscape, enabling full-blown drug resistance.
In Figure 6B, SPEN encodes a hormone transcriptional repressor. In normal breast cells, SPEN binds in a ligand-independent manner and negatively regulates the transcription of targets. In our data, two types of SPEN nonsense mutations were detected in the resistant cells (chr1: 16,257,217C→A in the DNA resulting in Tyr→stop codon in the protein; chr1: 16,257,524G→T in the DNA resulting in Glu→stop codon in the protein), resulting in a resistance effect probably by the re-activation of targets. Those targets include several DNB genes, including CEBPA, CREB1, FOSL2, HSPA13, IER3, JUN, MMD, S100P, TAPBP, and TP53.
In Figure 6C, the mutant gene FBF1 is a Fas-binding factor. The Fas cell surface receptor belongs to the TNF receptor family of cell death-inducing molecules. In the JNK and p38 MAP kinase pathways, Fas can activate p38 by successively activating DAXX, ASK1, and MKK3/MKK6. As a mitogen-activated protein kinase, p38 catalyzes the phosphorylation of certain transcription factors and induces the apoptosis by regulating the expression of a number of genes. Among those transcription factors, two DNB genes, ELK4 and DDIT3, are involved. In our data, a donor site mutation (chr17: 73,922,367 GT→GG) of FBF1 was identified in the resistant cells. This change could result in the production of a non-functional protein, which may be associated with the inhibition of apoptosis.
In Figure 6D, as a GTPase-activating protein, ARFGAP1 can regulate the activity of ARF1. ARF1 is a small guanine nucleotide-binding protein that plays a role in vesicular trafficking as an activator of phospholipase D (PLD1). PLD1 catalyzes the hydrolysis of phosphatidylcholine to yield phosphatidic acid (PA) and choline. As a biosynthetic precursor, PA can activate Raf-1, resulting in the activation of MAPK signaling cascades. The gene MAPK1 encoding ERK is upregulated (P = 7.07e−55) in the resistant cells. The activation of ERK increases cell growth and proliferation by regulating some of the DNB genes downstream, including CEBPA, H19, JUN, etc. In our data, a mutation at the 3′UTR region following the 14th exon of ARFGAP1 (chr20: 61,921,141C→A) was detected.
In Figure 6E, XIAP is an X-linked inhibitor of apoptosis. Two sites in the region of 3′UTR following the 7th exon were identified to have mutations (chrX: 123,046,680A→G; chrX: 123,046,689A→G). XIAP inhibits at least two members of the caspase family of cell death proteases, CASP3 and CAS7. The subsequent repression of PARP results in low synthesis levels of poly(ADP-ribose) and the occurrence of apoptosis. One of the first-order neighbors of DNB in the STRING network, PARP2, is one of the genes encoding the PARP protein. In Figure 6F–I, it can be seen that from KEGG pathway analysis, the interplay between mutant and DNB genes is involved in many signaling pathways, which may induce the proliferation, cellular senescence, and dysfunction of cell cycle.
Therefore, mutant ZBTB17, SPEN, FBF1, ARFGAP1, XIAP, etc., by directly or indirectly interacting with DNB genes upstream or downstream, may contribute to the development of tamoxifen resistance in MCF-7 cells. In particular, the mutant genes regulate the downstream DNBs, which subsequently cause changes in the transcriptional landscape before and after the acquisition of resistance, thereby enabling full-blown drug resistance.
The predictive ability of DNBs and drug-driven mutations in clinical prognosis
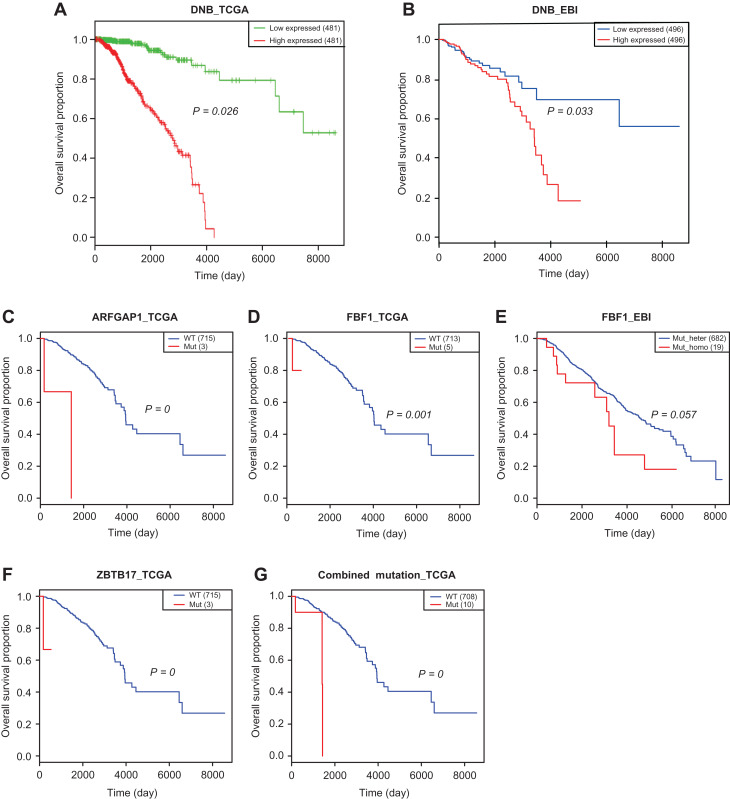
To validate the roles of the identified DNB genes and mutant genes in clinical prognosis, an integrative analysis using the expression profile, mutation profile, and clinical information from two independent datasets, TCGA and EBI, was applied towards the general cohorts with breast cancer. The Kaplan–Meier survival analysis was used to assess the predictive ability of the DNBs and the drug-driven mutations regarding the clinical outcomes of ER-positive breast cancer patients. The results showed that a higher level of the combined DNB score was significantly associated with poor survival in both the EBI (Figure 7A) and TCGA datasets (Figure 7B). Survival analyses for the individual DNB members are given in Supplementary Figure S5, which shows the significant functional effects of those DNB genes on disease progression. Moreover, among the five mutant genes above, the mutants ARFGAP1, FBF1, and ZBTB17 were identified to be significantly associated with poor overall survival in patients in the TCGA dataset (Figure 7C, D, and F). Notably, in the EBI dataset, a homozygous gene mutation in FBF1 was associated with poor survival compared with a heterozygous gene mutation in FBF1 (Figure 7E). Combining the mutations above, patients with ARFGAP1, FBF1, or ZBTB17 mutations were associated with a poor survival (Figure 7G). These results validated the functional effects of the DNB genes along with the associated mutated genes on disease progression. The survival analyses for the individual DNB members are shown in Supplementary Figure S5; based on these data, the collective group of DNB genes clearly performs better in predicting prognosis.
Figure 7.
Survival analysis results of DNB genes and mutational genes on two independent datasets. (A and B) The higher level of the combined expression index of DNBs is associated with the poorer survival in the TCGA dataset (A) and the EBI dataset (B). (C and D) Mutant ARFGAP1 (C) or FBF1 (D) is associated with the poor survival in the TCGA dataset. (E) In the EBI dataset, homozygous gene mutation in FBF1 is associated with the poor survival comparing with heterozygous gene mutation in FBF1. (F) Mutant ZBTB17 is associated with the poor survival. (G) The patients with ARFGAP1, FBF1, or ZBTB17 mutation are associated with the poor survival. The survival analysis results on individual DNB genes are given in Supplementary Figure S5. Survival analyses validate that DNB genes and mutant genes are associated with poor survival of breast cancer patients.
Discussion
Based on the time-course transcriptome data of the tamoxifen-treated MCF-7 breast cancer cells, we identified the critical cellular state associated with the acquisition of tamoxifen resistance by applying the data-driven DNB approach.
The DNB method is based on network and nonlinear theories, and is aimed at detecting the early-warning signals of a critical transition. By signaling the onset of the critical transition at an early stage, the DNB method provides a solution to the problem caused by the acquisition of drug resistances, and can thus guide the allocation of resources to better manage effective treatment. In our study, we used the time-course data obtained from an in vitro cell culture system, but the method is applicable to clinical datasets as well. In fact, although patients progress into the critical state before beginning to acquire drug resistance, it is difficult to determine such a stage using traditional biomarkers because there is no significant difference between the resistance and pre-resistance states in terms of clinical phenotypes, particularly for breast cancer. This fact, lamentably, is the barricade of reaching the early diagnosis of drug resistance in clinics. However, the DNB method provides a new way of using crucial regulators to explore the underlying mechanism of the acquisition of drug resistance, thus achieving an early adjustment of treatment. This approach to understanding drug resistance is one main contribution and value in the potential applications of the DNB method from a long-term point of view. This method can be used to simplify the process by which clinicians identify and screen patients using the omics data obtained from their non-invasive clinical samples for detecting drug resistance during cancer therapies.
In addition, based on both the TCGA and EBI datasets, the identified DNB molecules were validated using survival analyses, which demonstrated clear differences between the DNB-overexpressed and DNB-underexpressed samples. Indeed, an indicative early-warning signal was revealed by the DNB method around the 4th week during continual tamoxifen exposure, coincident with the experimental observation, which validates the effectiveness of the DNB method in one aspect and, more importantly, provides a clue to explain the underlying mechanism of the progression to the drug-tolerance stage. In clinics, the occurrence of such an early-warning sign may help screen out patients with the possibility of resistance to a specific drug, and thus achieve better management of effective treatment. Indeed, in a dynamic way, the DNB method reveals the existence of the pre-resistance state during continual exposure to a specific drug; however, such a state cannot be shown by individual molecular variables due to the ‘dynamics and network’ nature of these variables. Therefore, the benefits from using the DNB method in signaling the pre-resistance state make the identification and management of high-risk patients more effective. Our model was validated specifically with both the TCGA and EBI clinical samples, most of which had underexpressed DNB features and had relatively longer survival expectations, allowing the generalization of our method to this type of understudied drug-specific monitoring. In addition, rather than the correlation analysis (including both direct and indirect correlations) in DNB, we can further adopt the direct association analysis (Riggins et al., 2007; Musgrove and Sutherland, 2009) for DNB to improve the sensitivity.
In the molecular network perspective, the study reveals some interesting details during the process of drug resistance development in breast cancer cells. First, following the appearance of mutation genes, the DNB module in the molecular network changes significantly at the 4th week (Figure 4D), resulting in the global alteration of gene expression at the 5th week (Figure 3A) and triggering the ectopic re-activation of the cell cycle and DNA replication together with other functions. This may provide new insights into the underlying mechanism of drug resistance acquisition. Second, by considering that a higher level of the DNB score was significantly associated with poor survival (Figure 7A and B), the identified DNB genes may serve as markers for prognosis.
Our predictive method may greatly benefit the scheduling of treatments for complex diseases in which patients are exposed to considerably different drugs and may become drug resistant. Two common reasons for healthcare provider burnout are the prevalent use of complicated, error-prone devices and the rapid accumulation of patient data that must be processed in a timely and effective manner.
Materials and methods
Cell culture
The human breast adenocarcinoma MCF-7 cell line was purchased from American Type Culture Collection and propagated in Dulbecco’s Modified Eagle’s Media (Gibco) supplemented with 10% fetal bovine serum and antibiotics (100 units/ml penicillion, and 100 μg/ml streptomycin, Nacalai Tesque). For the chronological study of tamoxifen resistance, cells were seeded at 1 × 106 cells/100-mm dish in medium containing or lacking 1 μM tamoxifen (Sigma-Aldrich) and were continuously cultured for 3 months. The tamoxifen-untreated control cells were passaged with trypsin-EDTA (0.05% trypsin and 0.53 mM EDTA, Nacalai Tesque) once a week when they reached to confluence. The tamoxifen-treated (TamR) cells were passaged after one week when the cells reached to 70%–80% confluency. During weeks 2–4 of tamoxifen treatment, the cells grew slowly; therefore, only the medium was changed once a week instead of passaging the cells. From week 5, the tamoxifen-treated cells were passaged once a week in the same way as the control cells until week 12.
RNA extraction and cryopreservation
Cells were exposed or unexposed to tamoxifen for the indicated time periods in sextuplicate. Control cells from the 1st week to the 12th week and tamoxifen-treated cells of the 1st week and from the 5th week to the 12th week were re-plated at a density of 2 × 106 cells/100-mm dish and grown for 2 days to reach the log phase (~50% confluent). For the 2-, 3-, and 4-week tamoxifen-treated cells, the medium was changed instead of re-plating the cells, and the cultures were grown for 2 days. Therefore, the RNA samples from the 2-, 3-, and 4-week tamoxifen-treated cells were prepared from cultures in log phase but at a different confluency (20%–80% confluent). The tamoxifen exposure period was defined as up to the day of sample preparation. Quintuplicate samples (quadruplicate for the 2-week control sample) were used for total RNA extraction with Qiashredder (QIAGEN) and an RNeasy mini kit (QIAGEN). RNA concentration and integrity was evaluated using a Bioanalyzer 2100 (Agilent). For the validation study, the remaining cells were trypsinized, re-suspended in Cell Banker I (TaKaRa) freezing medium, and frozen in liquid nitrogen.
mRNA sequencing
mRNA (1 μg) was used for poly (A+) mRNA sequencing using the TruSeq Stranded mRNA Sample Prep Kit (or mRNA-Seq Sample Preparation Kit) (Illumina) according to the manufacturer’s protocol. One hundred-base pair-end reads or 36-base single-end reads were obtained using a HiSeq2500 (Illumina) instrument and analyzed using the analysis software provided by Illumina. The reads that were mapped to the ribosomal RNA were removed. For each time point, five samples were sequenced for the tamoxifen-treated (TamR) and untreated (control) cells.
Cell growth assay
MCF-7 cells were seeded at 1 × 106 cells per 100-mm dish in medium containing or lacking 1 μM tamoxifen (Sigma-Aldrich) as described above. Both the tamoxifen-treated and untreated cells were passaged after a week with trypsin-EDTA (0.05% trypsin and 0.53 mM EDTA, Nacalai Tesque) regardless of the culture confluency. Quantities of 2 × 105 cells/well were plated on 6-well plates in tamoxifen-containing (1 μM) medium or without tamoxifen and grown for the designated time period. The assay was performed with triplicates.
Cell transformation assay
Pseudopodia is a maker of aggressive cell phenotypes, which are likely to migrate (Cardone et al., 2005). We counted the numbers of cells with elongated edges (pseudopodia) in five image areas in a picture of cultured cells and calculated the percentage of cells with pseudopodia in the total number of cells in the area.
RNA-seq data processing
The RNA-seq reads were mapped to the hg19 reference genome using STAR (v2.4.0j) (Dobin et al., 2013). The read count for each gene was calculated using HTSeq (v0.6.1) (Anders et al., 2015). The RPKM values were quantified using Cufflinks (v2.1.1) (Trapnell et al., 2010). Differential gene expression analyses were performed using the DESeq2 (Love et al., 2014) package of Bioconductor in the R statistical software (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html).
Variants for each sample were called by GATK (v3.4.0) (McKenna et al., 2010) using standard hard filtering parameters according to the GATK Best Practices recommendations (DePristo et al., 2011; Van der Auwera et al., 2013).
Hierarchical clustering, heatmap visualization, and GSEA
We calculated the SD of each gene across tamoxifen-treated samples obtained over a period of 12 weeks and used the gene expression profiles of 120 individual samples with genes that had an SD ≥3 under a condition of tamoxifen treatment to generate a hierarchical clustering and heatmap with the ‘pheatmap’ package in R. The GSEA (Subramanian et al., 2005) was employed to determine the KEGG pathways (Kanehisa and Goto, 2000) enriched by a pre-ranked list of all genes, which were sorted by the statistical significance of the differential expression defined by DESeq2.
DNB method for detecting the tipping point
The process of acquiring drug resistance can be modeled by three states or stages (Figure 1) similar to disease progression (Chen et al., 2012; Liu et al., 2012; Sa et al., 2016; Li et al., 2017; Yang et al., 2018): the non-resistance state, which is a stable state with high resilience and robustness to perturbations; the pre-resistance state, which is the tipping point just before the catastrophic shift into the irreversible resistance state and is thus characterized by low resilience and robustness due to its critical dynamics, but is still reversible to the non-resistance state with appropriate treatments; and the resistance state, which is another stable state that acquires endocrine resistance generally with high resilience and robustness and is thus usually very difficult to return to the non-resistance state even with advanced treatments. Clearly, it is of great importance to predict the pre-resistance state, which not only holds the key to elucidating the molecular mechanisms of irreversible drug resistance at the dynamics and network levels but can also directly apply to the re-optimization of anti-resistance strategies from the clinical and therapeutic viewpoint.
However, different from the resistance state, it is a difficult task to identify the pre-resistance state or the early-warning signs of pre-resistance because there are generally no significant differences between the non-resistance state and the pre-resistance state in terms of the molecular signatures and clinical phenotypes (Chen et al., 2012), which leads to the failure of traditional biomarkers. In contrast, the DNB method was developed to quantitatively identify the tipping point or pre-disease state during disease progression based on the observed data. Theoretically, when a biological system is near the critical point, there exists a dominant group defined as the DNB molecules, which satisfy the following three conditions based on the observed data (Chen et al., 2012):
The correlation (PCCin) between any pair of members in the DNB group rapidly increases;
The correlation (PCCout) between one member of the DNB group and any other non-DNB molecule rapidly decreases;
The standard deviation (SDin) or coefficient of variation for any member in the DNB group drastically increases.
In other words, the above conditions can be approximately stated as: the appearance of a strongly fluctuating and highly correlated group of molecules implies the imminent transition to the resistance state. Then, these three conditions are adopted to quantify the tipping point as the early-warning signals of diseases, and further, the identified dominant group of molecules consists of DNB members. The DNB theory has been applied to a number of analyses of disease progression and biological processes to predict the critical states as well as their driven factors (Chen et al., 2012; Li et al., 2014; Mojtahedi et al., 2016; Richard et al., 2016; Lesterhuis et al., 2017; Sa et al., 2016; Li et al., 2017; Yang et al., 2018). In this work, by considering the acquisition of drug resistance as a nonlinear dynamics process, we further applied the DNB method to reveal the tipping point of endocrine resistance and the regulatory factors of this resistance. To quantify the critical state, the following criterion IDNB was used as the signal of the critical point by combining the above three statistical conditions:
Thus, from the observed data of a sample, whenever there is a group of molecules appearing with a high IDNB score, this group of molecules is the DNB group of molecules and the state of this sample is considered to be near the tipping point. Therefore, from the observed data (e.g. omics data) of each sample, we can identify the DNB members and further quantify whether or not this sample is near the critical state using the IDNB score.
To further reliably identify the critical state, we developed a new method called the landscape DNB, which explores both the local and global gene expression data as well as the network structure, and the detailed algorithm is provided in the Supplementary Information.
Drug-driven mutation detection
GATK (The Genome Analysis Toolkit, https://software.broadinstitute.org/gatk/) was used to call mutations from our sequencing data. For each mutation site detected by GATK, we compared its genotype in the TamR samples with the genotype respectively in three conditions: in control samples at the same time point, in reference genome and in MCF-7 cell line, to determine the mutation type. By the multiple comparisons of the genotypes above, the mutations were further classified as drug-driven mutation, random mutation, and MCF-7 mutation (Supplementary Figure S6). We defined those drug-driven mutations that started from around the tipping point and continually existed at least the following four time points as drug resistance-associated mutations.
Survive analysis
Clinical survival data of ER-positive breast cancer patients and their corresponding expression and mutation data were obtained from the TCGA dataset and EBI (EGAS00000000083) dataset. We carry out the survival analysis on the collection of DNB genes through a linear regression model
where is the expression value of the ith DNB gene and is a risk coefficient which is estimated by Cox regression model, a proportional hazards regression model, which allows analyzing the effect of several risk factors on survival. The analysis was established in R (http://cran.r-project.org) using the survival package.
Based on the status of a mutant gene, patients were divided into two groups with the first group harboring mutant gene and the second group harboring wild-type gene. Survival analysis between two groups was also assessed using survival package in R.
Availability of data and material
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA000580, which are publicly accessible at http://bigd.big.ac.cn/gsa. The source codes of the algorithm are provided at https://github.com/rabbitpei/drug-resistance-of-breast-cancer.
Supplementary Material
Acknowledgements
We thank Dr Shigeyuki Magi from the Laboratory for Integrated Cellular Systems, RIKEN Center for Integrative Medical Sciences (IMS) for help with the statistical analysis.
Edited by Jiarui Wu
Funding
This work was supported by grants from the National Key R&D Program of China (2017YFA0505500), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13040700), the National Natural Science Foundation of China (11771152, 91529303, 31771476, 31571363, 31771469, 91530320, 61134013, 81573023, 81501203, and 11326035), Pearl River Science and Technology Nova Program of Guangzhou (201610010029); FISRT, Aihara Innovative Mathematical Modeling Project from Cabinet Office, Japan; Fundamental Research Funds for the Central Universities (2017ZD095); JSPS KAKENHI (15H05707); Grant-in-Aid for Scientific Research on Innovative Areas (3901) and SPS KAKENHI (15KT0084, 17H06299, 17H06302, and 18H04031); RIKEN Epigenome and Single Cell Project Grants to M.O.-H. This work was performed in part under the International Cooperative Research Program of Institute for Protein Research, Osaka University (ICRa-17-01 to L.C. and M.O.-H.).
Conflict of interest
none declared.
References
- Anders S., Pyl P.T., and Huber W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.A., Bhat-Nakshatri P., Patel N.M., et al. (2001). Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J. Biol. Chem. 276, 9817–9824. [DOI] [PubMed] [Google Scholar]
- Cardone R.A., Bagorda A., Bellizzi A., et al. (2005). Protein kinase A gating of a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module regulates invasion in breast cancer cell lines. Mol. Biol. Cell 16, 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Liu R., Li Y., et al. (2016). Detecting critical state before phase transition of complex biological systems by hidden Markov model. Bioinformatics 32, 2143–2150. [DOI] [PubMed] [Google Scholar]
- Chen L., Liu R., Liu Z.P., et al. (2012). Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton C.J., Massarweh S., Huang S., et al. (2008). Development of resistance to targeted therapies transforms the clinically associated molecular profile subtype of breast tumor xenografts. Cancer Res. 68, 7493–7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A., Banks E., Poplin R., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee J.M., Robertson J.F., Ellis I.O., et al. (2001). Phosphorylation of ERK1/2 mitogen-activated protein kinase is associated with poor response to anti-hormonal therapy and decreased patient survival in clinical breast cancer. Int. J. Cancer 95, 247–254. [DOI] [PubMed] [Google Scholar]
- He D., Liu Z.P., Honda M., et al. (2012). Coexpression network analysis in chronic hepatitis B and C hepatic lesions reveals distinct patterns of disease progression to hepatocellular carcinoma. J. Mol. Cell Biol. 4, 140–152. [DOI] [PubMed] [Google Scholar]
- Hirata Y., Bruchovsky N., and Aihara K. (2010). Development of a mathematical model that predicts the outcome of hormone therapy for prostate cancer. J. Theor. Biol. 264, 517–527. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., and Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlden J.M., Hutcheson I.R., Jones H.E., et al. (2003). Elevated levels of epidermal growth factor receptor/c-erbB2 heterodimers mediate an autocrine growth regulatory pathway in tamoxifen-resistant MCF-7 cells. Endocrinology 144, 1032–1044. [DOI] [PubMed] [Google Scholar]
- Lesterhuis W.J., Bosco A., Millward M.J., et al. (2017). Dynamic versus static biomarkers in cancer immune checkpoint blockade: unravelling complexity. Nat. Rev. Drug Discov. 16, 264–272. [DOI] [PubMed] [Google Scholar]
- Li M., Zeng T., Liu R., et al. (2014). Detecting tissue-specific early warning signals for complex diseases based on dynamical network biomarkers: study of type 2 diabetes by cross-tissue analysis. Brief. Bioinform. 15, 229–243. [DOI] [PubMed] [Google Scholar]
- Li M., Li C., Liu W., et al. (2017). Dysfunction of PLA2G6 and CYP2C44-associated network signals imminent carcinogenesis from chronic inflammation to hepatocellular carcinoma. J. Mol. Cell Biol. 9, 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng Q., Bao C., et al. (2015). Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt B., Esteller R., Echauz J., et al. (2001). Epileptic seizures may begin hours in advance of clinical onset: a report of five patients. Neuron 30, 51–64. [DOI] [PubMed] [Google Scholar]
- Liu R., Li M., Liu Z.P., et al. (2012). Identifying critical transitions and their leading biomolecular networks in complex diseases. Sci. Rep. 2, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Wang X., Aihara K., et al. (2014. a). Early diagnosis of complex diseases by molecular biomarkers, network biomarkers, and dynamical network biomarkers. Med. Res. Rev. 34, 455–478. [DOI] [PubMed] [Google Scholar]
- Liu R., Yu X., Liu X., et al. (2014. b). Identifying critical transitions of complex diseases based on a single sample. Bioinformatics 30, 1579–1586. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massarweh S., Osborne C.K., Creighton C.J., et al. (2008). Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 68, 826–833. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSharry P.E., Smith L.A., and Tarassenko L. (2003). Prediction of epileptic seizures: are nonlinear methods relevant? Nat. Med. 9, 241–242. [DOI] [PubMed] [Google Scholar]
- Mojtahedi M., Skupin A., Zhou J., et al. (2016). Cell fate decision as high-dimensional critical state transition. PLoS Biol. 14, e2000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove E.A., and Sutherland R.L. (2009). Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 9, 631–643. [DOI] [PubMed] [Google Scholar]
- Ou K.W., Hsu K.F., Cheng Y.L., et al. (2010). Asymptomatic pulmonary nodules in a patient with early-stage breast cancer: Cryptococcus infection. Int. J. Infect. Dis. 14, e77–e80. [DOI] [PubMed] [Google Scholar]
- Richard A., Boullu L., Herbach U., et al. (2016). Single-cell-based analysis highlights a surge in cell-to-cell molecular variability preceding irreversible commitment in a differentiation process. PLoS Biol. 14, e1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggins R.B., Schrecengost R.S., Guerrero M.S., et al. (2007). Pathways to tamoxifen resistance. Cancer Lett. 256, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa R., Zhang W., Ge J., et al. (2016). Discovering a critical transition state from nonalcoholic hepatosteatosis to nonalcoholic steatohepatitis by lipidomics and dynamical network biomarkers. J. Mol. Cell Biol. 8, 195–206. [DOI] [PubMed] [Google Scholar]
- Saini K.S., Taylor C., Ramirez A.J., et al. (2012). Role of the multidisciplinary team in breast cancer management: results from a large international survey involving 39 countries. Ann. Oncol. 23, 853–859. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z., Liu R., Zheng L., et al. (2015). Cerebrospinal fluid protein dynamic driver network: at the crossroads of brain tumorigenesis. Methods 83, 36–43. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G.A., Carneiro M.O., Hartl C., et al. (2013). From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas J.G., Winkler T., Musch G., et al. (2005). Self-organized patchiness in asthma as a prelude to catastrophic shifts. Nature 434, 777–782. [DOI] [PubMed] [Google Scholar]
- Yang B., Li M., Tang W., et al. (2018). Dynamic network biomarker indicates pulmonary metastasis at the tipping point of hepatocellular carcinoma. Nat. Commun. 9, 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T., Zhang C.C., Zhang W., et al. (2014). Deciphering early development of complex diseases by progressive module network. Methods 67, 334–343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA000580, which are publicly accessible at http://bigd.big.ac.cn/gsa. The source codes of the algorithm are provided at https://github.com/rabbitpei/drug-resistance-of-breast-cancer.