Abstract
Structural changes in the corpus callosum have been reported in schizophrenia; however, the underlying molecular mechanism remains unclear. As the corpus callosum is high in lipid content, we analyzed the lipid contents of the corpora callosa from 15 patients with schizophrenia and 15 age- and sex-matched controls using liquid chromatography coupled to tandem mass spectrometry and identified lipid combinations associated with schizophrenia. Real-time quantitative polymerase chain reaction analyses using extended samples (schizophrenia, n = 95; control, n = 91) showed low expression levels of lipid metabolism-related genes and their potential upstream transcription factors in schizophrenia. Subsequent pathway analysis identified a gene regulatory network where nuclear factor of activated T cells 2 (NFATC2) is placed most upstream. We also observed low gene expression levels of microglial markers, inflammatory cytokines, and colony-stimulating factor 1 receptor (CSF1R), which is known to regulate the density of microglia, in the corpus callosum in schizophrenia. The interactions between CSF1R and several genes in the presently identified gene network originating from NFATC2 have been reported. Collectively, this study provides evidence regarding lipid abnormalities in the corpora callosa of patients with schizophrenia and proposes the potential role of impaired “NFATC2-relevant gene network-microglial axis” as its underlying mechanism.
Keywords: arachidonic acid, gene expression, liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS), microglia, postmortem brain
Schizophrenia is a major psychiatric illness, which features positive and negative symptoms and cognitive deficits. Although the etiology of schizophrenia is still unclear, structural changes in the white matter have been repeatedly observed in affected patients (Woodruff et al. 1995; Downhill Jr et al. 2000; Flynn et al. 2003; Arnone et al. 2008; Maniega et al. 2008; Collinson et al. 2014; Lener et al. 2015; Cetin-Karayumak et al. 2019). The molecular mechanism(s) underlying these white matter changes are unclear.
Structural and functional abnormalities involving lipids have attracted much attention from researchers (Horrobin et al. 1991; Glen et al. 1994; Yao et al. 2000; Schmitt et al. 2004; McNamara et al. 2007; Kale et al. 2008; Schwarz et al. 2008; Peters et al. 2012; Taha et al. 2013; Ghosh et al. 2017; Matsumoto et al. 2017). However, previous postmortem brain studies regarding lipids have mainly focused on the gray matter (Horrobin et al. 1991; Yao et al. 2000; Schmitt et al. 2004; McNamara et al. 2007; Taha et al. 2013; Matsumoto et al. 2017). Recent studies have demonstrated that the contents of specific lipid species are altered in the white matter of patients with schizophrenia (Schwarz et al. 2008; Ghosh et al. 2017). This is consistent with an earlier study that reported that polyunsaturated fatty acid (PUFA) concentrations in the erythrocyte membrane are associated with white-matter integrity in early-phase psychosis (Peters et al. 2012).
Based on these lines of evidence and the fact that the corpus callosum is the largest white-matter structure in the brain and has a high myelin content, which is mainly composed of lipids (approximately 75–80% of its dry weight) (O'Brien and Sampson 1965; Rumsby 1978), we hypothesized that abnormal lipid homeostasis may underlie the pathological changes in the corpus callosum of schizophrenia. We previously reported no significant differences in the contents of individual fatty acids in total phospholipid fractions extracted from the corpora callosa of patients with schizophrenia and control subjects (Hamazaki et al. 2017). In this study, using the same sample set, we performed a more detailed lipid analysis by leveraging a newly established liquid chromatography coupled to tandem mass spectrometry (LC–MS/MS)-based assay, focusing on the structural diversity in both the polar heads and fatty acid side chains of phospholipids and sulfatide (SUL; also known as 3-O-sulfogalactosylceramide). SUL is specifically abundant in the myelin sheath and harbors a fatty acid linked to a sphingoid base (Halder et al. 2007). The results provide mechanistic insights into the formation of white matter structural abnormalities in schizophrenia.
Methods and Materials
Chemicals
LC–MS grade acetonitrile and 2-propanol were purchased from Thermo Fisher Scientific K.K. (Yokohama, Japan). All lipid standards were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). More detailed information on the lipid standards is provided in the Supplementary Material.
Postmortem Brain Samples
Postmortem brain samples were provided by the Victorian Brain Bank at the Florey Institute for Neuroscience and Mental Health. All experimental procedures were approved by the Ethics Committee of RIKEN. Blocks of the anterior portion of corpus callosum (just posterior of the genu) and Brodmann area 8 (BA8) were sectioned and rapidly frozen at −80°C at the Florey Institute; the samples were then delivered to the RIKEN Center for Brain Science. Demographic data on the postmortem brain tissues used in lipid analysis (schizophrenia and control; both n = 15) and in gene expression analysis (schizophrenia, n = 95; control, n = 91) are shown in Table 1.
Table 1.
Demographic data for control and schizophrenia brains
| Sample set 1a | Sample set 2b | |||||
|---|---|---|---|---|---|---|
| Con (n = 15) | SZ (n = 15) | P value | Con (n = 91) | SZ (n = 95) | P value | |
| Sex (male/female) | 8/7 | 8/7 | – | 71/20 | 70/25 | – |
| Age (years, mean ± SD) | 57.27 ± 12.96 | 57.87 ± 13.67 | 0.927 | 47.88 ± 16.58 | 45.92 ± 17.37 | 0.393 |
| PMI (hours, mean ± SD) | 43.11 ± 17.74 | 42.93 ± 12.81 | 0.798 | 42.08 ± 13.89 | 41.71 ± 13.87 | 0.920 |
| Brain pH (mean ± SD) | 6.34 ± 0.23 | 6.18 ± 0.27 | 0.059 | 6.33 ± 0.21 | 6.26 ± 0.25 | 0.050 |
| History (years, mean ± SD) | – | 24.64 ± 14.25 | – | – | 18.41 ± 14.38 | – |
| CPz.eq (mean ± SD) | – | 706.3 ± 457.8 | – | – | 620.5 ± 575.2 | – |
| Suicide (n) | 0 | 3 | – | 1 | 44 | – |
aSamples for lipid analysis.
bSamples for gene expression analyses.
Con, control; CPz.eq, chlorpromazine equivalents; SZ, schizophrenia.
Lipid Extraction
Lipids were extracted from the postmortem brains using a modified Bligh and Dyer method (Bligh and Dyer 1959). The methodological details are provided in the Supplementary Material.
LC–MS/MS Analysis
To quantitatively evaluate the contents of diverse lipid species, we established an LC–MS/MS method using a hybrid triple quadrupole/linear ion trap MS (QTRAP 4500, ABSciex, Tokyo, Japan) coupled to an Agilent 1100 Series high performance liquid chromatography system (Agilent Technologies, Santa Clara, CA, USA) by referring to the method of Castro-Perez et al. (2010). Using this method, we detected five phospholipid classes (phosphatidylcholine [PC], phosphatidylethanolamine [PE], phosphatidylinositol [PI], phosphatidylserine [PS], and phosphatidylglycerol [PG]), one lysophospholipid (lysophosphatidylcholine [LPC]) class, and one sphingolipid class (SUL). In total, 121 lipid species were detected in the human corpus callosum samples. Individual lipid classes in each brain sample were separated on an Atlantis T3 Column (particle size: 3 μm, 2.1 × 150 mm, Waters, Milford, MA). After sample injection (10 μL), the composition of the mobile phase was maintained at 55% A (acetonitrile/water (40:60, v/v) containing 10 mM ammonium acetate and 0.1% acetic acid) and 45% B (acetonitrile/isopropanol (10:90, v/v) containing 10 mM ammonium acetate and 0.1% acetic acid) for 3.5 min. For the next 32.5 min, the gradient was increased in a linear fashion to 100% B and was held at this composition for 3.5 min. Then, the system was returned to 55% A and 45% B, and the column was re-equilibrated for an additional 6.5 min before the next run. The flow rate was 115 μL/min. MS/MS data acquisition was performed in the multiple reaction monitoring (MRM) mode. The optimized MRM parameters are shown in Supplementary Table S1. Individual lipid species were quantified by referencing the spiked internal standard and the calibration curve of each lipid standard.
Data Analyses of Lipidomic Data
Differences in lipid levels between the schizophrenia and control groups were statistically evaluated by Mann–Whitney U test (two-tailed) with false discovery rate (FDR) correction (two-stage linear step-up method of Benjamini et al. (2006)).
To categorize the samples, we performed multivariate analysis. First, we pretreated the lipidomic data by autoscaling (mean-centering and dividing by the standard deviation [SD] of each variable). Then, we performed a partial least squares-discriminant analysis (PLS-DA) to evaluate the lipid contents for their ability to discriminate between schizophrenia and control subjects. Next, we performed cluster analysis by generating a heat map using lipids with PLS-DA variable importance in projection (VIP) scores > 1.0, based on the Euclidean distance measure and Ward clustering algorithm (Ward Jr 1963).
The relevant lipids that distinguished the two groups were selected using the results of univariate (Mann Whitney U test, P < 0.05) and multivariate analyses (PLS-DA VIP score > 1.0). The predictive accuracy of the selected lipids as the variables was evaluated by the receiver operating characteristic (ROC) curve analysis. Fisher’s exact test (two-tailed) was used to test the enrichment of specific side chains in the selected lipid species. Correlations between the amounts of lipids and confounding factors (age, postmortem interval (PMI), brain pH, chlorpromazine equivalent dose and illness duration) were estimated using Spearman’s rank correlation coefficient. The P values of the correlations were applied to the FDR method (two-stage linear step-up method of Benjamini et al. (2006)) to account for multiple comparisons.
The data processing and the statistical analysis were performed using GraphPad Prism 7 software (GraphPad Software, San Diego, CA) and MetaboAnalyst 4.0 (http://www.metaboanalyst.ca/faces/home.xhtml).
Gene Expression Analysis
Target gene expression was measured by real-time quantitative polymerase chain reaction using TaqMan assays as previously described (Ide et al. 2019; Ohnishi et al. 2019). TaqMan probes for the genes of interest (Applied Biosystems, Foster City, CA, USA) are listed in Supplementary Table S2. GAPDH was used as an internal control. Outliers were defined as datapoints ±2 SDs from the mean and were removed from statistical analyses. Data were statistically evaluated by the Mann–Whitney U test (two-tailed) with FDR correction (two-stage linear step-up method of Benjamini et al. (2006)) or Dunn’s multiple comparison for post hoc testing after a Kruskal–Wallis test. The correlations among the expression levels of altered genes in schizophrenia and individual confounding factors were assessed using the Spearman’s rank correlation coefficient. The P values of the correlations were applied to the FDR method (two-stage linear step-up method of Benjamini et al. (2006)) to account for multiple comparisons. Analysis of covariance (ANCOVA) was conducted to examine differences in gene expression levels between patients with schizophrenia and controls while considering the covariates such as brain pH and PMI. Statistical analyses were performed using GraphPad Prism 7 software and the EZR software package (Kanda 2013).
Identification of Potential Transcription Factors Controlling Dysregulated Lipid Metabolism-Related Genes in the Corpus Callosum
To identify the potential upstream transcription factors that could control the expression of genes dysregulated in the corpus callosum, we performed in silico analysis to identify common transcription factors regulating the genes. We arbitrarily defined the promoters of the candidate genes as the regions spanning 1000 bp upstream and 100 bp downstream of the transcriptional start site, which included 1) FANTOM5 defined promoter regions (http://fantom.gsc.riken.jp/5/), 2) CpG islands, 3) DNase I hypersensitivity regions, and 4) regions with H3K4me3 marks, derived from ENCODE (https://www.encodeproject.org/). We next predicted the putative transcription factor binding sites in the promoter regions of the selected genes using TRANSFAC (TRANSFAC matrix table, Release 2017.3; http://genexplain.com/transfac/) with default parameters. We then selected the transcription factors that bind the target promoter sequences of at least five lipid metabolism-related genes of interest in the direction of transcription.
Exploration of the Schizophrenia-Related Gene Network
We generated a molecular interaction map containing the genes of interest using Ingenuity Pathway Analysis (IPA) software (QIAGEN, Hilden, Germany). To investigate whether any polymorphisms in the genes of interest are associated with schizophrenia, we used the Oxford Brain Imaging Genetics Server (version 2.0; http://big.stats.ox.ac.uk/) (Elliott et al. 2018).
Results
Quantitative Analysis of Corpus Callosum Lipids Using LC–MS/MS
Using our newly established LC–MS/MS system, we analyzed the corpus callosum samples from patients with schizophrenia and unaffected controls (both n = 15). These same samples were examined in our previous study (Hamazaki et al. 2017), in which we reported that none of the individual fatty acid contents in the total phospholipid fractions differed between the disease and control groups. Using our assay system, we quantified 121 lipid species, and among them, the levels of 18 phospholipid species and 2 SULs were lower (Mann–Whitney U test, P < 0.05) in patients with schizophrenia compared to those in controls (Supplementary Table S3). However, after applying FDR to adjust for multiple comparisons, the differences became insignificant (Supplementary Table S3). The results were consistent with our previous study, in which no significant differences were found in the contents of fatty acids in the total phospholipid fractions extracted from the same subjects (Hamazaki et al. 2017).
In contrast, PLS-DA followed by cluster analysis using the lipidomic data demonstrated that PLS-DA 2D and 3D score plots partially separated the schizophrenia and control groups (Supplementary Fig. S1A and B). A heat map of lipids with PLS-DA VIP scores > 1.0 discriminated seven patients from the other subjects, based on lipid content patterns (left seven columns, Fig. 1A). These preliminary data suggest that this distinct cluster represents a subgroup of schizophrenia patients in terms of the lipid pathology.
Figure 1.
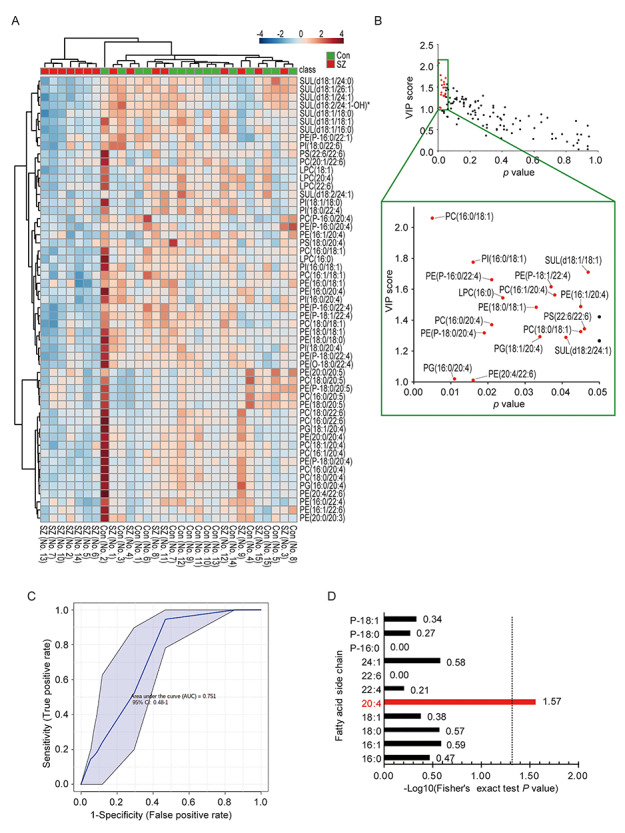
Analysis of lipidomic data from patients with schizophrenia and controls. (A) Cluster analysis of lipids with a PLS-DA VIP scores > 1.0. Each column represents a sample, and each row represents a lipid. The color indicates the relative lipid contents of each group. Green and red marks represent controls (Con) and patients with schizophrenia (SZ), respectively. *The existence of other molecular species at the same m/z and retention time. The possibility that control no. 2 was an outlier could not be ruled out. (B) Selection of lipids that can help differentiate between the schizophrenia and control samples. The x-axis and y-axis represent the P values of Mann–Whitney U test and VIP score, respectively. Red marks indicate selected lipids (Mann–Whitney U test, P < 0.05 and PLS-DA VIP score > 1.0) which differentiate between schizophrenia and control. (C) ROC curve to plot the performance of the combination of selected 17 lipids. The x-axis and y-axis represent 1-specificity and sensitivity, respectively. The AUC is shown with 95% confidence limits. (D) Enrichment analysis of the fatty acid side chain(s) of selected lipids that can discriminate between the schizophrenia and control samples. The x-axis represents the log10 of the P values of enriched terms, calculated by Fisher’s exact test.
Considering these results, we surmised that the unbalanced combination of multiple lipids may affect the pathology of a subset of schizophrenia patients, although the impact of individual lipids was small. To find the combination of lipids associated schizophrenia, we used the results of both univariate (Mann–Whitney U test, P < 0.05) and multivariate analyses (PLS-DA VIP score > 1.0) and identified 17 lipid species that can help discriminate between schizophrenia and control samples (Fig. 1B). The predictive accuracy of the variables was evaluated by ROC curve analysis. The result showed that an area under curve (AUC) was 0.751 (95% CI: 0.48–1) (Fig. 1C), indicating good accuracy. Next, we tested for enrichment of any specific side chains in the 17 lipid species and detected arachidonic acid (AA) to be significantly enriched (Fig. 1D). These results suggest that the combination of specific lipids, especially AA-containing lipids, may be associated with the pathology of schizophrenia.
Expression Levels of Lipid Metabolism-Related Genes in the Corpus Callosum
Lipid homeostasis is maintained by complicated metabolic pathways, through de novo synthesis, remodeling, and degradation (Supplementary Fig. S2A–C). Based on our data analysis, we hypothesized that lipid metabolism-related genes, especially those encoding enzymes and transporters involved in AA-related metabolic pathways, are dysregulated in schizophrenia. To test this hypothesis, we measured the mRNA levels of the relevant genes in an extended cohort of corpus callosum samples (patients with schizophrenia, n = 95; unaffected controls, n = 91; Table 1). The selection criteria for the genes analyzed were as follows (Supplementary Table S2 for each gene descriptions): 1) genes encoding enzymes that mediate the insertion of fatty acyl-CoA into lipids (GPAM, GPAT4, AGPAT3, AGPAT5, LPCAT2, CERS1, and CERS2), 2) genes encoding enzymes that mediate the release of fatty acids from phospholipids (PLA2G4A), 3) genes encoding AA metabolism-related enzymes (ELOVL2, ELOVL4, ELOVL5, FADS1, PTGS1, PTGS2, ALOX5, ALOX12, and ALOX15), and 4) genes encoding intracellular fatty acid chaperone proteins expressed in the brain (FABP3, FABP5, and FABP7) (Shimamoto et al. 2014).
Lower expression levels of the nine lipid metabolism-related genes (GPAT4, LPCAT2, ELOVL2, PLA2G4A, PTGS1, PTGS2, ALOX5, ALOX15, and FABP3) were detected in the corpora callosa of patients with schizophrenia compared with controls (Mann–Whitney U test, P < 0.05) (Table 2). After applying FDR analysis to adjust for multiple comparisons, the differences in these genes remained significant or showed a lower trend (six genes [ALOX5, ELOVL2, GPAT4, LPCAT2, PLA2G4A and PTGS1] showed significantly low [q < 0.05] and three genes [PTGS2, FABP3 and ALOX15] showed lower trends [q < 0.1] in schizophrenia). Although positive correlations between brain pH and the expression levels of GPAT4, LPCAT2, and PTGS1 were detected in both the schizophrenia and control groups (Supplementary Table S4), ANCOVA excluded potential contributions of brain pH (Supplementary Fig. S3). Low expression levels of these genes may explain the impaired AA metabolism in the corpus callosum of patients with schizophrenia because 1) ELOVL fatty acid elongase 2 (ELOVL2; encoded by ELOVL2), cyclooxygenase 1 (COX1; encoded by PTGS1), cyclooxygenase 2 (COX2; encoded by PTGS2), arachidonate 5-lipoxygenase (ALOX5; encoded by ALOX5), and arachidonate 15-lipoxygenase (ALOX5; encoded by ALOX15) catalyze AA conversion, 2) lysophosphatidylcholine acyltransferase 2 (LPCAT2; encoded by LPCAT2) exhibits a high affinity for arachidonoyl-CoA (Shindou et al. 2007), and 3) fatty acid-binding protein (FABP3; encoded by FABP3) preferentially binds to omega-6 PUFAs such as AA (Veerkamp et al. 1999; Liu et al. 2010).
Table 2.
Expression levels of lipid metabolism-related genes in the corpus callosum
| Gene | Con | SZ | Fold-change | P value§ | FDR q value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | n | Mean | SD | n | (SZ/Con) | |||
| FADS1 | 0.979 | 0.246 | 87 | 0.938 | 0.255 | 90 | 0.96 | 0.2057 | 0.2160 |
| ELOVL2 | 1.038 | 0.326 | 87 | 0.898 | 0.262 | 90 | 0.86 | 0.0030** | 0.0074†† |
| ELOVL4 | 0.878 | 0.313 | 85 | 0.806 | 0.255 | 92 | 0.92 | 0.1896 | 0.2144 |
| ELOVL5 | 1.011 | 0.254 | 88 | 1.015 | 0.233 | 91 | 1.00 | 0.9616 | 0.7067 |
| ALOX5 | 1.060 | 0.428 | 85 | 0.764 | 0.430 | 91 | 0.72 | <0.0001**** | <0.0001†††† |
| ALOX12 | 0.876 | 0.501 | 80 | 0.947 | 0.668 | 82 | 1.08 | 0.9049 | 0.7001 |
| ALOX15 | 0.816 | 0.425 | 86 | 0.694 | 0.377 | 90 | 0.85 | 0.0327* | 0.0686‡ |
| PTGS1 | 1.100 | 0.427 | 88 | 0.766 | 0.415 | 94 | 0.70 | <0.0001**** | <0.0001†††† |
| PTGS2 | 0.821 | 0.523 | 85 | 0.716 | 0.548 | 91 | 0.87 | 0.0424* | 0.0693‡ |
| PLA2G4A | 0.987 | 0.316 | 85 | 0.811 | 0.289 | 89 | 0.82 | <0.0001**** | 0.0004††† |
| GPAM | 0.979 | 0.179 | 84 | 0.949 | 0.172 | 89 | 0.97 | 0.4391 | 0.4035 |
| GPAT4 | 1.099 | 0.128 | 84 | 1.012 | 0.165 | 91 | 0.92 | 0.0002*** | 0.0007††† |
| AGPAT3 | 1.067 | 0.109 | 85 | 1.061 | 0.119 | 92 | 0.99 | 0.5985 | 0.4888 |
| AGPAT5 | 0.982 | 0.136 | 84 | 0.951 | 0.136 | 90 | 0.97 | 0.1094 | 0.1462 |
| LPCAT2 | 1.086 | 0.262 | 86 | 0.915 | 0.255 | 90 | 0.84 | <0.0001**** | 0.0004††† |
| CERS1 | 1.000 | 0.129 | 81 | 1.039 | 0.177 | 91 | 1.04 | 0.1321 | 0.1618 |
| CERS2 | 1.025 | 0.277 | 88 | 0.977 | 0.277 | 93 | 0.95 | 0.2638 | 0.2585 |
| FABP3 | 0.977 | 0.406 | 86 | 0.844 | 0.306 | 90 | 0.86 | 0.0390* | 0.0693‡ |
| FABP5 | 0.870 | 0.259 | 85 | 0.855 | 0.273 | 91 | 0.98 | 0.5542 | 0.4792 |
| FABP7 | 0.892 | 0.341 | 87 | 0.818 | 0.340 | 91 | 0.92 | 0.0802 | 0.1178 |
| FAAH | 0.935 | 0.233 | 86 | 0.970 | 0.258 | 92 | 1.04 | 0.2992 | 0.1571 |
| MGLL | 1.301 | 0.226 | 85 | 1.173 | 0.200 | 92 | 0.90 | 0.0001*** | 0.0001††† |
Values were normalized to GAPDH.*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, †q < 0.05, †††q < 0.001, ††††q < 0.0001, ‡q < 0.1, §Mann–Whitney U test
We also examined the expression levels of fatty acid amide hydrolase (FAAH), which releases AA from N-arachidonoylethanolamine (anandamide), and monoglyceride lipase (MGLL), which releases AA from 2-arachidonoylglycerol (2-AG; Supplementary Fig. S2B) (Cairns et al. 2016). Although FAAH did not change, MGLL was significantly lower in patients with schizophrenia compared with that in controls (Table 2). These results suggest that 2-AG metabolism may also be dysregulated in schizophrenia. Taken together, these data support the idea that lipid homeostasis mediated by fatty acid insertion into phospholipids, AA production from phospholipids and 2-AG, intracellular AA transport and AA degradation are dysregulated in schizophrenia.
Interestingly, the expression of FABP7 was significantly lower in patients with schizophrenia who died by suicide compared with nonsuicidal controls (P = 0.0014) and nonsuicidal schizophrenia cases (P = 0.0049; Supplementary Fig. S4). This raises the possibility that low FABP7 expression might be associated with the impulse to commit suicide.
Transcription Factors Regulating the Expression of Lipid Metabolism-Related Genes
To explore the potential causes for the lower levels of the 10 lipid metabolism-related genes (Table 2) in patients with schizophrenia, we examined the transcription factors with the potential to bind the promotor regions of >5 of the target genes. As a result, we identified 14 candidate transcription factors (Table 3). Of these, the levels of KLF4, KLF6, IKZF1, MZF1, NFATC2, and TFAP2A were lower in patients with schizophrenia compared to those in controls, whereas ZBTB14 was higher (Table 4, See Supplementary Table S2 for each gene description). The differences remained significant after applying FDR to adjust for multiple comparisons (Table 4) The potential effect of confounding factor (Supplementary Table S5) was excluded by ANCOVA (Supplementary Fig. S5). We found several correlations between genes encoding transcription factors and those involved in lipid metabolism (Supplementary Fig. S6), supporting roles for these transcription factors in the regulation of lipid metabolism-related genes.
Table 3.
Candidate transcription factors that potentially bind to the promotor regions of all or more than 5 lipid metabolism-related genes
| Gene symbol | Description | ALOX5 | ALOX15 | ELOVL2 | FABP3 | GPAT4 | LPCAT2 | PLA2G4A | PTGS1 | PTGS2 | MGLL | No. of genes regulated by TF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KLF6 | Kruppel like factor 6 | 24 | 16 | 9 | 5 | 11 | 3 | 1 | 14 | 10 | 16 | 10/10 |
| GTF2IRD1 | GTF2I repeat domain-containing 1 | 4 | 6 | 3 | 1 | 1 | 1 | 0 | 1 | 2 | 4 | 9/10 |
| TFAP2A | Transcription factor AP-2 alpha | 4 | 2 | 1 | 1 | 1 | 2 | 0 | 1 | 2 | 2 | 9/10 |
| CHURC1 | Churchill domain containing 1 | 8 | 2 | 3 | 2 | 0 | 2 | 0 | 2 | 3 | 2 | 8/10 |
| KLF4 | Kruppel-like factor 4 | 2 | 6 | 2 | 1 | 0 | 0 | 1 | 2 | 5 | 4 | 8/10 |
| AHR | Aryl hydrocarbon receptor | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 1 | 2 | 3 | 8/10 |
| NFATC2 | Nuclear factor of activated T cells 2 | 2 | 0 | 0 | 4 | 1 | 1 | 2 | 0 | 5 | 4 | 7/10 |
| ING4 | Inhibitor of growth family member 4 | 4 | 2 | 3 | 2 | 2 | 0 | 0 | 1 | 0 | 1 | 7/10 |
| ZBTB14 | Zinc finger and BTB domain containing 14 | 3 | 1 | 7 | 0 | 3 | 0 | 0 | 1 | 7 | 5 | 7/10 |
| ZNF333 | Zinc finger protein 333 | 1 | 0 | 0 | 0 | 3 | 2 | 5 | 2 | 13 | 4 | 7/10 |
| CRX | Cone-rod homeobox | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 4 | 0 | 6/10 |
| MZF1 | Myeloid zinc finger 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 0 | 2 | 6/10 |
| IKZF1 | IKAROS family zinc finger 1 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 5/10 |
| MYOG | Myogenin | 4 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 5/10 |
The numbers indicate the number of putative binding sites in the promotor region of each gene.
TF, transcription factor.
Table 4.
Expression levels of transcription factors in the corpus callosum
| Gene | Con | SZ | Fold-change | P value§ | FDR q value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | n | Mean | SD | n | (SZ/Con) | |||
| KLF6 | 0.949 | 0.283 | 86 | 0.841 | 0.246 | 89 | 0.89 | 0.0079** | 0.0167† |
| GTF2IRD1 | 0.984 | 0.229 | 91 | 0.972 | 0.187 | 88 | 0.99 | 0.9673 | 0.6250 |
| TFAP2A | 0.994 | 0.265 | 90 | 0.908 | 0.245 | 93 | 0.91 | 0.0353* | 0.0423† |
| CHURC1 | 0.952 | 0.160 | 86 | 0.940 | 0.140 | 92 | 0.99 | 0.6190 | 0.4773 |
| KLF4 | 0.964 | 0.520 | 89 | 0.714 | 0.420 | 90 | 0.74 | 0.0004*** | 0.0010†† |
| ING4 | 0.956 | 0.214 | 89 | 0.946 | 0.218 | 90 | 0.99 | 0.7131 | 0.4992 |
| MZF1 | 1.025 | 0.253 | 86 | 0.868 | 0.263 | 91 | 0.85 | 0.0003*** | 0.0010†† |
| AHR | 0.972 | 0.251 | 88 | 0.915 | 0.240 | 90 | 0.94 | 0.1383 | 0.1291 |
| NFATC2 | 0.946 | 0.318 | 85 | 0.836 | 0.359 | 91 | 0.88 | 0.0101* | 0.0170† |
| ZBTB14 | 0.992 | 0.144 | 84 | 1.044 | 0.173 | 90 | 1.05 | 0.0347* | 0.0423† |
| ZNF333 | 1.047 | 0.223 | 88 | 1.030 | 0.237 | 91 | 0.98 | 0.6251 | 0.4773 |
| IKZF1 | 1.064 | 0.443 | 86 | 0.748 | 0.417 | 90 | 0.70 | <0.0001**** | <0.0001†††† |
| MYOG | 0.797 | 0.382 | 86 | 0.711 | 0.413 | 88 | 0.89 | 0.0510 | 0.0536 |
Values were normalized to GAPDH.
* P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, †q < 0.05, †††q < 0.001, ††††q < 0.0001, §Mann–Whitney U test
Notably, we also found correlations between the identified transcription factors (Fig. 2A and Supplementary Fig. S7), suggesting the existence of a transcription factor network. To identify a hierarchy of genes, we analyzed interactions among the genes of interest and the putative transcription factors using IPA. Nuclear factor of activated T cells 2 (NFATC2) was placed upstream of four other transcription factor genes (KLF4, KLF6, IKZF1, and TFAP2A) and seven lipid metabolism genes (PLA2G4A, FABP3, PTGS1, PTGS2, ALOX5, ALOX15, and MGLL; Fig. 2B), suggesting that NFATC2 may act as a master regulator of the network relevant for schizophrenia in terms of white-matter pathology.
Figure 2.
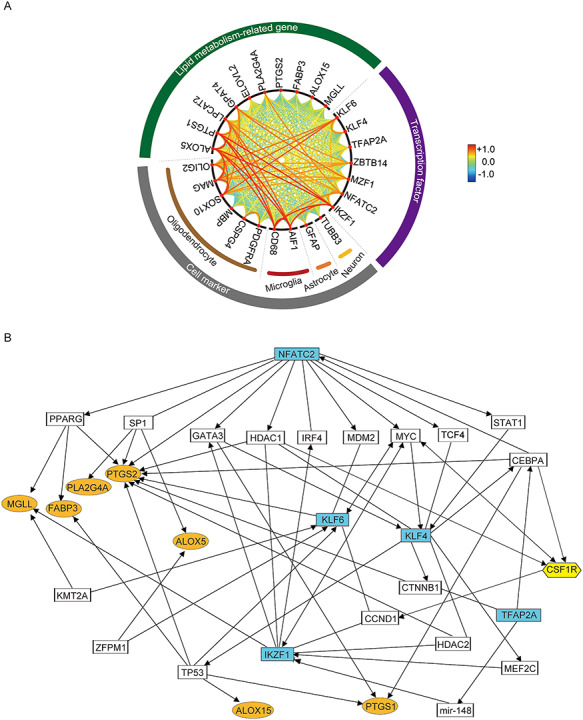
Interactions among identified schizophrenia-related genes. (A) Correlations between lipid metabolism-related genes, transcription factors, and cellular markers are displayed in a circle. The larger the absolute value of the correlation coefficient between two genes, the more the line connecting the genes is placed in front and in red color. (B) Predicted schizophrenia-related gene expression network by ingenuity pathway analysis. Transcription factors, lipid metabolism-related genes, and CSF1R dysregulated in schizophrenia are highlighted in blue, orange, and yellow, respectively. Each interaction is supported by at least one literature reference.
Cell Populations in the Corpus Callosum in Schizophrenia
We next analyzed whether the gene expression changes observed were elicited by changes in the cell populations in the corpus callosum of schizophrenia. We examined the expression levels of several cell type-specific markers: tubulin beta 3 class III (TUBB3) for neurons, glial fibrillary acidic protein (GFAP) for astrocytes, platelet-derived growth factor receptor alpha (PDGFRA), chondroitin sulfate proteoglycan 4 (CSPG4), SRY-box transcription factor 10 (SOX10), oligodendrocyte transcription factor 2 (OLIG2), myelin basic protein (MBP) and myelin-associated glycoprotein (MAG) for oligodendrocytes, and allograft inflammatory factor 1 (AIF1, also known as ionized calcium-binding adapter molecule 1 [IBA1]) and CD68 molecule (CD68) for microglia. Intriguingly, AIF1 and CD68 were downregulated in patients with schizophrenia compared with those in controls, while no changes were observed for the other marker genes (Table 5). Although correlations between brain pH and the expression levels of AIF1 and CD68 were detected (Supplementary Table S6), the potential effect of brain pH was negligible by ANCOVA (Supplementary Fig. S8A and B). AIF1 is expressed in microglia across their different morphological states, whereas CD68 is highly expressed in round/activated microglia (Hendrickx et al. 2017). Therefore, these results suggest that activated microglia (and steady-state microglia) maybe reduced in the corpus callosum in schizophrenia. The expression levels of ALOX5, LPCAT2, and PTGS1 were strongly correlated with those of the microglial markers (Fig. 2A and Supplementary Fig. S9), suggesting that the lower gene expression can be explained, at least in part, by the reduction of microglial cells.
Table 5.
Expression levels of cell marker genes in the corpus callosum
| Gene | Con | SZ | Fold-change | P value§ | FDR q value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | n | Mean | SD | n | (SZ/Con) | |||
| AIF1 | 1.076 | 0.514 | 88 | 0.659 | 0.434 | 93 | 0.61 | <0.0001**** | <0.0001†††† |
| CD68 | 1.135 | 0.509 | 89 | 0.736 | 0.443 | 93 | 0.65 | <0.0001**** | <0.0001†††† |
| TUBB3 | 0.806 | 0.375 | 84 | 0.710 | 0.296 | 91 | 0.88 | 0.1127 | 0.3157 |
| GFAP | 0.908 | 0.311 | 86 | 0.880 | 0.331 | 92 | 0.97 | 0.6037 | 0.5634 |
| PDGFRA | 0.932 | 0.298 | 87 | 0.875 | 0.324 | 92 | 0.94 | 0.2796 | 0.5634 |
| CSPG4 | 1.081 | 0.321 | 87 | 1.141 | 0.399 | 93 | 1.06 | 0.4418 | 0.5634 |
| SOX10 | 0.954 | 0.253 | 88 | 0.926 | 0.233 | 91 | 0.97 | 0.4554 | 0.5634 |
| OLIG2 | 1.042 | 0.288 | 86 | 1.010 | 0.266 | 90 | 0.97 | 0.5997 | 0.5634 |
| MBP | 1.270 | 0.671 | 90 | 1.360 | 0.724 | 89 | 1.07 | 0.5712 | 0.5634 |
| MAG | 1.019 | 0.271 | 88 | 1.018 | 0.279 | 91 | 1.00 | 0.8782 | 0.7377 |
Values were normalized to GAPDH.
**** P < 0.0001, ††††q < 0.0001, §Mann–Whitney U test.
One of the molecules that regulate the density of microglia is colony-stimulating factor 1 receptor (CSF1R) (Erblich et al. 2011). We examined the expression levels of CSF1R and found that the expression level of CSF1R was low in the corpus callosum of patients with schizophrenia (Fig. 3A), even after ANCOVA to consider the potential effects of cofounding factors (brain pH; Supplementary Table S6, Supplementary Fig. S8C). The expression level of CSF1R was strongly correlated with those of the microglial markers (Fig. 3B and C). Of note, pathway analysis demonstrated that CSF1R interacts with the several genes in the schizophrenia-related gene network (Fig. 2B), suggesting that the gene network including CSF1R is closely related to aberration of microglia and abnormal lipid metabolism in schizophrenia.
Figure 3.
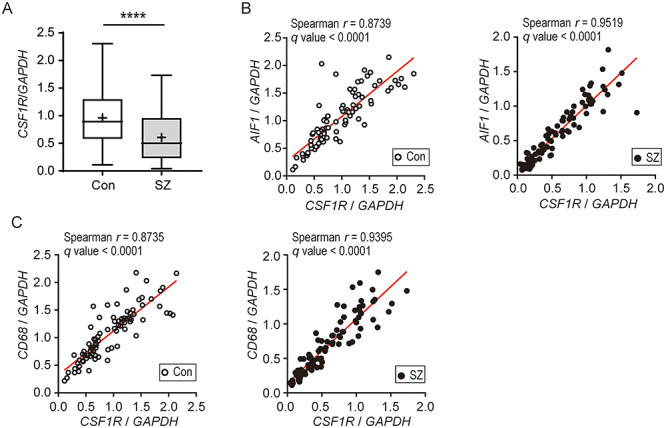
Expression levels of CSF1R and correlations between the expression levels of CSF1R and microglial markers in the corpora callosa of patients with schizophrenia and healthy controls (A) Box and whisker plots of the expression levels of CSF1R (Con, n = 85; SZ, n = 90). GAPDH was used as an internal control. The horizontal line in the box represents the median value, and the + represents the mean. Whiskers represent the 25th and 75th percentiles, and dots represent outliers. ****P < 0.0001. (B, C) Spearman’s correlation coefficients describing the relationships between the expression levels of CSF1R and microglial markers, AIF1 (B) and CD68 (C). Open circles (left panel) and closed circles (right panel) indicate controls and patients with schizophrenia, respectively.
Recent studies have demonstrated the roles of cross-talk between oligodendrocytes and microglia in oligodendrocyte proliferation, differentiation, and myelination activity (Hamilton and Rome 1994; Pasquini et al. 2011; Peferoen et al. 2014; Domingues et al. 2016; Hagemeyer et al. 2017; Lloyd et al. 2017). This cross-talk is mediated by cytokines, chemokines, growth factors, and exosomes (Hamilton and Rome 1994; Pasquini et al. 2011; Peferoen et al. 2014; Domingues et al. 2016; Lloyd et al. 2017). Gene expression analysis of inflammatory cytokines (IL1B, IL6, TNF, and TGFB1; see Supplementary Table S2 for each gene description) revealed that the expression levels of IL1B, TNF, and TGFB1 were significantly lower in patients with schizophrenia compared with those in controls (Table 6), although the difference of TNF between the two groups became nonsignificant after ANCOVA to consider the potential effect of cofounding factor (brain pH) (Supplementary Table S6, Table 6). The expression levels of IL1B and TGFB1 were highly positively correlated with the expression of microglial marker genes (Supplementary Fig. S10A). Although inflammation is a major topic in schizophrenia research, these results are consistent with our recent studies, in which we did not observe signs of inflammation but instead found that anti-inflammatory and antioxidative reactions were enhanced in postmortem brain samples from patients with schizophrenia (Toyoshima et al. 2016; Ide et al. 2019). Intriguingly, it is known that NFATC2, a candidate master regulator of schizophrenia-related transcription factor network, modulates microglial cytokine secretion (Nagamoto-Combs and Combs 2010; Manocha et al. 2017), and its expression level was positively correlated with the expression levels of cytokines (Supplementary Fig. S10B). In addition, the expression level of CSF1R was also positively correlated with the expression levels of cytokines (Supplementary Fig. S10C). Several positive correlations were detected among the levels of schizophrenia-related genes (lipid metabolism-related genes and transcription factor genes) and the levels of the oligodendrocyte markers OLIG2, SOX10, and MAG (Fig. 2A). These lines of evidence suggest that impaired microglia-oligodendrocyte cross-talk following to the dampened microglial activities leads to qualitative aberration of the oligodendrocyte/myelin sheath (Fig. 4).
Table 6.
Expression levels of inflammation cytokines in the corpus callosum
| Gene | Con | SZ | Fold-change | Mann–Whitney U test P value | FDR q value | ANCOVA with brain pH as covariate | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | n | Mean | SD | n | (SZ/Con) | p value | |||
| IL1B | 0.591 | 0.720 | 90 | 0.383 | 0.386 | 91 | 0.65 | 0.0002*** | 0.0002††† | – |
| TNF | 0.816 | 0.533 | 87 | 0.672 | 0.478 | 91 | 0.82 | 0.0402* | 0.0281† | 0.1282 |
| IL6 | 0.584 | 0.629 | 89 | 0.910 | 1.125 | 91 | 1.56 | 0.2692 | 0.1413 | – |
| TGFB1 | 1.023 | 0.320 | 85 | 0.766 | 0.327 | 91 | 0.75 | <0.0001**** | <0.0001†††† | – |
Values were normalized to GAPDH.
* P < 0.05, ***P < 0.001, ****P < 0.0001, †q < 0.05, †††q < 0.001, ††††q < 0.0001.
Figure 4.
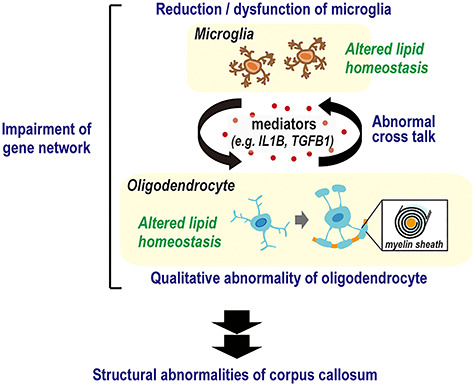
Potential mechanism of corpus callosum abnormality development in schizophrenia. In schizophrenia, reduced and dysfunctional microglial cells result in abnormal cross-talk with oligodendrocytes via mediators such as IL1B and TGFB1. This leads to transcriptional perturbation caused by impaired schizophrenia-related gene network including NFATC2 and CSF1R, resulting in abnormal lipid metabolism. This disturbs the normal properties and functions of oligodendrocytes and the myelin sheath, causing structural and functional deficits in the corpus callosum.
Gene Expression Analyses in the Frontal Cortex of Schizophrenia
To confirm whether the impaired schizophrenia-related gene network and aberrant population and activities of microglial cells observed in the corpora callosa of patients with schizophrenia can be generalized in the brain of patients with schizophrenia, we performed gene expression analysis using frontal cortex (BA8) samples. The lower levels of AIF1 and IL1B were also observed in the BA8 of patients with schizophrenia (Table 7). After applying FDR to adjust for multiple comparisons, the differences remained significant or showed a reducing trend (AIF1 showed significantly low [q < 0.05] and IL1B showed lower trends [q < 0.1] in schizophrenia). The potential effect of confounding factor (Supplementary Table S7) was excluded by ANCOVA (Supplementary Fig. S11). In contrast, there were no differences in the expression levels of the other schizophrenia-related genes identified in the corpora callosa of patients with schizophrenia between the two groups (Table 7). The results suggest that the reduction of specific subpopulation of microglia (e.g., AIF1+/CD68+ microglia), which is related to the impairment of schizophrenia-related gene network, maybe involved in the corpus callosum-specific abnormalities in schizophrenia, whereas the reduction of AIF1+ microglia may affect both frontal cortex and corpus callosum in schizophrenia.
Table 7.
Expression levels of transcription factors, microglial markers, inflammation cytokines, and CSF1R in the BA8
| Genes | Con | SZ | Fold-change | P value§ | FDR q value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | n | Mean | SD | n | (SZ/Con) | |||
| KLF6 | 1.079 | 0.309 | 90 | 1.053 | 0.286 | 91 | 0.976 | 0.5209 | 0.7520 |
| KLF4 | 0.846 | 0.616 | 87 | 0.978 | 0.875 | 92 | 1.156 | 0.4466 | 0.7369 |
| TFAP2A | 0.954 | 0.576 | 88 | 0.949 | 0.535 | 90 | 0.995 | 0.9093 | 0.8832 |
| MZF1 | 0.973 | 0.149 | 89 | 1.039 | 0.223 | 91 | 1.067 | 0.0421* | 0.1620 |
| ZBTB14 | 1.006 | 0.130 | 87 | 0.997 | 0.159 | 89 | 0.991 | 0.5957 | 0.7644 |
| IKZF1 | 1.115 | 0.485 | 89 | 1.013 | 0.521 | 91 | 0.909 | 0.0813 | 0.1878 |
| NFATC2 | 0.969 | 0.313 | 89 | 1.042 | 0.485 | 91 | 1.076 | 0.8211 | 0.8832 |
| AIF1 | 1.112 | 0.437 | 87 | 0.885 | 0.480 | 92 | 0.795 | 0.0009*** | 0.0100† |
| CD68 | 1.020 | 0.412 | 87 | 0.920 | 0.472 | 91 | 0.902 | 0.0632 | 0.1824 |
| IL1B | 0.760 | 0.954 | 89 | 0.675 | 1.320 | 91 | 0.888 | 0.0105* | 0.0604‡ |
| TGFB1 | 1.007 | 0.315 | 88 | 1.078 | 0.493 | 92 | 1.071 | 0.9176 | 0.8832 |
| CSF1R | 0.862 | 0.368 | 87 | 0.826 | 0.427 | 90 | 0.958 | 0.3978 | 0.7369 |
Values were normalized to GAPDH.
* P < 0.05, ***P < 0.001, ****P < 0.0001, †q < 0.05, ‡q < 0.1, §Mann–Whitney U test.
Discussion
In the present study, we performed lipid analysis of the corpus callosum using a newly established LC–MS/MS assay system and identified 17 lipid species that could help discriminate between the schizophrenia and control samples. Importantly, AA was enriched in the side chain of the lipids. As an underlying mechanism of the lipid pathology, we identified a candidate gene network in which NFATC2 is placed most upstream. Furthermore, we found that the expression levels of microglial markers, inflammatory cytokines, and colony-stimulating factor 1 receptor (CSF1R), which is known to regulate the density of microglia (Erblich et al. 2011), were low in the corpora callosa of patients with schizophrenia. Of note, interactions between CSF1R and several genes in the NFATC2-relevant gene network were observed. These results suggest that impairment of signaling through microglia and the schizophrenia-related gene network including NFATC2 and CSF1R plays an important role in the lipid pathology observed in the corpora callosa of patients with schizophrenia (Fig. 4).
Although it remains unclear whether the debilitation of the NFATC2-relevant gene network is the primary cause of the observed pathology, previous studies endorse the possibility. In a genome-wide association study of brain imaging phenotypes and other traits/diseases from the UK Biobank (http://big.stats.ox.ac.uk) (Elliott et al. 2018), we found that an NFATC2 variant (rs190024650; p value = 5.6 × 10−7) and a variant in the vicinity of GATA binding protein 3 gene (GATA3; rs191185553; p value = 7.5 × 10−7), a target of NFATC2 (Scheinman and Avni 2009), were associated with schizophrenia. In addition, a previous study (Ren et al. 2017) reported that an NFATC2 variant (rs193091397) was associated with reduced fractional anisotropy values in the white matter. NFATC2 [also known as nuclear factor of activated T cell (NFAT)1] is a member of the NFAT family, which in humans comprises five family members. Upon their dephosphorylation by calcineurin, NFAT proteins translocate from the cytoplasm to the nucleus, where they are transcriptionally active (Hogan et al. 2003). The calcineurin/NFAT signaling pathway plays critical roles in brain development, regulating myelination (Weider et al. 2018), axon outgrowth (Graef et al. 2003), and synaptogenesis (Yoshida and Mishina 2005; Kim et al. 2014; Kipanyula et al. 2016). Our previous genetic association study indicated that PPP3CC, which encodes the calcineurin A γ-subunit, and genes encoding early growth response (EGR) proteins (EGR2, EGR3 and EGR4) are associated with schizophrenia (Yamada et al. 2007). Notably, Egr2 and Egr3 levels are regulated by calcineurin through the activation of NFAT-mediated transcription (Rengarajan et al. 2000). Recently, Torshizi et al. (2019) reported that transcription factor 4, an NFATC2 target molecule (Chevrier et al. 2011), may act as a master regulator of a gene network that confers susceptibility to schizophrenia.
To the best of our knowledge, no reports have showed altered expression levels of schizophrenia-related genes examined in this study in animal models of schizophrenia. However, previous studies using genetically modified animals demonstrated the association between some currently identified schizophrenia-related genes and psychiatric-relevant behavioral abnormalities, which can be summarized as follows: 1) Nfatc2/Nfatc4 double-knockout mice displayed increased social interaction, increased locomotor activity, and decreased anxiety-related behavior relative to control mice (Arron et al. 2006), 2) aged Csf1r heterozygous mice showed cognitive deficits, sensorimotor deficits, depression, and anxiety-like behaviors (Chitu et al. 2015), 3) Fabp3 knockout mice showed impairment of social interaction (Shimamoto et al. 2014) and post-traumatic stress disorder-like behaviors such as cognitive deficits, hyperlocomotion and impaired fear extinction (Yabuki et al. 2018), and 4) COX-2 deficient knock-in mice showed increased hyperactivity, anxiety, repetitive behavior, motor deficit, and social abnormality (Wong et al. 2019). These results raise the possibility of the debilitation of genes in the currently identified schizophrenia-related network as a primary molecular mechanism of the pathology of schizophrenia.
The present study revealed that the expression level of AIF1 was low in both corpora callosa and frontal cortex from patients with schizophrenia compared with controls. In contrast, the expression level of CD68 was low in the corpora callosa but not in the prefrontal cortex of patients with schizophrenia. Furthermore, the differences in the expression levels of other schizophrenia-related genes seen in the corpora callosa from patients with schizophrenia were not detected in the frontal cortex. AIF1 is expressed in microglia across their different morphological states, whereas CD68 is highly expressed in round/activated microglia (Hendrickx et al. 2017). It is known that microglia in the white matter represent a unique population that are phenotypically and functionally different from other regional counterparts (Tan et al. 2019). This evidence suggests that the reduction in the specific subpopulation of microglia (e.g., activated CD68+/AIF1+ microglia), which is associated with the schizophrenia-related gene network, maybe involved in the corpus callosum (white matter)-specific abnormalities in schizophrenia.
Microglia contribute to oligodendrocyte proliferation, differentiation, and myelination activity by producing various factors, such as chemokines, cytokines, and growth factors (Hamilton and Rome 1994; Pasquini et al. 2011; Peferoen et al. 2014; Domingues et al. 2016; Hagemeyer et al. 2017; Lloyd et al. 2017). Our study revealed low levels of microglial markers and the inflammatory cytokines IL1B and TGFB1 in schizophrenia. In addition, low expression levels of genes related to the survival of microglia and neuroinflammation such as NFATC2 and CSF1R (Manocha et al. 2017; Weider et al. 2018; Coleman et al. 2020; Henry et al. 2020) were detected in the corpora callosa from patients with schizophrenia. It has been reported that Il1b knockout mice showed failure to remyelinate properly in a cuprizone model of demyelination and remyelination (Mason et al. 2001). TGF-b1 is also known to contribute to the remyelination of the central nervous system (Hamaguchi et al. 2019). This evidence suggests that impaired microglia-oligodendrocyte cross-talk following dampened microglial activities leads to qualitative aberration of the oligodendrocyte/myelin sheath (Fig. 4).
Although inflammation is a major topic in schizophrenia research, these results are consistent with those of our recent studies. Using the same postmortem brain set analyzed in this study, we recently reported enhanced anti-inflammatory/antioxidant processes involving excess hydrogen sulfide production as an antioxidant in the brains of patients with schizophrenia (Ide et al. 2019), in contrast to the prevailing neuroinflammation theory (Miller et al. 2011; De Picker et al. 2017). AA was enriched in the side chains of lipid species that discriminate between schizophrenia and control samples. This may represent an abnormal substrate availability for the generation of free AA and AA-derived bioactive molecules, such as prostaglandins. In addition, the expression of genes for enzymes that produce inflammatory mediators from AA, such as COX1/2 and ALOX5/15, were low in patients with schizophrenia. These results are collectively consistent with anti-inflammatory conditions in a subset of brains from patients with schizophrenia.
The ability to identify subgroups of patients with schizophrenia characterized by biological features is crucial for personalized treatment. In this study, we observed a potential schizophrenia subgroup during cluster analysis of corpus callosum lipid content patterns. The shared clinical characteristics of this subgroup of patients remain to be addressed. Suicide is a major cause of death among patients with schizophrenia, and recently, Lee et al. reported an association between white-matter alterations and suicide attempts in patients with schizophrenia or schizophreniform disorder (Lee et al. 2016). Our data showed that FABP7 expression was significantly low in the corpora callosa of suicide completers among patients with schizophrenia compared with nonsuicidal controls and nonsuicidal schizophrenia cases. Fatty acid-binding protein 7 (FABP7, encoded by FABP7) is a fatty acid chaperone protein expressed in astrocytes and oligodendrocyte progenitor cells (Sharifi et al. 2011), and it preferentially binds to omega-3 PUFAs (Xu et al. 1996; Balendiran et al. 2000; Liu et al. 2010), which enhance oligodendrocyte differentiation (Pu et al. 2013; Bernardo et al. 2017). Our prior study revealed the existence of rare functional variants of FABP7 in a schizophrenia cohort (Shimamoto et al. 2014). Disturbed FABP7 function can increase the fragmentation of sleep (Gerstner et al. 2017), and a relationship between sleep quality and suicide has been reported (Keshavan et al. 1994; Pigeon et al. 2012; Li et al. 2016).
This study has several limitations. First, we were not able to rule out the effects of diet, cigarette smoking, diseases such as liver disease and hyperlipidemia, or alcohol intake on lipid contents and the expression of schizophrenia-related genes in the brain, as this information was not available. Second, our sample size was small. Future replication studies using larger cohorts with more detailed clinical information will be required to validate these results. It should be noted that a high-throughput method will be necessary for large-scale lipid analysis because the LC–MS/MS method used in this study is complicated and time-consuming. Third, we quantified major species of PC, PE, PI, PS, PG, LPC, and SUL; however, these constitute a limited portion of the lipids contained in the corpus callosum. Quantification of other lipids will be required to comprehensively understand abnormal lipid metabolism in schizophrenia.
In summary, this study provides evidence for abnormal lipid metabolism in the corpus callosum in schizophrenia and proposes roles of reduced number and malfunction of microglial cells in this pathology. Impaired signaling through the schizophrenia-related gene network and microglial cells could lead to abnormal cross-talk between microglia and oligodendrocytes in the white matter structures of patients with schizophrenia. Further studies are warranted to enhance our understanding of the mechanisms regulating this pathology, including genetic and epigenetic aspects.
Notes
The authors thank members of the RIKEN Center for Sustainable Resource Science for LC–MS maintenance. We are also deeply grateful to Dr Yugi Katsuyuki for advice on bioinformatic analysis of the lipidomic data. Conflicts of Interest: None declared.
Funding
The Japan Society for the Promotion of Science KAKENHI (grant number 16K21622 and 19K17122 to C.S.); the Ministry of Education, Culture, Sports, Science and Technology (a Grant-in-Aid for Scientific Research on Innovative Areas: grant number JP18H05435 to T.Y.); the Japan Agency for Medical Research and Development (AMED) (AMED-CREST program: grant number JP19gm0910004 and the Strategic Research Program for Brain Sciences from AMED: grant number JP19dm0107083 to T.Y.).
Declaration
The funding agencies had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplementary Material
References
- Arnone D, McIntosh A, Tan G, Ebmeier K. 2008. Meta-analysis of magnetic resonance imaging studies of the corpus callosum in schizophrenia. Schizophr Res. 101:124–132. [DOI] [PubMed] [Google Scholar]
- Arron JR, Winslow MM, Polleri A, Chang C-P, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK. 2006. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 441:595. [DOI] [PubMed] [Google Scholar]
- Balendiran GK, Schnutgen F, Scapin G, Borchers T, Xhong N, Lim K, Godbout R, Spener F, Sacchettini JC. 2000. Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J Biol Chem. 275:27045–27054. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D. 2006. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 93:491–507. [Google Scholar]
- Bernardo A, Giammarco M, De Nuccio C, Ajmone-Cat M, Visentin S, De Simone R, Minghetti L. 2017. Docosahexaenoic acid promotes oligodendrocyte differentiation via PPAR-γ signalling and prevents tumor necrosis factor-α-dependent maturational arrest. Biochim Biophys Acta. 1862:1013–1023. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 37:911–917. [DOI] [PubMed] [Google Scholar]
- Cairns EA, Baldridge WH, Kelly ME. 2016. The endocannabinoid system as a therapeutic target in glaucoma. Neural Plast. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Perez JM, Kamphorst J, DeGroot J, Lafeber F, Goshawk J, Yu K, Shockcor JP, Vreeken RJ, Hankemeier T. 2010. Comprehensive LC-MS E lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J Proteome Res. 9:2377–2389. [DOI] [PubMed] [Google Scholar]
- Cetin-Karayumak S, Di Biase MA, Chunga N, Reid B, Somes N, Lyall AE, Kelly S, Solgun B, Pasternak O, Vangel M. 2019. White matter abnormalities across the lifespan of schizophrenia: a harmonized multi-site diffusion MRI study. Mol Psychiatry. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, Gat-Viks I, Tonti E, DeGrace MM, Clauser KR et al. 2011. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 147:853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V, Gokhan S, Gulinello M, Branch CA, Patil M, Basu R, Stoddart C, Mehler MF, Stanley ER. 2015. Phenotypic characterization of a Csf1r haploinsufficient mouse model of adult-onset leukodystrophy with axonal spheroids and pigmented glia (ALSP). Neurobiol Dis. 74:219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman LG, Zou J, Crews FT. 2020. Microglial depletion and repopulation in brain slice culture normalizes sensitized proinflammatory signaling. J Neuroinflammation. 17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson SL, Gan SC, Woon PS, Kuswanto C, Sum MY, Yang GL, Lui JM, Sitoh YY, Nowinski WL, Sim K. 2014. Corpus callosum morphology in first-episode and chronic schizophrenia: combined magnetic resonance and diffusion tensor imaging study of Chinese Singaporean patients. Br J Psychiatry. 204:55–60. [DOI] [PubMed] [Google Scholar]
- De Picker LJ, Morrens M, Chance SA, Boche D. 2017. Microglia and brain plasticity in acute psychosis and schizophrenia illness course: a meta-review. Front Psych. 8:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues HS, Portugal CC, Socodato R, Relvas JB. 2016. Oligodendrocyte, astrocyte, and microglia crosstalk in myelin development, damage, and repair. Front Cell Dev Biol. 4:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downhill JE Jr, Buchsbaum MS, Wei T, Spiegel-Cohen J, Hazlett EA, Haznedar MM, Silverman J, Siever LJ. 2000. Shape and size of the corpus callosum in schizophrenia and schizotypal personality disorder. Schizophr Res. 42:193–208. [DOI] [PubMed] [Google Scholar]
- Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, Marchini J, Smith SM. 2018. Genome-wide association studies of brain imaging phenotypes in UK biobank. Nature. 562:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. 2011. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn S, Lang D, Mackay A, Goghari V, Vavasour I, Whittall K, Smith G, Arango V, Mann J, Dwork A. 2003. Abnormalities of myelination in schizophrenia detected in vivo with MRI, and post-mortem with analysis of oligodendrocyte proteins. Mol Psychiatry. 8:811. [DOI] [PubMed] [Google Scholar]
- Gerstner JR, Perron IJ, Riedy SM, Yoshikawa T, Kadotani H, Owada Y, Van Dongen HPA, Galante RJ, Dickinson K, Yin JCP et al. 2017. Normal sleep requires the astrocyte brain-type fatty acid binding protein FABP7. Sci Adv. 3:e1602663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Dyer RA, Beasley CL. 2017. Evidence for altered cell membrane lipid composition in postmortem prefrontal white matter in bipolar disorder and schizophrenia. J Psychiatr Res. 95:135–142. [DOI] [PubMed] [Google Scholar]
- Glen AI, Glen EM, Horrobin DF, Vaddadi KS, Spellman M, Morse-Fisher N, Ellis K, Skinner FS. 1994. A red cell membrane abnormality in a subgroup of schizophrenic patients: evidence for two diseases. Schizophr Res. 12:53–61. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. 2003. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 113:657–670. [DOI] [PubMed] [Google Scholar]
- Hagemeyer N, Hanft K-M, Akriditou M-A, Unger N, Park ES, Stanley ER, Staszewski O, Dimou L, Prinz M. 2017. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 134:441–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder RC, Jahng A, Maricic I, Kumar V. 2007. Mini review: immune response to myelin-derived sulfatide and CNS-demyelination. Neurochem Res. 32:257–262. [DOI] [PubMed] [Google Scholar]
- Hamaguchi M, Muramatsu R, Fujimura H, Mochizuki H, Kataoka H, Yamashita T. 2019. Circulating transforming growth factor-β1 facilitates remyelination in the adult central nervous system. Elife. 8:e41869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki K, Maekawa M, Toyota T, Dean B, Hamazaki T, Yoshikawa T. 2017. Fatty acid composition of the postmortem corpus callosum of patients with schizophrenia, bipolar disorder, or major depressive disorder. Eur Psychiatry. 39:51–56. [DOI] [PubMed] [Google Scholar]
- Hamilton SP, Rome LH. 1994. Stimulation of in vitro myelin synthesis by microglia. Glia. 11:326–335. [DOI] [PubMed] [Google Scholar]
- Hendrickx DA, Eden CG, Schuurman KG, Hamann J, Huitinga I. 2017. Staining of HLA-DR, Iba1 and CD68 in human microglia reveals partially overlapping expression depending on cellular morphology and pathology. J Neuroimmunol. 309:12–22. [DOI] [PubMed] [Google Scholar]
- Henry RJ, Ritzel RM, Barrett JP, Doran SJ, Jiao Y, Leach JB, Szeto GL, Wu J, Stoica BA, Faden AI. 2020. Microglial depletion with CSF1R inhibitor during chronic phase of experimental traumatic brain injury reduces neurodegeneration and neurological deficits. J Neurosci. 40:2960–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205–2232. [DOI] [PubMed] [Google Scholar]
- Horrobin DF, Manku MS, Hillman H, Iain A, Glen M. 1991. Fatty acid levels in the brains of schizophrenics and normal controls. Biol Psychiatry. 30:795–805. [DOI] [PubMed] [Google Scholar]
- Ide M, Ohnishi T, Toyoshima M, Balan S, Maekawa M, Shimamoto-Mitsuyama C, Iwayama Y, Ohba H, Watanabe A, Ishii T. 2019. Excess hydrogen sulfide and polysulfides production underlies a schizophrenia pathophysiology. EMBO Mol Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale A, Joshi S, Naphade N, Sapkale S, Raju MS, Pillai A, Nasrallah H, Mahadik SP. 2008. Opposite changes in predominantly docosahexaenoic acid (DHA) in cerebrospinal fluid and red blood cells from never-medicated first-episode psychotic patients. Schizophr Res. 98:295–301. [DOI] [PubMed] [Google Scholar]
- Kanda Y. 2013. Investigation of the freely available easy-to-use software ‘EZR’for medical statistics. Bone Marrow Transplant. 48:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavan MS, Reynolds C, Montrose D, Miewald J, Downs C, Sabo E. 1994. Sleep and suicidality in psychotic patients. Acta Psychiatr Scand. 89:122–125. [DOI] [PubMed] [Google Scholar]
- Kim M-S, Shutov LP, Gnanasekaran A, Lin Z, Rysted JE, Ulrich JD, Usachev YM. 2014. Nerve growth factor (NGF) regulates activity of nuclear factor of activated T-cells (NFAT) in neurons via the phosphatidylinositol 3-kinase (PI3K)-Akt-glycogen synthase kinase 3β (GSK3β) pathway. J Biol Chem. 289:31349–31360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipanyula MJ, Kimaro WH, Etet PF. 2016. The emerging roles of the calcineurin-nuclear factor of activated T-lymphocytes pathway in nervous system functions and diseases. J Aging Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-J, Kim B, Oh D, Kim M-K, Kim K-H, Bang SY, Choi TK, Lee S-H. 2016. White matter alterations associated with suicide in patients with schizophrenia or schizophreniform disorder. Psychiatry Res: Neuroimag. 248:23–29. [DOI] [PubMed] [Google Scholar]
- Lener MS, Wong E, Tang CY, Byne W, Goldstein KE, Blair NJ, Haznedar MM, New AS, Chemerinski E, Chu K-W. 2015. White matter abnormalities in schizophrenia and schizotypal personality disorder. Schizophr Bull. 41:300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SX, Lam SP, Zhang J, Yu MWM, Chan JWY, Chan CSY, Espie CA, Freeman D, Mason O, Wing Y-K. 2016. Sleep disturbances and suicide risk in an 8-year longitudinal study of schizophrenia-spectrum disorders. Sleep. 39:1275–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RZ, Mita R, Beaulieu M, Gao Z, Godbout R. 2010. Fatty acid binding proteins in brain development and disease. Int J Dev Biol. 54:1229–1239. [DOI] [PubMed] [Google Scholar]
- Lloyd AF, Davies CL, Miron VE. 2017. Microglia: origins, homeostasis, and roles in myelin repair. Curr Opin Neurobiol. 47:113–120. [DOI] [PubMed] [Google Scholar]
- Maniega SM, Lymer G, Bastin M, Marjoram D, Job D, Moorhead T, Owens D, Johnstone E, McIntosh A, Lawrie S. 2008. A diffusion tensor MRI study of white matter integrity in subjects at high genetic risk of schizophrenia. Schizophr Res. 106:132–139. [DOI] [PubMed] [Google Scholar]
- Manocha GD, Ghatak A, Puig KL, Kraner SD, Norris CM, Combs CK. 2017. NFATc2 modulates microglial activation in the AβPP/PS1 mouse model of Alzheimer’s disease. J Alzheimers Dis. 58:775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JL, Suzuki K, Chaplin DD, Matsushima GK. 2001. Interleukin-1β promotes repair of the CNS. J Neurosci. 21:7046–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto J, Nakanishi H, Kunii Y, Sugiura Y, Yuki D, Wada A, Hino M, Niwa SI, Kondo T, Waki M, et al. 2017. Decreased 16:0/20:4-phosphatidylinositol level in the post-mortem prefrontal cortex of elderly patients with schizophrenia. Sci Rep 7:45050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara RK, Jandacek R, Rider T, Tso P, Hahn CG, Richtand NM, Stanford KE. 2007. Abnormalities in the fatty acid composition of the postmortem orbitofrontal cortex of schizophrenic patients: gender differences and partial normalization with antipsychotic medications. Schizophr Res. 91:37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BJ, Buckley P, Seabolt W, Mellor A, Kirkpatrick B. 2011. Meta-analysis of cytokine alterations in schizophrenia: clinical status and antipsychotic effects. Biol Psychiatry. 70:663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamoto-Combs K, Combs CK. 2010. Microglial phenotype is regulated by activity of the transcription factor, NFAT (nuclear factor of activated T cells). J Neurosci. 30:9641–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien JS, Sampson EL. 1965. Lipid composition of the normal human brain: gray matter, white matter, and myelin. J Lipid Res. 6:537–544. [PubMed] [Google Scholar]
- Ohnishi T, Balan S, Toyoshima M, Maekawa M, Ohba H, Watanabe A, Iwayama Y, Fujita Y, Tan Y, Hisano Y. 2019. Investigation of betaine as a novel psychotherapeutic for schizophrenia. EBioMedicine. 45:432–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquini LA, Millet V, Hoyos HC, Giannoni J, Croci D, Marder M, Liu F-T, Rabinovich GA, Pasquini JM. 2011. Galectin-3 drives oligodendrocyte differentiation to control myelin integrity and function. Cell Death Differ. 18:1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peferoen L, Kipp M, Valk P, Noort JM, Amor S. 2014. Oligodendrocyte-microglia cross-talk in the central nervous system. Immunology. 141:302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BD, Machielsen MW, Hoen WP, Caan MW, Malhotra AK, Szeszko PR, Duran M, Olabarriaga SD, Haan L. 2012. Polyunsaturated fatty acid concentration predicts myelin integrity in early-phase psychosis. Schizophr Bull. 39:830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigeon WR, Pinquart M, Conner K. 2012. Meta-analysis of sleep disturbance and suicidal thoughts and behaviors. J Clin Psychiatry. [DOI] [PubMed] [Google Scholar]
- Pu H, Guo Y, Zhang W, Huang L, Wang G, Liou AK, Zhang J, Zhang P, Leak RK, Wang Y. 2013. Omega-3 polyunsaturated fatty acid supplementation improves neurologic recovery and attenuates white matter injury after experimental traumatic brain injury. J Cereb Blood Flow Metab. 33:1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Wang Q, Lei W, Zhang C, Li Y, Li X, Li M, Deng W, Huang C, Du F. 2017. The common variants implicated in microstructural abnormality of first episode and drug-naïve patients with schizophrenia. Sci Rep. 7:11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J, Mittelstadt PR, Mages HW, Gerth AJ, Kroczek RA, Ashwell JD, Glimcher LH. 2000. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity. 12:293–300. [DOI] [PubMed] [Google Scholar]
- Rumsby MG. 1978. Organization and structure in central-nerve myelin. Biochem Soc Trans. 6:448–462. [DOI] [PubMed] [Google Scholar]
- Scheinman EJ, Avni O. 2009. Transcriptional regulation of GATA3 in T helper cells by the integrated activities of transcription factors downstream of the interleukin-4 receptor and T cell receptor. J Biol Chem. 284:3037–3048. [DOI] [PubMed] [Google Scholar]
- Schmitt A, Wilczek K, Blennow K, Maras A, Jatzko A, Petroianu G, Braus DF, Gattaz WF. 2004. Altered thalamic membrane phospholipids in schizophrenia: a postmortem study. Biol Psychiatry. 56:41–45. [DOI] [PubMed] [Google Scholar]
- Schwarz E, Prabakaran S, Whitfield P, Major H, Leweke FM, Koethe D, McKenna P, Bahn S. 2008. High throughput lipidomic profiling of schizophrenia and bipolar disorder brain tissue reveals alterations of free fatty acids, phosphatidylcholines, and ceramides. J Proteome Res. 7:4266–4277. [DOI] [PubMed] [Google Scholar]
- Sharifi K, Morihiro Y, Maekawa M, Yasumoto Y, Hoshi H, Adachi Y, Sawada T, Tokuda N, Kondo H, Yoshikawa T et al. 2011. FABP7 expression in normal and stab-injured brain cortex and its role in astrocyte proliferation. Histochem Cell Biol. 136:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto C, Ohnishi T, Maekawa M, Watanabe A, Ohba H, Arai R, Iwayama Y, Hisano Y, Toyota T, Toyoshima M, et al. 2014. Functional characterization of FABP3, 5 and 7 gene variants identified in schizophrenia and autism spectrum disorder and mouse behavioral studies. Hum Mol Genet 23:6495–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindou H, Hishikawa D, Nakanishi H, Harayama T, Ishii S, Taguchi R, Shimizu T. 2007. A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells cloning and characterization of acetyl-CoA: lyso-PAF acetyltransferase. J Biol Chem. 282:6532–6539. [DOI] [PubMed] [Google Scholar]
- Taha AY, Cheon Y, Ma K, Rapoport SI, Rao JS. 2013. Altered fatty acid concentrations in prefrontal cortex of schizophrenic patients. J Psychiatr Res. 47:636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y-L, Yuan Y, Tian L. 2019. Microglial regional heterogeneity and its role in the brain. Mol Psychiatry. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torshizi AD, Armoskus C, Zhang H, Forrest MP, Zhang S, Souaiaia T, Evgrafov OV, Knowles JA, Duan J, Wang K. 2019. Deconvolution of transcriptional networks identifies TCF4 as a master regulator in schizophrenia. BioRxiv. 133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima M, Akamatsu W, Okada Y, Ohnishi T, Balan S, Hisano Y, Iwayama Y, Toyota T, Matsumoto T, Itasaka N. 2016. Analysis of induced pluripotent stem cells carrying 22q11. 2 deletion. Transl Psychiatry. 6:e934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerkamp JH, Moerkerk HT, Prinsen CF, Kuppevelt TH. 1999. Structural and functional studies on different human FABP types. Mol Cell Biochem. 192:137–142. [PubMed] [Google Scholar]
- Ward JH., Jr 1963. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 58:236–244. [Google Scholar]
- Weider M, Starost LJ, Groll K, Küspert M, Sock E, Wedel M, Fröb F, Schmitt C, Baroti T, Hartwig AC. 2018. Nfat/calcineurin signaling promotes oligodendrocyte differentiation and myelination by transcription factor network tuning. Nat Commun. 9:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CT, Bestard-Lorigados I, Crawford DA. 2019. Autism-related behaviors in the cyclooxygenase-2-deficient mouse model. Genes Brain Behav. 18:e12506. [DOI] [PubMed] [Google Scholar]
- Woodruff PW, McManus IC, David AS. 1995. Meta-analysis of corpus callosum size in schizophrenia. J Neurol Neurosurg Psychiatry. 58:457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LZ, Sanchez R, Sali A, Heintz N. 1996. Ligand specificity of brain lipid-binding protein. J Biol Chem. 271:24711–24719. [DOI] [PubMed] [Google Scholar]
- Yabuki Y, Takahata I, Matsuo K, Owada Y, Fukunaga K. 2018. Ramelteon improves post-traumatic stress disorder-like behaviors exhibited by fatty acid-binding protein 3 null mice. Mol Neurobiol. 55:3577–3591. [DOI] [PubMed] [Google Scholar]
- Yamada K, Gerber DJ, Iwayama Y, Ohnishi T, Ohba H, Toyota T, Aruga J, Minabe Y, Tonegawa S, Yoshikawa T. 2007. Genetic analysis of the calcineurin pathway identifies members of the EGR gene family, specifically EGR3, as potential susceptibility candidates in schizophrenia. Proc Natl Acad Sci. 104:2815–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JK, Leonard S, Reddy RD. 2000. Membrane phospholipid abnormalities in postmortem brains from schizophrenic patients. Schizophr Res. 42:7–17. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Mishina M. 2005. Distinct roles of calcineurin-nuclear factor of activated T-cells and protein kinase A-cAMP response element-binding protein signaling in presynaptic differentiation. J Neurosci. 25:3067–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


