Abstract
The presence of heterogeneity/subgroups in infants and older populations against single-domain brain or behavioral measures has been previously characterized. However, few attempts have been made to explore heterogeneity at the brain–behavior relationship level. Such a hypothesis posits that different subgroups of infants may possess qualitatively different brain–behavior relationships that could ultimately contribute to divergent developmental outcomes even with relatively similar brain phenotypes. In this study, we aimed to explore such relationship-level heterogeneity and delineate the subgrouping structure of newborns with differential brain–behavior associations based on a typically developing sample of 81 infants with 3-week resting-state functional magnetic resonance imaging scans and 4-year intelligence quotient (IQ) measures. Our results not only confirmed the existence of relationship-level heterogeneity in newborns but also revealed divergent developmental outcomes associated with two subgroups showing similar brain functional connectivity but contrasting brain–behavior relationships. Importantly, further analyses unveiled an intriguing pattern that the subgroup with higher 4-year IQ outcomes possessed brain–behavior relationships that were congruent to their functional connectivity pattern in neonates while the subgroup with lower 4-year IQ not, providing potential explanations for the observed IQ differences. The characterization of heterogeneity at the brain–behavior relationship level may not only improve our understanding of the patterned intersubject variability during infancy but could also pave the way for future development of heterogeneity-inspired, personalized, subgroup-specific models for better prediction.
Keywords: brain–behavior relationship, infant subgrouping, intersubject variability, relational heterogeneity, resting-state functional connectivity
Introduction
One of the most important goals of human neuroscience research lies in the delineation of robust brain–behavior relationships to derive brain-based biomarkers for behavioral prediction. This research is especially important for infants as subtle brain changes during infancy may cascade into major deviations later in life (Tau and Peterson 2010; Gao et al. 2017; O’Donnell and Meaney 2017; Monk et al. 2019). However, the difficulty of detecting robust brain–behavior relationships in this population is well recognized and the reported associations are often weak to moderate (Alcauter et al. 2014, 2015b). On the one hand, technological limitations may partly underlie these difficulties and future development of more advanced imaging/image analysis methods may help derive more sensitive biomarkers with tighter behavioral associations. However, another reason may relate to the conventional way of characterizing brain–behavior relationships based on the homogeneity assumption. This is because instead of homogeneity, many recent observations suggest heterogeneity of brain–behavior relationships across different populations or even within an otherwise “homogeneous” population. For example, two parallel studies conducted by Elton and colleagues found that beyond categorical differences in brain functional connectivity, children with attention-deficit hyperactivity disorder (ADHD) (Elton et al. 2014) and autism spectrum disorder (Elton et al. 2016) also show contrasting brain–behavioral relationships compared with normal controls. Moreover, sex-related differences in brain–intelligence quotient (IQ) relationships in a cohort of control adults (Jiang et al. 2019) and socioeconomic status-related differences in brain-reading skills correlations in a typically developing children sample were also reported (Noble et al. 2006). Therefore, previous reports suggest that there might be heterogeneity at the brain–behavioral relationship level even within an otherwise homogeneous population.
The benefits of exploring and delineating such brain–behavior relationship heterogeneities in infants are at least 2-fold. First, it would greatly improve our understanding of the intersubject variability pattern during this critical period of development (Gao et al. 2014). If confirmed, the existence of such relationship-level heterogeneity would indicate mechanistic differences and imply that even with similar brain phenotypes, different infants may go on to develop divergent behavioral outcomes, given differential brain–behavioral mechanisms. Therefore, such delineation goes beyond the conventional single-domain characterization of heterogeneity and could offer greater insights into potential brain mechanistic differences. Second, a subgrouping structure could be derived based on the detected brain–behavior relationship heterogeneity, which may offer novel ways for brain-based predictions of later developmental outcomes through deriving subgroup-specific prediction models. Such heterogeneity-inspired, personalized, subgroup-specific prediction approaches, compared with those based on generic whole-group models, may better utilize the enriched information embedded in the infant brain for better prediction.
In this paper, we sought to take the first step and determine whether there exists brain–behavior relationship heterogeneity in a relatively “homogenous” control infant sample (i.e., all with full-term birth, <24-h stay at a neonatal intensive care unit (NICU), and no maternal mental disorder diagnosis, N = 81). For each infant subject, we have a successful newborn resting-state functional magnetic resonance imaging (rsfMRI) scan and a 4-year cognitive outcome measure (i.e., 4YR IQ). The correlation structures between functional connections measured at ~3 weeks of age and 4YR IQ scores were examined to determine if there exist significantly different brain–behavior relationships in different subgroups of infants at the connection level. We further examined if a unified subgrouping structure could be defined at the whole-brain level. We hypothesize positive answers for both questions. Moreover, we further expect that the derived subgroups would also show differences in their 4-year developmental outcomes, given their differences in brain–behavior mechanisms. Our results confirmed all three hypotheses and provided strong support for the existence of relationship-level heterogeneity in neonates. More interestingly, when comparing between brain–behavior relationships and neonatal functional connectivity patterns, an intriguing pattern emerged that the subgroup with higher 4YR IQ showed brain–behavior relationships that were largely congruent with their neonatal functional connectivity pattern (i.e., reflecting their pre−/perinatal functional connectivity growth directions), particularly within the default-mode (Raichle et al. 2001), frontoparietal control (Vincent et al. 2008), and limbic networks, while the subgroup with lower 4YR IQ not. These findings greatly improved our understanding of the intersubject variability at the brain–behavioral relationship level and lay the foundation for future developments of subgroup-specific models for behavioral prediction.
Materials and Methods
Participants and Imaging
The infant participants were from the University of North Carolina (UNC) at Chapel Hill Early Brain Development Study, characterizing early childhood brain and behavior development (Gao et al. 2017; Gilmore et al. 2018). A total of 81 neonate subjects with passed-quality control 3-week rsfMRI scans and successful 4YR IQ measures were retrospectively identified and included in this study. All included infants were full term (gestational age ≥37 weeks), no maternal psychiatric disorders diagnosis, and <24-h stay at an NICU. Note our original sample have both twins and singletons, but the final sample only included randomly selected one of each twin pair to minimize between-subject correlation. This sample was chosen to demonstrate the potential presence of brain–behavior relationship heterogeneity within an otherwise homogenous infant sample. Demographic information of the sample was summarized in Table 1. The study protocols were approved by both the UNC at Chapel Hill and Cedars-Sinai Institutional Review Boards.
Table 1.
Summary of demographic information
| Neonates | |
|---|---|
| Infants with 4YR IQ measures |
N = 81 IQ: 110.22 ± 12.22 |
| Gestational age at birth (days) | 273.33 ± 9.43 |
| Postnatal age at scan (days) | 23.41 ± 11.37 |
| Birth weight (grams) | 3216.2 ± 558.9 |
| Sex (male/female) | N: 34/47 |
| Twin (twin/single birth) | N: 25/56 |
The magnetic resonance imaging (MRI) data were acquired using two scanners: a 3 T Siemens Allegra scanner with circular polarization head coil (69 neonatal scans) and a 3 T Siemens Tim Trio with 32-channel head coil (12 neonatal scans) with the same MRI protocols. Different scanners were included as a control variable in subgroup comparisons. Functional images were acquired with a T2*-weighted echo planar imaging sequence: repetition time/echo time (TR/TE) = 2000 ms/32 ms, 33 slices, voxel size = 4 mm3, 150 volumes. Structural images were acquired using a 3D MPRAGE sequence: TR/TE = 1820 ms/4.38 ms, inversion time = 1100 ms, voxel size = 1 mm3. Infant subjects were fed, swaddled, and fitted with ear protection prior to imaging. All subjects were in a natural sleep state during the imaging session.
Image Preprocessing
Functional images were preprocessed using the FMRIB’s Software Libraries (FSL) (Smith et al. 2004) and Analysis of Functional NeuroImages (Cox 1996), including discarding the first three volumes, slice timing and head motion correction, bandpass filtering (0.01–0.08 Hz), and nuisance signal regression. The 24 motion-related parameters (six motion correction parameters, derivative, and their quadratic terms), signals from the white matter, cerebrospinal fluid (plus their derivative and quadratic terms), and the global signal were included as nuisance signals. All nuisance signals were band-pass filtered (0.01–0.08 Hz) before regression to match the frequency of the blood oxygen level-dependent signal. Despite of the already short acquisition time (5 min), rigorous additional motion correction steps through scrubbing were employed to mitigate the widely reported motion-related artifacts (Power et al. 2012, 2014). Specifically, volumes with global signal changes >0.5% and/or frame-wise displacements (FD) >0.3 mm (Power et al. 2012) were excluded (plus one before and two after). After this procedure, different subjects had different numbers of volumes left and to avoid introducing another potential variability, we chose to uniformly cut the length of remaining data of all subjects to be 90 volumes (i.e., 3 min) as a compromise between sample size and data length. Finally, the images were spatially smoothed with Gaussian kernel (full width at half maximum = 6 mm). The amount of scrubbed volumes and residual frame-wise FD were correlated with behavior scores and gestational age at scan. None of these tests yield significance. After functional images preprocessing, all functional images were registered to the age-specific anatomical template space for neonate (Shi et al. 2011) using the combined transformation field from a two-step registration, namely an affine transformation from individual functional images to anatomical images and a nonlinear registration from individual anatomical images to the target images. Spatial transformations were performed in FSL.
Behavioral Measures of IQ
Behavioral data of IQ were collected using the Stanford–Binet Intelligence Scales, 5th edition (Roid 2003). The Stanford–Binet is a series of tasks administered individually in a structured setting. These scales were designed to assess intelligence across the lifespan (appropriate for individuals aged 2 through 85 years), specifically focusing on five major domains, including fluid, knowledge, quantitative, visual–spatial, and working memory. In the current study, the abbreviated IQ (ABIQ) score was used as an estimate of general cognitive ability. ABIQ is calculated from performance on two scales: nonverbal fluid reasoning and verbal knowledge. The ABIQ score provides a quick estimate of a child’s general cognitive ability, and as it requires the administration of only two subtests, it is easier to obtain than the full-scale IQ, especially for 4-year-old children. The ABIQ score has shown strong test–retest (r = 0.87) reliability. The Stanford–Binet scales also have strong inter-rater reliability (ranging from 0.74 to 0.97 across all scales).
Experimental Design and Statistical Analysis
Detection of Significant Brain–Behavior Relationships as a Whole Group
To determine the existence of significant brain–behavioral relationships as a whole group (i.e., homogenous relationships), anatomical automatic labeling (AAL)-based functional connectivity matrix (Rushe et al. 2001) was calculated for each of the 81 newborns and correlations between each functional connection and 4YR IQ scores were calculated. Gestational age at birth, postnatal age at scan, birth weight, sex, twin status, and scanner were included as control variables. Specifically, after imaging preprocessing, the infant age-specific AAL atlas (Shi et al. 2011) was used to extract reginal rsfMRI time series for the calculation of pair-wise correlation values among 90 cortical regions (i.e., a 90 × 90 correlation matrix for each subject). After Fisher Z-transform, connection strengths were correlated with 4YR IQ scores within the full sample of 81 newborns to detect significant one-group brain–behavioral relationships after controlling for the six control variables as described above. To improve robustness of this brain–behavioral correlation detection, a nonparametric permutation test was conducted to detect significant relationships. Specifically, 10 000 randomizations on the 4YR behavior IQ were conducted and the probability of a randomized squared correlation value larger than the true correlation was defined as the P value. P < 0.05 based on this nonparametric test was defined to be significant.
Detection of Heterogenous Brain–Behavior Relationships
To determine the existence of heterogenous brain–behavior relationships for each of the 4005 connections, we designed an algorithm to examine if splitting the whole sample into two subgroups could significantly improve the whole-group brain–behavior relationship model as introduced above. The algorithm was designed to optimize two subgroups {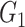 ,
,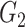 } within the original group
} within the original group 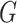 so that the linear relationships {
so that the linear relationships {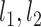 } within each of the two subgroups explain the maximum amount of variance. To achieve this, we designed an iterative algorithm, minimizing the sum of absolute values of residuals from {
} within each of the two subgroups explain the maximum amount of variance. To achieve this, we designed an iterative algorithm, minimizing the sum of absolute values of residuals from {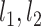 } fitting of all the data points, which is conceptually similar as k-means clustering. The linear regression model was defined as follows:
} fitting of all the data points, which is conceptually similar as k-means clustering. The linear regression model was defined as follows: 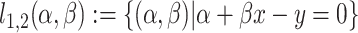 , in which intercept:
, in which intercept: 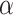 and slope:
and slope: 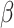 were estimated from data
were estimated from data 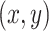 using ordinary least squares (OLS) regression.
using ordinary least squares (OLS) regression.
To start the algorithm, two lines were defined as the initialization of {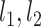 }: One fitting the whole group
}: One fitting the whole group 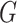 and the other orthogonal with the first one, both crossing the origin.
and the other orthogonal with the first one, both crossing the origin. 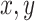 were normalized into Z-score before fitting. Then, the point
were normalized into Z-score before fitting. Then, the point 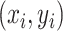 -to-line {
-to-line {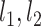 } distance was defined as follows:
} distance was defined as follows: 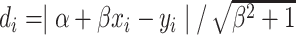 . Based on the distance, point
. Based on the distance, point 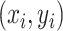 could be assigned into
could be assigned into 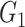 if
if 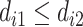 or into
or into 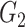 if
if 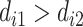 , yielding subgroups {
, yielding subgroups {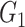 ,
,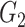 }. After all points got an assignment, {
}. After all points got an assignment, {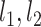 } were refitted based on the newly assigned point memberships in each subgroup and another iteration started.
} were refitted based on the newly assigned point memberships in each subgroup and another iteration started. 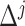 was defined as the Euler distance between the updated slopes {
was defined as the Euler distance between the updated slopes {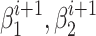 } and the previous slopes {
} and the previous slopes {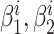 } (
} (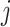 represented iterations). These steps were repeated until
represented iterations). These steps were repeated until 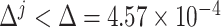 (equivalent to that the angular distance was <1.5°). Final outputs resulted in optimized subgroup {
(equivalent to that the angular distance was <1.5°). Final outputs resulted in optimized subgroup {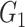 ,
,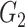 } at individual connection. The pseudocode of the iterative algorithm was listed below.
} at individual connection. The pseudocode of the iterative algorithm was listed below.
Algorithm Dataset Fitting
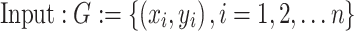 |
 |
 |
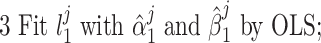 |
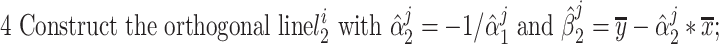 |
 |
 |
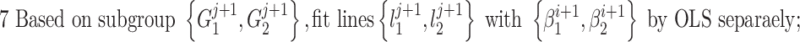 |
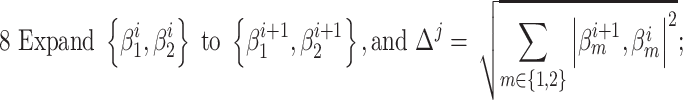 |
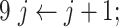 |
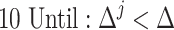 |
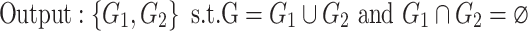 |
Note even with a random distribution, splitting a whole sample into two subgroups may improve the linear fitting. To avoid this “false positive” scenario and only detect those connections that show significantly more improvement after splitting than random distributions, we employed a consistent nonparametric random permutation test to define significance of subgrouping for each connection. Specifically, the nonparametric permutation test was conducted by randomly permuting the behavioral scores of all subjects 10 000 times and rerunning the subgrouping algorithm, as described above, for each random permutation. Therefore, 10 000 pairs of “randomized” {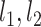 } were estimated during this process to serve as the null distribution and a P value was assigned for each connection as the percentage of times the averaged R2 of the two randomized subgrouping lines from the 10 000 permutations surpass the corresponding true averaged R2 of the subgrouping based on the original data. This testing ensured that we only detect connections that showed a significantly better subgrouping structure than random organizations. P < 0.05 based on this nonparametric test was defined to be significant.
} were estimated during this process to serve as the null distribution and a P value was assigned for each connection as the percentage of times the averaged R2 of the two randomized subgrouping lines from the 10 000 permutations surpass the corresponding true averaged R2 of the subgrouping based on the original data. This testing ensured that we only detect connections that showed a significantly better subgrouping structure than random organizations. P < 0.05 based on this nonparametric test was defined to be significant.
Subgrouping Structure of Brain–Behavior Relationship at Whole-Brain Level
We further explored if a unified whole-brain level subgrouping structure can be derived from the individual connection-level subgrouping classifications. To do this, the subgrouping characteristics of each subject for each of the detected connections showing significant two-group fittings were calculated as the difference values of dot-to-lines distances of each subject’s data point to the two regression lines. These connection-specific values were concatenated to form a subject-level subgrouping profile vector across all detected connections showing significant subgrouping as described above. This vector thus represented each individual subject’s subgrouping membership profile across all detected “heterogenous” connections and was used to calculate an across-subject correlation matrix reflecting the between-subject similarity of their individual connection-level subgrouping profiles. Based on this similarity matrix, k-means clustering was conducted to determine if there are subgroups (i.e., cohorts showing significantly higher within-cohort similarity than between-cohort similarity) that possessed similar subgrouping profiles across all heterogenous connections. The optimal k was defined based on the “Davies–Bouldin” criterion (Davies and Bouldin 1979), which calculated the ratio of within-cluster and between-cluster distances. In order to assess the robustness of this optimal number, 100 repetitions of k-means clustering were conducted.
After clustering, the defined whole-brain level subgroups were compared against their 4YR IQ outcomes and six control variables including gestational age at birth, postnatal age at scan, birth weight, and ratios of male/female, twin/singleton, and two scanners. The within-subgroup brain–behavior relationships were also calculated and compared. To further compare the spatial distribution of within-subgroup connection-level relationships at network level, the 90 AAL regions were assigned to eight functional networks (Yeo et al. 2011) based on spatial overlapping (i.e., winner-take-all approach to assign each region to the network with the highest level of overlap in volume) to group all connections as either within one (both nodes within one network) or between two networks (each node belonging to a different network). Finally, to test the robustness of our results against the choice of brain parcellations, the Cedars–UNC neonate-specific functional brain atlas (Shi et al. 2018) was used and the above-mentioned comparisons of within-subgroup functional connectivity and brain–behavior relationships were repeated to explore if findings can be replicated across different template choices. The overall workflow of our methods is presented in Supplementary Figure S1.
Results
Heterogenous Brain–Behavioral Relationships at the Connection Level
The group mean whole-brain correlation matrix is shown in Figure 1a. Regions within the same functional networks were grouped together, and it was apparent that most networks were already well synchronized, although primary ones were synchronized at a higher degree than high-order ones, which is consistent with previous findings (Gao et al. 2011, 2015). Based on the nonparametric permutation test, there were 343 connections (8.56%, Fig. 1b) detected showing significant homogenous/one-group connectivity–IQ relationships (nonparametric P < 0.05). These one-group relationships were expectedly weak (i.e., average R2 = 0.072 with range between 0.046 and 0.165, Fig. 1c, upper row). On the other hand, 334 connections (8.34%, Fig. 1b) were detected to show two-group/heterogenous brain–behavior relationships based on our nonparametric test (nonparametric P < 0.05). As expected, the subgroup-level linear brain–behavioral relationships for the 334 heterogenous connections from the two-group fittings were much stronger (i.e., average R2 = 0.501 with range between 0.410 and 0.661, Fig. 1c, bottom row) compared with their corresponding one-group correlations (average R2 = 0.039, P < 1 × 10−10, Fig. 1c, middle row) and the 343 significant single-group correlations (P < 1 × 10−10, Fig. 1c, upper row). Interestingly, there were 114 connections showing both significant one-group and two-group correlations, indicating that despite of significant correlations with 4YR IQ as a whole group, there were still detectable slope differences in their brain–behavioral relationships. Consistently, when directly testing these 114 connections, the two-group fittings significantly outperformed one-group fittings in terms of variance explained (P < 1 × 10−10, Supplementary Fig. S2), suggesting subgroups were preferred to model these connections than one-group models. Three examples of homogeneous/one-group relationships, heterogenous/two-group relationships, and overlapping relationships are shown in Figure 1d.
Figure 1.
The homogenous (one-group) and heterogenous (two-group) brain–behavior relationships between neonate functional connectivity (NEO FC) and 4-year IQ (4YR IQ). (a) The group mean FC matrix of the 81 neonates, grouped by eight functional networks (i.e., visual [VIS], sensorimotor [SM], dorsal attention [DA], ventral attention [VA], limbic system [LIM], frontoparietal control [FP], default-mode [DMN], and subcortical [SUB] network); (b) The number and percentage of significant homogenous (one-group) and heterogenous (two-group) connections among all connections; (c) The mean variance explained (i.e., mean R2) of the 343 homogenous connections by the one-group fitting (R2_1_1; upper bar), the mean variance explained of the 334 heterogenous connections by one-group fitting (R2_1_2, middle bar), and mean variance explained of the 334 heterogenous connections by the two-subgroup fitting (R2_2_2, bottom bar); “****” indicates P < 1.0 × 10−10 for group comparison; (d) Three exemplar scatter plots of the connections showing one-group relationships in left column, two-group relationships in middle column, and connections showing both significant one-group and two-group relationships in the right column. x-axis represents the functional connectivity (Z-scores) in neonate and y-axis represents the 4-year IQ (Z-scores).
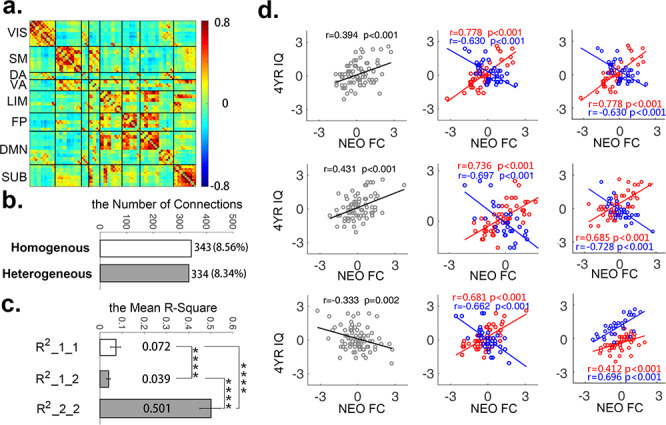
The spatial distributions of the two types of connections were shown in glass brains in Figure 2a and summarized based on their network assignments in Fig. 2b. Lists of all regions of interest (ROI)-to-ROI connections that showed one−/two-group relationships were presented in Supplementary Tables S1 and S2, respectively. Overall, both the homogenous and heterogenous connections featured a brain-wide distribution spreading across all eight functional networks.
Figure 2.
The distribution of the detected homogenous and heterogenous connections. (a) The spatial distribution of the detected homogenous (N = 343) and heterogenous (N = 334) connections in glass brains. (b) The network-level distribution of homogenous and heterogenous connections. The connections between each network and the rest seven networks were summarized as “between-network” in this plot. The numbers in y-axis represent the normalized number of connections (normalized against the number of nodes within each network) showing significant relationships normalized against the total possible number of within−/between-network connections. VIS, visual; SM, sensorimotor; DA, dorsal attention; VA, ventral attention; LIM, limbic system; FP, frontoparietal control; DMN, default-mode; and SUB, subcortical network.
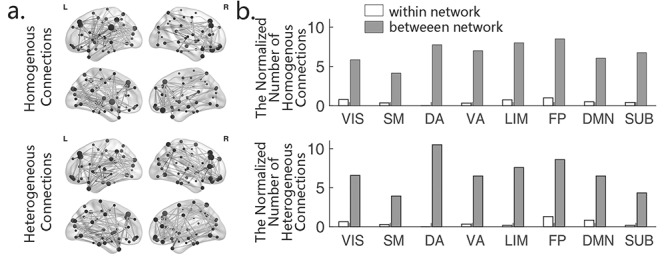
A Robust Whole-Brain Level Subgrouping Structure Derived from the Connection-Level Subgrouping Profiles
To explore the possibility of defining a whole-brain consistent subgrouping structure, k-means clustering was conducted based on individual connection-level subgrouping profiles. Based on the Davies–Bouldin criterion (Davies and Bouldin 1979), the 81 neonatal subjects were clustered into two subgroups (N = 27/54, Fig. 3a) at the whole-brain level. The results demonstrated significantly higher within-subgroup similarity than between-subgroup similarity (P < 0.001) in their connection-level subgrouping profiles, supporting the existence of whole-brain level subgroups (Fig. 3b,c). In fact, subgroup 1 (S1) and subgroup 2 (S2) showed primarily positive within-group similarities, but largely negative correlations between their connection-level subgrouping profiles. In contrast, when we tried to perform similar whole-brain clustering based on the homogenous connections as a control analysis, the k-means clustering was not converging and no subgroups were detected. Examples of the connection-level subgrouping profiles from pairs of subjects either within the same subgroup or from different subgroups are shown in Supplementary Figure S3.
Figure 3.
Whole-brain level subgrouping structure based on the individual connection-level subgrouping profiles. (a) The optimal number of clusters (i.e., k = 2); (b) Similarity of connection-level subgrouping profiles within and between the two defined whole-brain level subgroups (S1 and S2); (c) Statistical comparisons of within- and between-subgroups (S1 and S2) similarity of connection-level subgrouping profiles; (d) Statistical comparisons of 4YR IQ and six control variables (i.e., gestational age at birth [GA, in days], postnatal age at scan [PA, in days], birth weight [BW, in grams], percentages of two sexes, twin/singleton, and two scanners) and two motion parameters (the residual frame-wise displacement [rFD] and the number of scrubbed TRs [nSC]) between S1 and S2. ***P < 0.001.
When examined against control variables including gestational age at birth, postnatal age at scan, and birth weight, the two subgroups did not show any significant differences (Fig. 3d). Similarly, the ratios of male/female, twin/singleton, motion parameters, and the number of datasets acquired in scanner 1/scanner 2 were not different between the two subgroups either. However, when examining behavioral outcomes, S1 and S2 showed a significant difference in 4YR IQ with S1 showing higher IQ than S2 (P < 0.001, Fig. 3d, left panel).
When examining functional connections alone, the two subgroups showed highly consistent whole-brain functional connectivity patterns and none of the connections showed significant difference between S1 and S2 (Fig. 4a, first column). Therefore, as expected, the two subgroups featured similar brain functional connectivity but different behavioral outcomes. Subsequent examination of within-subgroup brain–behavioral relationships revealed highly contrasting brain–behavior relationships between the two subgroups (Fig. 4a, second column). In fact, the two sets of brain–behavior relationships from S1 and S2 showed a significant negative relationship (r = −0.116, P < 1.0 × 10−10).
Figure 4.
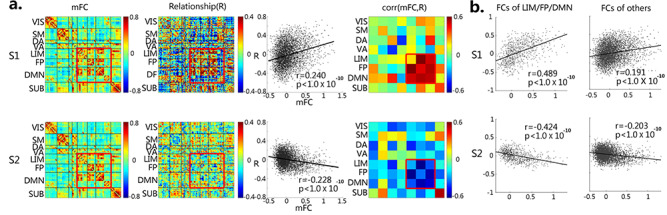
Contrasting brain–behavior relationships between the two subgroups. (a) First column: Mean functional connectivity patterns for S1 and S2 (visual [VIS], sensorimotor [SM], dorsal attention [DA], ventral attention [VA], limbic system [LIM], frontoparietal control [FP], default-mode [DMN] and subcortical [SUB] network); second column: Unthresholded brain–behavior correlation patterns for S1 and S2 (thresholded matrices at P < 0.05 are shown in Supplementary Fig. S5, the left column); third column: Scatter plots between mean functional connectivity patterns (column 1) and mean brain–behavior relationship patterns (column 2) for S1 and S2, respectively; fourth column: Column 3 relationships in network format. Red boxes highlight the tri-network set (i.e., the LIM, FP, and DMN); (b) First column: Scatter plots of column 3 relationships within the DMN–LIM–FP set; second column: Scatter plots of column 3 relationships outside of the DMN–LIM–FP set.
A closer look revealed that the brain–behavior relationship pattern in S1 qualitatively resembled its own neonatal functional connectivity pattern, but S2 failed to demonstrate such similarity. Quantitatively, when correlating between these two patterns, S1 showed highly significant positive correlations (r = 0.240, P < 1.0 × 10−10), while S2 demonstrated a negative correlation (r = −0.228, P < 1.0 × 10−10, Fig. 4a, third column), suggesting that S1’s brain–behavior relationships were more congruent with its own functional connectivity developmental status at the neonatal stage while S2 not. When further decomposing the whole-brain pattern into different functional networks, the tri-network set including the limbic, the frontoparietal control, and the default-mode networks (red box in Fig. 4a, fourth column) stood out and showed the most significant positive correlations in S1 (r = 0.489, P < 1.0 × 10−10, compared with the rest showing r = 0.191), but the most negative correlations in S2 (r = −0.424, P ≤ 1.0 × 10−10, compared with the rest showing r = −0.203, Fig. 4b). When further examining these patterns using a different neonate-specific functional atlas, the observed within-group functional connectivity and brain–behavior relationship patterns remained highly consistent (Supplementary Fig. S4).
Finally, although the two detected subgroups did not differ in their scanner distributions (Fig. 3d), the use of different scanners may represent a source of potential heterogeneity. Therefore, we repeated our analysis based only on subjects from the Allegra scanner (N = 69) and highly consistent results were obtained (Supplementary Fig. S6). Specifically, based on this one-scanner sample, there were 296/243 connections showing significant one−/two-group relationships (Supplementary Fig. S6a). After the same whole-brain subgrouping analysis based on these connection-level subgrouping profiles, a similar two-group subgrouping structure was again detected to be optimal (Supplementary Fig. S6b), and 91.3% of subjects (i.e., 63 out of 69) were detected to be within the same subgroup as they did in the original analysis based on the 81 subjects. When the within-subgroup brain–behavior relationship analysis was repeated, a highly consistent pattern with that shown in Figure 4 was observed; subjects in S1 showed brain–behavior relationships that were largely consistent with their neonatal functional connectivity pattern, but S2 not (Supplementary Fig. S6c). Moreover, the tri-network set of the limbic, the frontoparietal control, and the default-mode network also showed the most positive/negative correlations in S1 and S2, respectively (Supplementary Fig. S6c, red boxes).
Discussion
In this study, we demonstrated the presence of brain–behavior relationship heterogeneity in newborns, not only at connection level but also at the whole brain level. In particular, two subgroups of newborns were detected with similar brain functional connectivity patterns but different 4YR IQ outcomes and contrasting brain–behavior relationships. More intriguingly, the subgroup with higher 4YR IQ showed brain–behavior relationships that were largely congruent to its neonatal functional connectivity pattern while the subgroup with lower 4YR IQ performances not. These findings add new perspectives to our current understanding of intersubject variability during infancy and may inspire future developments of better brain-based prediction models for behavioral outcomes.
First, the detection of 334 heterogenous brain–behavioral relationships within a relatively homogenous neonate sample (i.e., all full term, <24-h NICU stay, and no parental disorder diagnosis) confirmed the presence of relational heterogeneities in human neonates with respect to their 4YR IQ outcomes (Fig. 1). Moreover, a robust subgrouping structure at the whole-brain level was also detectable showing much higher within-subgroup similarity than between-subgroup similarity based on their connection-level subgrouping profiles (Fig. 3). In fact, while their within-subgroup clustering profiles were positively correlated, their between-group profiles were largely negatively correlated, indicating highly contrasting brain–behavioral relationships. These findings suggest that infants within S1 and S2 likely employ divergent brain mechanisms for related domains of IQ development. Importantly, when examined against different control variables including gestation age at birth, postnatal age at scan, birth weight, sex, twin status, motion parameters, and scanner used to acquire data, no significant differences were observed (Fig. 3) between the two subgroups, indicating that the observed divergence in brain–behavioral relationships were not driven by these control variables.
Given their early appearance with minimal environmental exposures, the genetically driven prenatal processes likely contributed to such relational predispositions (Wang et al. 2018). However, adverse prenatal environmental risk factors, especially those directly interacting with fetal neural development (e.g., prenatal drug exposure [Grewen et al. 2015; Salzwedel et al. 2015, 2016], maternal obesity [Salzwedel et al. 2019a] /depression [Graham et al. 2015; Qiu et al. 2015], etc.), may have also influenced the emergence of such heterogeneity, either independently or interacting with genetic factors (Gao et al. 2019). Future studies explicitly monitoring these factors are needed to test these interactions. Regardless, our findings of heterogenous relationships between neonatal brain functional connectivity measures and 4-year cognitive outcomes in a relatively homogenous neonate sample support the critical importance of individualized/personalized perinatal brain mechanisms in long-term outcomes. More broadly, this finding is also consistent with one of the most notorious observations in human mental disorder research; when comparing a group of patients with mental disorders with normal controls, the differences in their brains, either structurally or functionally, are often subtle and can only be detected at the group level. However, individual patients often have dramatic and sometimes extreme behavioral manifestations that are easily discernable at individual level (Lombardo et al. 2015; Elton et al. 2016). Findings in this study suggest that it is likely not only categorical differences in brain metrics but also qualitative differences in brain–behavioral mechanisms that may have contributed to these mismatches. Overall, the observed heterogeneity suggests that future efforts exploring brain–behavior relationships in infants and older populations should not only focus on whole-group homogenous ones but also pay attention to subgroups with divergent relationships. Notably, pioneering previous studies have demonstrated the presence of such brain–behavior relationship-level heterogeneity against clinical diagnosis (e.g., ADHD [Elton et al. 2014], autism spectrum disorder [Elton et al. 2016], Turner’s syndrome [Xie et al. 2015], cerebral small vessel disease [Su et al. 2018]), sex (Jiang et al. 2019), and socioeconomic status (Noble et al. 2006), which is conceptually consistent with the current study. However, a major difference between most previous studies and the current one relates to the data-driven nature of the relationship-level heterogeneity detection approach employed in this study. As a result, the current approach does not rely on a prior hypothesis regarding the underlying dimension of heterogeneity. Instead, it data-drivenly detects heterogeneity within the examined population in question and could theoretically detect heterogeneity that either exists within an otherwise homogenous population or cuts across different dimensions. Therefore, applications of the currently proposed new concept and methods in future studies may provide novel insights in either subtyping within a diagnosis or detecting heterogeneity/homogeneity that cuts across different clinical diagnosis and/or other known phenotypes, a concept in line with the Research Domain Criteria framework (Ross and Margolis 2019).
A further analysis of the relationships between the brain–behavior associations and the neonatal functional connectivity patterns within S1 and S2 revealed intriguing patterns of how different brain–behavioral mechanisms may have contributed to the observed IQ differences. The current results revealed that S1’s brain–behavior relationships are highly consistent with its own functional connectivity development pattern at the neonatal stage while S2’s not. This observation suggests that through the employment of a set of brain–behavior mechanisms that are “conforming” to their own functional connectivity growth directions till the neonatal stage, infants in S1 are more likely to develop better behavioral outcomes. However, with much weaker or even opposite relationships between the two domains, infants in S2 would likely not be able to similarly “take advantage of” existing pre−/perinatal functional connectivity growth for better IQ outcomes later in development. Therefore, findings in this study not only revealed brain–behavior relational heterogeneity in neonates but also revealed a potential guiding principle for better behavioral outcomes, which underscores the importance of “alignment” between brain–behavior relationships and initial brain growth directions.
At network level, the tri-network set including the default-mode network (Raichle et al. 2001), the frontoparietal control network (Vincent et al. 2008), and the limbic network stands out to be the networks showing the strongest positive correlations between brain–behavior relationships and neonatal functional connectivity patterns in S1 (Fig. 4; Supplementary Fig. S4). Although to a lesser extent, these three networks also seem to show the most negative/weakest correlations in S2. Previous reports have consistently documented the early development and fast synchronization of the default-mode (Gao et al. 2009) and limbic networks (Alcauter et al. 2015a; Salzwedel et al. 2019b) in neonates, suggesting their critical importance in the early orchestration of functional circuits critical for self-referential, arousal, salience detection, and emotional regulations (Fransson 2005; Di Martino et al. 2008; Roy et al. 2009). Notably, the two networks, especially the default-mode network, are often observed to show anticorrelations with externally driven networks, including the frontoparietal control network, in adults studies, and this relationship has been promoted to be one of the guiding principles in adult brain functional organization (Fox et al. 2005; Fornito et al. 2012). Our previous findings also revealed the fast development of this between-network relationship during the first year of life, which may underlie the improvement of attention and self-awareness functions in infants (Gao et al. 2013). Overall, the three networks, including the default-mode, the limbic, and the frontoparietal control network, are likely involved in the critical development of salience detection/attention and socioemotional functions during infancy that pave the foundation for later intellectual and cognitive development (Atzil et al. 2018). The observation that these three networks stand out to show most significant brain–behavior associations and drive the positive correlations between brain–behavior relationships and neonatal functional connectivity patterns in the subgroup with superior 4YR IQ outcomes is thus not surprising and reinforces their fundamental importance in early brain and behavioral development.
There are several limitations in this study that deserves further consideration. The first one relates to the predefined number of two when delineating the subgrouping structure at the connection level. This number was chosen considering the relatively limited sample size (N = 81), so future studies with larger sample sizes are needed to explore if more subgroups exist at the connection level. As an initial try, we did perform an additional set of analysis to explore if more whole-brain level subgroups could be detected if we define three subgroups at the connection level. Our results showed that even based on the 262 connections showing significant three-group relationships at the connection level (Supplementary Fig. S7a), a two-subgroup whole-brain clustering structure was still optimal (Supplementary Fig. S7b). In fact, 72.8% of the subjects overlapped between the new subgrouping structure and the original one. However, the newly detected subgroups showed lower levels of within-group homogeneity (especially for S2, P < 1.0 × 10−10, Supplementary Fig. S7b) and less separation between the two subgroups (P < 1.0 × 10−10, Supplementary Fig. S7b) compared with our original results; thus we deemed this subgrouping results suboptimal to our main results. Therefore, it is likely that a larger sample size and more subjects with enhanced heterogeneity profiles (e.g., including preterm babies and/or other at-risk babies) are needed to define more subgroups in this population and this represents a future direction. Second, we chose to use the AAL template (Shi et al. 2011) because of its wide application in neuroimaging studies, which makes it easier for important future replication studies. However, we do recognize that different templates may affect the results. Therefore, we tested a second functionally defined, neonate-specific functional atlas (Shi et al. 2018) and highly consistent patterns between S1 and S2 were observed (Supplementary Fig. S4), supporting the robustness of the current findings against different template choices. Third, our sample consists of data acquired from two scanners, although we did not observe any difference in scanner distribution between the two detected subgroups (Fig. 3d), this may represent a source of potential heterogeneity in our original sample. Therefore, we repeated our analysis based only on subjects from the Allegra scanner (N = 69) and highly consistent results were obtained (Supplementary Fig. S6), supporting the robustness of the reported results against potential scanner-related differences. A fourth limitation relates to the relatively short rsfMRI scan time (i.e., 5 min) and data included in the analysis after motion scrubbing (i.e., 3 min). However, when we compared the functional connectivity matrices resulting from 3- and 4-min data in a subsample of 58 subjects with at least 4-min data remaining after scrubbing (Supplementary Fig. S8), the two group mean matrices showed highly consistent patterns, the individual connection values were highly correlated (r = 0.994, P < 1.0 × 10−10), and no connection showed significant difference even at the uncorrected P < 0.05 level. Nevertheless, future studies with longer rsfMRI scans are needed to independently validate the current findings. Finally, a general limitation in infant rsfMRI study relates to the lack of objective sleeping stage monitoring, which prevented us from investigating whether brain stage differences may have contributed to the identified relationship-level heterogeneity and subsequent subgrouping structures. Simultaneous electroencephalography-fMRI recordings could potentially shed more light into this confound, but great challenges remain in its practical applications in scanning naturally sleeping infants. Future efforts are needed to address this limitation.
Overall, in this study, we found two subgroups of neonates that do not differ in single-domain brain functional connectivity measures but significantly contrast each other in their brain–behavioral relationships and 4YR IQ outcomes, providing strong support for the presence of relational heterogeneity even in a relatively homogenous neonatal sample. Importantly, the higher performance group of S1 demonstrated brain–behavior mechanisms that were consistent with its own pre−/perinatal functional connectivity growth directions, which likely had contributed to the observed higher IQs while the lower-performance group of S2 showed no such relationships. Intriguingly, the tri-network set of the default-mode, the limbic, and the frontoparietal control network dominated these relationships, reinforcing the importance of these networks in early brain development (Gao et al. 2009; Alcauter et al. 2015a; Salzwedel et al. 2019b). These findings significantly improved our understandings of the intersubject variability during infancy. If validated, the finding of relational heterogeneity in this study may inspire novel brain-based prediction approaches through personalized, subgroup-specific prediction models, which would likely better utilize the rich and differential mechanistic information embedded in the newborn brain for better prediction.
Notes
Conflict of Interest: None declared.
Supplementary Material
Funding
This work was supported by National Institutes of Health (R34DA050255, R01DA042988 and R01DA043678 to W.G., R01MH064065 and R01HD05300 to J.H.G.) and Cedars-Sinai Precision Health Initiative Awards to W.G.
References
- Alcauter S, Lin W, Smith JK, Gilmore JH, Gao W. 2015a. Consistent anterior-posterior segregation of the insula during the first 2 years of life. Cereb Cortex. 25:1176–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcauter S, Lin W, Smith JK, Goldman BD, Reznick JS, Gilmore JH, Gao W. 2015b. Frequency of spontaneous BOLD signal shifts during infancy and correlates with cognitive performance. Dev Cogn Neurosci. 12:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcauter S, Lin W, Smith JK, Short SJ, Goldman BD, Steven Reznick J, Gilmore JH, Gao W. 2014. Development of thalamocortical connectivity during infancy and its cognitive correlations. J Neurosci. 34:9067–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzil S, Gao W, Fradkin I, Barrett LF. 2018. Growing a social brain. Nat Hum Behav. 2:624–636. [DOI] [PubMed] [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 29:162–173. [DOI] [PubMed] [Google Scholar]
- Davies DL, Bouldin DW. 1979. A cluster separation measure. IEEE Trans Pattern Anal Mach Intell. PAMI-1:224–227. [PubMed] [Google Scholar]
- Di Martino A, Scheres A, Margulies DS, Kelly AMC, Uddin LQ, Shehzad Z, Biswal B, Walters JR, Castellanos FX, Milham MP. 2008. Functional connectivity of human striatum: a resting state fMRI study. Cereb Cortex. 18:2735–2747. [DOI] [PubMed] [Google Scholar]
- Elton A, Alcauter S, Gao W. 2014. Network connectivity abnormality profile supports a categorical-dimensional hybrid model of ADHD. Hum Brain Mapp. 35:4531–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton A, Di Martino A, Hazlett HC, Gao W. 2016. Neural connectivity evidence for a categorical-dimensional hybrid model of autism spectrum disorder. Biol Psychiatry. 80:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Harrison BJ, Zalesky A, Simons JS. 2012. Competitive and cooperative dynamics of large-scale brain functional networks supporting recollection. Proc Natl Acad Sci U S A. 109:12788–12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. 2005. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 102:9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P. 2005. Spontaneous low-frequency BOLD signal fluctuations: an fMRI investigation of the resting-state default mode of brain function hypothesis. Hum Brain Mapp. 26:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Alcauter S, Elton A, Hernandez-Castillo CR, Smith JK, Ramirez J, Lin W. 2015. Functional network development during the first year: relative sequence and socioeconomic correlations. Cereb Cortex. 25:2919–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Elton A, Zhu H, Alcauter S, Smith JK, Gilmore JH, Lin W. 2014. Intersubject variability of and genetic effects on the brain’s functional connectivity during infancy. J Neurosci. 34:11288–11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Gilmore JH, Giovanello KS, Smith JK, Shen D, Zhu H, Lin W. 2011. Temporal and spatial evolution of brain network topology during the first two years of life. PLoS One. 6:e25278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Gilmore JH, Shen D, Smith JK, Zhu H, Lin W. 2013. The synchronization within and interaction between the default and dorsal attention networks in early infancy. Cereb Cortex. 23:594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Grewen K, Knickmeyer RC, Qiu A, Salzwedel A, Lin W, Gilmore JH. 2019. A review on neuroimaging studies of genetic and environmental influences on early brain development. Neuroimage. 185:802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Lin W, Grewen K, Gilmore JH. 2017. Functional connectivity of the infant human brain: plastic and modifiable. Neuroscientist. 23:169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Zhu H, Giovanello KS, Smith JK, Shen D, Gilmore JH, Lin W. 2009. Evidence on the emergence of the brain’s default network from 2-week-old to 2-year-old healthy pediatric subjects. Proc Natl Acad Sci. 106:6790–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JH, Knickmeyer RC, Gao W. 2018. Imaging structural and functional brain development in early childhood. Nat Rev Neurosci. 19:123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AM, Pfeifer JH, Fisher PA, Carpenter S, Fair DA. 2015. Early life stress is associated with default system integrity and emotionality during infancy. J Child Psychol Psychiatry Allied Discip. 56:1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewen K, Salzwedel AP, Gao W. 2015. Functional connectivity disruption in neonates with prenatal marijuana exposure. Front Hum Neurosci. 9:601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Calhoun VD, Fan L, Zuo N, Jung R, Qi S, Lin D, Li J, Zhuo C, Song M et al. . 2019. Gender differences in connectome-based predictions of individualized intelligence quotient and sub-domain scores. Cereb Cortex. 30:888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MV, Pierce K, Eyler LT, CarterBarnes C, Ahrens-Barbeau C, Solso S, Campbell K, Courchesne E. 2015. Different functional neural substrates for good and poor language outcome in autism. Neuron. 86:567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk C, Lugo-Candelas C, Trumpff C. 2019. Prenatal developmental origins of future psychopathology: mechanisms and pathways. Annu Rev Clin Psychol. 15:317–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble KG, Wolmetz ME, Ochs LG, Farah MJ, McCandliss BD. 2006. Brain-behavior relationships in reading acquisition are modulated by socioeconomic factors. Dev Sci. 9:642–654. [DOI] [PubMed] [Google Scholar]
- O’Donnell KJ, Meaney MJ. 2017. Fetal origins of mental health: the developmental origins of health and disease hypothesis. Am J Psychiatry. 174:319–328. [DOI] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 59:2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. 2014. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 84:320–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Anh TT, Li Y, Chen H, Rifkin-Graboi A, Broekman BFP, Kwek K, Saw SM, Chong YS, Gluckman PD et al. . 2015. Prenatal maternal depression alters amygdala functional connectivity in 6-month-old infants. Transl Psychiatry. 5:e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. 2001. A default mode of brain function. Proc Natl Acad Sci U S A. 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roid GH. 2003. Stanford-Binet intelligence scales. 5th ed. Itasca (IL): Riverside Publishing. [Google Scholar]
- Ross CA, Margolis RL. 2019. Research domain criteria: strengths, weaknesses, and potential alternatives for future psychiatric research. Mol Neuropsychiatry. 5:218–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AK, Shehzad Z, Margulies DS, Kelly AMC, Uddin LQ, Gotimer K, Biswal BB, Castellanos FX, Milham MP. 2009. Functional connectivity of the human amygdala using resting state fMRI. Neuroimage. 45:614–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushe TM, Rifkin L, Stewart AL, Townsend JP, Roth SC, Wyatt JS. 2001. Neuropsychological outcome at adolescence of very preterm birth and its relation to brain structure. Dev Med Child Neurol. 43:226–233. [DOI] [PubMed] [Google Scholar]
- Salzwedel AP, Gao W, Andres A, Badger TM, Glasier CM, Ramakrishnaiah RH, Rowell AC, Ou X. 2019a. Maternal adiposity influences neonatal brain functional connectivity. Front Hum Neurosci. 12:514. doi: 10.3389/fnhum.2018.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel AP, Grewen KM, Goldman BD, Gao W. 2016. Thalamocortical functional connectivity and behavioral disruptions in neonates with prenatal cocaine exposure. Neurotoxicol Teratol. 56:16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel AP, Grewen KM, Vachet C, Gerig G, Lin W, Gao W. 2015. Prenatal drug exposure affects neonatal brain functional connectivity. J Neurosci. 35:5860–5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzwedel AP, Stephens RL, Goldman BD, Lin W, Gilmore JH, Gao W. 2019b. Development of amygdala functional connectivity during infancy and its relationship with 4-year behavioral outcomes. Biol Psychiatry Cogn Neurosci Neuroimaging. 4:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Salzwedel AP, Lin W, Gilmore JH, Gao W. 2018. Functional brain parcellations of the infant brain and the associated developmental trends. Cereb Cortex. 28:1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, Shen D. 2011. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One. 6:e18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE et al. . 2004. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 23:S208–S219. [DOI] [PubMed] [Google Scholar]
- Su N, Liang X, Zhai FF, Zhou LX, Ni J, Yao M, Tian F, Zhang SY, Jin ZY, Cui LY et al. . 2018. The consequence of cerebral small vessel disease: linking brain atrophy to motor impairment in the elderly. Hum Brain Mapp. 39:4452–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tau GZ, Peterson BS. 2010. Normal development of brain circuits. Neuropsychopharmacology. 35:147–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Kahn I, Snyder AZ, Raichle ME, Buckner RL. 2008. Evidence for a frontoparietal control system revealed by intrinsic functional connectivity. J Neurophysiol. 100:3328–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shen M, Guillaume B, Chong YS, Chen H, Fortier MV, Meaney MJ, Qiu A. 2018. FKBP5 moderates the association between antenatal maternal depressive symptoms and neonatal brain morphology. Neuropsychopharmacology. 43:564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Zhang Z, Zhao Q, Zhang J, Zhong S, Bi Y, He Y, Pan H, Gong G. 2015. The effects of X chromosome loss on neuroanatomical and cognitive phenotypes during adolescence: a multi-modal structural MRI and diffusion tensor imaging study. Cereb Cortex. 25:2842–2853. [DOI] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR et al. . 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


