Abstract
Objective
The development of machine learning (ML) algorithms to address a variety of issues faced in clinical practice has increased rapidly. However, questions have arisen regarding biases in their development that can affect their applicability in specific populations. We sought to evaluate whether studies developing ML models from electronic health record (EHR) data report sufficient demographic data on the study populations to demonstrate representativeness and reproducibility.
Materials and Methods
We searched PubMed for articles applying ML models to improve clinical decision-making using EHR data. We limited our search to papers published between 2015 and 2019.
Results
Across the 164 studies reviewed, demographic variables were inconsistently reported and/or included as model inputs. Race/ethnicity was not reported in 64%; gender and age were not reported in 24% and 21% of studies, respectively. Socioeconomic status of the population was not reported in 92% of studies. Studies that mentioned these variables often did not report if they were included as model inputs. Few models (12%) were validated using external populations. Few studies (17%) open-sourced their code. Populations in the ML studies include higher proportions of White and Black yet fewer Hispanic subjects compared to the general US population.
Discussion
The demographic characteristics of study populations are poorly reported in the ML literature based on EHR data. Demographic representativeness in training data and model transparency is necessary to ensure that ML models are deployed in an equitable and reproducible manner. Wider adoption of reporting guidelines is warranted to improve representativeness and reproducibility.
Keywords: demographic data; machine learning; electronic health record; clinical decision support, bias, transparency
INTRODUCTION
The ubiquity of electronic health records (EHRs) has facilitated the development of machine learning (ML) models to assist with clinical decision-making for diagnosis, treatment, and prognosis.1 The use of ML models in clinical practice has increased in recent years, and specific models have approached the performance of expert clinicians in specialties such as pathology and radiology.2–4 However, questions remain as to whether these models will generalize more broadly and deliver benefit to diverse populations.5,6 As ML tools proliferate across clinical settings, it is important to understand potential demographic biases underlying model development.
Recently, reports have questioned whether ML models in healthcare might perpetuate discrimination if trained on historical data—which is often poorly representative of broader populations.7 Representativeness may be a larger problem in countries with significant health disparities on the basis of demographics, such as the United States (US). If ML models are not trained on populations for which they will be applied, these advances may further perpetuate disparities and fail to demonstrate external validity in broader patient communities.8 Recent evidence highlights this issue, particularly in the US, where ML-drive decision support often falls short for nonwhite populations, likely due to a lack of diversity in the training data.8,9
Another challenge in the EHR ML literature is the reproducibility of results.10–13 This may be further exacerbated by the lack of clarity related to model development and evaluation. Indeed, recent studies showed an alarming lack of reproducibility using the same data, suggesting that public sharing of model code could enhance reproducibility.14,15 Furthermore, the paucity of shared code means that many ML models are not validated on external health systems.13 Recently, 2 evaluations of ML models on EHR data highlighted important gaps related to these issues, including limitations such as single-center data collection and inadequate reporting of missing data as well as concerns regarding the clinical impact of models.16,17 Promoting access to ML models’ code and details about the training data is crucial to advance the applicability and generalizability of ML in biomedicine.
The goal of the current study is to evaluate the reporting of demographic data in ML models using EHRs and the availability of information needed for reproducibility. Our work specifically focuses on whether studies disclose and/or include the demographic variables in the models, validation protocols, and reproducibility aspects of the models. To identify and provide best practices for designing useful ML studies, the transparent reporting and documentation of key demographic variables as well as reproducibility and generalizability safeguards are necessary to ensure the equitable implementation of these technologies.
MATERIALS AND METHODS
Literature selection
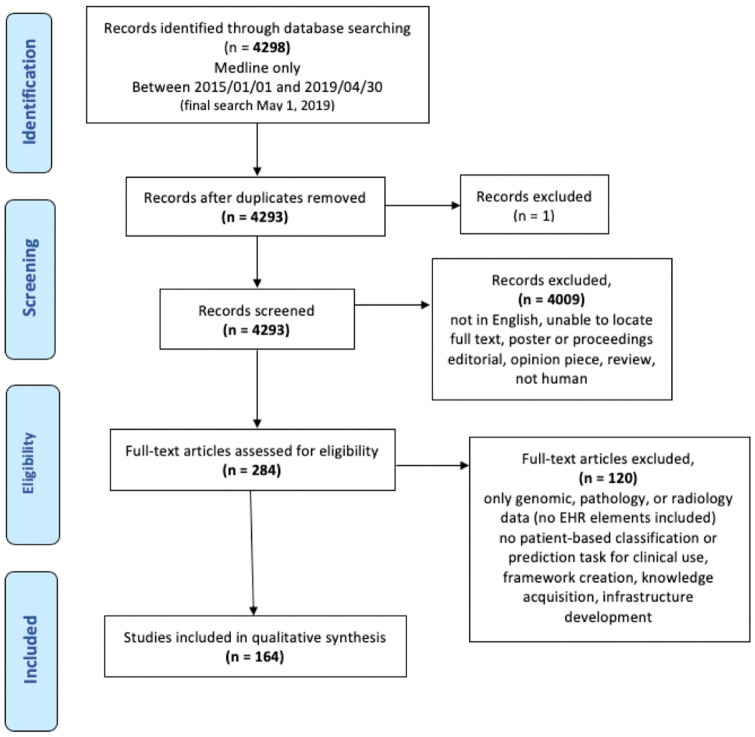
We systematically searched PubMed (Medline) for studies applying ML models to support clinical decision-making using EHR data—specifically structured and unstructured EHR data (eg, laboratory tests, vitals, medications, diagnosis codes, clinical notes, etc.). We limited our search to papers published between January 1, 2015 and April 30, 2019 (final search completed on May 1, 2019). We included English-language studies using EHR data as their primary data source for development of a prediction or classification model. Papers were excluded if they used only imaging or genomic data that were not linked to patient demographics. Studies using statistical regression models, such as logistic regression without any train/test splitting or cross validation, were also excluded because they are generally constructed based on theory and assumptions, do not learn automatically from data, and hence are not designed as a machine learning model framework. The complete search strategy and inclusion/exclusion criteria are shown in Figure 1.18 Four reviewers (SB, EMC, RS, JALV) independently reviewed the studies (each study was assessed by 2 reviewers), reaching a consensus on all eligible studies after 2 adjudication meetings. Inclusion and exclusion criteria are highlighted in Figure 1. Our search strategy included MeSH terms and keywords that appeared in the title or abstract. Our complete query can be found in the Supplementary Material S1.
Figure 1.
Study selection flowchart.
Data collection, extraction, and analysis
Eligible articles were downloaded, and the full text was independently assessed by the same 2 reviewers who performed the first selection. A randomly selected subset of 20 articles was screened by all reviewers, and disagreements were discussed in a focus group meeting with an ML expert.
Of the 4298 retrieved articles, 164 matched the inclusion criteria and were further analyzed. Papers were evaluated on 5 aspects: 1) study design, which includes sample size, clinical setting, disease condition; 2) reporting of demographic variables, specifically gender, age, socioeconomic status, race/ethnicity, and any other sensitive demographic indicators; 3) comparisons of overall demographics of the samples evaluated in type 2 diabetes mellitus (T2DM) studies to National Health and Nutrition Examination Survey (NHANES) population to provide a use case demonstrating US population representation; 4) model validation setting, including internal and external validation; and 5) data sharing and code open-sourcing.
A standardized form was used for data extraction from each study, including authors, year, journal type (clinical, biomedical informatics, and other), clinical setting (inpatient, outpatient, emergency department, intensive care unit), clinical condition (eg, oncology, cardiovascular disease, diabetes) and sample size. This information was collected from the abstract, main text, and any supplementary material available.
Evaluation of demographic variables
Demographic variables were evaluated across 3 domains: 1) variables reported for the study population; 2) variables included as features in the model; and 3) representativeness of the training data to the target population. All categories were presented as frequencies and percentages.
Training data characteristics
Comparison with NHANES population
In studies where race/ethnicity and gender were reported, we combined populations from all US studies to generate the average demographic statistics of all ML studies included in this report and compared it to the NHANES population. For the calculation of gender distributions, we excluded 9 studies focused on breast cancer (n = 3), obstetric patients (n = 4) and systemic lupus erythematosus (n = 2). Age distribution among studies was not compared due to the inconsistent reporting formats (mean, median or frequencies). P values were calculated using the 1 sample test for proportions.
T2DM use case
To investigate whether the training data of ML models used is representative of the target population, we evaluated papers focused on type 2 Diabetes Mellitus (T2DM) because data exist regarding the real distribution of T2DM in the US population.19 We compared the overall race/ethnicity and gender percentages of T2DM studies included in our study to the T2DM patients within the NHANES population using sample weights to represent overall population distribution.
Validation protocols
Validation protocols of the papers were evaluated based on the following definitions. Internal validation (including cross-validation) is defined as the performance of the model in verifying that the conclusions drawn from a subset of the collected data are consistent in the remaining data of same origin.20 External validation of the models uses unseen data that are entirely separate from the data used for model development, verifying that the conclusions drawn from the collected data were based on a new dataset of different origin to assess the model’s generalizability.21
Data sharing and code sourcing
Evaluation of data and code availability was performed in 2 steps: 1) we searched for the following keywords: “GitHub,” “GitLab,” “Bitbucket,” “open source,” “available,” “availability,” “data,” and “code” in the full text; 2) wherever none of the keywords were found, we manually reviewed the article text for any mention of data or code availability. In addition, missing data reporting in the studies were evaluated as reported or/and imputed.
RESULTS
A total of 164 studies were eligible for inclusion in the analysis (Supplementary Table S1). Of these, 74 (45%) articles were from clinical journals, 42 (26%) from medical informatics journals and 48 (30%) from other journals. The number of ML papers using EHR data increased over the course of the 5-year inclusion period. Studies originated from EHRs in 16 different countries, with the majority conducted in the US (n = 121, 74%) and China (n = 13, 8%). The sample size in the studies ranged between 73 and 4 637 297 patients, with a median of 1237. The studies reviewed differed in their targeted age groups. A majority of studies, 133 (81%), were conducted in adult populations, whereas 15 (9%) addressed pediatric cohorts, and 13 (8%) included both adults and children. Two (1%) studies focused only on geriatric patients and 1 (0.6%) study focused on neonates.
A large number of studies (n = 57, 35%) used EHR data from inpatient settings exclusively; followed by 36 (22%) studies using outpatient data only; 15 (9%) using intensive care unit (ICU) data only; and 10 (6%) emergency department data only. Additionally, the included studies considered a variety of clinical conditions and diseases with cardiovascular disease being the most frequently predicted condition covered by 23 (14%) of the articles and followed by sepsis in 10 (6%) articles.
Demographic variable reporting
The reporting of sensitive demographic variables is summarized in Table 1. There was heterogeneity in whether, and how, the distribution of demographic variables was defined and reported. For instance, age distribution of a cohort was reported either as a range, median or mean. Relevant demographic variables were absent from several papers, yet a large number of studies reported gender (n = 124, 76%) and age (n = 129, 79%) distributions. However, race/ethnicity and socioeconomic status were not reported in 105 (64%) and 150 (92%) studies, respectively. The distribution of other sensitive demographic variables was rarely reported.
Table 1.
Demographic variables reported in the studies
| Variables N (%) | Distribution reported |
Distribution not reported |
||||
|---|---|---|---|---|---|---|
| Total | Not included in the model | Included in the model | Total | Not included in the model | Included in the model | |
| Gender | 124 (76) | 30 (18) | 94 (57) | 40 (24) | 20 (12) | 20 (12) |
| Age | 129 (79) | 16 (10) | 113 (69) | 35 (21) | 12 (7) | 23 (14) |
| Race/ethnicity | 59 (36) | 18 (11) | 41 (25) | 105 (64) | 86 (52) | 19 (12) |
| Socio-economic status | 14 (8) | 1 (1) | 13 (8) | 150 (92) | 145 (88) | 5 (3) |
| Other sensitive demographic variables | ||||||
| Marital status | 9 (6) | – | 9 (6) | 155 (95) | 149 (91) | 6 (4) |
| Zip code | – | – | 164 (100) | 160 (98) | 4 (2) | |
| Employment status | 1 (1) | – | 1 (1) | 163 (99) | 160 (98) | 3 (2) |
| Education level | 2 (1) | – | 2 (1) | 162 (99) | 160 (98) | 2 (1) |
| Religion | 1 (1) | – | 1 (1) | 163 (99) | 161 (98) | 2 (1) |
| Childhood status | – | – | 1 (1) | 163 () | 1 (1) | |
| Housing status | – | – | 1 (1) | 163 () | 1 (1) | |
Sensitive variables as model inputs
The inclusion of demographic variables as model inputs varied across studies (Table 1). Age and gender were often included as model features, regardless of whether their distribution was reported. For instance, 23 studies (14%) did not report the age distribution of the training dataset, yet age was declared as an input feature in their model. Race/ethnicity was reported and included as a feature in 41 (25%) studies and socioeconomic status in 13(8%) studies. Other sensitive variables were rarely included as features in the model.
Training data characteristics
Comparison with the NHANES population
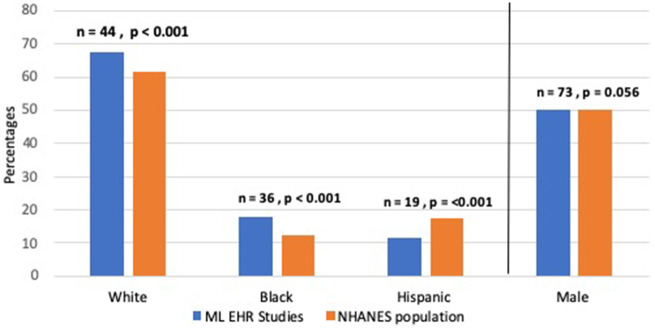
Figure 2 shows the overall percentages of gender and race/ethnicity in US papers (121, 74%). The percentage of male subjects varied between 27% and 71% with a mean of 50%, which is close to the population rate of 49% in NHANES (P = .056). Race/ethnicity percentages show slight differences compared to NHANES; the percentage of White subjects was higher in ML studies vs NHANES (68% versus 62%, respectively P < .001). Yet Black subjects were overrepresented (18% versus 12%, respectively P < .001) and Hispanics underrepresented in ML models compared to NHANES (11% versus 18%, respectively P < .001).
Figure 2.
Percentages of gender and race/ethnicity variables in ML studies compared to the NHANES population.
T2DM use case
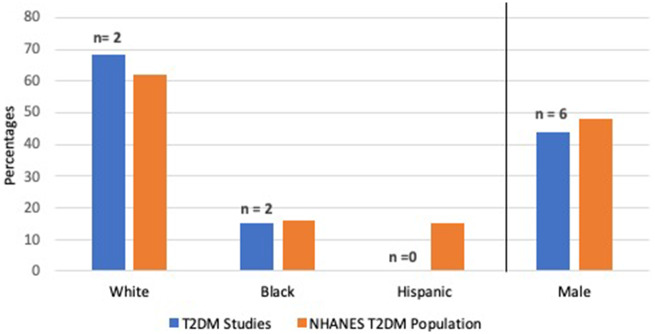
Figure 3 shows the overall percentages of gender and race/ethnicity in articles related to T2DM that were US-based (7 articles, 5%). Gender distribution was reported in 6 (86%) studies whereas race/ethnicity was reported in 2 (29%) studies, although neither of these included the percentage of Hispanic T2DM patients. Due to the small number of studies we did not provide P values for comparisons.
Figure 3.
Percentages of 2 demographic variables in ML studies on T2DM cohorts compared to the NHANES T2DM population.
Validation protocols
The generalizability of clinical studies can be tested via internal validation and/or external validation. In 147 (88%) studies, only internal validation was performed; the remaining 12% of studies showed results for external validation. In addition, in 10 (6%) studies, the validation results for all the data without splitting it into subgroups (train, test, or leave-one-out) were given.
Data and code availability
Among 164 studies, 12 (7%) used a publicly available dataset, 8 (5%) shared their data in an anonymized (deidentified) format and 1 study shared an example subset of the real-world data. Among studies using publicly available datasets, 9 (6%) used the MIMIC dataset, a freely available dataset containing deidentified health data associated with over 55 000 intensive care unit admissions.22 Among all studies, 82 (50%) reported missing data, of which 37 (22%) used imputation techniques (eg, mean/median imputation or regression imputation) while 8 (5%) excluded those cases with missing data. In 27 (17%) studies, authors open-sourced their code through online repositories such as GitHub or GitLab.
DISCUSSION
The proliferation of ML models offers novel opportunities for improvement in clinical decision-making. However, demographic descriptions of training data are poorly reported. This study demonstrates that key demographic variables such as race/ethnicity and socioeconomic status are often not reported in the clinical ML literature. Moreover, ML models were rarely validated in external populations and authors rarely share the code associated with published studies. This work highlights the need for improved reporting standards along with code-sharing across the EHR research community.
In the studies included in this report, data essential to interpret the outcome and intended target populations of ML models were inconsistently described. When demographic information was reported, we observed trends similar to previous analyses,23 with an unbalanced distribution of race/ethnicity. While we do not expect every clinical cohort to match the general population, differences in population distributions must be transparent, particularly when applications are developed with the intention to deploy in populations outside their training population. Transparency of demographic representation in the training data is essential for end users intending to use the model to guide clinical decisions.21,24
To assess population representativeness of ML models’ training population, direct access to study data is often necessary, as is sometimes done for randomized controlled trials.19,22 Other approaches include qualitative analyses based on consensus of authors for each target population.25 However, this approach also requires disclosure of demographic data and inclusion/exclusion criteria. In an attempt to appraise ML models sample representative of the population to which they will be applied, we evaluated T2DM models as an example. We found only 2 of 7 articles include population demographics and, in these papers, White and Black populations included in the training data were similar to the US T2DM population. However, neither study reported Hispanics in their training data. This raises concern given Hispanics higher prevalence and complication rates for T2DM compared to Whites.26 It is unclear how T2DM models trained without representation of Hispanics would fair more broadly in nationwide healthcare systems to benefit diverse populations. The inability to quantify sample representativeness in ML models demonstrates a strong need for a framework to evaluate sample representativeness to ensure appropriate downstream applications of the ML models.
It is important to remember that EHRs were not intended for secondary analyses; missing and inaccurate data are common.27 Clearly, many sensitive variables, such as socioeconomic or housing status may not be available in EHRs, hence the low reporting we found on these variables is not surprising. In addition, missing data can substantially affect the results of predictive models and in our analyses half of the studies included reported missingness and of those, 22% used imputation techniques to address missingness. While not all data can be imputed, a clear statement of missingness and imputation methods should be provided. While imperfect for secondary analyses, EHRs are a dominant data source for clinical decision support. Their unbiased application is dependent on the representativeness of the training data to the applied population, a measure that can only be determined through the thoughtful comparison of sensitive variables across populations.
The performance of any predictive model attempting to influence clinical decision-making broadly depends on its reliability and generalizability to other settings and populations than those represented in the training data.21,28 However, many of the studies screened exclusively used data from a single facility, and the models were rarely validated in external populations. This compounds the aforementioned nonrepresentativeness of the majority of models and may enhance the risk of generalization failure if these models were utilized in more diverse patient cohorts. Indeed, lack of data sharing or a deidentified EHR data source for researchers to validate and evaluate the models could help mitigate this problem. However, data are not being shared across systems or entities, in part due to a lack of interoperability and in part due to a lack of appropriate incentives for data sharing.29 There is a need to develop strategies and policies to facilitate data sharing and regulations are moving in this direction, but clarification on what data can and should be made available to improve the applicability of ML models is needed.23,30
To facilitate broader dissemination of ML technologies, published models should strive to provide their source code where possible. Wiens et al31 presented a framework for accelerating the translation of ML-based interventions in healthcare and suggested that it is good practice to share code, packages, and inputs, as well as supporting documentation. The lack of models available for open-source review also presents a risk that the code underlying the model may be tailored specifically to attributes of the training data and therefore could not be extrapolated effectively to other settings.
Our work suggests that reporting guidelines must be adapted to studies proposing new ML models to clearly elaborate on the presence or absence of sensitive demographic variables31,32 as we and others have highlighted.10,33,34 Additionally, efforts need to be made to explicitly report the representativeness of training data to the applied population. When feasible, all models developed need to be externally validated on datasets containing relevant variables and outcomes of interest. Finally, similar to preregistration of clinical trials serving to ensure their accountability, adopters, journals, regulators, and other stakeholders should push for open-sourcing of code as a prerequisite to its broad scale acceptability.
This empirical assessment of ML studies has certain limitations. First, the studies included were limited to journal articles published in English and indexed only in Medline. A more robust search for articles disseminated in other sources, such as ACM digital library and IEEE eXplore, could extend the scope of this work and merit further investigation. Second, some studies included may be proof of concept and not intended for implementation in a clinical setting. However, the plurality of the articles included in this study were published in clinical journals with a goal to use their approaches in clinical decision-making. Third, though we aimed to include studies that would be relevant to the general populations when reporting descriptive statistics, caution should be employed in interpreting our results. Some study populations may genuinely deviate from the general population and the goal of representativeness needs to be carefully defined on a case by case basis. Finally, lack of sufficient reporting leaves substantial ambiguity for some variables considered in these studies. Better reporting standards would address this ambiguity.
CONCLUSION
Our empirical assessment of a large number of studies provides insights into whether demographic data has been sufficiently considered and/or appropriately managed when developing ML models. We found demographic characteristics of training data are poorly reported in the ML EHR literature. Additionally, external model validation and code open-sourcing is infrequently performed. These factors limit the generalizability and reproducibility of ML research. Wider adoption of reporting guidelines with an explicit focus on demographic inclusion and equity is critical for the safe and equitable deployment of ML models in practice.
FUNDING
Research reported in this publication was supported by Stanford’s Presence Center’s AI in Medicine: Inclusion & Equity Initiative. The content is solely the responsibility of the authors and does not necessarily represent the official views of Stanford University.
AUTHOR CONTRIBUTIONS
MGS and THB conceived the project. THB and JPAI directed the project. SB, MGS, EMC, and THB chose the variables of interest, decided on the inclusion and exclusion criteria for participation in the study, and SB, EMC, SR, and JALV collected the data. SB and THB analyzed and evaluated the data and take responsibility for both the integrity of the data and the accuracy of the data analysis. SB drafted the original article and all authors reviewed and approved the manuscript. THB takes responsibility for the integrity of the data and the accuracy of the data analysis and provided administrative, technical, and material support.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
MGS is currently an employee of Google Health; however, this project represents prior work at Stanford. All other authors have no conflict of interest to declare.
Supplementary Material
REFERENCES
- 1. Rothman B, Leonard JC, Vigoda MM.. Future of electronic health records: implications for decision support. Mt Sinai J Med 2012; 79 (6): 757–68. [DOI] [PubMed] [Google Scholar]
- 2. Zhang Z, Chen P, McGough M, et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat Mach Intell 2019; 1 (5): 236–45. [Google Scholar]
- 3. Rajpurkar P, Irvin J, Ball RL, et al. Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 2018; 15 (11): e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018; 2 (3): 158–64. [DOI] [PubMed] [Google Scholar]
- 5. Saria S, Butte A, Sheikh A.. Better medicine through machine learning: what’s real, and what’s artificial? PLoS Med 2018; 15 (12): e1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G.. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med 2018; 178 (11): 1544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Char DS, Shah NH, Magnus D.. Implementing machine learning in health care—addressing ethical challenges. N Engl J Med 2018; 378 (11): 981–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cahan EM, Hernandez-Boussard T, Thadaney-Israni S, Rubin DL.. Putting the data before the algorithm in big data addressing personalized healthcare. NPJ Digit Med 2019; 2: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adamson AS, Smith A.. Machine learning and health care disparities in dermatology. JAMA Dermatol 2018; 154 (11): 1247–8. [DOI] [PubMed] [Google Scholar]
- 10. Moons KGM, Wolff RF, Riley RD, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med 2019; 170 (1): W1–33. [DOI] [PubMed] [Google Scholar]
- 11. Cowley LE, Farewell DM, Maguire S, Kemp AM.. Methodological standards for the development and evaluation of clinical prediction rules: a review of the literature. Diagn Progn Res 2019; 3 (1): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Munafò MR, Nosek BA, Bishop DVM, et al. A manifesto for reproducible science. Nat Hum Behav 2017; 1 (1): 0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vollmer S, Mateen BA, Bohner G, et al. Machine learning and AI research for patient benefit: 20 critical questions on transparency, replicability, ethics and effectiveness. arXiv preprint arXiv: 181210404; 2018. [DOI] [PMC free article] [PubMed]
- 14. Sanchez-Pinto LN, Luo Y, Churpek MM.. Big data and data science in critical care. Chest 2018; 154 (5): 1239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Price WN. Medical Malpractice and Black-Box Medicine. In: Cohen I, Lynch H, Vayena E, Gasser U, eds. Big Data, Health Law, and Bioethics. Cambridge: Cambridge University Press; 2018: 295–306. [Google Scholar]
- 16. Goldstein BA, Navar AM, Pencina MJ, Ioannidis JP.. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc 2017; 24 (1): 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiao C, Choi E, Sun J.. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc 2018; 25 (10): 1419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol 2009; 62 (10): e1–34. [DOI] [PubMed] [Google Scholar]
- 19. He Z, Wang S, Borhanian E, Weng C.. Assessing the collective population representativeness of related type 2 diabetes trials by combining public data from ClinicalTrials.gov and NHANES. Stud Health Technol Inform 2015; 216: 569–73. [PMC free article] [PubMed] [Google Scholar]
- 20. Steyerberg EW, Harrell FE, Borsboom GJ, Eijkemans MJ, Vergouwe Y, Habbema JD.. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol 2001; 54 (8): 774–81. [DOI] [PubMed] [Google Scholar]
- 21. Riley RD, Ensor J, Snell KI, et al. External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ 2016; 353: i3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He Z, Ryan P, Hoxha J, et al. Multivariate analysis of the population representativeness of related clinical studies. J Biomed Inform 2016; 60: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Obermeyer Z, Powers B, Vogeli C, Mullainathan S.. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019; 366 (6464): 447–53. [DOI] [PubMed] [Google Scholar]
- 24. Sendak MP, Gao M, Brajer N, Balu S.. Presenting machine learning model information to clinical end users with model facts labels. NPJ Digit Med 2020; 3: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kennedy-Martin T, Curtis S, Faries D, Robinson S, Johnston J.. A literature review on the representativeness of randomized controlled trial samples and implications for the external validity of trial results. Trials 2015; 16 (1): 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention. Hispanic/Latino Americans and type 2 diabetes In: Centers for Disease Control and Prevention, ed. Atlanta, GA: US Department of Health & Human Services, HHS; 2019. https://www.cdc.gov/diabetes/library/features/hispanic-diabetes.html Accessed May 2, 2020. [Google Scholar]
- 27. Hersh WR, Weiner MG, Embi PJ, et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care 2013; 51 (8 Suppl 3): S30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou H, Della PR, Roberts P, Goh L, Dhaliwal SS.. Utility of models to predict 28-day or 30-day unplanned hospital readmissions: an updated systematic review. BMJ Open 2016; 6 (6): e011060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holmgren AJ, Patel V, Adler-Milstein J.. Progress in interoperability: measuring US hospitals’ engagement in sharing patient data. Health Aff (Millwood) 2017; 36 (10): 1820–7. [DOI] [PubMed] [Google Scholar]
- 30. Hripcsak G, Duke JD, Shah NH, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 2015; 216: 574–8. [PMC free article] [PubMed] [Google Scholar]
- 31. Wiens J, Saria S, Sendak M, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019; 25 (9): 1337–40. [DOI] [PubMed] [Google Scholar]
- 32. Benchimol EI, Smeeth L, Guttmann A, et al. RECORD Working Committee. The Reporting of studies Conducted using Observational Routinely collected health Data (RECORD) statement. PLoS Med 2015; 12 (10): e1001885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagendran M, Chen Y, Lovejoy CA, et al. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ 2020; 368: m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hernandez-Boussard T, Bozkurt S, Ioannidis J, Shah N.. MINIMAR: MINimum Information for Medical AI Reporting—developing reporting standards for artificial intelligence in healthcare. J Am Med Inform Assoc 2020. doi: 10.1093/jamia/ocaa088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Among 164 studies, 12 (7%) used a publicly available dataset, 8 (5%) shared their data in an anonymized (deidentified) format and 1 study shared an example subset of the real-world data. Among studies using publicly available datasets, 9 (6%) used the MIMIC dataset, a freely available dataset containing deidentified health data associated with over 55 000 intensive care unit admissions.22 Among all studies, 82 (50%) reported missing data, of which 37 (22%) used imputation techniques (eg, mean/median imputation or regression imputation) while 8 (5%) excluded those cases with missing data. In 27 (17%) studies, authors open-sourced their code through online repositories such as GitHub or GitLab.