Abstract
Objective
In applying machine learning (ML) to electronic health record (EHR) data, many decisions must be made before any ML is applied; such preprocessing requires substantial effort and can be labor-intensive. As the role of ML in health care grows, there is an increasing need for systematic and reproducible preprocessing techniques for EHR data. Thus, we developed FIDDLE (Flexible Data-Driven Pipeline), an open-source framework that streamlines the preprocessing of data extracted from the EHR.
Materials and Methods
Largely data-driven, FIDDLE systematically transforms structured EHR data into feature vectors, limiting the number of decisions a user must make while incorporating good practices from the literature. To demonstrate its utility and flexibility, we conducted a proof-of-concept experiment in which we applied FIDDLE to 2 publicly available EHR data sets collected from intensive care units: MIMIC-III and the eICU Collaborative Research Database. We trained different ML models to predict 3 clinically important outcomes: in-hospital mortality, acute respiratory failure, and shock. We evaluated models using the area under the receiver operating characteristics curve (AUROC), and compared it to several baselines.
Results
Across tasks, FIDDLE extracted 2,528 to 7,403 features from MIMIC-III and eICU, respectively. On all tasks, FIDDLE-based models achieved good discriminative performance, with AUROCs of 0.757–0.886, comparable to the performance of MIMIC-Extract, a preprocessing pipeline designed specifically for MIMIC-III. Furthermore, our results showed that FIDDLE is generalizable across different prediction times, ML algorithms, and data sets, while being relatively robust to different settings of user-defined arguments.
Conclusions
FIDDLE, an open-source preprocessing pipeline, facilitates applying ML to structured EHR data. By accelerating and standardizing labor-intensive preprocessing, FIDDLE can help stimulate progress in building clinically useful ML tools for EHR data.
Keywords: machine learning, electronic health records, preprocessing pipeline
INTRODUCTION
To date, researchers have successfully leveraged electronic health record (EHR) data and machine learning (ML) tools to build patient risk stratification models for many adverse outcomes, including healthcare–associated infections,1–3 sepsis and septic shock,4,5 acute respiratory distress syndrome,6 and acute kidney injury,7,8 among others.9 Though these works take advantage of ML techniques, prior to applying ML, substantial effort must be devoted to preprocessing. EHR data are messy, often consisting of high-dimensional, irregularly sampled time series with multiple data types and missing values. Transforming EHR data into feature vectors suitable for ML techniques requires many decisions, such as what input variables to include, how to resample longitudinal data, and how to handle missing data, among many others.
Currently, EHR data preprocessing is largely ad hoc and can vary widely between studies. For example, on the same task of predicting in-hospital mortality, Silva et al10 (in PhysioNet/CinC challenge 2012) used 41 input variables, while Harutyunyan et al11 used 17 input variables. To handle missing values, Purushotham et al12 used mean imputation, whereas Harutyunyan et al11 used prespecified “normal” values. This heterogeneity in the steps preceding the application of ML makes it difficult to meaningfully compare different ML algorithms and ensure reproducibility. To this end, researchers have proposed preprocessing pipelines, such as MIMIC-Extract.13 However, such pipelines make assumptions that do not necessarily generalize to new data sets. As the role of ML in health care expands, there is an increasing need for the systematization of generalizable preprocessing techniques for EHR data.
In an effort to speed up and standardize the preprocessing of EHR data, we propose FIDDLE (Flexible Data-Driven Pipeline), which systematically transforms structured EHR data into representations that can be used as inputs to ML algorithms. Our proposed approach is largely data-driven and incorporates good practices from the literature. FIDDLE was designed to work out of the box with reasonable default settings, but it also allows users to customize certain arguments and incorporate task-specific domain knowledge. While it applies broadly to structured clinical data, we evaluated FIDDLE through a proof-of-concept experiment in the context of MIMIC-III and the eICU Collaborative Research Database: 2 different but widely used large-scale EHR data sets that represent health data collected in intensive care units (ICUs) throughout the United States.14,15 We demonstrate the pipeline’s utility and flexibility across a variety of clinically important prediction tasks and several common ML algorithms. The code for FIDDLE and all analyses is open source (https://gitlab.eecs.umich.edu/MLD3/FIDDLE). Though FIDDLE is not a one-size-fits-all solution to preprocessing and further work is needed to test the limits of its generalizability, it can help accelerate ML research applied to EHR data. By reducing the time and effort spent on labor-intensive data preprocessing steps, FIDDLE streamlines the process. Moreover, it provides an easily shareable and reproducible baseline, presenting researchers with a quick and reasonable starting point.
MATERIALS AND METHODS
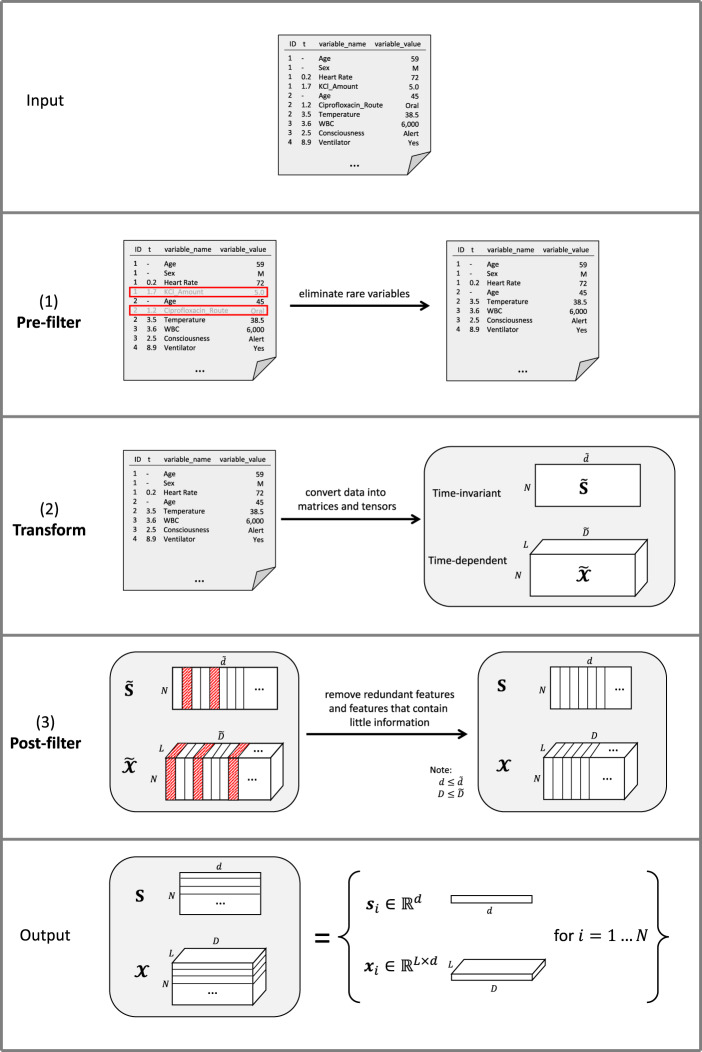
FIDDLE is an open-source preprocessing pipeline for structured data extracted from the EHR (Figure 1). Preprocessing EHR data for ML presents numerous challenges (Table 1), many of which are not unique to EHR data and arise in other settings. In the subsections below, we describe how FIDDLE tackles these challenges in the context of EHR data.
Figure 1.
Overview of FIDDLE. Given formatted input data and user-defined arguments, FIDDLE processes data in 3 stages: (1) pre-filter, (2) transform, and (3) post-filter. So long as the units are consistent, timestamps in the t column may be recorded at any level of granularity (eg, seconds, minutes, hours, days, visits, etc.). In this sample input file, we consider time in hours. A row with [1, 0.2, Heart Rate, 72] corresponds to a patient with ID = 1 with a heart rate = 72 bpm recorded at t = 0.2 h. In (1) pre-filter, FIDDLE eliminates rare variables. In (2) transform, FIDDLE transforms data into tensors containing time-invariant and time-dependent features. In (3) post-filter, FIDDLE removes redundant features and features that are likely uninformative. The output consists of binary vectors and , describing the features for each ID. bpm: beats per minute; FIDDLE: Flexible Data-Driven Pipeline: ID: unique identifier; KCl: potassium chloride; WBC: white blood cell.
Table 1.
Challenges in preprocessing EHR data and FIDDLE’s solution
| Challenges | Example | Solutions in FIDDLE |
|---|---|---|
| Some data have associated timestamps, while others do not |
|
Handle time-invariant and time-dependent data separately.2,16,17 |
Data have heterogeneous types
|
|
Different representations for each value type |
| Data are sparse and irregularly sampled, and different variables can have different frequencies of recording |
|
|
After resampling the data according to some temporal granularity () we might have:
|
|
|
High-dimensional feature space
|
Data extracted from the EHR typically contain hundreds, if not thousands, of variables, including medications, labs, CPT codes, etc. |
Note: bpm: beats per minute; CPT: current procedure terminology; EHR: electronic health record; FIDDLE: Flexible Data-Driven Pipeline; ICD-9: International Classification of Diseases, Ninth Edition; ICU: intensive care unit; IV: intravenous.
Input and output
FIDDLE takes as input tabular data with 4 columns—ID, t, variable_name, and variable_value—where ID is a unique identifier for each example and t is the time of recording, measured relative to a fiducial marker (eg, time of admission, t = 0). Generally, an ID may have multiple rows representing recordings of different variables at different times. When t is null, the pipeline assumes a time-invariant value recorded once (eg, baseline variables like “age” and “sex”). Each variable_name uniquely encodes the name of a variable (eg, “heart rate” and “white blood cell count”) and is consistent across all IDs. The variable_value column may contain numbers (eg, heart rate of “72” beats per minute) or strings (eg, “abdominal pain”) and cannot be null. Each variable_name can be automatically classified as either numerical or categorical, based on the associated variable_value type. A user can always override the type of a variable_name to be either numerical, categorical, or hierarchical (eg, International Classification of Diseases [ICD]/current procedure terminology [CPT] codes, where the user can specify which level[s] of the hierarchy to consider). We do not make assumptions regarding the completeness of the data; it is likely that not every ID will have a value associated with every variable_name.
Given data in the format described above and a set of user-defined arguments (Table 2), FIDDLE generates feature vectors based on data within the observation period. This feature representation can be used to make predictions at regarding whether an outcome will occur after . Tables 2 and 3 summarize FIDDLE’s inputs and outputs. More specifically, FIDDLE outputs , a set of features for each example , where contains time-invariant features and contains time-dependent features. Here, refers to the number of examples (unique IDs in the data table), is the number of time-steps after “windowing” (ie, resampling) the observation period into time bins of size . The dimensionalities of the time-invariant and time-dependent features are denoted by and , respectively. To generate these feature vectors, FIDDLE processes the formatted data in 3 steps—(1) pre-filter, (2) transform, and (3) post-filter—as illustrated in Figure 1 and described below.
Table 2.
Summary of notation in user-defined arguments of FIDDLE
| Argument | Description |
|---|---|
| A positive number specifying the time of prediction; is the observation period to consider when processing time-dependent data. The unit of could be minutes, hours, days, etc., and must be the same as the unit of . | |
| A positive number specifying the temporal granularity (eg, hourly vs daily) at which to resample the time-dependent data. The unit of must be the same as the unit of . | |
| A value between 0 and 1 specifying the threshold for the pre-filter step. | |
| A value between 0 and 1 specifying the threshold for the post-filter step. | |
| A positive number specifying the threshold, in terms of the average number of measurements per time window, at which we deem a variable “frequent” (for which summary statistics will be calculated). | |
| A set of statistics functions (eg, min, max, mean). Each function takes as input 1 or more recordings within a time bin and outputs a single summary statistic. These functions are only applicable to “frequent” variables, as determined by . | |
| discretize | A Boolean flag (default value: True) specifying whether features with numerical values are kept as raw values or discretized into binary features. |
| discretization_encoding | A string specifying how numerical values are encoded into binary features after discretization. Possible values are: “one-hot” (default) and “ordinal.” This argument is ignored and should not be used when discretize=False. |
Note: FIDDLE: Flexible Data-Driven Pipeline.
Table 3.
Symbols used to describe FIDDLE’s implementation
| Symbol | Shape | Description |
|---|---|---|
| – | The number of examples. | |
| – | The number of time bins, calculated as . | |
| , | – | The number of input variables that are time-invariant/time-dependent after the pre-filter step. |
| , | – | The dimensionalities of time-invariant / time-dependent features after the transform step and before the post-filter step. |
| , | – | The final dimensionalities of time-invariant / time-dependent features. |
| Data tables containing values of raw time-invariant/time-dependent values after the pre-filter step. | ||
| Tensors containing the time-invariant/time-dependent features for all examples, after the transform step and before the post-filter step. | ||
| Tensors containing the final time-invariant/time-dependent features for all examples. | ||
Note: FIDDLE: Flexible Data-Driven Pipeline.
Processing steps
Pre-filter
First, rows with timestamp t outside the observation period are eliminated and variables that occur rarely are removed. Specifically, all rows with a variable_name that appears in of examples are filtered out. That is, if a specific drug is administered to only 1% of examples and , then rows/observations involving this drug are removed. This step speeds up downstream analyses, though aggressive filtering could result in a potential loss of useful information.
Transform
Processing continues based on the timestamp types. If the timestamp t of a variable_name is null, then data corresponding to that variable_name are processed as “time-invariant”; otherwise, those data are processed as “time-dependent” to capture dynamics and longitudinal patterns.
Time-invariant data
All time-invariant data are concatenated into a table of shape , where each row corresponds to a single ID and each column pertains to a time-invariant variable. If a variable is not available for a particular ID, then that row-column pair is set to null.
Time-dependent data
All time-dependent data are concatenated into a tensor of shape , similar to above but with an additional temporal dimension . Here, we provide a high-level overview of the processing steps; additional details are available in the code and in Supplementary Appendix 1. Each slice in the tensor corresponds to a single ID and contains the values of all processed time-dependent variables over the time bins. The pipeline processes “non-frequent” and “frequent” variables differently (a numeric variable is considered “frequent” if recorded times on average over all examples and all time bins). Each “non-frequent” variable is simply represented by its most recent recorded value for every time bin (null if not available). To encode information within each time bin, each “frequent” variable is mapped to processed variables: (1) value, (2) mask, (3) delta time, and (4) summary statistics resulting from . Missing values are handled with carry-forward imputation,26 as it makes fewer assumptions about the data and is potentially more feasible to implement in real time.24,25 As EHR data are assumed “missing not at random,” the imputed values are tracked by a “mask” (indicating presence) and “delta time” (the number of time bins since the previous non-imputed measurement).27,28 Finally, all processed time-dependent variables are concatenated together, resulting in dimensions for “non-frequent” variables and “frequent” variables.
The pipeline then discretizes categorical variables into binary features using a one-hot encoding.2,3,6 If the “discretize” option is set to true, numerical variables are first quantized based on quintiles19 and then mapped to binary features using a one-hot or an ordinal encoding (as specified by the user); otherwise, each numerical variable is used as a feature (raw values are used to preserve interpretability; standardization/normalization might be necessary depending on the ML algorithm). Missing entries (null) are mapped to 0s in all categories/bins. The final output of this step is a matrix and a tensor , where and are the dimensions of time-invariant/time-dependent features after discretization.
Post-filter
After the data are transformed, features that are equal to 1 (or 0) in of examples are removed (where is small: eg, 0.01).30–33 This removes features that are unlikely to be informative. Each group of duplicated features (ie, features that are pairwise perfectly correlated) are then combined into a single feature. This produces a matrix and a tensor , with time-invariant features and time-dependent features where and .
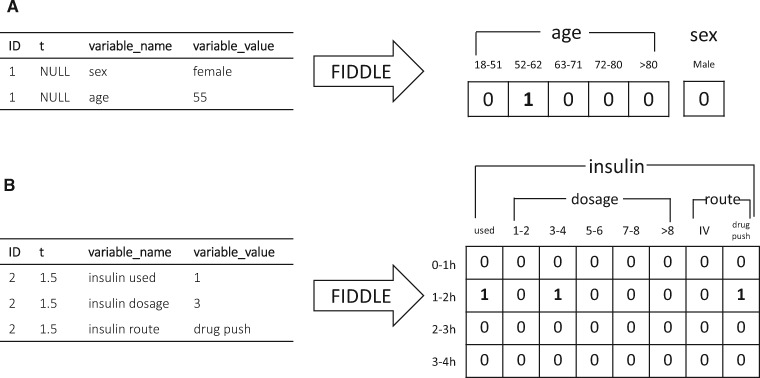
These preprocessing steps produce a data representation that can be used as input to ML algorithms. Figure 2 illustrates how FIDDLE can transform formatted EHR data into feature vectors, providing examples for both time-invariant and time-dependent data. Additional details of FIDDLE and guidelines on argument settings are described in Supplementary Appendix 1.
Figure 2.
Examples of FIDDLE input and output for time-invariant and time-dependent data. In this example, each ID represents a patient (an example). Timestamps are recorded in hours. Only the subset of input/output relevant for illustration is shown. The bins for numerical variables and the categories for categorical variables are automatically determined from the entire input data table (not shown). (A) Time-invariant input data and output features for Patient 1. Patient 1 is female with an age of 55. The feature “sex = female” is dropped in the post-filter step because it is perfectly correlated with “sex = male.” (B) Time-dependent input data and output features for Patient 2. At t = 1.5 h, Patient 2 had an insulin administration of 3 units via drug push. No imputation in 2–4 h is done, since the 3 variables related to insulin are not considered “frequent,” resulting in 0 s in the output features for the corresponding time bins. FIDDLE: Flexible Data-Driven Pipeline; ID: unique identifier; IV: intravenous.
Experiments
To demonstrate that FIDDLE generalizes across data sets and produces useful features, we consider its use across a number of different prediction tasks. We performed a proof-of-concept experiment where we applied FIDDLE to 2 different EHR data sets, training and evaluating various ML models for a set of clinically relevant prediction tasks. Here, the goal was not to train state-of-the-art models, but produce a reasonable representation from which one could rapidly iterate. We measured the predictive performance of the learned models as a proxy for the utility of FIDDLE as a feature preprocessing pipeline.
Data
In our experiments, we used the MIMIC-III database14 and the eICU Collaborative Research Database,15 interpreting each ICU visit as a unique example. Tables in the 2 data sets encompass many different aspects of patient care: demographics, physiological measurements, laboratory measurements, medications, fluid output, microbiology, and so forth. These tables contain both observations and interventions, and can have numerical or categorial values. Additional information about the data extraction process can be found in Supplementary Appendix 2.
From MIMIC-III,14 we focused on 17,710 patients (23,620 ICU visits) monitored using the iMDSoft MetaVision system (2008–2012) for its relative recency over the Philips CareVue system (2001–2008), thus representing more up-to-date clinical practices. Each ICU visit is identified by a unique ‘ICUSTAY_ID', for which we extracted data from 10 structured tables (Table 4).
Table 4.
Summary of MIMIC-III tables used in our analysis
| MIMIC-III | ||
|---|---|---|
| Table name | Description | Example variables |
| PATIENTS | Information on unique patients | Age, Sex |
| ADMISSIONS | Information on unique hospitalizations |
Admission type Admission location |
| ICUSTAYS | Information on unique ICU stays |
Care unit Ward ID Admission-to-ICU time |
| CHARTEVENTS | Charted data, including vital signs, and other information relevant to patients’ care |
Heart rate Pain location Daily weight |
| LABEVENTS | Laboratory test results from the hospital database |
Lactate WBC |
| INPUTEVENTS_MV | Fluid intake administered, including dosage and route (eg, oral or intravenous) |
NaCl 0.45% Whole blood |
| OUTPUTEVENTS | Fluid output during the ICU stay |
OR urine Stool |
| PROCEDUREEVENT_MV | Patients’ procedures during the ICU stay |
CT scan X-ray |
| MICROBIOLOGYEVENTS | Microbiology specimen from hospital database | Sputum |
| DATETIMEEVENTS | Documentation of dates and times of certain events |
Last dialysis Pregnancy due |
Note: We used all structured tables that pertain to patient health.
CT: computed tomography; ICU: intensive care unit; ID: unique identifier; OR: operating room; WBC: white blood cell.
The eICU15 database consists of data from 139,367 patients (200,859 ICU visits) who were admitted to 200 different ICUs located throughout the United States in 2014 and 2015. Each ICU visit is identified by a unique ‘patientunitstayid.' Similar to above, we extracted data from 18 structured tables that pertain to patient health (Table 5).
Table 5.
Summary of eICU tables used in our analysis
| eICU | ||
|---|---|---|
| Table name | Description | Example variables |
| patient | Information on unique patients, hospitalizations, and ICU stays |
Age, Sex Hospital/ward ID |
|
vitalPeriodic vitalAperiodic |
Vital signs measured through bedside monitors or invasively |
Temperature End Tidal CO2 |
|
lab customLab |
Laboratory tests |
CPK troponin - I |
|
medication infusionDrug intakeOutput |
Active medication orders, the intake of drug through infusions, and intake/output of fluids |
Morphine dosage Dialysis total |
| microLab | Microbiology cultures taken from patients |
Culture site (wound) Organism |
|
note nurseAssessment nurseCare nurseCharting |
Documentation of physician/nurse assessment |
Abdominal pain Psychological status Respiratory rate |
| pastHistory | Relevant past medical history |
Transplant AIDS |
| physicalExam | Results of physical exam (structured) |
Blood pressure Verbal score |
|
respiratoryCare respiratoryCharting |
Respiratory care data |
Airway position Vent details |
| treatment | Structured data documenting specific, active treatments | Thrombolytics |
Note: We used all structured tables that pertain to patient health.
AIDS: acquired immunodeficiency syndrome; CPK: creatine phosphokinase; ICU: intensive care unit; ID: unique identifier.
The code to extract and format MIMIC-III and eICU data is provided in our implementation. When mapping data from raw database tables to the appropriate format as input to FIDDLE, we worked closely with a critical care physician (MWS) to ensure the mapping was appropriate (details are in Supplementary Appendix 2).
Clinical outcomes
In our evaluation of FIDDLE, we trained ML models to predict in-hospital mortality, acute respiratory failure (ARF), and shock.35,36 Interpreting each ICU stay as an example, we developed pragmatic outcome definitions (see Supplementary Appendix 3) based on the clinical experience of a critical care physician (MWS). In contrast to previous definitions based on ICD diagnosis codes,11 we focused on clinical data indicating the onset of events (eg, mechanical ventilation and administration of vasopressors), since records of ICD codes do not indicate the time of onset and may correspond poorly to the actual diagnoses.37,38 Our clinical-based definitions for these 2 decompensation tasks more accurately reflect the timing of outcomes. For ARF and shock, we defined onset time as the earliest time the outcome criteria (Supplementary Appendix 3) were met.
Applying FIDDLE to MIMIC-III and eICU
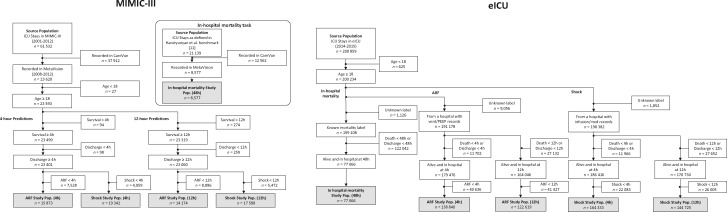
We defined 5 prediction tasks, each with a distinct study cohort (Figure 3; Supplementary Appendix 3). In all analyses, we excluded neonates and children (age <18) because their physiology and risk factors differ from adults.11,39 We did not attempt to exclude patients with treatment limitations (eg, those who may be placed on comfort measures), given the difficulty in identifying this status reliably across data sets. While this allows us to compare with previous work,11,13 it could ultimately make the prediction tasks easier and limit the clinical utility of the learned models. For in-hospital mortality, we used hours to predict whether the outcome would occur after following existing work.11 For ARF and shock, we used both hours and hours. Examples (ICU stays) with an event onset time before , or discharges and deaths before , were excluded. For the eICU data, we also excluded examples for which the ground truth labels could not be reliably determined due to a lack of sufficient documentation.15 Specifically, for ARF and shock, we excluded entire hospitals without any relevant ventilation records or vasopressor records, respectively.
Figure 3.
Harutyunyan et al11 definitions of the study cohorts. For each data set (MIMIC-III and eICU), we defined 5 prediction tasks, each with a distinct study cohort: in-hospital mortality at 48 h, ARF at 4 h, ARF at 12 h, shock at 4 h, and shock at 12 h. ARF: acute respiratory failure; ICU: intensive care unit; PEEP: positive end-expiratory pressure.
These 5 tasks correspond to a single prediction based on a fixed look-back period; in the Supplementary Material, we further demonstrate how FIDDLE applies to (1) multiple predictions over the prediction window using a sequence-to-sequence long short-term memory network model (Supplementary Appendix 8.1), and (2) predicting the 90-day post-discharge mortality of MIMIC-III patients using clinical data in ICUs and ICD codes at discharge (Supplementary Appendix 8.2). These additional experiments illustrate how FIDDLE can be applied more broadly and even handle hierarchical values.
When applying FIDDLE to the 5 cohorts on MIMIC-III, we used the following user-defined arguments (Tables 2 and 3): ; ; ; (, , ); discretize=True; and discretization_encoding = “one-hot.” These settings were determined based on how often the variables were recorded and the class balance within each cohort, in line with our recommendations in Supplementary Appendix 1. On the eICU cohorts, because of the large sample size and feature space, we used more aggressive filtering thresholds (mortality: ; 4-hour tasks: ; 12-hour tasks: ). To understand the effect of user-defined arguments on the utility of the features generated by FIDDLE, we also tested FIDDLE using (1) different filtering thresholds, ; (2) temporal granularities, (3) a continuous versus one-hot encoding versus ordinal encoding representation; and (4) carry-forward imputation versus median imputation versus no imputation (results are reported in Supplementary Appendix 7.5).
Model training and evaluation
We used the features generated by FIDDLE directly as input to 4 classification algorithms: penalized logistic regression (LR), random forest (RF), 1-dimensional convolutional neural networks (CNN), and long short-term memory networks (LSTM), adapting the features depending on the model type. For models expecting flat input (LR and RF), we flattened the time-dependent features and concatenated them with the time-invariant features , resulting in a feature vector of shape . For models expecting sequential input (CNN and LSTM), we repeated the time-invariant features at every time-step of 16 resulting in a feature matrix of shape .
We randomly assigned each patient to either the train or the test partition, and then split each study cohort into train and test sets (containing ICU stays) accordingly. Hyperparameters (Supplementary Appendix 5) were selected using the training/validation data and a random search40 with a budget of 50, maximizing the average area under the receiver operating characteristics curve (AUROC). Due to the large sample size, deep models (CNN and LSTM) on the eICU data were given a budget of 5. Models were evaluated on the held-out test sets in terms of the receiver operating characteristic curve (ROC), precision-recall curve (PR), and calibration performance. We also measured the area under the ROC and PR curves (AUROC and AUPR, respectively). Empirical 95% confidence intervals (CIs) were estimated using 1000 bootstrapped samples of the test set. When comparing model performance, statistical significance was determined using a resampling test on the same 1000 bootstraps,41 with a Bonferroni correction for multiple hypothesis testing where appropriate.42
All experiments were implemented in Python 3.6,43 Scikit-learn,44 and Pytorch.45 The code for FIDDLE and all analyses is open-source and available online, along with documentation and further usage notes (Supplementary Appendix 1).
Baseline MIMIC-Extract
Increasingly, ML researchers in the clinical domain are sharing preprocessing code, leading to improved reproducibility.11–13 However, there exist limited efforts in developing generalizable tools for EHR feature extraction. Most closely aligned with our goal is MIMIC-Extract,13 a recently developed data extraction and preprocessing pipeline that transforms EHR data from MIMIC-III into data frames used for common ML models. Specific to MIMIC-III, it includes outlier detection and the aggregation of semantically equivalent features. However, due to this specificity, it does not readily port to other data sets (eg, the eICU data). In our experiments, for each prediction task on MIMIC-III we compared FIDDLE to MIMIC-Extract following Wang et al’s13 implementation (see Supplementary Appendix 6 for details).
RESULTS
FIDDLE applied to MIMIC-III and the eICU data: study cohorts and extracted features
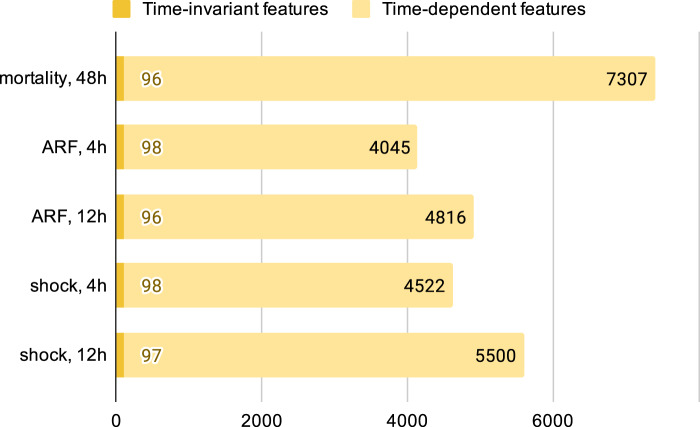
The MIMIC-III study cohorts varied in size from 8,577 to 19,342 examples, whereas eICU varied from 77,066 to 164,333 examples (Supplementary Appendix 3). The formatted input tables contained up to 320 million rows. Applied to MIMIC-III and the eICU data, FIDDLE produced feature vectors that varied in dimension from 4,143 to 7,403 and from 2,528 to 7,084, respectively. Later prediction times resulted in more time-dependent features, but the number of time-invariant features was approximately the same across all tasks (Figure 4). The transform step identified 6 important vital signs as “frequent” variables: heart rate, respiratory rate, temperature, systolic blood pressure, diastolic blood pressure, and peripheral oxygen saturation. Since we set , this means these variables were recorded more than once per hour on average across patients.
Figure 4.
Dimensionality of feature vectors for each prediction task on MIMIC-III. After applying FIDDLE to the MIMIC-III study cohorts, an ICU visit is represented by time-invariant features and time-dependent features, both of which are high-dimensional. Though the number of time-invariant features is similar across tasks, the number of time-dependent features varies because more data (likely corresponding to more variables) are collected for a later prediction time. FIDDLE: Flexible Data-Driven Pipeline; ICU: intensive care unit.
Examples of features generated by FIDDLE on the task of ARF prediction (12 hours) from MIMIC-III are displayed in Tables 6 and 7. Time-invariant features correspond to age, sex, ICU locations, and different sources of hospital admission (Table 6), whereas time-dependent features cover a diverse range of descriptors, including vital signs, medications, laboratory results, and so forth. Much of the information in the EHR is retained; for example, drug administrations are described by both dosage and route (Table 7).
Table 6.
Examples of time-invariant features extracted by FIDDLE on the 12-hour ARF cohort for MIMIC-III
| Time-invariant features |
|---|
| Age in Q1 (18–51) |
| Age in Q2 (52–62) |
| Age in Q3 (63–71) |
| Age in Q4 (72–80) |
| Age in Q5 (>80) |
| Sex = Female |
| ICU Location ID = 12 |
| ICU Location ID = 15 |
| ICU Location ID = 23 |
| ICU Location ID = 33 |
| ICU Location ID = 52 |
| ICU Location ID = 57 |
| Hospital admission source: clinic referral |
| Hospital admission source: transfer from hospital |
| Hospital admission source: from emergency room |
Note: ARF: acute respiratory failure; FIDDLE: Flexible Data-Driven Pipeline; ICU: intensive care unit; ID: unique identifier; Q, quintile.
Table 7.
Examples of time-dependent features extracted by FIDDLE on the 12-hour ARF cohort for MIMIC-III
| Time-dependent features |
|---|
| At 0–1 h, insulin dosage in Q1 (≤2 units) |
| At 0–1 h, insulin dosage in Q2 (>2 units, ≤3 units) |
| At 0–1 h, insulin dosage in Q3 (>3 units, ≤4 units) |
| At 0–1 h, insulin dosage in Q4 (>4 units, ≤8 units) |
| At 0–1 h, insulin dosage in Q5 (>8 units) |
| At 0–1 h, insulin route = intravenous |
| At 0–1 h, insulin route = drug push |
| At 1–2 h, insulin dosage in Q1 (≤2 units) |
| At 1–2 h, insulin dosage in Q2 (>2 units, ≤3 units) |
| At 1–2 h, insulin dosage in Q3 (>3 units, ≤4 units) |
| At 1–2 h, insulin dosage in Q4 (>4 units, ≤8 units) |
| At 1–2 h, insulin dosage in Q5 (>8 units) |
| At 1–2 h, insulin route = intravenous |
| At 1–2 h, insulin route = drug push |
Note: ARF: acute respiratory failure; FIDDLE: Flexible Data-Driven Pipeline; Q, quintile.
Applied to each cohort of MIMIC-III, FIDDLE extracted feature vectors in approximately 30–150 minutes, depending on the size of input data and argument settings. In contrast, MIMIC-Extract took 8 hours in total (including database operations, etc), but the feature-processing stage alone took 1–2 hours. Due to the larger sample sizes of eICU cohorts, processing took longer compared to MIMIC-III (∼10 hours using FIDDLE). In Supplementary Appendix 4, we report detailed results on the runtime, the number of variables remaining at each step, and the final feature dimensionalities for each task.
Assessing the utility of FIDDLE
We measure the utility of FIDDLE by evaluating the performance of ML models trained/tested on features generated by FIDDLE. We summarize the results in Tables 8 and 9; extended results and comparisons to additional baselines are reported in Supplementary Appendix 7.
Table 8.
Summary of performance on MIMIC-III for all FIDDLE-based models, compared to MIMIC-Extract
| Task |
In-hospital mortality, 48 h n = 1264 |
ARF, 4 h n = 2358 |
ARF, 12 h n = 2093 |
Shock, 4 h n = 2867 |
Shock, 12 h n = 2612 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | |
| MIMIC-Extract | LR | 0.859 (0.830–0.887) | 0.445 (0.358–0.540) | 0.777 (0.752–0.803) | 0.604 (0.561–0.648) | 0.723 (0.683–0.759) | 0.250 (0.200–0.313) | 0.796 (0.771–0.821) | 0.505 (0.454–0.557) | 0.748 (0.712–0.784) | 0.242 (0.193–0.310) |
| RF | 0.852 (0.821–0.882) | 0.448 (0.359–0.537) | 0.821 (0.799–0.843) | 0.660 (0.617–0.698) | 0.747 (0.709–0.782) | 0.289 (0.235–0.356) | 0.824 (0.801–0.845) | 0.541 (0.488–0.588) | 0.778 (0.742–0.812) | 0.307 (0.248–0.369) | |
| CNN | 0.851 (0.820–0.879) | 0.439 (0.353–0.529) | 0.788 (0.763–0.814) | 0.633 (0.591–0.672) | 0.722 (0.684–0.758) | 0.258 (0.207–0.320) | 0.798 (0.773–0.824) | 0.520 (0.471–0.572) | 0.741 (0.704–0.778) | 0.247 (0.198–0.317) | |
| LSTM | 0.837 (0.803–0.867) | 0.441 (0.358–0.523) | 0.796 (0.770–0.822) | 0.634 (0.590–0.675) | 0.700 (0.661–0.736) | 0.229 (0.184–0.286) | 0.801 (0.778–0.825) | 0.513 (0.463–0.562) | 0.753(0.717–0.791) | 0.248 (0.199–0.313) | |
| FIDDLE | LR | 0.856(0.821–0.888) | 0.444(0.357–0.545) | 0.817(0.792–0.839) | 0.657(0.614–0.696) | 0.757(0.720–0.789) | 0.291(0.236–0.354) | 0.825(0.803–0.846) | 0.548(0.501–0.595) | 0.792(0.758–0.824) | 0.274(0.227–0.338) |
| RF | 0.814(0.780–0.847) | 0.357(0.279–0.448) | 0.817(0.795–0.839) | 0.652(0.608–0.690) | 0.760(0.726–0.793) | 0.317(0.255–0.382) | 0.809(0.786–0.833) | 0.516(0.467–0.566) | 0.773(0.740–0.806) | 0.288(0.231–0.355) | |
| CNN | 0.886(0.854–0.916) | 0.531(0.434–0.629) | 0.827(0.803–0.848) | 0.666(0.626–0.705) | 0.768(0.733–0.800) | 0.294(0.238–0.361) | 0.831(0.811–0.851) | 0.541(0.493–0.589) | 0.791(0.758–0.823) | 0.295(0.239–0.361) | |
| LSTM | 0.868(0.835–0.897) | 0.510(0.411–0.597) | 0.827(0.801–0.846) | 0.664(0.623–0.703) | 0.771(0.737–0.802) | 0.326(0.267–0.397) | 0.824(0.803–0.845) | 0.541(0.497–0.587) | 0.792(0.759–0.823) | 0.314(0.251–0.386) | |
Note: Reported as AUROC and AUPR with 95% CIs in parentheses on the respective held-out test set for the 5 prediction tasks. For each task (column), the bolded results are the best-performing model for either MIMIC-Extract or FIDDLE.
ARF: acute respiratory failure; AUROC: area under the receiver operating characteristics curve; AUPR: area under the precision-recall curve; CI: confidence interval; CNN: convolutional neural networks; FIDDLE: Flexible Data-Driven Pipeline; LR: logistic regression; LSTM: long short-term memory networks; RF: random forest.
Table 9.
Summary of performance on eICU for all FIDDLE-based models
| Task | In-hospital mortality, 48 h n = 11 542 |
ARF, 4 h n = 20 749 |
ARF, 12 h n = 18 275 |
Shock, 4 h n = 24 647 |
Shock, 12 h n = 21 642 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | AUROC | AUPR | AUROC | AUPR |
| FIDDLE-LR | 0.824(0.812–0.836) | 0.401(0.374–0.428) | 0.810(0.799–0.821) | 0.269(0.246–0.293) | 0.778(0.763–0.794) | 0.201(0.178–0.225) | 0.846(0.836–0.855) | 0.338(0.314–0.360) | 0.797(0.782–0.811) | 0.187(0.168–0.210) |
| FIDDLE-RF | 0.787(0.774–0.800) | 0.340(0.314–0.366) | 0.792(0.779–0.803) | 0.236(0.217–0.258) | 0.749(0.734–0.764) | 0.166(0.149–0.187) | 0.810(0.800–0.820) | 0.279(0.258–0.298) | 0.768(0.753–0.783) | 0.152(0.136–0.171) |
| FIDDLE-CNN | 0.845(0.834–0.855) | 0.433(0.404–0.461) | 0.828(0.817–0.839) | 0.276(0.252–0.300) | 0.799(0.784–0.813) | 0.212(0.190–0.236) | 0.854(0.846–0.863) | 0.351(0.327–0.374) | 0.813(0.800–0.826) | 0.200(0.180–0.223) |
| FIDDLE-LSTM | 0.841(0.830–0.852) | 0.435(0.408–0.464) | 0.833(0.822–0.844) | 0.296(0.272–0.322) | 0.803(0.788–0.817) | 0.216(0.194–0.241) | 0.853(0.844–0.861) | 0.356(0.332–0.379) | 0.816(0.802–0.828) | 0.199(0.178–0.223) |
Note: Reported as AUROC and AUPR with 95% CI on the respective held-out test set for the 5 prediction tasks.
ARF: acute respiratory failure; AUROC: area under the receiver operating characteristics curve; AUPR: area under the precision-recall curve; CI: confidence interval; CNN: convolutional neural networks; FIDDLE: Flexible Data-Driven Pipeline; LR: logistic regression; LSTM: long short-term memory networks; RF: random forest.
FIDDLE across tasks and data sets
We hypothesized that the generated features are useful across the 2 data sets and the 5 prediction tasks (involving 3 outcomes and different prediction times). Comparing model performance across MIMIC-III and eICU, the ML models performed similarly on each task (Tables 8 and 9). Compared to MIMIC-Extract, FIDDLE-based models achieved similar performance, despite the lack of data set–specific curation (Table 8).
FIDDLE-generated features as input to different ML algorithms
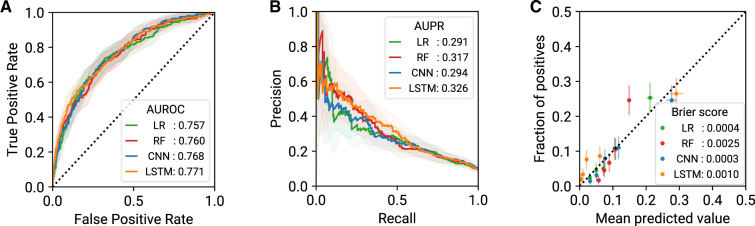
To evaluate the generalizability of FIDDLE features across ML algorithms, we compared the performance of the 4 ML algorithms on the same prediction task. On MIMIC-III, for predicting ARF at 12 hours, all 4 ML models achieved good discriminative performance (Figure 5A and B) and good model calibration (Figure 5C). Trends were similar across the other 4 tasks (Table 8; Supplementary Appendix 7.2) and on eICU (Table 9; Supplementary Appendix 7.2), supporting our claim that FIDDLE-generated features are useful for common ML algorithms.
Figure 5.
Model performance (with 95% CI) for prediction of ARF at t = 12 h on MIMIC-III, evaluated on the held-out test set (n = 2093). On this task, all 4 FIDDLE-based models exhibited similarly good discriminative and calibration performance. (A) ROC curves and AUROC scores. (B) PR curves and AUPR scores. (C) Calibration plots and Brier scores. ARF: acute respiratory failure; AUROC: area under the receiver operating characteristics curve; AUPR: area under the precision-recall curve; CI: confidence interval; CNN: convolutional neural networks; FIDDLE: Flexible Data-Driven Pipeline; LR: logistic regression; LSTM: long short-term memory networks; PR: precision-recall curve; RF: random forest; ROC: receiver operating characteristics curve.
DISCUSSION
When applying ML to EHR data, researchers often fall back on easily extracted, hand-selected features, because more comprehensive preprocessing can be time intensive. In this work, we developed FIDDLE as an open-source tool to streamline this process. In our proof-of-concept experiments, features generated by FIDDLE led to good predictive performance across different outcomes, prediction times, and classification algorithms, with AUROCs comparable to those of MIMIC-Extract applied to MIMIC-III. Furthermore, we demonstrated that FIDDLE readily applies to other EHR data sets, such as the eICU Collaborative Research Database, provided that the data are appropriately formatted. FIDDLE has the potential to greatly speed up EHR data preprocessing, aiding ML practitioners who work with health data.
The proposed approach is largely data-driven; for example, variable discretization depends on the underlying data distribution. Such an approach relies less on domain knowledge compared to common alternatives, such as the manual specification of ranges for normal/abnormal values. With enough data, data-driven approaches can help reduce human effort and save time. Though largely data-driven, FIDDLE still allows users to tailor the pipeline to their cohort/task. This kind of flexibility is critical to many applications of ML in health care. For example, users can set the temporal granularity at which the input is considered. Moreover, given the open-source nature of the pipeline, researchers can build upon FIDDLE and adapt it to their task requirements by modifying specific components (as illustrated in Supplementary Appendix 7.5), such as using different imputation methods for missing data, increasing the number of quantile bins, or incorporating additional clinical expertise. While FIDDLE is by no means the single best way to preprocess data for all use cases, it facilitates reproducibility and the sharing of preprocessing code (oftentimes overlooked or not fully described in the literature).
FIDDLE helps to address many limitations in existing work. In contrast to previous studies that have focused on a small set of hand-selected variables,10–12,46,47 FIDDLE allowed us to consider nearly all available structured data in MIMIC-III and eICU, producing features that capture a rich representation of a patient’s physiological state and longitudinal history. These extracted features enable ML models to leverage the potentially high-dimensional patterns in the data. Specifically, while MIMIC-Extract13 and FIDDLE share similarities in their goals, there are notable differences. Unlike FIDDLE, which produces feature vectors that can be used as input to ML algorithms, MIMIC-Extract outputs several data frames (potentially containing null/missing values) that require further preprocessing. Additionally, MIMIC-Extract assumes a fixed resampling rate and performs clinical groupings of variables. These decisions limit the generalizability of MIMIC-Extract to tasks at different time scales or other data sets. In contrast, FIDDLE relies on fewer assumptions and can be applied to any EHR data set that meets the required format. Beyond MIMIC-Extract, alternative techniques to incorporate all available data in an EHR system exist. For example, Rajkomar et al39 proposed a representation learning framework for EHR data in the Fast Healthcare Interoperability Resources (FHIR) format. Their learned embeddings, however, lack the interpretability of FIDDLE-generated features. The feature representation of FIDDLE facilitates debugging, as illustrated in Supplementary Appendix 7.4. Moreover, their approach is not open-source; the open-source nature of FIDDLE makes it readily accessible to researchers.
It is worth noting that FIDDLE only streamlines the preprocessing of data extracted from the EHR; to obtain a usable ML model starting from data collected at the bedside requires many more steps beyond preprocessing. To this end, researchers have proposed other EHR data pipelines that solve problems complementary to FIDDLE’s goal (Supplementary Appendix 9). However, before FIDDLE can be integrated into an EHR system,48 adaptations must be made to generate features in a prospective setting (for example, by storing the feature transformation functions and filters). Going forward, such integrations might consider applying FIDDLE on top of interoperable data formats, such as FHIR,49 Observational Medical Outcomes Partnership Common Data Model,50 and The National Patient-Centered Clinical Research Network (PCORnet®).51
As a pipeline, FIDDLE has limitations. First, FIDDLE processes all numerical variables identically, which may be inappropriate in certain settings. Though FIDDLE does process “frequent” variables through user-defined summary statistics, future versions could allow a user to specify different summary statistics for different groups of variables (eg, “most recent” for vital signs, “total” for bodily fluids like urine output). Second, FIDDLE considers only the structured contents in the EHR. For the unstructured contents, such as imaging and nursing notes, techniques from computer vision and natural language processing could be used to generate a set of embeddings that can then be incorporated. Finally, FIDDLE does not attempt to harmonize data across institutions. How to transfer models or feature representations across different institutions remains an open problem.52,53
Though a data-driven approach, like FIDDLE, can help speed up ML analyses, it does not eliminate the critical need for model checking. In our experiments, we used FIDDLE to generate high-dimensional feature vectors (ie, d > 1000). In contrast to a hypothesis-driven approach that starts with a small set of hand-selected variables, FIDDLE can leverage the entire structured contents of the EHR. In doing so, the model may take advantage of variables specific to a particular hospital or even unintended short-cuts in the data. For example, in our experiments with MIMIC-III, the models identified patients with a code status of “do not resuscitate” as “lower risk” for ARF. Given our treatment-based definition of ARF, the model was able to capitalize on the code status feature, improving predictive performance in this subset of patients, but not necessarily improving the clinical utility of the model. Carefully reviewing the learned model and validating it in ways that mimic the clinical use case remains necessary. While FIDDLE does not allow one to skip these critical steps, by saving the extracted feature names, it can help one in the debugging process. Finally, when integrating a model into hospital operations, there is a significant maintenance cost associated with each included variable. A tradeoff arises between improvements in performance and the “technical debt” that comes with including more variables in deployed ML models.54 To address this, researchers may consider either (1) tuning the filtering threshold in FIDDLE to be more aggressive, or (2) applying downstream feature selection approaches (eg, filtering, wrapper, or embedded methods).55
CONCLUSION
In summary, FIDDLE can help ML researchers preprocess data extracted from the EHR. By accelerating and standardizing the labor-intensive preprocessing steps, FIDDLE can help stimulate progress in building clinically useful ML tools. We have made FIDDLE open source, available online to the research community. We hope that FIDDLE will be useful to other researchers; ultimately, once the community starts using the tool, we will be able to collectively refine and build on it together.
FUNDING
This work was supported by the Michigan Institute for Data Science (MIDAS); the National Science Foundation award number IIS-1553146; the National Heart, Lung, and Blood Institute grant number R25HL147207; and the National Library of Medicine grant number R01LM013325. The views and conclusions in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the Michigan Institute for Data Science; the National Science Foundation; the National Heart, Lung and Blood Institute; nor the National Library of Medicine.
AUTHOR CONTRIBUTORS
ST, JW, MWS, and DK designed and conceptualized the overall study. ST, PD, and YS implemented the preprocessing pipeline. ST performed the cohort extraction, data cleaning, model development, and model evaluation. PD and YS contributed in conducting experiments and evaluating the results. MWS provided valuable insights regarding clinical motivation and interpretation. ST, PD, and YS drafted the manuscript. All authors reviewed the manuscript critically for scientific content, and all authors approved the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST
None declared.
Supplementary Material
REFERENCES
- 1. Wiens J, Horvitz E, Guttag JV.. Patient risk stratification for hospital-associated C. diff as a time-series classification task. In: proceedings of the twenty-sixth annual conference on neural information processing systems (NeurIPS); December 2–6, 2012: 467–76; Lake Tahoe, Nevada. [Google Scholar]
- 2. Oh J, Makar M, Fusco C, et al. A generalizable, data-driven approach to predict daily risk of Clostridium difficile infection at two large academic health centers. Infect Control Hosp Epidemiol 2018; 39 (4): 425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li BY, Oh J, Young VB, Rao K, Wiens J.. Using machine learning and the electronic health record to predict complicated Clostridium difficile infection. Open Forum Infect Dis 2019; 6 (5). doi: 10.1093/ofid/ofz186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Desautels T, Calvert J, Hoffman J, et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform 2016; 4 (3): e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Henry KE, Hager DN, Pronovost PJ, Saria S.. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med 2015; 7 (299): 299ra122. [DOI] [PubMed] [Google Scholar]
- 6. Zeiberg D, Prahlad T, Nallamothu BK, Iwashyna TJ, Wiens J, Sjoding MW.. Machine learning for patient risk stratification for acute respiratory distress syndrome. PLOS One 2019; 14 (3): e0214465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koyner JL, Carey KA, Edelson DP, Churpek MM.. The development of a machine learning inpatient acute kidney injury prediction model. Crit Care Med 2018; 46 (7): 1070–77. [DOI] [PubMed] [Google Scholar]
- 8. Tomašev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 2019; 572 (7767): 116–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miotto R, Li L, Kidd BA, Dudley JT.. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016; 6 (1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Silva I, Moody G, Scott DJ, Celi LA, Mark RG.. Predicting in-hospital mortality of ICU patients: the PhysioNet/computing in cardiology challenge 2012. Comput Cardiol 2010; 39: 245–48. [PMC free article] [PubMed] [Google Scholar]
- 11. Harutyunyan H, Khachatrian H, Kale DC, Ver Steeg G, Galstyan A.. Multitask learning and benchmarking with clinical time series data. Sci Data 2019; 6 (1): 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Purushotham S, Meng C, Che Z, Liu Y.. Benchmarking deep learning models on large healthcare datasets. J Biomed Inform 2018; 83: 112–34. [DOI] [PubMed] [Google Scholar]
- 13. Wang S., McDermott MBA., Chauhan G., Ghassemi M., Hughes MC., Naumann T. MIMIC-Extract: a data extraction, preprocessing, and representation pipeline for MIMIC-III. In: proceedings of the Association for Computing Machinery Conference on Health, Inference, and Learning; July 23–24, 2020; Toronto, Canada.
- 14. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O.. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data 2018; 5 (1): 180178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fiterau M, Bhooshan S, Fries J, et al. ShortFuse: biomedical time series representations in the presence of structured information. In: proceedings of the 2nd Machine Learning for Healthcare Conference; August 18–19, 2017; Boston, MA. [PMC free article] [PubMed]
- 17. Wu J, Roy J, Stewart WF.. Prediction modeling using EHR data: challenges, strategies, and a comparison of machine learning approaches. Med Care 2010; 48 (6): S106–13. [DOI] [PubMed] [Google Scholar]
- 18. Hardy MA. Regression with dummy variables. Thousand Oaks, CA: SAGE; 1993. [Google Scholar]
- 19. Cochran WG. The effectiveness of adjustment by subclassification in removing bias in observational studies. Biometrics 1968; 24 (2): 295–313. [PubMed] [Google Scholar]
- 20. Collins GS, Ogundimu EO, Cook JA, Manach YL, Altman DG.. Quantifying the impact of different approaches for handling continuous predictors on the performance of a prognostic model. Stat Med 2016; 35 (23): 4124–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. World Health Organization. The ICD-10 classification of mental and behavioural disorders: clinical descriptions and diagnostic guidelines. Wkly Epidemiol Rec 1992; 67 (30): 227–27. [Google Scholar]
- 22. Zhang Y. A hierarchical approach to encoding medical concepts for clinical notes. In: proceedings of the ACL-08: HLT Student Research Workshop; June 15–20, 2008; Columbus, Ohio.
- 23. Sherman E, Gurm H, Balis U, Owens S, Wiens J. Leveraging clinical time-series data for prediction: a cautionary tale. In: proceedings of the AMIA Annual Symposium; November 4–8, 2017; Washington, DC. [PMC free article] [PubMed]
- 24. Little R, Rubin D.. Statistical analysis with missing data. New York, NY: Wiley; 1987. [Google Scholar]
- 25. Nemati S, Ghassemi MM, Clifford GD. Optimal medication dosing from suboptimal clinical examples: A deep reinforcement learning approach. In: proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); August 16–20, 2016; Orlando, FL. [DOI] [PubMed]
- 26. Churpek MM, Adhikari R, Edelson DP.. The value of vital sign trends for detecting clinical deterioration on the wards. Resuscitation 2016; 102: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Che Z, Purushotham S, Cho K, Sontag D, Liu Y.. Recurrent neural networks for multivariate time series with missing values. Sci Rep 2018; 8 (1): 6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rubin DB. Inference and missing data. Biometrika 1976; 63 (3): 581–92. [Google Scholar]
- 29. Kuhn M. Building predictive models in R using the caret package. J Stat Soft 2008; 28 (5): 1–26. [Google Scholar]
- 30. Tang J, Alelyani S, Liu H.. Feature selection for classification: a review. In: Aggarwal CC, ed. Data Classification: Algorithms and Applications; Boca Raton, FL: CRC Press; 2014: 37–64. [Google Scholar]
- 31. Wang L, Wang Y, Chang Q.. Feature selection methods for big data bioinformatics: a survey from the search perspective. Methods 2016; 111: 21–31. [DOI] [PubMed] [Google Scholar]
- 32. Mitra P, Murthy CA, Pal SK.. Unsupervised feature selection using feature similarity. IEEE Trans Pattern Anal Machine Intell 2002; 24 (3): 301–12. [Google Scholar]
- 33. Yu L, Liu H.. Efficient feature selection via analysis of relevance and redundancy. J Mach Learn Res 2004; 5: 1205–24. [Google Scholar]
- 34. Solorio-Fernández S, Carrasco-Ochoa JA, Martínez-Trinidad JF.. A review of unsupervised feature selection methods. Artif Intell Rev 2020; 53 (2): 907–42. [Google Scholar]
- 35. Oh J, Wang J, Tang S, Sjoding MW, Wiens J. Relaxed parameter sharing: effectively modeling time-varying relationships in clinical time-series. In: proceedings of the 4th Machine Learning for Healthcare Conference; August 8–10, 2019; Ann Arbor, MI.
- 36. Zhang Y, Jarrett D, van der Schaar M. Stepwise model selection for sequence prediction via deep kernel learning. In: proceedings of the 23rd International Conference on Artificial Intelligence and Statistics (AISTATS); August 26–28, 2020.
- 37. O'Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM.. Measuring diagnoses: ICD code accuracy. Health Serv Res 2005; 40 (5p2): 1620–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiens J, Saria S, Sendak M, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019; 25 (9): 1337–40. [DOI] [PubMed] [Google Scholar]
- 39. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1 (1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bergstra J, Bengio Y.. Random search for hyper-parameter optimization. J Machine Learn Res 2012; 13: 281–305. [Google Scholar]
- 41. LaFleur BJ, Greevy RA.. Introduction to permutation and resampling-based hypothesis tests. J Clin Child Adolesc Psychol 2009; 38 (2): 286–94. [DOI] [PubMed] [Google Scholar]
- 42. Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika 1986; 73 (3): 751–4. [Google Scholar]
- 43.Van Rossum G, Drake FL. Python Language Reference, version 3.6. https://www.python.org/ Accessed September 2018.
- 44. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res 2011; 12: 2825–30. [Google Scholar]
- 45. Paszke A, Gross S, Chintala S. Pytorch version 1.0. https://pytorch.org/ Accessed September 2018.
- 46. Pirracchio R. Mortality prediction in the ICU based on MIMIC-II results from the super ICU learner algorithm (SICULA) project. In: Secondary Analysis of Electronic Health Records. Cham, Switzerland: Springer; 2016: 295–313. [PubMed] [Google Scholar]
- 47. Johnson AEW, Pollard TJ, Mark RG. Reproducibility in critical care: a mortality prediction case study. In: proceedings of the 2nd Machine Learning for Healthcare Conference; August 18–19, 2017; Boston, MA.
- 48. Sendak MP, Balu S, Schulman KA.. Barriers to achieving economies of scale in analysis of EHR data. Appl Clin Inform 2017; 8 (3): 826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bender D, Sartipi K. HL7 FHIR: an agile and RESTful approach to healthcare information exchange. In: proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems; June 20–22, 2013; Porto, Portugal.
- 50. Stang PE, Ryan PB, Racoosin JA, et al. Advancing the science for active surveillance: rationale and design for the observational medical outcomes partnership. Ann Intern Med 2010; 153 (9): 600–06. [DOI] [PubMed] [Google Scholar]
- 51. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS.. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc 2014; 21 (4): 578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wiens J, Guttag J, Horvitz E.. A study in transfer learning: leveraging data from multiple hospitals to enhance hospital-specific predictions. J Am Med Inform Assoc 2014; 21 (4): 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wiens J, Shenoy ES.. Machine learning for healthcare: on the verge of a major shift in healthcare epidemiology. Clin Infect Dis 2018; 66 (1): 149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sculley D, Holt G, Golovin D, et al. Machine learning: the high interest credit card of technical debt. Montréal, QC, Canada: SE4ML (Software Engineering for Machine Learning) (NeurIPS 2014 Workshop); 2014.
- 55. Chandrashekar G, Sahin F.. A survey on feature selection methods. Comput Electr Eng 2014; 40 (1): 16–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.