Abstract
Skin cutaneous melanoma is characterized by significant heterogeneity in its molecular, genomic and immunologic features. Whole transcriptome RNA sequencing data from The Cancer Genome Atlas of skin cutaneous melanoma (n = 328) was utilized. CIBERSORT was used to identify immune cell type composition, on which unsupervised hierarchical clustering was performed. Analysis of overall survival was performed using Kaplan–Meier estimates and multivariate Cox regression analyses. Membership in the lymphocyte:monocytelow, monocytehigh and M0high cluster was an independently poor prognostic factor for survival (HR: 3.03; 95% CI: 1.12–8.20; p = 0.029) and correlated with decreased predicted response to immune checkpoint blockade. In conclusion, an M0-macrophage-enriched, lymphocyte-to-monocyte-ratio-low phenotype in the primary melanoma tumor site independently characterizes an aggressive phenotype that may differentially respond to treatment.
Keywords: : cutaneous melanoma, immunotherapy, lymphocytes, macrophages, monocytes, RNA sequencing, tumor immune microenvironment
Practice points.
Melanoma is a deadly skin cancer that is increasing in prevalence yearly. Although most patients with melanoma experience good outcomes, many still do not.
Here the authors use publicly available gene sequencing data from a large cohort of patients with melanoma to better understand how the tumor immune microenvironment might influence disease progression and response to therapies.
The authors found that a subset of patients with tumors containing a low lymphocyte-to-monocyte ratio and high undifferentiated macrophage content in the primary tumor site had significantly decreased overall survival compared with other groups.
Additionally, this same group demonstrated lower predicted rates of response to immune checkpoint blockade therapy and increased rates of metastasis compared with other groups in the total cohort.
The authors' study may provide a tool to help risk-stratify patients with melanoma and may lead to the identification of new therapeutic targets for these patients in the future.
Melanoma is the most aggressive form of skin cancer known, and its incidence continues to rise [1,2]. It is estimated that by the end of 2020 there will be 100,350 new cases and 6850 deaths as a result of melanoma [3]. Several factors have been demonstrated to be predictive of poor prognosis in patients with melanoma; these include sex, age, ulceration, mitotic rate and Breslow tumor thickness [4–6]. Fortunately, most patients who present at an early, localized stage are potentially curable. By contrast, patients who present with advanced melanoma have a poor prognosis, with a 5-year survival rate of 10% [7]. There remains a subset of patients, however, who present with localized disease but still demonstrate a poor overall survival due to recurrence and disease progression [4–7]. It is for this reason that it is of paramount importance to clinicians to identify novel and efficient prognostic markers for patients with malignant melanoma.
It has become well known in recent years that inflammatory responses play key roles in the pathways of tumor initiation, development and progression [8,9]. For this reason, hematologic parameters of the inflammatory response have recently become more well studied and have been shown to demonstrate prognostic value in various cancers [10,11]. These hematologic indexes include markers such as C-reactive protein [11,12], platelet-to-lymphocyte ratio [13,14] and neutrophil-to-lymphocyte ratio [15–17]. However, given the limited prevalence of publicly available whole exome tumor sequencing data, there is a paucity of data on inflammatory parameters within the primary tumor allowing for exploration of the tumor immune microenvironment (TIME).
Recently, it has been found that a decreased peripheral lymphocyte-to-monocyte (L:M) ratio may be a poor prognostic indicator in various cancers [18–23]. The prognostic value of the L:M ratio may be explained by the fact that lymphopenia is a surrogate marker of weak immune response, whereas an elevated monocyte count may stand as a surrogate marker of high tumor burden. Monocytes comprise a large population of innate immune cells that circulate in the bloodstream and traffic to tissues during a steady state, with increased circulation rates during inflammation. These cells play key roles in supporting tissue homeostasis, initiating and propagating host responses to pathogens and resolving immune responses before excessive tissue damage occurs [24]. Recently, monocytes have emerged as important regulators of cancer development and progression, with different subsets appearing to have opposing roles in enabling tumor growth and preventing metastatic spread of cancerous cells [25]. Monocytes also serve as a primary source of long-lived tumor-associated macrophages (TAMs) and dendritic cells that shape the tumor microenvironment [26]. Even more interestingly, it has previously been demonstrated that this ratio may predict immunotherapy response. In 2017, Failing et al. demonstrated that a decreased lymphocyte-to-macrophage ratio (<1.7) in the peripheral blood may in fact predict a poor response to pembrolizumab in metastatic melanoma patients [27]. In this analysis, the authors leverage data from The Cancer Genome Atlas (TCGA) to explore the landscape of TIME in melanoma as well as the prognostic impact of the L:M ratio in whole-exome sequenced tumor specimens, rather than in the peripheral blood, using bulk tumor sequencing data.
Methods
Study design
The authors conducted a retrospective analysis of patients with a histological diagnosis of skin cutaneous melanoma (SKCM) in the TCGA database. Samples came from institutions across Australia, Brazil, Canada, Georgia, Germany, Moldova, Romania, Russia, the United States, Vietnam and Yemen. Each patient with clinical and genomic data was independently reviewed by two authors (NK Jairath and M Farha). Since all data utilized are available to the public, approval from the institutional review board was not required for this analysis. All analyses were performed using R 3.6.3 software (R Foundation for Statistical Computing, Vienna, Austria) for statistical computing.
Clustering based on immune cell subpopulations
The CIBERSORT in silico flow cytometry tool was used to quantify the relative levels of distinct immune cell types in the TCGA SKCM dataset [28]. A mixture file containing RNA sequencing by expression maximization gene expression data from the samples in the TCGA SKCM dataset was downloaded from the cBioPortal and formatted according to the guidelines outlined in the CIBERSORT manual (Palo Alto, CA, USA) [29–31]. The LM22 signature gene file was used as a reference point for comparison. LM22 contains 547 genes that accurately distinguish 22 mature human hematopoietic populations and activation states, including seven T-cell types, naive and memory B cells, plasma cells, natural killer cells and myeloid subsets. Lymphocytes in this analysis included naive and memory B cells; CD8 T cells; naive, resting memory and activated memory CD4 T cells; follicular helper T cells; regulatory T cells; γδ T cells; and natural killer cells. The LM22 file was constructed from the gene expression profiles of those cell types measured on the Affymetrix U133A Plus2 and Illumina Expression BeadChip (HumanHT-12 v4) platforms (Illumina Dx, CA, USA). By default, CIBERSORT estimates the relative fraction of each cell type in the sample, such that the sum of all relative fractions for each of the 22 cell subsets is equal to 1 for the sample.
The ComplexHeatmap package was downloaded from Bioconductor (31) (R Foundation for Statistical Computing). Unsupervised hierarchical clustering was performed by individual patient sample using lymphocyte and monocyte relative infiltration values as well as ratios of macrophage subtypes to total macrophages and L:M ratio as clustering criteria. Clustering was performed on both x- and y-axes to distinguish high versus low expression values of both clusters and cell types. The clustering distance metric was set to maximum distance between rows, and the clustering method was Ward's minimum variance. The dataset was divided into four clusters based on the immune cell subpopulation distribution. Sample identifiers corresponding to each cluster were extracted for further analysis.
Survival analysis of clusters
The primary outcome in the authors' analysis was overall survival (OS), defined as the time from pathologic diagnosis to death or loss to follow-up, as defined by the TCGA study group. Demographic and treatment details, including BRAF, NF1 and RAS mutations; tumor mutational burden; percent necrosis in the pathologic sample; percent tumor content (percentage of cells in the sample that were malignant); Breslow depth; occurrence of metastatic disease; age at diagnosis; lymphocyte score (a surrogate for distribution and density of lymphocytic infiltration in the tumor sample); and timing and dose of therapies, including chemotherapy, immunotherapy and radiation, were obtained using the TCGAbiolinks web tool (R Foundation for Statistical Computing version 3.6.2). Patients still alive were censored at the time of last follow-up. Patients were separated into two groups for Kaplan–Meier analysis: those with evidence of distant metastases or regional lymph node metastasis at diagnosis (metastasis) and those for whom the tumor had not yet metastasized (primary tumor). Each endpoint was assessed using the Kaplan–Meier method, and survival curves were compared using the Mantel–Cox log-rank test. Survival analysis was carried out using the survminer R package (R Foundation for Statistical Computing). Log-rank p-value and risk tables are displayed on each figure.
Retrieving raw count data
The TCGAbiolinks package was downloaded from Bioconductor (31) (R Foundation for Statistical Computing version 3.6.2) [32–34]. The Genomic Data Commons query function was used to retrieve Illumina HiSeq RNA data from primary tumors in the TCGA SKCM dataset, and the data were downloaded using the Genomic Data Commons download function (National Cancer Institute, MD, USA). Raw count data were normalized, and low count genes were filtered according to the default 25% quantile across all samples. The table of normalized and filtered raw count data was extracted for use in downstream analysis.
Statistical analysis
Clinical, pathologic and molecular characteristics were compared between clusters using statistical methods in R using a chi-square test. The Kaplan–Meier method was used to compare endpoints across clusters with the log-rank test. To determine the influence of immunologic and clinicopathologic covariates on OS, a multivariate Cox regression analysis was performed. Statistical significance was set at p < 0.05.
Prediction of immunotherapy response
Understanding the relevance of the immune composition of tumors in predicting response to immune checkpoint blockade (ICB) is essential. However, most publicly available transcriptome data do not have corresponding ICB treatment data. To overcome this limitation, the Tumor Immune Dysfunction and Exclusion (TIDE) tool was utilized (Dana Farber Cancer Institute, MA, USA). TIDE is a web-based tool that uses a gene expression signature to predict response to ICB [35]. Z-score-transformed whole transcriptome (RNA sequencing by expression maximization) data were downloaded from the cBioPortal and input into the tool. TIDE performance has been validated in melanoma and non-small-cell lung cancer datasets, making it an ideal choice for the authors' study. Output included response prediction, with lower TIDE score corresponding to better immunotherapy response, and other gene signatures associated with immune dysfunction, including IFN-γ response, microsatellite instability, CD274 (regulatory T-cell marker) and T-cell dysfunction.
Results
Time clustering & baseline demographics
All patients with SKCM and available clinical and sequencing data in the SKCM TCGA database (n = 328) were clustered based on relative intratumoral lymphocyte, monocyte, M0 macrophage, M1 macrophage and M2 macrophage concentrations in each individual sample. Patients were divided into four clusters: cluster 1 (n = 85, 26%, lymphocyte:monocytelow, monocytehigh and M0high), cluster 2 (n = 116, 35%, lymphocyte:monocytelow, monocytemid, M0Low), cluster 3 (n = 101, 31%, lymphocyte:monocytemid, monocytelow, M0low) and cluster 4 (n = 26, 8%, lymphocyte:monocytehigh, monocytelow and M0low). Full, unsupervised hierarchical clustering for 22 immune cell subsets can be seen in Figure 1A. The absolute immune cell infiltration in each cluster was determined by the CIBERSORT absolute immune score output and displayed in Figure 1B. Pairwise t-tests were conducted to confirm significant differences among cell types between clusters. All clusters demonstrated significantly different levels of each type of immune cell infiltrate (p < 0.05). There were no significant differences between median age, sex, site of primary tumor, specimen site or median follow-up between clusters. Full demographic information can be found in Table 1.
Figure 1. . Immune cell composition of clusters.
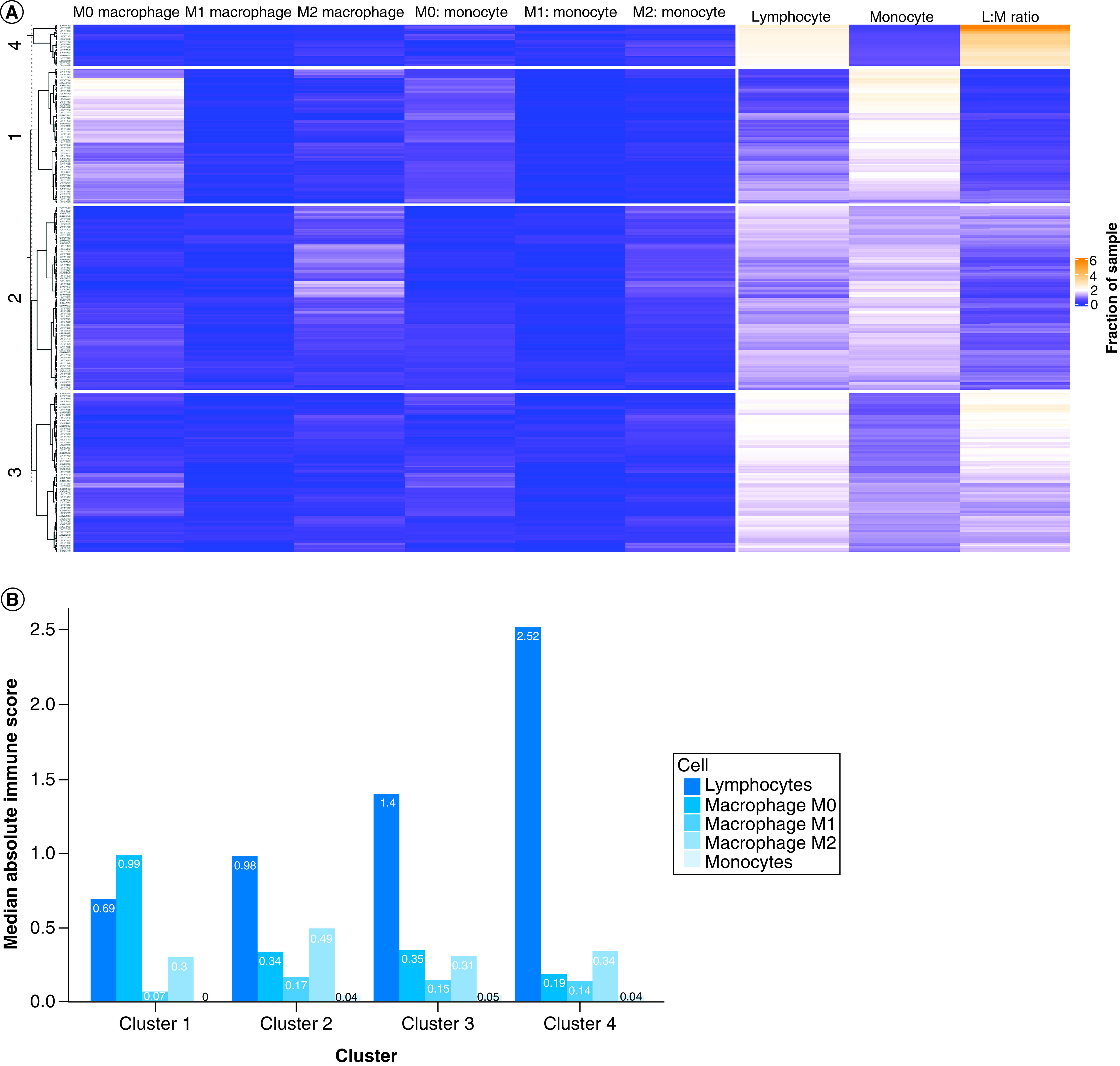
(A) Heatmap detailing the immune microenvironment landscape of melanoma in The Cancer Genome Atlas. (B) Bar plot representing the median absolute immune infiltration separated by cell type and cluster as calculated by CIBERSORT.
LM: Lymphocyte-to-monocyte.
Table 1. . Demographics.
| Patient and tumor characteristics | Cluster 1 (n = 85) | Cluster 2 (n = 116) | Cluster 3 (n = 101) | Cluster 4 (n = 26) |
|---|---|---|---|---|
| Median age at diagnosis, years | 54 | 57 | 57 | 58 |
| Sex | ||||
| – Male | 50 | 62 | 57 | 18 |
| – Female | 35 | 54 | 44 | 8 |
| Site of primary tumor | ||||
| – Trunk | 45 | 58 | 59 | 11 |
| – Extremities | 31 | 41 | 36 | 12 |
| – Head and neck | 7 | 13 | 1 | 3 |
| – Other | 2 | 4 | 3 | 0 |
| Specimen site | ||||
| – Primary tumor | 9 | 13 | 14 | 3 |
| – Regional skin or soft tissue | 18 | 20 | 12 | 3 |
| – Regional lymph node | 38 | 39 | 66 | 18 |
| – Distant metastasis | 13 | 13 | 8 | 2 |
| – Not recorded | 5 | 0 | 1 | 0 |
| – Median follow-up, days | 1060 | 1010 | 1136 | 1133 |
TIME cluster prognostic effect
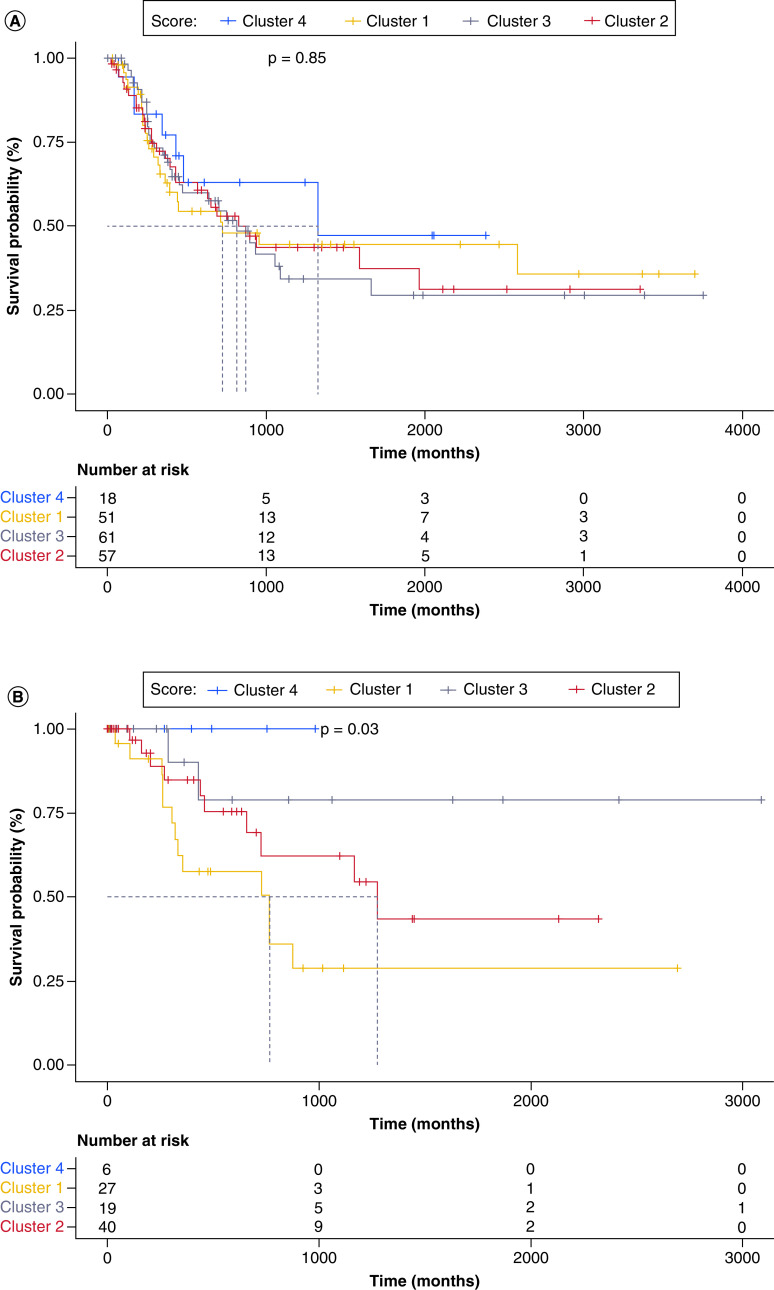
The median follow-up time for all patients was 40 months and was not significantly different across clusters (35, 34, 38 and 38 months for clusters 1–4, respectively). The primary endpoint of the study, OS, was analyzed for each cluster using Kaplan–Meier estimates and compared using the log-rank test. OS was 21.2 months in cluster 1, 35.2 months in cluster 2, 38.1 months in cluster 3 and 55.5 months in cluster 4. Patients were separated into localized (sample curated from primary tumor or regional skin or soft tissue) or metastatic (distant metastasis or regional lymph node) collection site to understand whether TIME of the primary site might guide tumor behavior. There were no significant differences between clusters in the metastatic site cohort (p = 0.85) (Figure 2A). The cluster with the poorest performance in the localized site SKCM cohort was cluster 1, as demonstrated by pairwise comparisons, which expressed an lymphocyte:monocytelow, monocytehigh and M0High phenotype (p = 0.03) (Figure 2B).
Figure 2. . Kaplan–Meier curves demonstrating the overall survival of all four clusters in The Cancer Genome Atlas skin cutaneous melanoma cohort.
(A) Kaplan–Meier curve demonstrating survival difference between clusters based on collection from distant metastatic or regional lymph node site. (B) Kaplan–Meier curve demonstrating survival difference between clusters based on collection from primary tumor or regional skin or soft tissue site.
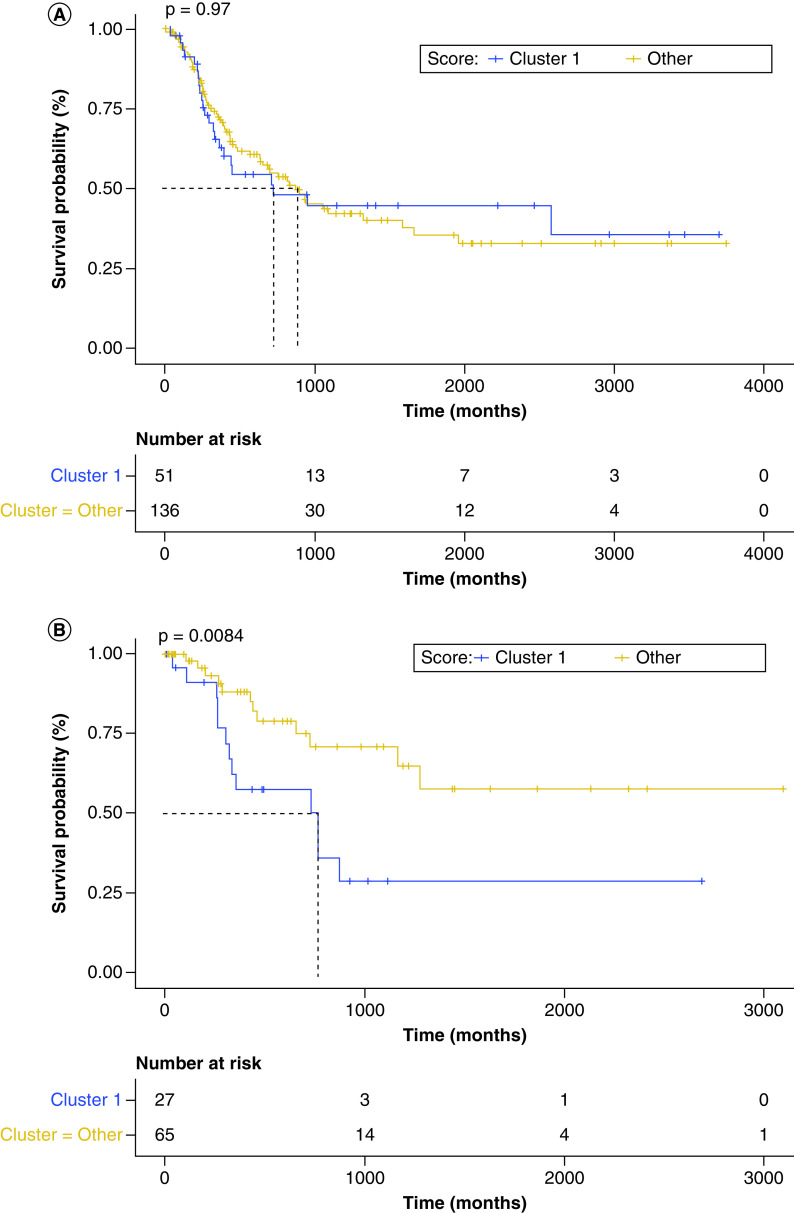
When stratifying patients by tumor specimen curated from a localized site versus a metastatic site and grouping clusters to isolate the poorly performing cluster 1, the metastatic group demonstrated no significant difference between the lymphocyte:monocytelow, M0-enriched group (cluster 1) and the other clusters in the analysis (HR: 0.9919; 95% CI: 0.6145–1.601; p = 0.974) (Figure 3A). However, when examining only the localized samples, the lymphocyte:monocytelow, M0-enriched group (cluster 1) demonstrated significantly poorer survival compared with the other groups (HR: 2.804; 95% CI: 1.262–6.234; p = 0.0114) (Figure 3B). These results indicate a poorer OS in patients who have localized tumors with M0-macrophage-enriched and lymphocyte:monocytelow phenotype.
Figure 3. . Survival differences between clusters.
(A) Kaplan–Meier curve displaying overall survival, which demonstrates no difference between cluster 1 (L:Mlow and M0-enriched) and the remaining clusters (grouped) for patients in whom the tissue site sequenced was a distant metastasis or regional lymph node (HR: 0.9919; 95% CI: 0.6145–1.601; p = 0.974). (B) Kaplan–Meier curve displaying overall survival, which demonstrates a significant difference between cluster 1 (L:Mlow and M0-enriched) and the remaining clusters (grouped) for patients in whom the tissue site sequenced was a distant metastasis or regional lymph node. Cluster 1 demonstrated a poorer prognosis (HR: 2.804; 95% CI: 1.262–6.234; p = 0.0114).
LM: Lymphocyte-to-monocyte.
Multivariate survival analysis
After establishing that SKCM with an M0-macrophage-enriched, L:Mlow phenotype in the primary tumor site offers the poorest prognosis of the immune subpopulations, a multivariate analysis was performed to determine the independent prognostic impact of TIME clusters on OS in the entire cohort (Figure 4). Only patients with complete clinical data were included in the multivariate analysis. Cluster 1, with cluster 4 as a reference (HR: 3.03; 95% CI: 1.12–8.20; p = 0.029), was independently prognostic for the endpoint of OS. Stage II disease (HR: 11.89; 95% CI: 2.70–52.30; p = 0.001), stage III disease (HR: 31.41; 95% CI: 7.35–134.20; p < 0.001) and stage IV disease (HR: 72.05; 95% CI: 11.13–466.40; p < 0.001) at diagnosis also correlated with significantly worse prognosis in a predictable pattern when compared with the reference stage I, as did Breslow depth (HR: 1.32 per mm increase; 95% CI: 1.07–1.60; p = 0.009). BRAF V600K mutation status also trended toward poorer prognosis (HR: 2.47; 95% CI: 0.86–7.10; p = 0.093) but did not reach statistical significance.
Figure 4. . Multivariate Cox regression analysis demonstrating the independent prognostic value of important clinical and genetic factors, including sex, age, stage, Breslow depth, BRAF status and cluster designation.
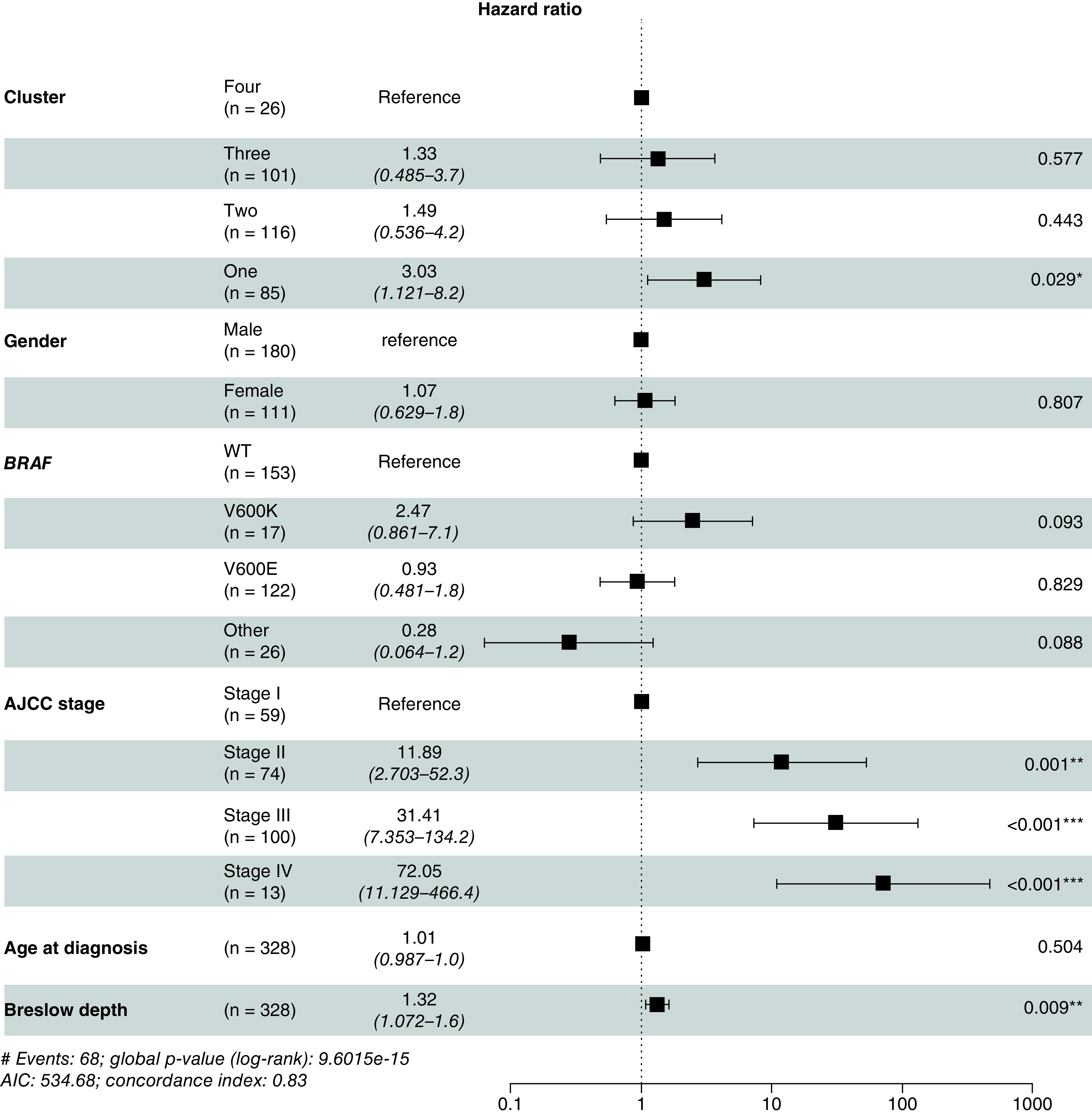
Cluster 1 was independently prognostic of overall survival (HR: 3.02; 95% CI: 1.30–7.02; p = 0.01).
*P < 0.05; **P < 0.01 and ***P < 0.001.
AIC: Akaike Information Criterion; AJCC: American Joint Commission on Cancer; HR: Hazard ratio.
TIDE ICB response, clinical & molecular characteristics
Using the TIDE tool described earlier as well as clinical and molecular data collected by the TCGA study group, the authors sought to characterize the poorly performing M0-macrophage-enriched, L:Mlow cluster 1 by predicted response to ICB as well as known poor prognosticators and clinical outcomes. Cluster 1 demonstrated increased rates of metastasis (51 vs 40, 41 and 42% for clusters 2–4, respectively); decreased predicted response to immunotherapy (19 vs 34, 35 and 38% in clusters 2–4, respectively); increased rate of BRAF mutations (64 vs 51, 44 and 44% in clusters 2–4, respectively); increased percent necrosis in the pathologic tumor sample (5.8 vs 2.7, 3.2 and 0.8% in clusters 2–4, respectively); increased median Breslow depth (4.5 vs 3, 3.2 and 2.5 mm in clusters 2–4, respectively); and decreased lymphocyte score, a surrogate for distribution and density of lymphocytic infiltration in the tumor sample (2.2 vs 2.8, 3 and 3.9 for clusters 2–4, respectively). Notably, tumor mutational burden and tumor content were similar across clusters, and cluster 1 demonstrated few RAS mutations and a decreased microsatellite instability score when compared with the other clusters. Complete representation of these data can be seen in Table 2.
Table 2. . Molecular landscape, clinical factors and markers of immune response by cluster in The Cancer Genome Atlas.
| Tumor molecular, clinical and immune characteristics | Cluster 1 (n = 85) | Cluster 2 (n = 116) | Cluster 3 (n = 101) | Cluster 4 (n = 26) |
|---|---|---|---|---|
| BRAF, % | 64 | 51 | 44 | 44 |
| NF1, % | 19 | 16 | 16 | 4 |
| RAS, % | 21 | 34 | 31 | 27 |
| TMB | 457 | 482 | 399 | 299 |
| Necrosis, %† | 5.8 | 2.7 | 3.2 | 0.8 |
| Tumor content, %‡ | 87 | 84 | 85 | 82 |
| Lymphocyte score§ | 2.2 | 2.8 | 3 | 3.9 |
| Breslow depth, median | 4.5 | 3 | 3.2 | 2.5 |
| Any metastatic disease, % | 51 | 40 | 41 | 42 |
| Metastatic recurrence, % | 42 | 34 | 35 | 38 |
| TIDE ICB response, % | 19 | 34 | 35 | 27 |
| TIDE raw score | 0.54 | 0.24 | -0.6 | 0.35 |
| IFN-γ response score | -0.27 | 0.06 | 0.05 | 0.52 |
| MSI score | 0.41 | 0.54 | 0.49 | 0.51 |
| CD274 score | 0.06 | 0.03 | -0.03 | 0.05 |
| T-cell dysfunction-- | -0.15 | -0.17 | -0.06 | 0.62 |
Reported as a mean value; remaining continuous variables reported as median values.
Represents percentage of malignant cells in the tumor sample.
Reported as calculated by the TCGA study group.
ICB: Immune checkpoint blockade; MSI: Microsatellite instability; TCGA: The Cancer Genome Atlas; TIDE: Tumor immune dysfunction and exclusion; TMB: Tumor mutational burden.
Discussion
With the prevalence of melanoma continuing to rise and the persistence of a subgroup of patients with a poor prognosis despite appropriate therapies, it is of increasing importance that novel predictors of therapeutic response and therapeutic targets are developed. In this analysis, the authors explore the landscape of TIME in melanoma and identify unique clusters based on the prevalence of lymphocytes and monocytes within tumor samples. The authors demonstrate that a low lymphocyte:monocyte ratio in the primary tumor specimen with high M0 macrophage enrichment confers worse prognosis in SKCM. Additionally, this group displays fewer predicted responders to immunotherapy, concordant with results presented from analysis involving peripheral blood ratios [27], poorly prognostic clinical characteristics (high tumor mutational burden, necrosis, low lymphocyte score, high percentage of BRAF mutants) and high rates of metastatic disease and recurrence. Together, these results suggest that a large undifferentiated macrophage pool in the setting of low lymphocytic infiltration in the primary tumor site may serve to create a favorable environment for tumor progression. It is also possible that undifferentiated macrophages exert an immunosuppressive effect on the tumor, leading to poor lymphocytic infiltration and thus poorer OS.
Peripheral blood lymphocyte:monocyte ratio was first found to be a prognostic factor in hematological malignancies [36,37]. Subsequently, a higher L:M ratio has been shown to be associated with improved OS in over a dozen different solid tumors [38]. Most of these studies have been published since 2014, which exemplifies the recent interest in the prognostic value of the lymphocyte:monocyte ratio. Lymphocyte:monocyte ratio derived from primary or metastatic tumor sequencing has yet be to investigated. It is as yet unclear what the actual mechanisms are behind the correlation of low lymphocyte:monocyte ratio with poor outcome in cancer patients. It is possible this could be mediated through tumor-infiltrating immune cells, which play a critical role in tumor growth suppression and enhancement. Of particular importance are tumor-infiltrating lymphocytes and TAMs. These immune cells are found in tumors and have been found to be important prognostic factors in various cancers [39]. It is thought that tumor-infiltrating lymphocytes are involved in cellular and humoral anti-tumor immune responses that themselves are involved in tumor control. Moreover, high tumor-infiltrating lymphocyte numbers are associated with improved clinical outcomes [40–42]. Additionally, it has been demonstrated that lymphopenia correlated with OS in a prospectively collected series of patients with metastatic breast cancer, non-Hodgkin lymphoma and soft tissue sarcoma [43]. A low lymphocyte count might result in an inadequate immune response in the control of the tumor, which may help to explain why a low lymphocyte:monocyte ratio correlated with poorer OS and increased rates of metastasis, necrosis and Breslow depth at diagnosis. With regard to melanoma, a recent study by Osella-Abate et al. demonstrated that increased expression of regulatory T-cell markers, known to be associated with anergy and blunted CD8 T-cell response, led to decreased tumor regression compared with groups that demonstrated decreased regulatory T-cell infiltration [44]. This suggests that more targeted approaches to specific lymphocyte populations may prove beneficial to achieving melanoma tumor regression.
TAMs are regarded as key contributors to the crosstalk between tumor and stromal cells, orchestrating key events necessary for cancer progression, including skewing adaptive responses, cell growth, angiogenesis and extracellular matrix remodeling, changes that all lead to a pre-metastatic niche [45,46]. Further subclassification of TAMs is necessary, as their polarization influences their behavior. At a basic level, macrophages are separated into the M1 subtype, which is pro-inflammatory, anti-fibrotic and activated by lipopolysaccharides, TNF and IFN-γ, and the M2 subtype, which is anti-inflammatory, pro-fibrotic and stimulated by IL-4 and IL-13 [47,48]. Given the dynamic nature of the tumor microenvironment and the numerous stimuli within it [47], emerging classification paradigms describe TAMs on a continuum of many subtypes or as a mixed phenotype that is consistent with neither M1 nor M2 phenotypes [49]. Regardless of the phenotype, all TAMs participate in some degree of immunosuppression [50].
In ovarian cancer and glioblastoma, transcriptomic profiling has demonstrated that M0 macrophages do not fit into the canonical M1 or M2 model, but M0 macrophages do have high expression of M2 markers and a transcriptional profile more similar to M2 macrophages [51,52]. Ultimately, M0s may represent another type of TAM or an incompletely differentiated M2 [51]. M0 macrophages have been found to be one of the cell subsets most strongly associated with poor outcome in breast cancer [53], prostate cancer [54] and lung adenocarcinoma [55], whereas reduced M0 content has been associated with better prognosis in bladder cancer [56]. In a comprehensive analysis of digestive system cancers, M0 macrophages were among the most prevalent immune cell fractions, with M0-enriched clusters associated with decreased recurrence-free survival and worse prognostic immune score [57]. It is possible that the presence of these tumor-promoting cells offered more prognostic significance in the primary tumor sample in the authors' current study because of their promotion of immunosuppression, leading to aggressive phenotype and a pre-metastatic niche, which translated to poorer survival. In patients who have already metastasized, prognosis will likely already be poorer, and the relative intratumoral immune cell concentrations at these sites thereby offer little in the way of stratifying prognosis.
With the advent of pembrolizumab, nivolumab and ipilimumab, the outlook for treatment of advanced melanoma has improved significantly. Patients treated with pembrolizumab in the KEYNOTE-001 study were found to have a median progression-free survival of 4 months and a median OS of 23 months [58]. However, these medications fail to induce a response in a subset of patients, they can cause immune-related adverse events and they are quite expensive. Therefore, finding biomarkers to help predict which patients are more likely to benefit from treatment is an important area of investigation. The authors' study found that the cluster of patients with a decreased lymphocyte:monocyte ratio and M0 macrophage enrichment demonstrated worse OS in the primary tumor and in a multivariate analysis accounting for other poor clinical prognostic factors, such as stage, Breslow depth and BRAF status. Monocytes are recruited into tumors and promote tumor progression [59]. IL-10 is an immunosuppressive cytokine produced mainly by monocytes. In metastatic melanoma, high IL-10 levels have been correlated with worse survival [60]. Treatment with ipilimumab, nivolumab or an ipilimumab/nivolumab combination in patients caused significant changes in gene expression in CD3+ T cells but relatively fewer changes in monocytes [61]. These properties of monocytes may explain the authors' finding that patients with higher baseline M0 macrophage content (and therefore lower L:M ratio) demonstrated poor prognosis and fewer predicted responders to immunotherapy.
The authors' analysis is a snapshot in time, reflecting when the tumor was resected and sequenced. As a result, the dynamic influences on macrophage polarization and the changing tumor microenvironment were not captured. Additionally, bulk tumor specimen analysis does not capture the immune contexture that is critical to macrophage behavior. Single-cell approaches may address some of these issues; however, they are subject to a bias toward more highly expressed genes, they require optimally preserved clinical specimens and their high cost limits profiling of large numbers of patients [62]. For these reasons, in a clinical decision-making context, an exclusively single-cell approach is not feasible. Thus, it is important to use single-cell approaches to augment bulk tumor profiling from databases such as TCGA by validating findings from larger-scale analyses of bulk specimens.
Conclusion
The authors' study has built on the previous literature by characterizing immune clusters within SKCM. With this study, the authors characterize the immune microenvironment landscape of melanoma as it relates to intratumoral lymphocyte and macrophage concentrations and thereby define a subset of patients with melanoma who display poorer overall outcomes, including decreased OS and fewer ICB responders, based on primary tumor biopsy findings. This subset of patients may benefit from more aggressive treatment earlier in the disease course. Additionally, the authors demonstrate a technique to conduct a comprehensive bioinformatical analysis using publicly available whole exome sequencing data to characterize the tumor immune microenvironment in SKCM and how it relates to clinical outcomes. These results shed light on a potential risk stratification tool and provide an impetus to further explore the TIME of melanoma, especially as it relates to the primary tumor site, and how it may influence disease progression and define novel therapeutic targets in the future.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit https://osf.io/3xk6u/?view_only=8e316039e80842209742245fc31a8eb2
Author contributions
This article was conceptualized by NK Jairath, M Farha, R Jairath, P W Harms, L C Tsoi and T Tejasvi; methodology contribution by NK Jairath, M Farha, R Jairath, P W Harms, L C Tsoi and T Tejasvi; study validation by NK Jairath, M Farha, R Jairath, P W Harms, L C Tsoi and T Tejasvi; formal analysis by M Farha and NK Jairath; investigation by NK Jairath and M Farha; resources provided by NK Jairath and M Farha; data curation by NK Jairath and M Farha. Original draft was prepared by NK Jairath, M Farha, R Jairath, P W Harms, L C Tsoi and T Tejasvi; review and editing by NK Jairath, M Farha, R Jairath, P W Harms, L C Tsoi and T Tejasvi; visualization by NK Jairath, R Jairath and M Farha. Supervision by P W Harms, L C Tsoi and T Tejasvi; project administration: T Tejasvi; funding acquisition: NK Jairath.
Financial & competing interests disclosure
This research was funded by NIH T35 grant no. HL007690-35 (NK Jairath). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
Since all data utilized in this retrospective analysis are available to the public, approval from the institutional review board was not required.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Davey RJ, van der Westhuizen A, Bowden NA. Metastatic melanoma treatment: combining old and new therapies. Crit. Rev. Oncol. Hematol. 98, 242–253 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Eggermont AM, Spatz A, Robert C. Cutaneous melanoma. Lancet 383(9919), 816–827 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 70(1), 7–30 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Balch CM, Gershenwald JE, Soong SJ. et al. Multivariate analysis of prognostic factors among 2,313 patients with stage III melanoma: comparison of nodal micrometastases versus macrometastases. J. Clin. Oncol. 28(14), 2452–2459 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson JF, Soong SJ, Balch CM. et al. Prognostic significance of mitotic rate in localized primary cutaneous melanoma: an analysis of patients in the multi-institutional American Joint Committee on Cancer melanoma staging database. J. Clin. Oncol. 29(16), 2199–2205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balch CM, Soong SJ, Gershenwald JE. et al. Age as a prognostic factor in patients with localized melanoma and regional metastases. Ann. Surg. Oncol. 20(12), 3961–3968 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garbe C, Eigentler TK, Keilholz U, Hauschild A, Kirkwood JM. Systematic review of medical treatment in melanoma: current status and future prospects. Oncologist 16(1), 5–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coussens LM, Werb Z. Inflammation and cancer. Nature 420(6917), 860–867 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature 454(7203), 436–444 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Templeton AJ, McNamara MG, Šeruga B. et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J. Natl Cancer Inst. 106(6), dju124 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Shrotriya S, Walsh D, Bennani-Baiti N, Thomas S, Lorton C. C-reactive protein is an important biomarker for prognosis, tumor recurrence and treatment response in adult solid tumors: a systematic review. PLoS One 10(12), e0143080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leuzzi G, Galeone C, Gisabella M. et al. Baseline C-reactive protein level predicts survival of early-stage lung cancer: evidence from a systematic review and meta-analysis. Tumori. 102(5), 441–449 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Gu X, Gao XS, Cui M. et al. Clinicopathological and prognostic significance of platelet to lymphocyte ratio in patients with gastric cancer. Oncotarget 7(31), 49878–49887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu X, Sun S, Gao XS. et al. Prognostic value of platelet to lymphocyte ratio in non-small cell lung cancer: evidence from 3,430 patients. Sci. Rep. 6, 23893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchioni M, Cindolo L, Autorino R. et al. High neutrophil-to-lymphocyte ratio as prognostic factor in patients affected by upper tract urothelial cancer: a systematic review and meta-analysis. Clin. Genitourin. Cancer 15(3), 343–349.e1 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Yin J, Qin Y, Luo YK, Feng M, Lang JY. Prognostic value of neutrophil-to-lymphocyte ratio for nasopharyngeal carcinoma: a meta-analysis. Medicine (Baltimore) 96(29), e7577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Sun Y, Peng P, Zhu S, Sun W, Zhang P. Prognostic and clinicopathologic significance of neutrophil-to-lymphocyte ratio in esophageal squamous cell carcinoma: evidence from a meta-analysis. Onco. Targets Ther. 10, 1165–1172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stotz M, Szkandera J, Stojakovic T. et al. The lymphocyte to monocyte ratio in peripheral blood represents a novel prognostic marker in patients with pancreatic cancer. Clin. Chem. Lab. Med. 53(3), 499–506 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Neofytou K, Smyth EC, Giakoustidis A. et al. The preoperative lymphocyte-to-monocyte ratio is prognostic of clinical outcomes for patients with liver-only colorectal metastases in the neoadjuvant setting. Ann. Surg. Oncol. 22(13), 4353–4362 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Lucca I, de Martino M, Hofbauer SL, Zamani N, Shariat SF, Klatte T. Comparison of the prognostic value of pretreatment measurements of systemic inflammatory response in patients undergoing curative resection of clear cell renal cell carcinoma. World J. Urol. 33(12), 2045–2052 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Hutterer GC, Sobolev N, Ehrlich GC. et al. Pretreatment lymphocyte-monocyte ratio as a potential prognostic factor in a cohort of patients with upper tract urothelial carcinoma. J. Clin. Pathol. 68(5), 351–355 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Szkandera J, Gerger A, Liegl-Atzwanger B. et al. The lymphocyte/monocyte ratio predicts poor clinical outcome and improves the predictive accuracy in patients with soft tissue sarcomas. Int. J. Cancer 135(2), 362–370 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Stotz M, Pichler M, Absenger G. et al. The preoperative lymphocyte to monocyte ratio predicts clinical outcome in patients with stage III colon cancer. Br. J. Cancer 110(2), 435–440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11(11), 762–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna RN, Cekic C, Sag D. et al. Patrolling monocytes control tumor metastasis to the lung. Science 350(6263), 985–990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat. Rev. Cancer 16(7), 447–462 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Failing JJ, Yan Y, Porrata LF, Markovic SN. Lymphocyte-to-monocyte ratio is associated with survival in pembrolizumab-treated metastatic melanoma patients. Melanoma Res. 27(6), 596–600 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Newman AM, Steen CB, Liu CL. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37(7), 773–782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2(5), 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, Aksoy BA, Dogrusoz U. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6(269), pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32(18), 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Colaprico A, Silva TC, Olsen C. et al. TCGAbiolinks: an r/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44(8), e71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva TC, Colaprico A, Olsen C. et al. Analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Res. 5, 1542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mounir M, Lucchetta M, Silva TC. et al. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 15(3), e1006701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang P, Gu S, Pan D. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24(10), 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li ZM, Huang JJ, Xia Y. et al. Blood lymphocyte-to-monocyte ratio identifies high-risk patients in diffuse large B-cell lymphoma treated with R-CHOP. PLoS One 7(7), e41658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porrata LF, Ristow K, Colgan JP. et al. Peripheral blood lymphocyte/monocyte ratio at diagnosis and survival in classical Hodgkin's lymphoma. Haematologica 97(2), 262–269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishijima TF, Muss HB, Shachar SS, Tamura K, Takamatsu Y. Prognostic value of lymphocyte-to-monocyte ratio in patients with solid tumors: a systematic review and meta-analysis. Cancer Treat. Rev. 41(10), 971–978 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Man YG, Stojadinovic A, Mason J. et al. Tumor-infiltrating immune cells promoting tumor invasion and metastasis: existing theories. J. Cancer 4(1), 84–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahmoud SM, Paish EC, Powe DG. et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 29(15), 1949–1955 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br. J. Cancer 105(1), 93–103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortes C, Mastroeni S, Mannooranparampil TJ. et al. Tumor-infiltrating lymphocytes predict cutaneous melanoma survival. Melanoma Res. 25(4), 306–311 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Ray-Coquard I, Cropet C, Van Glabbeke M. et al. Lymphopenia as a prognostic factor for overall survival in advanced carcinomas, sarcomas, and lymphomas. Cancer Res. 69(13), 5383–5391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osella-Abate S, Conti L, Annaratone L. et al. Phenotypic characterisation of immune cells associated with histological regression in cutaneous melanoma. Pathology 51(5), 487–493 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Solinas G, Germano G, Mantovani A, Allavena P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J. Leukoc. Biol. 86(5), 1065–1073 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol. 22(2), 231–237 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Xue J, Schmidt SV, Sander J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40(2), 274–288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Capece D, Fischietti M, Verzella D. et al. The inflammatory microenvironment in hepatocellular carcinoma: a pivotal role for tumor-associated macrophages. Biomed Res. Int. 2013, 187204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torroella-Kouri M, Silvera R, Rodriguez D. et al. Identification of a subpopulation of macrophages in mammary tumor-bearing mice that are neither M1 nor M2 and are less differentiated. Cancer Res. 69(11), 4800–4809 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Laoui D, Van Overmeire E, Movahedi K. et al. Mononuclear phagocyte heterogeneity in cancer: different subsets and activation states reaching out at the tumor site. Immunobiology 216(11), 1192–1202 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Zhang Q, Li H, Mao Y. et al. Apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferation and migration of ovarian cancer cells by activating the ERK signaling pathway. Int. J. Mol. Med. 45(1), 10–22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabrusiewicz K, Rodriguez B, Wei J. et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 1(2), e85841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PLoS Med. 13(12), e1002194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jairath NK, Farha MW, Srinivasan S. et al. Tumor immune microenvironment clusters in localized prostate adenocarcinoma: prognostic impact of macrophage enriched/plasma cell non-enriched subtypes. J. Clin. Med. 9(6), 1973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Wu S, Yang Y, Zhao M, Zhu G, Hou Z. The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed. Pharmacother. 95, 55–61 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Li W, Zeng J, Luo B. et al. High expression of activated CD4+ memory T cells and CD8+ T cells and low expression of M0 macrophage are associated with better clinical prognosis in bladder cancer patients. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 36(2), 97–103 (2020). [PubMed] [Google Scholar]
- 57.Yang S, Liu T, Cheng Y, Bai Y, Liang G. Immune cell infiltration as a biomarker for the diagnosis and prognosis of digestive system cancer. Cancer Sci. 110(12), 3639–3649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ribas A, Hamid O, Daud A. et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA 315(15), 1600–1609 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel) 6(3), 1670–1690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torisu-Itakura H, Lee JH, Huynh Y, Ye X, Essner R, Morton DL. Monocyte-derived IL-10 expression predicts prognosis of stage IV melanoma patients. J. Immunother. 30(8), 831–838 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Das R, Verma R, Sznol M. et al. Combination therapy with anti-CTLA-4 and anti-PD-1 leads to distinct immunologic changes in vivo. J. Immunol. 194(3), 950–959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suvà ML, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol. Cell 75(1), 7–12 (2019). [DOI] [PubMed] [Google Scholar]