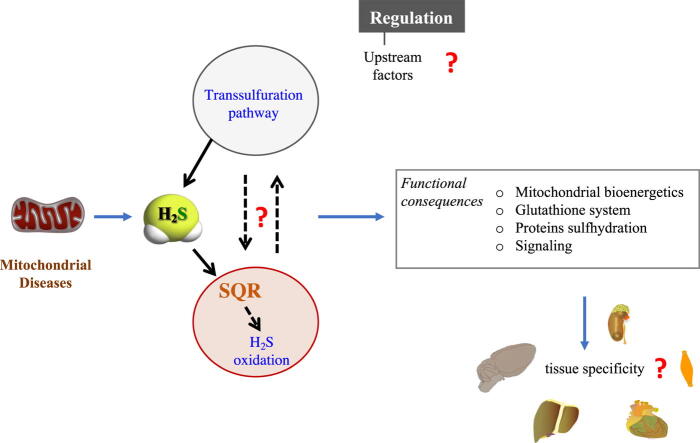
Graphical abstract
Keywords: Coenzyme Q, Mitochondria, ROS, Oxidative stress, Glutathione
Abstract
Background
Mitochondrial disorders are genetic diseases for which therapy remains woefully inadequate. Therapy of these disorders is particularly challenging partially due to the heterogeneity and tissue-specificity of pathomechanisms involved in these disorders. Abnormalities in hydrogen sulfide (H2S) metabolism are emerging as novel mechanism in mitochondrial dysfunction. However, further studies are necessary to understand the effects, protective or detrimental, of these abnormalities, and their relevance, in mitochondrial diseases.
Aim of Review
To review the recent evidences of derangement of the metabolism of H2S, at biosynthesis or oxidation levels, in mitochondrial dysfunction, focusing specifically on the alterations of H2S oxidation caused by primary Coenzyme Q (CoQ) deficiency.
Key Scientific Concepts of Review
Mitochondria play a key role in the regulation of H2S and GSH metabolism pathways. However, further studies are needed to understand the consequences of abnormalities of H2S and GSH synthesis on the oxidation pathway, and vice versa; and on the levels of H2S and GSH, their tissue-specific detrimental effects, and their role the role in mitochondrial diseases. Beside the known H2S pathways, additional, tissue-specific, enzymatic systems, involved in H2S production and elimination, might exist.
Introduction
Mitochondrial diseases are metabolic disorders, clinically heterogeneous, and characterized by tissue specificity [1]. The reasons of this tissue-specificity are not completely understood [1], but it may be related to tissue-specific pathological or compensatory mechanisms. Recent findings in in vitro and in vivo models of mitochondrial diseases indicate that mitochondrial dysfunction alters the metabolism of hydrogen sulfide (H2S), at biosynthesis or oxidation levels [2], [3], [4], [5], [6], [7], [8]. But whether, in the context of mitochondrial diseases, these abnormalities have functional, beneficial or detrimental relevance, and how modifications of the biosynthetic and catabolic pathways affect each other, and H2S levels, are still unclear. Here, we review these recent findings, focusing specifically on the alterations of H2S oxidation caused by primary Coenzyme Q (CoQ) deficiency [9], [10].
Impairment of H2S oxidation in CoQ deficiency
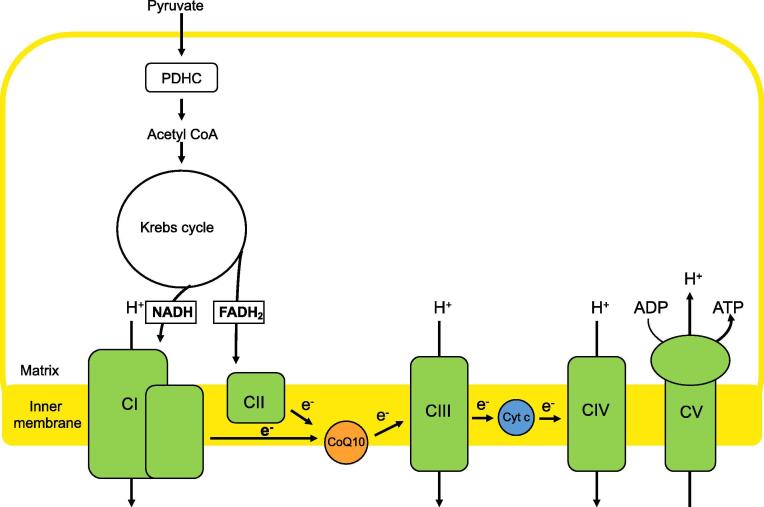
CoQ is a critical intermediate in the mammalian respiratory chain located in the mitochondrial inner membrane (MIM), as it transfers electrons from complexes I (CI) and II (CII) to complex III (CIII) in the course of producing ATP via oxidative phosphorylation (OxPhos) (Fig. 1) [11]. {Turunen, 2004 #8} Therefore, it is not surprising that mutations in CoQ synthesis can cause OxPhos disease [9], [10], presumably via (a) reduced ATP synthesis as a result of reduced electron flow and/or (b) via increases in CoQ-derived reactive oxygen species (ROS) that damage the OxPhos machinery [11]. In fact, there are several lines of in vitro and in vivo evidence that oxidative stress is a deleterious factor in the pathogenesis of CoQ deficiency [12], [13], [14], [15], [16], [17].
Fig. 1.
Schematic representation of CoQ in the mitochondria. CoQ transfers electrons from NADH-ubiquinoneoxidoreducatse (CI) to succinate dehydrogenase (CII) and CoQ-cytochrome c reductase (CIII) in the mitochondrial respiratory chain.
However, besides its role in the respiratory chain, CoQ participates in other metabolic pathways [11], including the conversion of sulfide (as H2S) and sulfite (as SO3) to thiosulfate (as SSO3) by sulfide-quinone oxidoreductase (SQR; SQOR; gene SQOR) (Fig. 1), the first reaction in the H2S oxidation pathway (Fig. 2) [18]. Importantly, electrons, in this redox reaction, that pass through CoQ are transferred to CIII and CIV, bypassing CI and CII. The ATP produced via this “SQR-driven respiration” is ordinarily a minor fraction of total ATP synthesis, but becomes significant in reducing environments [19], [20].
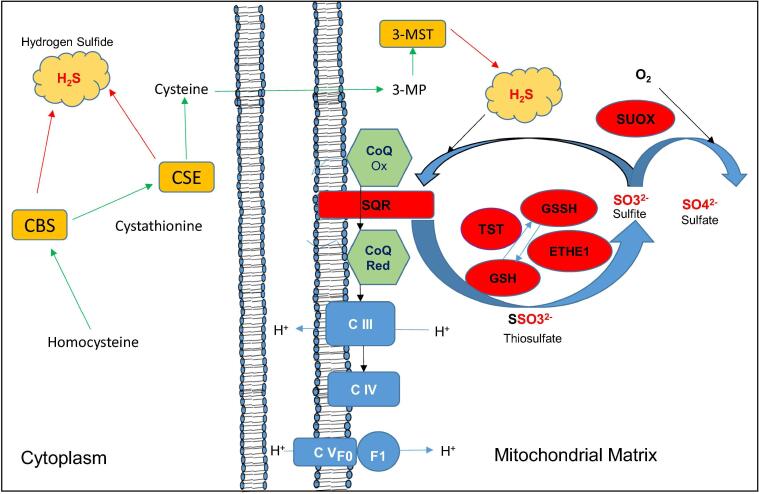
Fig. 2.
Transsulfuration and H2S oxidation pathways. In the transsulfuration pathway (in orange), the enzymes cystathionine β-synthase (CBS) and cystationine γ-ligase (CSE) use cysteine as a substrate for the synthesis of sulfides (H2S). The same cysteine is used by glutamate cysteine ligase (GCL) for the synthesis of GSH in the glutathione pathway (not shown). In the mitochondrial H2S oxidation (in red), sulfide-quinone oxidoreductase (SQR) converts sulfide into thiosulfate by transferring two electrons from H2S to CoQ. Thiosulfate is then converted into sulfite by thiosulfate sulfurtransferase (TST) and persulfide dioxygenase (ETHE1), a reaction that requires glutathione (GSH) as an electron acceptor. Excess sulfite is converted into sulfate by sulfite oxidase (SUOX). 3-MP = 3-mercaptopyruvate, 3-MST = 3-mercaptopyruvate sulfurtransferase, CoQ Ox = Coenzyme Q oxidized, CoQRed = Coenzyme Q reduced..
The initial evidence of this biological function of CoQ was identified in yeast ~ 20 years ago [21], [22], when studies in the fission yeast S. pombe revealed that strains with defects in CoQ biosynthetic genes, were unable to produce CoQ, accumulated H2S, and required cysteine and glutathione to grow on minimal medium [21], [22]. In these strains, grown in both rich and minimum media, levels of H2S are decreased by cysteine supplementation, suggesting that cysteine, one of the sulfur amino acids together with methionine, controls the production of H2S [21], [22].
More recent studies in mammalian cells and tissues, showed that CoQ deficiency severely decreases SQR levels and, as a consequence, impairs SQR-driven respiration, and H2S oxidation, and leads to accumulation of H2S [7], [8]. Specifically, defects of SQR-driven respiration and decreased levels of SQR protein, proportional to the decrease in the levels of CoQ, were found in fibroblasts from patients with CoQ deficiency due to various molecular defects in enzymes of CoQ biosynthesis [7], [8]. These abnormalities were rescued by CoQ supplementation, and recapitulated by genetic and pharmacological inhibition of CoQ synthesis, indicating that CoQ regulates SQR levels. In contrast, SQR, rather than CoQ levels, were responsible for the up-regulation of the down-stream H2S oxidation enzymes (Fig. 2) [7], [8].
Importantly, CoQ supplementation also increased levels of SQR in control cells [7], indicating that CoQ regulates H2S oxidation in physiological conditions, as well as in pathological states. This finding might have therapeutic implications for mitochondrial OxPhos defects other than CoQ deficiency, as deficiencies of CI and CII, which are localized up-stream SQR, and might be by-passed, by using SQR to feed electrons to complex III. For example, it may explain the results obtained by Vafai and colleagues, who developed a chemical screening platform to identify CI by bass factors, and found that naphthoquinones supplementation has therapeutic effects in CI deficient murine myoblasts [23].
H2S oxidation was found to be impaired also in three mouse models of human primary CoQ deficiency, which manifest clinically with nephrotic syndrome (NS), encephalopathy, and myopathy [7], [8], three of the most common phenotypes of human CoQ deficiency [10]. The models showed similarities in the biochemical and molecular phenotype, as well as differences, probably reflective of tissue specific pathomechanisms [7], [8].
In the mouse model of CoQ-deficient NS due to a missense mutation in Pdss2, the first and rate-limiting step in CoQ biosynthesis [24], [25], we observed that all organs had low CoQ levels and OxPhos deficiency, but remarkably, only kidney also had elevated reactive oxygen species (ROS) and markers of oxidative stress [5], [16], decreased levels of H2S oxidation enzymes, accumulation of H2S, and GSH depletion [5], [7]. Pdss2kd/kd mice have also low levels of plasma and urine thiosulfate, and increased blood C4-C6 acylcarnitines [5], [7], indicating a defect in short-chain fatty acid oxidation, due to the inhibition of the short- chain acyl-CoA dehydrogenases (SCAD), a known toxic effect of H2S accumulation [26].
Notably, increased ROS and low SQR were already evident in kidney of Pdss2kd/kd mice at age 1 month, before the onset of the disease [5]. Furthermore, administration of CoQ in these Pdss2kd/kd mice prolonged survival, from ~ 6 months in untreated mice to > 20 months in the CoQ-treated animals, prevented NS, as well as significantly reversed oxidative stress and sulfide derangement, acetylcarnitine profile, and GSH levels [5].
In Coq9R239X and Coq9Q95X mice, models of CoQ deficiency due to mutations in Coq9, SQR levels and function were decreased proportionally to residual CoQ levels in kidney, brain, and muscle [8]. Coq9 is required for the stability and function of Coq7, which is responsible for one of the three hydroxylations of CoQ benzoquinone ring [27], [28]. Coq9R239X mice develop a fatal mitochondrial encephalopathy, associated with oxidative stress and a defect of respiratory supercomplexes assembly in brain, while Coq9Q95X mice develop a late‐onset mild mitochondrial myopathy in females [27], [29]. Supplementation with ubiquinol-10 (the reduced form of CoQ) partially rescues the SQR depletion in muscle and kidney of Coq9R239X, parallel to increases in CoQ levels on those tissues [8]. In contrast to what observed in Pdss2kd/kd mice, tissues of Coq9R239X and Coq9Q95X did not show down-regulation of the enzymes down-stream of SQR; in fact, TST levels and activity were increased, as observed in CoQ deficient cells [8]. However, similar to Pdss2kd/kd mice, H2S levels were increased in kidney, while levels of total GSH were significantly decreased [8]. The presence of additional H2S biosynthetic pathways in the kidney, beside the transsulfuration pathway, might explains why H2S accumulates preferentially in this tissue [30], [31].
The consequences of H2S accumulation found in CoQ deficiency, partially recapitulate the biochemical and molecular abnormalities previously reported in Ethylmalonic aciduria encephalopathy (EE), an autosomal recessive disorder caused by mutations in the gene encoding for the mitochondrial enzyme ETHE1, a persulfide dioxygenase involved in the same H2S oxidation pathway as SQR (Fig. 2) [32]. Impairment of ETHE1 activity in human and mice causes chronic accumulation of H2S in tissues, and SCAD inhibition with accumulation of C4- and C5-acylcarnitines in plasma. Furthermore, it causes tissue-specific cytochrome c oxidase (COX) deficiency [32], [33], due to the formation of a covalent bond between H2S and the Fe atom coordinated by heme a. The chronic exposure and binding of H2S to COX causes accelerated degradation of its protein subunits thus reducing the amount of fully assembled and functionally active enzyme [34], [35]. COX activity, but not its protein levels, are decreased also in tissues of the first three patients described carrying mutations in SQR, a novel cause of Leigh syndrome [36]. Interestingly, there is no evidence of COX deficiency in CoQ deficiency, perhaps due to less severe accumulation of H2S. Fibroblasts from patients with EE have low levels of SQR and GSH, complicating the understanding of the mechanism underlying ETHE1 deficiency [37], [38].
Impairment of the glutathione pathway in H2S oxidation defects
H2S has been linked to ROS production through different mechanisms [39], [40]. One of the mechanisms that links H2S to oxidative stress is GSH depletion [39], [40], which is a well-known cause of ROS and oxidative stress, both present in mitochondrial diseases [41], [42], [43], [44], including CoQ and ETHE1 deficiencies [12], [13], [14], [15], [16], [45]. Studies in yeast indicate that levels of H2S are tightly regulated by the equilibrium of the transsulfuration and oxidation pathways (Fig. 2). Thus, increased levels of H2S (e.g. as a result of reduced SQR in CoQ deficiency) might operate in a negative feedback loop on the transsulfuration pathway. Because L-cysteine is also a key precursor of GSH biosynthesis, it is possible that as a consequence of reduced utilization of L-cysteine by the transsulfuration pathway, cysteine for GSH synthesis will also be reduced. Alternatively, H2S auto-oxidation of transsulfuration enzyme may generate reactive sulfur and oxygen radicals that deplete GSH. Thus, this reduction in the GSH levels could induce a reduction in GPx4 levels and the activities of glutathione peroxidase (GPx) and glutathione reductase (GRd), as observed in Coq9R239X mice [8]. Nevertheless, the glutathione system is not globally depleted in CoQ deficient fibroblasts, and Sqr depleted Hepa1c1c7 cells [8], indicating that GSH depletion is a tissue-specific, perhaps secondary effect of SQR depletion.
Interestingly, GSH, and GPX4 depletion, together with lipid ROS formation, characterize ferroptosis, a form of regulated non-apoptotic cell death [46]. GPX4 has been known to prevent ferroptosis by converting lipid hydroperoxides into non-toxic lipid alcohols [47]. Recently, also apoptosis-inducing factor mitochondrial 2 (AIFM2) has been identified as a potent ferroptosis-resistance factor, which co-operates with GPX4 and GSH to suppress phospholipid peroxidation and ferroptosis [48], [49]. AIFM2, re-named ferroptosis suppressor protein 1 (FSP1), is recruited to the plasma membrane where it functions as an oxidoreductase that reduces CoQ -using NAD(P)H- which halts the propagation of lipid peroxides [48], [49]. However, AIFM2 might block ferroptosis through a mechanism independent of CoQ [50]. CoQ deficiency causes cell death, which correlates with ROS levels, and oxidative stress [12], [13], [14], [15], but whether it causes specifically ferroptosis, has never been investigated.
Other mechanisms unrelated to GSH might be responsible for H2S-mediated oxidative stress; for example, through increased S-sulfhydration of proteins specifically involved in cell redox status. In fact, protein S-sulfhydration, a post-translational modification of protein cysteine residues, is important for regulation of various cell functions [51], [52], and is increased in CoQ deficient fibroblasts [7]. Nevertheless, in aging there are increased levels of ROS and oxidative stress, but protein S-sulfhydration is decreased [53].
Up regulation of the transsulfuration and gluthatione pathways in mitochondrial DNA defects
H2S is produced mostly from L-cysteine, that can be taken up with the diet, extracted from endogenous proteins, or synthesized endogenously via trans‐sulfuration of serine by L‐methionine, through the transsulfuration pathway (Fig. 2), which is expressed in all tissues [30].
Notably, L-cysteine, is also a key precursor of glutathione (GSH) biosynthesis, and its entry into that pathway is rate-limiting. Thus, mechanisms that regulate the availability of L-cysteine, likely affect both pathways, as demonstrated by metabolomics/transcriptomics approaches in different models of mitochondrial dysfunction. For example, studies on the effects of the 1-methyl-4-phenylpyridinium (MPP + ), whose neurotoxicity is mediated by different mechanisms, including CI inhibition, revealed increased GSH associated with upregulation of ATF-4 and the transsulfuration enzymes CTH and CBS. In cells stressed with MPP+, knockdown of ATF-4 or CTH, reduced GSH levels [54]. CI deficiency also causes GSH synthesis in a model of renal oncocytoma [55].
Up-regulation of the transsulfuration and GSH synthesis enzymes is found in mitochondrial DNA (mtDNA) depleted HEK-293 cells, and in mice with mtDNA replication defects due to mutations in the mitochondrial helicase Twinkle [3], [4]. These alterations are not isolated, but rather represent an aspect of a metabolic switch likely mediated by ATF-4 activation [3], [4], and the contribution of every metabolic pathway to the pathogenesis of human mitochondrial disease has not yet been investigated. Evidence of abnormalities of the transsulfuration pathway were also identified in patients with mtDNA maintenance/translation disorders. A similar pattern was observed also in plasma of patients with mitochondrial dysfunction, including mtDNA deletions, secondary to inclusion bodies myositis (IBM) [2], [56].
Up-regulation of the transsulfuration pathway and H2S accumulation are present in a variety of models of longevity and stress resistance associated with dietary restriction [57], and they might contribute to the beneficial effects of hypoxia shown in Ndufs4-/- mice [58], [59], possibly through SQR-driven respiration, or through protection against oxidative stress mediated by protein S-sulfhydration. Ndufs4-/- mice lack NADH: ubiquinone oxidoreductase iron-sulfur protein 4 (Ndufs4), and recapitulate the main findings of CI-related Leigh syndrome, the most common infantile mitochondrial encephalopathy. They develop a rapidly progressive encephalopathy, starting at ~ 40 days after birth, with > 90% mortality by 50 days of life [60]. Muscle, brain, and fibroblasts show evidence of oxidative stress and abnormal mitochondria [61], [62]. In this model, hypoxia has been shown to prevent and rescue neurological phenotype, and to prolong survival, but the mechanism is still unknown [58], [59].
Conclusions
In the last few years, abnormalities of H2S metabolism have been reported in a variety of models of mitochondrial dysfunction; however, the role of these abnormalities in mitochondrial diseases is still unknown. Further studies are needed to understand the relation of sulfide metabolism and mitochondrial diseases and the consequences of abnormalities of H2S and GSH synthesis on the oxidation pathway, and vice versa; and on the levels of H2S and GSH, and their tissue-specific detrimental effects. In this regard, we should consider not only the known, additional, tissue-specific, enzymatic pathways, that produce H2S beside the transsulfuration pathway, but also those other, still unknown systems, involved in H2S production and elimination, that might exist. Importantly, available data indicate that mitochondria play a key role in the regulation of those H2S pathways.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by NIH P01 HD080642 (CMQ), and Ministerio de Ciencia e inn (LCL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Biographies
Catarina M Quinzii, MD, is Associate professor of Neurology at CUMC, Columbia University, New York (US). She obtained her MD at the University of Milan, Milan (Italy). She has been working on mitochondrial diseases, particularly coenzyme Q10 deficiencies, for 15 years. She identified the first molecular defect of CoQ10 biosynthesis, the first molecular defect associated with secondary CoQ10 deficiency, and she studied the pathomechanisms of CoQ10 deficiencies in mammalian cells and animal models. Her group was among the firsts to unveil the role of H2S oxidation impairment in the pathogenesis of CoQ deficiency.
Luis C Lopez, PhD, is Associate professor at the University of Granada, Granada, Spain. After receiving his PhD in Biotechnology and Biomedicine at the University of Granada, he worked in the laboratories of Drs. DiMauro and Hirano at Columbia University, New York (USA). He became group leader in 2012 (“Ramón y Cajal” researcher). Since then, his group has generated and characterized the first two mouse models of mitochondrial encephalopathy and mitochondrial myopathy due to CoQ deficiency, discovering the function of COQ9, and identifying new pathomechanisms, such as the supercomplexes instability in symptomatic tissue, the indirect correlation in the efficacy of NMD and the severity of CoQ deficiency, and the disruption in the sulfide oxidation pathway.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Khan S., Ince-Dunn G., Suomalainen A., Elo L.L. Integrative omics approaches provide biological and clinical insights: examples from mitochondrial diseases. J Clin Invest. 2020;130(1):20–28. doi: 10.1172/JCI129202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J. Buzkova, J. Nikkanen, S. Ahola, A.H. Hakonen, K. Sevastianova, T. Hovinen, et al. Metabolomes of mitochondrial diseases and inclusion body myositis patients: treatment targets and biomarkers. EMBO Mol Med. 2018;10(12). doi: 10.15252/emmm.201809091. [DOI] [PMC free article] [PubMed]
- 3.X.R. Bao, S.E. Ong, O. Goldberger, J. Peng , R. Sharma, D.A. Thompson, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife. 2016;5. doi: 10.7554/eLife.10575 [DOI] [PMC free article] [PubMed]
- 4.Nikkanen J., Forsstrom S., Euro L., Paetau I., Kohnz R.A., Wang L. Mitochondrial DNA Replication Defects Disturb Cellular dNTP Pools and Remodel One-Carbon Metabolism. Cell Metab. 2016;23(4):635–648. doi: 10.1016/j.cmet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 5.G. Kleiner, E. Barca, M. Ziosi M, V. Emmanuele, Y Xu, A. Hidalgo-Gutierrez, et al. CoQ10 supplementation rescues nephrotic syndrome through normalization of H2S oxidation pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864(11):3708-22. doi: 10.1016/j.bbadis.2018.09.002 [DOI] [PMC free article] [PubMed]
- 6.Quinzii C.M., Luna-Sanchez M., Ziosi M., Hidalgo-Gutierrez A., Kleiner G., Lopez L.C. The Role of Sulfide Oxidation Impairment in the Pathogenesis of Primary CoQ Deficiency. Front Physiol. 2017;8:525. doi: 10.3389/fphys.2017.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziosi M., Di Meo I., Kleiner G., Gao X.H., Barca E., Sanchez-Quintero M.J. Coenzyme Q deficiency causes impairment of the sulfide oxidation pathway. EMBO Mol Med. 2017;9(1):96–111. doi: 10.15252/emmm.201606356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luna-Sanchez M., Hidalgo-Gutierrez A., Hildebrandt T.M., Chaves-Serrano J., Barriocanal-Casado E., Santos-Fandila A. CoQ deficiency causes disruption of mitochondrial sulfide oxidation, a new pathomechanism associated with this syndrome. EMBO Mol Med. 2017;9(1):78–95. doi: 10.15252/emmm.201606345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Awad A.M., Bradley M.C., Fernandez-Del-Rio L., Nag A., Tsui H.S., Clarke C.F. Coenzyme Q10 deficiencies: pathways in yeast and humans. Essays Biochem. 2018;62(3):361–376. doi: 10.1042/EBC20170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcazar-Fabra M., Trevisson E., Brea-Calvo G. Clinical syndromes associated with Coenzyme Q10 deficiency. Essays Biochem. 2018;62(3):377–398. doi: 10.1042/EBC20170107. [DOI] [PubMed] [Google Scholar]
- 11.Bentinger M., Tekle M., Dallner G. Coenzyme Q–biosynthesis and functions. Biochem Biophys Res Commun. 2010;396(1):74–79. doi: 10.1016/j.bbrc.2010.02.147. [DOI] [PubMed] [Google Scholar]
- 12.Quinzii C.M., Lopez L.C., Von-Moltke J., Naini A., Krishna S., Schuelke M. Respiratory chain dysfunction and oxidative stress correlate with severity of primary CoQ10 deficiency. FASEB J. 2008;22(6):1874–1885. doi: 10.1096/fj.07-100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinzii C.M., Lopez L.C., Gilkerson R.W., Dorado B., Coku J., Naini A.B. Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. FASEB J. 2010;24(10):3733–3743. doi: 10.1096/fj.09-152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez L.C., Quinzii C.M., Area E., Naini A., Rahman S., Schuelke M. Treatment of CoQ(10) deficient fibroblasts with ubiquinone, CoQ analogs, and vitamin C: time- and compound-dependent effects. PLoS ONE. 2010;5(7) doi: 10.1371/journal.pone.0011897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinzii C.M., Tadesse S., Naini A., Hirano M. Effects of inhibiting CoQ10 biosynthesis with 4-nitrobenzoate in human fibroblasts. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinzii C.M., Garone C., Emmanuele V., Tadesse S., Krishna S., Dorado B. Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J. 2013;27(2):612–621. doi: 10.1096/fj.12-209361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Corzo L., Luna-Sanchez M., Doerrier C., Ortiz F., Escames G., Acuna-Castroviejo D. Ubiquinol-10 ameliorates mitochondrial encephalopathy associated with CoQ deficiency. Biochim Biophys Acta. 2014;1842(7):893–901. doi: 10.1016/j.bbadis.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Jackson M.R., Melideo S.L., Jorns M.S. Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry. 2012;51(34):6804–6815. doi: 10.1021/bi300778t. [DOI] [PubMed] [Google Scholar]
- 19.Abou-Hamdan A., Guedouari-Bounihi H., Lenoir V., Andriamihaja M., Blachier F., Bouillaud F. Oxidation of H2S in mammalian cells and mitochondria. Methods Enzymol. 2015;554:201–228. doi: 10.1016/bs.mie.2014.11.042. [DOI] [PubMed] [Google Scholar]
- 20.Lagoutte E., Mimoun S., Andriamihaja M., Chaumontet C., Blachier F., Bouillaud F. Oxidation of hydrogen sulfide remains a priority in mammalian cells and causes reverse electron transfer in colonocytes. Biochim Biophys Acta. 2010;1797(8):1500–1511. doi: 10.1016/j.bbabio.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Uchida N., Suzuki K., Saiki R., Kainou T., Tanaka K., Matsuda H. Phenotypes of fission yeast defective in ubiquinone production due to disruption of the gene for p-hydroxybenzoate polyprenyl diphosphate transferase. J Bacteriol. 2000;182(24):6933–6939. doi: 10.1128/jb.182.24.6933-6939.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M., Wakitani S., Hayashi K., Miki R., Kawamukai M. High production of sulfide in coenzyme Q deficient fission yeast. BioFactors. 2008;32(1–4):91–98. doi: 10.1002/biof.5520320111. [DOI] [PubMed] [Google Scholar]
- 23.Vafai S.B., Mevers E., Higgins K.W., Fomina Y., Zhang J., Mandinova A. Natural Product Screening Reveals Naphthoquinone Complex I Bypass Factors. PLoS ONE. 2016;11(9) doi: 10.1371/journal.pone.0162686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng M., Jarett L., Meade R., Madaio M.P., Hancock W.W., George A.L., Jr Mutant prenyltransferase-like mitochondrial protein (PLMP) and mitochondrial abnormalities in kd/kd mice. Kidney Int. 2004;66(1):20–28. doi: 10.1111/j.1523-1755.2004.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng M., Falk M.J., Haase V.H., King R., Polyak E., Selak M. Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS Genet. 2008;4(4) doi: 10.1371/journal.pgen.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen C.B., Bross P., Winter V.S., Corydon T.J., Bolund L., Bartlett K. Misfolding, degradation, and aggregation of variant proteins. The molecular pathogenesis of short chain acyl-CoA dehydrogenase (SCAD) deficiency. J Biol Chem. 2003;278(48):47449–47458. doi: 10.1074/jbc.M309514200. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Corzo L., Luna-Sanchez M., Doerrier C., Garcia J.A., Guaras A., Acin-Perez R. Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum Mol Genet. 2013;22(6):1233–1248. doi: 10.1093/hmg/dds530. [DOI] [PubMed] [Google Scholar]
- 28.Lohman D.C., Forouhar F., Beebe E.T., Stefely M.S., Minogue C.E., Ulbrich A. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc Natl Acad Sci U S A. 2014;111(44):E4697–E4705. doi: 10.1073/pnas.1413128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luna-Sanchez M., Diaz-Casado E., Barca E., Tejada M.A., Montilla-Garcia A., Cobos E.J. The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol Med. 2015;7(5):670–687. doi: 10.15252/emmm.201404632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasinath B.S., Feliers D., Lee H.J. Hydrogen sulfide as a regulatory factor in kidney health and disease. Biochem Pharmacol. 2018;149:29–41. doi: 10.1016/j.bcp.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Shibuya N., Koike S., Tanaka M., Ishigami-Yuasa M., Kimura Y., Ogasawara Y. A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat Commun. 2013;4:1366. doi: 10.1038/ncomms2371. [DOI] [PubMed] [Google Scholar]
- 32.Di Meo I., Lamperti C. V. Tiranti V. Mitochondrial diseases caused by toxic compound accumulation: from etiopathology to therapeutic approaches. EMBO Mol Med. 2015;7(10):1257–1266. doi: 10.15252/emmm.201505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiranti V., Viscomi C., Hildebrandt T., Di Meo I., Mineri R., Tiveron C. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat Med. 2009;15(2):200–205. doi: 10.1038/nm.1907. [DOI] [PubMed] [Google Scholar]
- 34.Di Meo I., Fagiolari G., Prelle A., Viscomi C., Zeviani M., Tiranti V. Chronic exposure to sulfide causes accelerated degradation of cytochrome c oxidase in ethylmalonic encephalopathy. Antioxid Redox Signal. 2011;15(2):353–362. doi: 10.1089/ars.2010.3520. [DOI] [PubMed] [Google Scholar]
- 35.Tiranti V., Zeviani M. Altered sulfide (H(2)S) metabolism in ethylmalonic encephalopathy. Cold Spring Harb Perspect Biol. 2013;5(1) doi: 10.1101/cshperspect.a011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friederich M.W., Elias A.F., Kuster A., Laugwitz L., Larson A.A., Landry A.P. Pathogenic variants in SQOR encoding sulfide:quinone oxidoreductase are a potentially treatable cause of Leigh disease. J Inherit Metab Dis. 2020 doi: 10.1002/jimd.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmfeldt J., Vang S., Stenbroen V., Pavlou E., Baycheva M., Buchal G. Proteomics reveals that redox regulation is disrupted in patients with ethylmalonic encephalopathy. J Proteome Res. 2011;10(5):2389–2396. doi: 10.1021/pr101218d. [DOI] [PubMed] [Google Scholar]
- 38.Sahebekhtiari N., Fernandez-Guerra P., Nochi Z., Carlsen J., Bross P., Palmfeldt J. Deficiency of the mitochondrial sulfide regulator ETHE1 disturbs cell growth, glutathione level and causes proteome alterations outside mitochondria. Biochim Biophys Acta Mol Basis Dis. 2019;1865(1):126–135. doi: 10.1016/j.bbadis.2018.10.035. [DOI] [PubMed] [Google Scholar]
- 39.Eghbal M.A., Pennefather P.S., O'Brien P.J. H2S cytotoxicity mechanism involves reactive oxygen species formation and mitochondrial depolarisation. Toxicology. 2004;203(1–3):69–76. doi: 10.1016/j.tox.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 40.Truong D.H., Eghbal M.A., Hindmarsh W., Roth S.H., O'Brien P.J. Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab Rev. 2006;38(4):733–744. doi: 10.1080/03602530600959607. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi G., Cortopassi G. Oxidative stress in inherited mitochondrial diseases. Free Radic Biol Med. 2015;88(Pt A):10–17. doi: 10.1016/j.freeradbiomed.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pastore A., Petrillo S., Tozzi G., Carrozzo R., Martinelli D., Dionisi-Vici C. Glutathione: a redox signature in monitoring EPI-743 therapy in children with mitochondrial encephalomyopathies. Mol Genet Metab. 2013;109(2):208–214. doi: 10.1016/j.ymgme.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Enns G.M., Moore T., Le A., Atkuri K., Shah M.K., Cusmano-Ozog K. Degree of glutathione deficiency and redox imbalance depend on subtype of mitochondrial disease and clinical status. PLoS ONE. 2014;9(6) doi: 10.1371/journal.pone.0100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enns G.M., Cowan T.M. Glutathione as a Redox Biomarker in Mitochondrial Disease-Implications for. Therapy. J Clin Med. 2017;6(5). doi: 10.3390/jcm6050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Moura Alvorcem L., Britto R., Parmeggiani B., Glanzel N.M., da Rosa-Junior N.T., Cecatto C. Evidence that thiol group modification and reactive oxygen species are involved in hydrogen sulfide-induced mitochondrial permeability transition pore opening in rat cerebellum. Mitochondrion. 2019;47:141–150. doi: 10.1016/j.mito.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Jelinek A., Heyder L., Daude M., Plessner M., Krippner S., Grosse R. Mitochondrial rescue prevents glutathione peroxidase-dependent ferroptosis. Free Radic Biol Med. 2018;117:45–57. doi: 10.1016/j.freeradbiomed.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 47.Hayano M., Yang W.S., Corn C.K., Pagano N.C., Stockwell B.R. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016;23(2):270–278. doi: 10.1038/cdd.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bersuker K., Hendricks J.M., Li Z., Magtanong L., Ford B., Tang P.H. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doll S., Freitas F.P., Shah R., Aldrovandi M., da Silva M.C., Ingold I. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 50.Dai E., Zhang W., Cong D., Kang R., Wang J., Tang D. AIFM2 blocks ferroptosis independent of ubiquinol metabolism. Biochem Biophys Res Commun. 2020;523(4):966–971. doi: 10.1016/j.bbrc.2020.01.066. [DOI] [PubMed] [Google Scholar]
- 51.Mustafa A.K., Gadalla M.M., Sen N., Kim S., Mu W., Gazi S.K. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2(96)::ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paul B.D., Snyder S.H. H(2)S signalling through protein sulfhydration and beyond. Nat Rev Mol Cell Biol. 2012;13(8):499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]
- 53.J. Zivanovic, E. Kouroussis, J.B. Kohl, B. Adhikari, B. Bursac, S. Schott-Roux, et al. Selective Persulfide Detection Reveals Evolutionarily Conserved Antiaging Effects of S-Sulfhydration. Cell Metab. 2019;30(6):1152-70 e13. doi: 10.1016/j.cmet.2019.10.007 [DOI] [PMC free article] [PubMed]
- 54.Krug A.K., Gutbier S., Zhao L., Poltl D., Kullmann C., Ivanova V. Transcriptional and metabolic adaptation of human neurons to the mitochondrial toxicant MPP(+) Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopal R.K., Calvo S.E., Shih A.R., Chaves F.L., McGuone D., Mick E. Early loss of mitochondrial complex I and rewiring of glutathione metabolism in renal oncocytoma. Proc Natl Acad Sci U S A. 2018;115(27):E6283–E6290. doi: 10.1073/pnas.1711888115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tadiboyina V.T., Rupar A., Atkison P., Feigenbaum A., Kronick J., Wang J. Novel mutation in DGUOK in hepatocerebral mitochondrial DNA depletion syndrome associated with cystathioninuria. Am J Med Genet A. 2005;135(3):289–291. doi: 10.1002/ajmg.a.30748. [DOI] [PubMed] [Google Scholar]
- 57.Hine C., Harputlugil E., Zhang Y., Ruckenstuhl C., Lee B.C., Brace L. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell. 2015;160(1–2):132–144. doi: 10.1016/j.cell.2014.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferrari M., Jain I.H., Goldberger O., Rezoagli E., Thoonen R., Cheng K.H. Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc Natl Acad Sci U S A. 2017;114(21):E4241–E4250. doi: 10.1073/pnas.1621511114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.I.H. Jain, L. Zazzeron, O. Goldberger, E. Marutani, G.R. Wojtkiewicz, T. Ast, et al. Leigh Syndrome Mouse Model Can Be Rescued by Interventions that Normalize Brain Hyperoxia, but Not HIF Activation. Cell Metab. 2019;30(4):824-32 e3. doi: 10.1016/j.cmet.2019.07.006 [DOI] [PMC free article] [PubMed]
- 60.S.C. Johnson, M. E. Yanos, E.B. Kayser, A. Quintana, M. Sangesland, A. Castanza A, et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342(6165):1524-8. doi: 10.1126/science.1244360 [DOI] [PMC free article] [PubMed]
- 61.Valsecchi F., Grefte S., Roestenberg P., Joosten-Wagenaars J., Smeitink J.A., Willems P.H. Primary fibroblasts of NDUFS4(-/-) mice display increased ROS levels and aberrant mitochondrial morphology. Mitochondrion. 2013;13(5):436–443. doi: 10.1016/j.mito.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 62.de Haas R., Das D., Garanto A., Renkema H.G., Greupink R., van den Broek P. Therapeutic effects of the mitochondrial ROS-redox modulator KH176 in a mammalian model of Leigh Disease. Sci Rep. 2017;7(1):11733. doi: 10.1038/s41598-017-09417-5. [DOI] [PMC free article] [PubMed] [Google Scholar]