Graphical abstract
Keywords: Hydrogen sulfide, Toxicity, Sulfate-reducing bacteria, Dissimilatory sulfate reduction, Ulcerative colitis
Highlights
-
•
Species of Desulfovibrio genus are dominant among intestinal sulfate-reducing bacteria.
-
•
Hydrogen sulfide amounts are higher in feces of patients with ulcerative colitis.
-
•
The increased number of sulfate-reducing bacteria causes changes in the intestinal microbiota.
-
•
H2S can have a synergistic effect with acetate and can be involved in colitis development.
Abstract
Introduction
Increased numbers of sulfate-reducing bacteria (SRB) are often found in the feces of people and animals with inflammatory bowel disease. The final products of their metabolism are hydrogen sulfide and acetate, which are produced during dissimilatory sulfate reduction process.
Objectives
The aim of the study was to monitor processes concerning sulfate reduction microbial metabolisms, including: the main microbial genera monitoring and their hydrogen sulfide production in the intestines of healthy and not healthy individuals, phylogenetic analysis of SRB isolates, cluster analysis of SRB physiological and biochemical parameters, SRB growth kinetic parameters calculation, same as the application of the two-factor dispersion analysis for finding relationship between SRB biomass accumulation, temperature and pH. Feces samples from healthy people and patients with colitis were used for isolation of sulfate-reducing microbial communities.
Methods
Microbiological, biochemical, biophysical, molecular biology methods, and statistical processing of the results have been used for making an evaluation of gained results.
Results
Two dominant SRB morphotypes differed in colony size and quantitative ratio in the feces of healthy and colitis patients were observed and identified. In the feces of healthy people, 93% of SRB of morphotype I prevailed (Desulfovibrio) while morphotype II made only 7% (Desulfomicrobium); in the feces of patients with colitis, the ratio of these morphotypes was 99:1, respectively. Hydrogen sulfide concentrations are also higher in the feces of people with colitis and certain synergy effects exist among acetate produced by SRB.
Conclusions
The study results brought important findings concerning colony environments with developed colitis and these findings can lead to the development of possible risk indicators of ulcerative colitis prevalence.
Introduction
Hydrogen sulfide is a final product of sulfate-reducing bacteria (SRB) metabolism [1], [2], [3], [4]. High concentrations of this metabolite and SRB amount are often found in feces of people with bloody diarrhea, the mono- and polymicrobial infections in the gastrointestinal tract [5], [6], [7], [8]. SRB are heterogeneous group that metabolize organic compounds (serving them as energy and carbon sources) incompletely (to acetate) and completely (to CO2) [9], [10], [11]. SRB species and their quantitative composition on the intestinal mucosa surface differ from other microorganisms in the lumen [7], [12]. It is believed that SRB can cause frequent defecation, weight loss and increased intestinal permeability [8], [13], [14], [15], [16]. Such genera of SRB, Desulfovibrio, Desulfomicrobium, Lawsonia, and Bilophila, are the most often isolates found in the intestines of healthy and not healthy people, same as in animals [4], [17].
In low concentrations (not toxic amounts), hydrogen sulfide is found in the brain, heart, genitourinary, blood vessels, same as in the gastrointestinal tract [8], [18], [19], [20]. The first disadvantage of hydrogen sulfide higher amounts reflects on the butyrate consumption of colonocytes, since butyrate oxidation is inhibited [7], [13]. Studies that included experimental rats with developed intestinal ulceration indicated correlations between illness development and hydrogen sulfide concentrations in the colon. These statements are also supported by literature data that emphasize the connection with inflammatory bowel diseases and sulfate-reducing bacteria [4]. Certainly, hydrogen sulfide production and concentrations plays an important role in the colon [4], [21], [22], [23], [24], [25], [26]. The place in the colon is also important, since H2S production is higher in the distal intestine than in the proximal part of the colon [18]. SRB formed dense biofilms around ulcer [27], [28]. Other important factors that can be connected with the development of inflammatory bowel disease (IBD) include, intestinal lumen pH, immune status of the organism (host), and the availability of sulfate [1], [6]. Mucosa cells also have a more permeable epithelial barrier in the environment with increased H2S amounts [7], [8], [13], [20], [29]. IBD is an umbrella term used to describe disorders that involve chronic inflammation of the digestive tract [7], [13]. Types of IBD include ulcerative colitis (UC) and Crohn's disease. UC is a condition that causes long-lasting inflammation and sores (ulcers) in the innermost lining of the large intestine (colon) and rectum. Crohn's disease is the type of IBD characterized by inflammation of the lining of the digestive tract, which often spreads deep into affected tissues. Both UC and Crohn's disease, usually involve severe diarrhea, abdominal pain, and fatigue and weight loss. It should be noted that acetate is often used as chemically induced UC (in animal colitis models) [7]. It should be emphasized that the relationship between acetate produced by SRB and IBD development has not been fully described yet.
Certainly that SRB and their hydrogen sulfide production play an important role in the development of different IBD, especially UC. Consequently, studies providing new information about sulfate reducing bacteria, hydrogen sulfide production and their interactions with other microorganisms present in intestines, represent crucial steps toward a better understanding of IBD and especially preventive future treatments.
The aim of the research was to investigate processes considering sulfate-reducing bacteria environments, consisting out of four focuses: (a) monitoring the main microbial genera and hydrogen sulfide concentrations in the intestines of healthy people and with developed UC, (b) phylogenetic analysis of selected SRB isolates, (c) investigate SRB physiological and biochemical parameters and form a cluster analysis, (d) to calculate kinetic parameters of SRB growth, (e) to evaluate SRB biomass accumulation relationship with temperature and pH by two-factor dispersion analysis.
Material and methods
Experimental procedure
Bacterial culture and cultivation. The SRB isolated strains were identified based on physiological and biochemical properties and sequence analysis of 16S rRNA gene. The strains were kept in the collection of microorganisms at the Laboratory of Anaerobic Microorganisms of the Department of Experimental Biology at Masaryk University (Brno, Czech Republic). The bacteria were grown in modified liquid Postgate C medium [30], [31]. The medium was heated in boiling water for 30 min in order to obtain an oxygen-free medium, and cooled to +37 °C temperature. The final optimal pH 7.5 was provided by a sterile 1 M solution of NaOH (0.9 mL/l). The bacteria were grown for 72 h at 37 °C under anaerobic conditions. The tubes with strain were brim-filled with medium and closed to provide anaerobic conditions.
Samples. The ecological-trophic groups of microorganisms of intestinal microbiome were determined in the stool samples of the people. Determination of the number of microorganisms was carried out by tenfold dilutions, following the screening on elective nutrient media. To determine the number of SRB and the isolation of their pure cultures, isolated colonies were multiple replanted in the appropriate modified selective liquid and agar Postgate medium [31]. Pure bacterial cultures were isolated by the Koch method. Cultivation lasted for 10 days at a temperature of +37 °C under the fixed anaerobic conditions using boxes with oxygen absorbing generators (Genbox anaer Biomerieux, France). Phenotypic studies of pure SRB cultures were carried out by generally known methods [31], [32]. To determine the phylogenetic identity of SRB, the molecular and genetic identification, using analysis of 16S rRNA gene full sequences, was carried out.
Isolation and purification of DNA carried out using biomass of three daily cultures of SRB by QIAmp DNA Mini Kit (QIAGEN, Germany). Amplification of 16S rRNA gene fragments was performed by W.G. Weisburg et al. (1991) [33] and D.H. Pershing (2011) [34] using pairs of universal primer: 8FPL (5′-AGT-TTG-ATC-CTG-GCT-CAG-3′), 1492RPL (5′-GGT-TAC-CTT-GTT-ACG-ACT-T-3′), 8FPL (5′-AGT-TTG-ATC-CTG-GCT-CAG-3′) and 806R (5′-GGA-CTA-CCA-GGG-TAT-CTA-AT-3′) (Eurofins Scientific, Luxembourg). The purification of amplicons was done by the MinElute Gel Extraction Kit (QIAGEN). Sequencing was performed using the Genetic Analyzer (Life Technologies Corporation, USA) and reagents BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Analysis of the obtained sequences of 16S rRNA gene with homologous nucleotide sequences of the genes deposited in database GenBank, was performed by the BLASTN program (www.ncbi.nlm.nih.gov/nblast). The sequences of gene fragments of 16S rRNA SRB were deposited in the GenBank database as the numbers: KT881309, KT989311–KT989316.
Hydrogen sulfide determination. Hydrogen sulfide was measured spectrophotometrically. Calibration solutions were prepared in distilled water at concentrations of 12.5, 25, 50, and 100 µM sodium sulfide. The calibration curve has been constructed with the same process. 1 mL of the sample was added to 10 mL of 5 g/L aqueous solution of zinc acetate. Right after, 2 mL of 0.75 g/mL p-aminodimethylaniline in a solution of sulfuric acid (2 M) was added. The mixture stayed for 5 min at room temperature. After that, 0.5 mL of 12 g/L solution of ferric chloride dissolved in 15 mM sulfuric acid was added. After standing another 5 min at room temperature, the mixture was centrifuged 5000 × g at 23 °C. The absorbance of the mixture was determined to measure hydrogen sulfide at a wavelength of 665 nm by a spectrophotometer (Cecil Aquarius CE 7200 Double Beam Spectrophotometer) [35].
Statistical analysis. Using the experimental data, the basic statistical parameters (M – mean, SE – standard error, M ± SE) have been calculated [36]. Statistical analysis was done by SPSS 20 statistical software (IBM Corporation, Armonk, USA). Plots were built by software package Origin7.0 (www.origin-lab.com).
Results and discussion
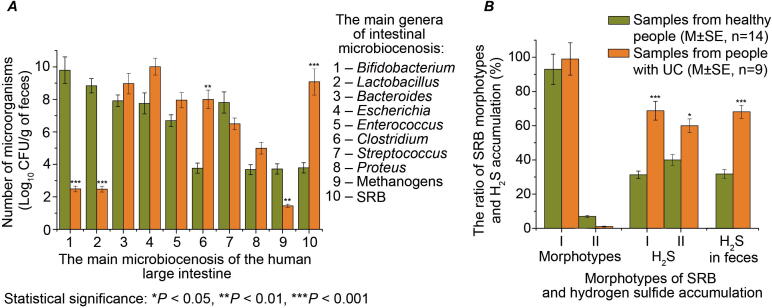
The results were obtained from monitoring intestinal microbial communities in healthy individuals and people with UC. The main genera ratio changes of autochthonous and facultative microbiota are shown in Fig. 1A. The autochthonous bacterial flora (Lactobacillus and Bifidobacterium genera) in patients with UC were 6 to 7 orders lower in comparison with healthy individuals. Also, facultative and conditionally pathogenic bacteria (Bacteroides, Escherichia, Enterococcus, and Proteus) were 1 to 4 orders higher in comparison with healthy individuals’ microbiota. The same trend of increased bacteria counts was observed among patients with UC in the content of sulfate-reducing bacteria and species of Clostridium genus. These differences were 4 to 5 orders higher than in healthy individuals. These findings were supported by the lower occurrence of methanogenic microorganisms (two orders decrease) since these bacteria compete for the substrate with SRB [12], [37], [38]. The different ratios of SRB morphotypes were found in the feces of healthy individuals and people with UC (Fig. 1B). Out of SRB morphotypes colonies, large colonies I (ø 2–3 mm vibrios cell shape) and small colonies II (ø to 1 mm, short rods) were detected. Vibrios SRB (93%) were dominated in the feces of healthy individuals and only 7% of SRB rod shaped was detected. The ratio of these morphotypes was 99:1 in the feces of patients with colitis. Also, the concentration of hydrogen sulfide was 2 times higher among individuals with UC. Indicative is that SRB cultured from patients with UC were capable to produce 1.5 to 2 times higher concentrations of hydrogen sulfide.
Fig. 1.
Changes in the number of the main microbial genera in the intestine of healthy people and patients with UC (A), the ratio of SRB morphotypes isolates and accumulation of hydrogen sulfide (B).
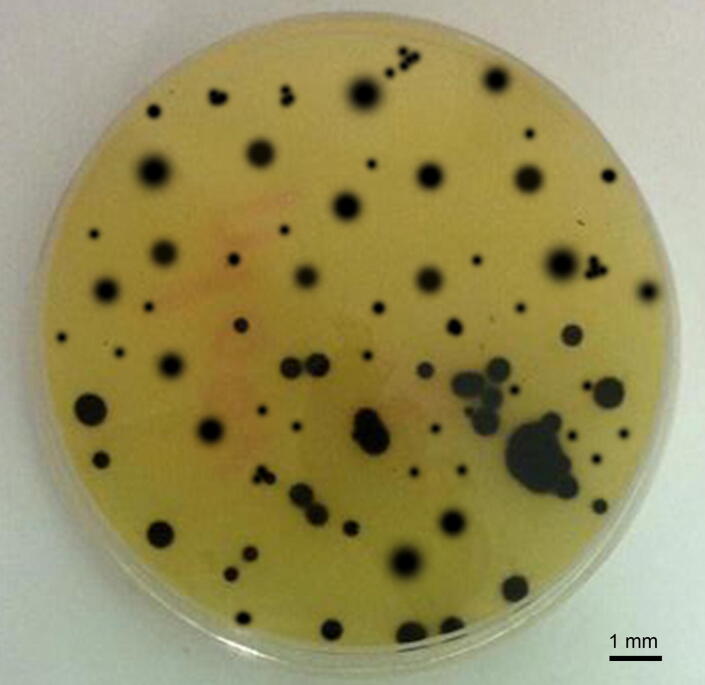
Hydrogen sulfide can be in ionized, non-volatile and volatile states (S2–, HS– and H2S) [39], [40]. It was also found that 95% of sulfide produced in the intestine is absorbed and the rest 5% is bounded with other compounds and found in feces [40]. H2S concentrations in feces also interact with other substances present in the intestinal lumen [41]. Certain studies are indicating that the total number of SRB in the feces of people with developed UC was not increased, but only their ration was changed and they started to be more aggressive and grow more rapidly in the culture medium. The level of H2S production in the feces of people with UC was also noticed to be increased by 28% [42]. In total, 157 SRB colonies were analyzed in our study; 79% (124 colonies) belonged to morphotype I, while 21% (33 colonies) belonged to morphotype II. There were not noticed differences in bacterial morphology between isolates found in the feces of healthy individuals and in the feces of people with developed UC (Fig. 2).
Fig. 2.
Isolated colonies of intestinal sulfate-reducing bacteria in modified Postgate agar medium.
In total, 20 isolates of bacteria (10 from healthy people and 10 from unhealthy people with UC) underwent further phenotypic identification. SRB strains were catalase positive and able to reduce nitrate to nitrite. They were not able to produce indole, but all of them were able to produce hydrogen sulfide in significant amounts. They metabolized pyruvate, malate, lactate, citrate, ethanol and glucose as sources of carbon and an electron donor. The affinity toward different electron acceptors and donors were checked for the both isolated morphotypes (Table 1).
Table 1.
Phenotypic properties of both SRB morphotypes.
| Compounds | Number of SRB isolates |
|
|---|---|---|
| Morphotype I: 124 isolates | Morphotype II: 33 isolates | |
| Electron acceptor | ||
| Sulfate | +++* | +++ |
| Sulfite | +++ | ++ |
| Sulfur | – | – |
| Thiosulfate | ++ | + |
| Fumarate | + | ++ |
| Nitrate | + | + |
| Electron donor | ||
| Laktate | +++ | +++ |
| Ethanol | ++ | + |
| H2 | +++ | + |
| Formic acid | – | – |
| Acetate | + | + |
| Pyruvate | + | + |
| Sukcinate | ++ | + |
| Malate | + | + |
*most intense growth: +++; intense growth: ++; less intense growth: +; no growth: –.
The growth of tested isolates was found in medium containing H2 and CO2 + acetate. Organic acids, alcohols and certain amino acids (alanine, aspartate, and glutamate) intestinal SRB are able to assimilate. Characteristics of SRB morphotypes (I and II) isolated from human intestines are shown in Table 2.
Table 2.
Characteristics of SRB morphotypes isolated from human intestine.
| Properties | Analyzed of SRB morphotypes |
||
|---|---|---|---|
| I (large) 124 colonies | II (small) 33 colonies | ||
| SRB (Log10, CFU/g) | Healthy | 3.76 ± 0.35 | 2.61 ± 0.23 |
| UC | 9.19 ± 0.93 | 7.32 ± 0.72 | |
| Cell shape | Vibrio | Short rods | |
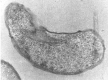 |
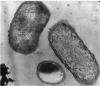 |
||
| Cell size (wide × lenght, µm) | 0.6–0.8 × 2.7–3.5 | 0.4–0.5 × 0.8–1.0 | |
| Gram staining | Negative | Negative | |
| Form/colonies edges | round/straight | round/straight | |
| Diametr of colonies (mm) | 2–3 | less than 1 | |
| Spore forming | no | no | |
| Pigment | Desulfoviridin | Desulforubidin | |
They had a chemolithoheterotrophic type of growth. SRB biomass of 3.89 ± 0.35 g/l was able to produce 3.23 ± 0.29 mM of hydrogen sulfide, after 6 days of cultivation (the achievement of stationary phase at +35 °C and pH 7.0–8.0). These conditions are in accordance with SRB intestinal environment. SRB produce hydrogen sulfide from dissimilation of sulfate, while acetate is the result of not complete lactate oxidation. According to cultural, morphological and biochemical findings (Bergey's Manual of Determinative Bacteriology), vibrio shaped SRB isolates (Vib-1, Vib-2, Vib-3, Vib-4, Vib-5, Vib-6, Vib-7, Vib-8, Vib-9, Vib-10) have been previously identified as Desulfovibrio genus, while Desulfomicrobium genus is defined as short rod shaped SRB (Rod-1, Rod-2, Rod-3, Rod-4, Rod-5, Rod-6, Rod-7, Rod-8, Rod-9, Rod-10).
Table 3 is showing the following results: lag phase length, generation time (Td) and maximum growth rate (µmax) of strains isolated from healthy and unhealthy individuals. Desulfovibrio strains from patients with UC had shorter generation time (1.73 ± 0.15 h) in comparison with Desulomicrobium genus (2.17 ± 0.23 h). The maximum measured growth rate among desulfovibrios was 0.058 ± 0.007 h−1. Desulfomicrobium bacterial strains isolated from healthy people were able to multiply faster (generation time: 1.93 ± 0.17 h; maximum growth rate: 0.052 ± 0.004 h−1. SRB kinetic parameters of growth did not differ significantly (p > 0.05) between results gained from healthy and unhealthy individuals. These findings are in accordance with previous studies [43].
Table 3.
The kinetic parameters of strains growth Desulfovibrio and Desulfomicrobium genus isolated from healthy people and patients with UC.
| SRB strains | Duration of lag phase (h) | Time of generation Td (h) | Maximal speed of the growth µmax (h-1) |
|---|---|---|---|
| Desulfovibrio isolated from | |||
| patients with UC | 5.21 ± 0.49 | 1.73 ± 0.15 | 0.058 ± 0.007 |
| healthy people | 5.46 ± 0.51 | 1.77 ± 0.19 | 0.056 ± 0.009 |
| Desulfomicrobium isolated from | |||
| patients with UC | 4.02 ± 0.35 | 2.17 ± 0.23 | 0.046 ± 0.002 |
| healthy people | 5.08 ± 0.46 | 1.93 ± 0.17 | 0.052 ± 0.004 |
The detailed identification of SRB was done by sequencing of 16 rRNA gene analysis. The nucleotide similarity compared with strains in GenBank is shown in Table 4. Desulfovibrio bacteria had a nucleotide sequence of 7 strains (Vib-1, Vib-2, Vib-3, Vib-4, Vib-7, Vib-8, Vib-10) that had homology of 99% with Desulfovibrio piger ATCC 29098 (NR 041778.1, deposited in the GenBank). Desulfovibrio sp. Vib-5, Vib-6 and Vib-9 had nucleotide sequence homology of 95%–96% and due to this homology it is not possible to determine their species membership, but only genera level (referent strain would be D. piger ATCC 29098) Rod-9 of Desulfomicrobium genus had nucleotide homology of 99% with Desulfomicrobium orale strain NY677 (AJ251629.1). Other 9 strains of Desulfomicrobium genus could not be determined due to lower homology. Desulfovibrio piger cultures have been deposited in the GenBank database under following numbers (KT989311), Vib-2 (KT989312), Vib-3 (KT989313), Vib-4 (KT989314), Vib-7 (KT881309), Vib-8 (KT989315), Vib-10 (KT989316).
Table 4.
The results of 16S rRNA gene sequence analysis of SRB strains isolated from human intestine.
| Strains and access number in GenBank | Length of the gene fragment (bp) | Reference strains in GenBank and accession number | Identity (%) |
|---|---|---|---|
| Vib-7 KT881309 | 1370 | D. piger ATCC 29098 NR 041778.1 | 99 |
| D. fairfieldensis ATCC700045 | 96 | ||
| D. desulfuricans Essex 6 NR 104990.1 | 95 | ||
| Vib-10 KT989316 | 734 | D. piger ATCC 29098 NR 041778.1 | 99 |
| D. desulfuricans Essex 6 NR 104990.1 | 95 | ||
| D. legallii strain H1 NR 108301.1 | 94 | ||
| Vib-1 KT989311 Vib-2 KT989312 Vib-3 KT989313 Vib-4 KT989314 Vib-8 KT989315 |
742 742 743 736 748 |
D. piger ATCC 29098 NR 041778.1 D. desulfuricans Essex 6 NR 104990.1 D. legallii strain H1 NR 108301.1 D. intestinalis KMS2 NR 026413.1 |
99 94–95 93–94 93–94 |
| Vib-5 | 748 | D. piger ATCC 29098 NR 041778.1 | 96 |
| D. desulfuricans Essex 6 NR 104990.1 | 95 | ||
| D. intestinalis KMS2 NR 026413.1 | 93 | ||
| Vib-6 | 722 | D. piger ATCC 29098 NR 041778.1 | 95 |
| Vib-9 | 742 | D. piger ATCC 29098 NR 041778.1 | 96 |
| Rod-9 | 1470 | D. orale NY677 AJ251629.1 | 99 |
| D. orale DSM 12838 CP014230.1 | 99 | ||
| D. orale JCM 17150 NR 113205.1 | 99 |
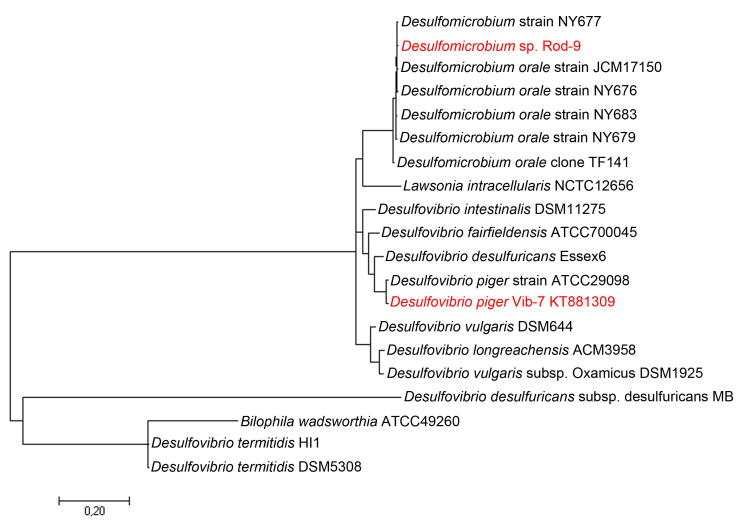
Phylogenetic analysis was done according to the sequence analysis of 16 rRNA and dendrogram was created, showing the affinity of D. piger Vib-7 and D. orale Rod-9 strains with other members of the Desulfovibrio and Desulfomicrobium genera (Fig. 3). Bacteria D. piger Vib-7 shared one cluster with D. piger ATCC 29098, D. desulfuricans Essex 6, D. fairfieldensis ATCC700045, D. intestinalis DSM 11275. These bacteria were isolated from humans’ and animals’ intestines. Desulfomicrobium orale Rod-9 formed one cluster with group D. orale strains.
Fig. 3.
Phylogenetic analysis dendrogram of relationship sequences of 16S rRNA gene of Desulfovibrio and Desulfomicrobium genera with SRB isolated from intestine. Comment: The dendrogram was constructed by Neighbor Joining methods, the scale indicates the genetic distance between the species.
The literature data indicate that D. piger belongs to SRB, found in the human intestine [17], [44]. Overall, Desulfovibrio genera are the most often found SRB in human feces with inflammatory bowel disease [2], [3], [5], [13], [14].
D. piger isolated and identified in the study were genetically similar (according to physiological and biochemical characteristics) to Desulfomonas pigra (isolated for the first time from the feces by W.E. Moore in 1976) [45] and later reclassified as D. piger [44]. Bacteria D. piger in human feces is represented in various collections ATCC 29098, HAQ-6, EBA23-28 [17]. Desulfomicrobium orale is often found in the oral cavity of people having gum problems, such as gum bleeding and periodontal disease [46]. In total, 20 strains of SRB isolated from the human intestine had a nucleotide sequence of SRB strains 99% homology with Desulfovibrio piger ATCC 29,098 (NR 041778.1), while Desulfomicrobium sp. strain Rod-9 belongs to D. orale NY677 species (AJ251629.1).
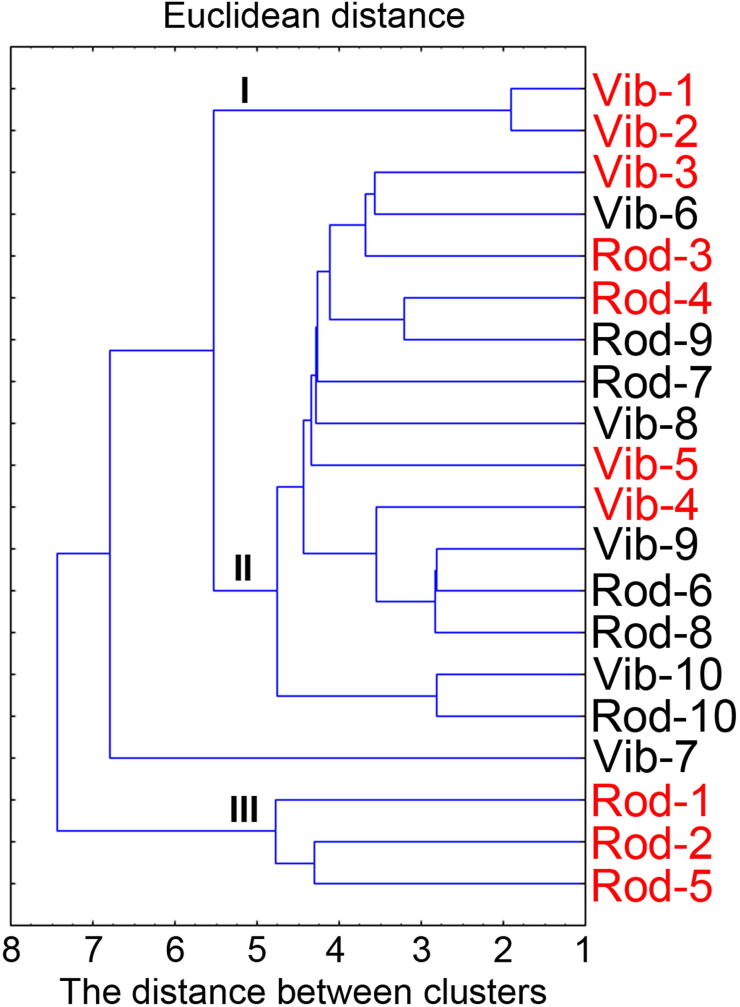
The role of SRB in the development of UC and dissimilatory sulfate reduction parameters (bacterial growth, sulfate and lactate consumption, hydrogen sulfide and acetate production) underwent statistical study, including multidimensional dataset. Cluster analysis was used to determine differences between bacterial growth and parameters of isolates from healthy and not healthy individuals. It can be seen from Fig. 4 that strains isolated from individuals with UC formed two clusters that are separated by Euclidean distance (I and III). These clusters are formed by Desulfovibrio piger (Vib-1, Vib-2) and Desulfomicrobium genus (Rod): Rod-1, 2, 5. These SRB strains belong to different genera. The cluster II was formed by not clearly defined subclusters (strains of Rod and Vib bacteria, both from healthy and unhealthy individuals). The ratio between bacteria isolates from healthy people and patients with the developed UC was 9:5. Different bacteria genera resulted to be the determining factor in the processes of dissimilatory sulfate reduction for Desulfovibrio genus leading to the symptoms of colitis, such as biomass accumulation and the production of hydrogen sulfide and acetate. These results are helping to better understand affinity between intestinal SRB.
Fig. 4.
Dendrogram of SRB affinity based on physiological and biochemical parameters of dissimilatory sulfate reduction. Comment: red color (here and thereafter) on the correlogram indicates the SRB strains isolated from samples of patients with colitis and black color from healthy people. Roman numerals indicate the grouping of bacterial isolates in clusters.
Dispersion analysis was carried out in order to combine the total impact of physical and chemical factors concerning the growth of Desulfovibrio and Desulfomicrobium genera isolated from healthy people and patients with UC (Table 5). The proportions of measured factors (temperature, pH and biomass accumulation) were possible to analyze together due to the application of two-factor dispersion. The influence share of temperature and pH were 60:38% and 50:50% of the Desulfovibrio genus and Desulfomicrobium genus, respectively. Biomass accumulation (not accounted growth factor) of Desulfomicrobium was 2.5 times higher in comparison with Desulfovibrio genus. Significant (p less than 0.05) statistical differences were found between variable factors averages. The bowel inflammation leads to the increased temperature that also consequently leads to more intensive growth of Desulfovibrio genus (the measurable effect of temperature was 60%). This is supported by the fact that Desulfovibrio genus is dominant bacteria during various inflammatory processes. The high concentrations of Desulfovibrio genus also immediately mean high production of hydrogen sulfide. pH is changed due to acetate production by SRB and these conditions support further complications.
Table 5.
Two-factor dispersion analysis of temperature and pH influence on SRB biomass accumulation.
| Factor of influence | Indicators of analysis of variance |
||||
|---|---|---|---|---|---|
| Strains of Desulfovibrio genus |
Strains of Desulfomicrobium genus |
||||
| UC | Healthy | UC | Healthy | ||
| Share of influence (η2, %) | t | 60.58 | 61.35 | 45.3 | 49.03 |
| pH | 38.39 | 37.18 | 51.21 | 48.28 | |
| Not included factors (%) | 1.03 | 1.47 | 3.45 | 2.69 | |
| Fisher coefficient (F practical) | t | 235.60 | 167.11 | 52.58 | 72.87 |
| pH | 149.30 | 101.29 | 59.39 | 71.77 | |
Comments: Fisher coefficient (F critical) for all parameters is 3, the reliability of influence (p) is 0.99.
The synergistic action of acetate can affect the mechanisms of hydrogen sulfide binding, its lumen distribution and withdrawal/absorption in the intestine different parts. These processes influence the biochemical and physiological parameters of intestinal cells that can lead to microbial changes in general. Enzyme rhodanese can be inhibited by acetate influence, increasing the permeability of histohematogenous barriers to hydrogen sulfide [7]. These synergic interactions between acetate and hydrogen sulfide influence the functionality of various important biological substrates, ionophores, membrane and cytoplasmic receptors. Certainly, their synergic effects can lead to increment of expressiveness level of their side effects and further development of inflammation and complications [39], [47].
Dispersion measurements are an appropriate technique for small sample analysis [36]. Dispersion analysis allowed to take into calculations percentages of factors (physical and chemical factors, same as not accountable factors) in one system and to establish the total impact (Table 3). Two-factor dispersion analysis gave an overview and opportunity to evaluate the proportion of these factors. SRB, including D. piger Vib-7, are the same as other intestinal bacteria sensitive to hydrogen sulfide and it is a limiting factor for them too [21]. In the lumen of the human large intestine, hydrogen sulfide is present in concentrations from 1.0 mM to 2.4 mM [20]. Fecal components have large capacity to bind sulfide, free sulfide (unbound) is present only in micromolar quantities [8], [18]. Hydrogen sulfide can be also produced out of endogenous sulfur-containing compounds, such as amino acids. Higher H2S concentrations can significantly inhibit cytochrome c oxidase (the terminal oxidase of the mitochondrial electron transport chain) and the consumption of mitochondrial oxygen. Colonocytes can metabolize H2S and in this way they can resist free luminal sulfide excessive concentrations [20]. The processes concerning excessive H2S in the large intestine have been studied scarcely.
H2S can interfere with the metabolism of colonic epithelial cells. Other microorganisms can produce H2S too, such as Clostridium species [37]. Quinone oxidoreductase, sulfur dioxygenase, and rhodanese are probably included in the processes of detoxification in the intestinal environment with high H2S concentrations. Though, metabolic pathways have not been still fully explained. Endogenously produced H2S should be also taken into consideration. Present literature data indicate that H2S is a pro- or antinociceptive agent in the large intestine. Human diet also influences H2S production in the intestine. On the other hand, sulfate concentrations present in the diet correspond with food types [47]. SRB is competing with methanogenic bacteria and this competition can lead to better conditions for SRB growth [48], [49].
Conclusions
Sulfate-reducing bacteria represent the important part of the intestinal microbiota. They can influence the intestinal environment by hydrogen sulfide and acetate production. Since their presence is connected with IBD, the processes around these bacteria are of high priority to be understood better. The results of the study clearly indicate higher SRB counts, especially Desulfovibrio, and higher hydrogen sulfide concentrations in patients with UC. The effect of hydrogen sulfide can be also supported by acetate accumulation, which is also produced by SRB in the process of incomplete oxidation of organic compounds during dissimilatory sulfate reduction. Both these compounds can lower the pH in the intestine and can be in the interaction (synergic effect) and increase aggressiveness of hydrogen sulfide. According to sequence analysis 16S rRNA genes (SRB isolates from healthy and unhealthy individuals) phylogenetic analysis was done and compared with the GenBank database. Cluster analysis differentiated strains according to physiological and biochemical parameters. Desulfovibrio strains (Vib-1, Vib-2 and Vib-3) and Desulfomicrobium strains (Rod-1, Rod-2 and Rod-5), isolated from patients with UC, formed one cluster and they were on opposite sides. SRB biomass accumulation was in correlations with pH and temperature; it can be concluded that for Desulfovibrio genus temperature affected more their growth, while Desulfomicrobium genus pH affected more the growth in patients with developed UC. The gained results of the research represent useful information for the development of possible UC occurrence risk indicators.
Compliance with ethics requirements
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This study was supported by Grant Agency of the Masaryk University (MUNI/A/0947/2019).
Author contribution
Ivan Kushkevych, Dani Dordević, Monika Vítězová wrote the article. All authors contributed to the conception, design and critically revised the manuscript.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Ivan Kushkevych, Email: ivan.kushkevych@gmail.com.
Monika Vítězová, Email: vitezova@sci.muni.cz.
References
- 1.Loubinoux J., Bronowicji J.P., Pereira I.A. Sulphate-reducing bacteria in human feces and their association with inflammatory diseases. FEMS Microbiol Ecol. 2002;40:107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 2.Kováč J., Vítězová M., Kushkevych I. Metabolic activity of sulfate-reducing bacteria from rodents with colitis. Open Med. 2018;13:344–349. doi: 10.1515/med-2018-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kushkevych I., Vítězová M., Fedrová P., Vochyanová Z., Paráková L., Hošek J. Kinetic properties of growth of intestinal sulphate-reducing bacteria isolated from healthy mice and mice with ulcerative colitis. Acta Vet Brno. 2017;86:405–411. [Google Scholar]
- 4.Gibson G.R., Cummings J.H., Macfarlane G.T. Growth and activities of sulphate-reducing bacteria in gut contents of health subjects and patients with ulcerative colitis. FEMS Microbiol Ecol. 1991;86:103–112. [Google Scholar]
- 5.McDougall R., Robson J., Paterson D., Tee W. Bacteremia caused by a recently described novel Desulfovibrio species. J Clin Microbiol. 1997;35:1805–1808. doi: 10.1128/jcm.35.7.1805-1808.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loubinoux J., Mory F., Pereira I.A., Le Faou A.E. Bacteremia caused by a strain of Desulfovibrio related to the provisionally named Desulfovibrio fairfieldensis. J Clin Microbiol. 2000;38:931–934. doi: 10.1128/jcm.38.2.931-934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowan F.E., Docherty N.G., Coffey J.C., O’Connell P.R. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br J Surg. 2009;96:151–158. doi: 10.1002/bjs.6454. [DOI] [PubMed] [Google Scholar]
- 8.Pitcher M.C., Cummings J.H. Hydrogen sulphide: A bacterial toxin in ulcerative colitis? Gut. 1996;39:1–4. doi: 10.1136/gut.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kushkevych I., Fafula R., Parak T., Bartoš M. Activity of Na+/K+-activated Mg2+-dependent ATP hydrolase in the cell-free extracts of the sulfate-reducing bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9 Acta Vet Brno. 2015;84:3–12. [Google Scholar]
- 10.Kushkevych I.V. Activity and kinetic properties of phosphotransacetylase from intestinal sulfate-reducing bacteria. Acta Biochem Pol. 2015;62:1037–1108. doi: 10.18388/abp.2014_845. [DOI] [PubMed] [Google Scholar]
- 11.Kushkevych I.V. Kinetic Properties of Pyruvate Ferredoxin Oxidoreductase of Intestinal Sulfate-Reducing Bacteria Desulfovibrio piger Vib-7 and Desulfomicrobium sp. Rod-9. Pol J Microbiol. 2015;64:107–114. [PubMed] [Google Scholar]
- 12.Gibson G.R., Macfarlane S., Macfarlane G.T. Metabolic interactions involving sulphate-reducing and methanogenic bacteria in the human large intestine. FEMS Microbiol Ecol. 1993;12:117–125. [Google Scholar]
- 13.Cummings J.H., Macfarlane G.T., Macfarlane S. Intestinal bacteria and ulcerative colitis. Curr Issues Intest Microbiol. 2003;4:9–20. [PubMed] [Google Scholar]
- 14.Kushkevych I., Dordević D., Vítězová M., Kollár P. Cross-correlation analysis of the Desulfovibrio growth parameters of intestinal species isolated from people with colitis. Biologia. 2018;73:1137–1143. [Google Scholar]
- 15.Kushkevych I., Dordević D., Vítězová M. Analysis of pH dose-dependent growth of sulfate-reducing bacteria. Open Med. 2019;14:66–74. doi: 10.1515/med-2019-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kushkevych I., Dordević D., Kollar P. Analysis of physiological parameters of Desulfovibrio strains from individuals with colitis. Open Life Sci. 2018;13:481–488. doi: 10.1515/biol-2018-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner DJ., Krieg NR, Staley JT, Garrity GM. Bergey’s manual of Systematic Bacteriology. The Proteobacteria, Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria. Second Edition. Printed in the United States of America, 2005; 2: 1388 p.
- 18.Attene-Ramos M.S., Wagner E.D., Plewa M.J., Gaskins H.R. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 19.Beauchamp R.O., Bus J.S., Popp J.A., Boreiko C.J., Andjelkovich D.A., Leber P. A critical review of the literature on hydrogen sulfide toxicity. CRC Crit Rev Toxicol. 1984;13:25–97. doi: 10.3109/10408448409029321. [DOI] [PubMed] [Google Scholar]
- 20.Blachier F., Davila A.M., Mimoun S. Luminal sulfide and large intestine mucosa: Friend or foe? Amino Acids. 2010;39:335–347. doi: 10.1007/s00726-009-0445-2. [DOI] [PubMed] [Google Scholar]
- 21.Kushkevych I., Dordević D., Vítězová M. Toxicity of hydrogen sulfide toward sulfate-reducing bacteria Desulfovibrio piger Vib-7. Arch Microbiol. 2019;201:389–397. doi: 10.1007/s00203-019-01625-z. [DOI] [PubMed] [Google Scholar]
- 22.Kushkevych I., Dordević D., Kollar P., Vítězová M., Drago L. Hydrogen sulfide as a toxic product in the small-large intestine axis and its role in IBD development. J Clin Med. 2019;8:1054. doi: 10.3390/jcm8071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kushkevych I., Vítězová M., Kos J., Kollár P., Jampilek J. Effect of selected 8-hydroxyquinoline-2-carboxanilides on viability and sulfate metabolism of Desulfovibrio piger. J Appl Biomed. 2018;16:241–246. [Google Scholar]
- 24.Kushkevych I., Kollar P., Suchy P., Parak T., Pauk K., Imramovsky A. Activity of selected salicylamides against intestinal sulfate-reducing bacteria. Neuroendocrinol Lett. 2015;36:106–113. [PubMed] [Google Scholar]
- 25.Kushkevych I., Kollar P., Ferreira A.L., Palma D., Duarte A., Lopes M.M. Antimicrobial effect of salicylamide derivatives against intestinal sulfate-reducing bacteria. J Appl Biomed. 2016;14:125–130. [Google Scholar]
- 26.Kushkevych I., Kos J., Kollar P., Kralova K., Jampilek J. Activity of ring-substituted 8-hydroxyquinoline-2-carboxanilides against intestinal sulfate-reducing bacteria Desulfovibrio piger. Med Chem Res. 2018;27:278–284. [Google Scholar]
- 27.Macfarlane S., Dillon J.F. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol. 2007;102:1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 28.Macfarlane S., Hopkins M.J., Macfarlane G.T. Bacterial growth and metabolism on surfaces in the large intestine. Microb Ecol Health Dis. 2000;2:64–72. [Google Scholar]
- 29.Beauchamp R.O., Bus J.S., Popp J.A. A critical review of the literature on hydrogen sulfide toxicity. Crit Rev Toxicol. 1984;13:25–97. doi: 10.3109/10408448409029321. [DOI] [PubMed] [Google Scholar]
- 30.Kováč J., Kushkevych I. Proceedings of the International PhD Students Conference MendelNet, Brno, Czech Republic, 6–7 November 2017. 2017. New modification of cultivation medium for isolation and growth of intestinal sulfate-reducing bacteria; pp. 702–707. [Google Scholar]
- 31.Postgate J.R. Cambridge University Press; Cambridge, UK: 1984. The Sulfate Reducing Bacteria. [Google Scholar]
- 32.Kushkevych I. Isolation and Purification of Sulfate-Reducing Bacteria. In: Microorganisms edited by: Dr. Miroslav Blumenberg. IntechOpen, London, UK.
- 33.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persing D.H. second ed. ASM Press; Publisher: 2011. Molecular Microbiology: Diagnostic Principles and Practice; p. 960. [Google Scholar]
- 35.Cline J.D. Spectrophotometric determination of hydrogen sulfide in natural water. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 36.Bailey N.T.J. Cambridge University Press; Cambridge, UK: 1995. Statistical Methods in Biology. [Google Scholar]
- 37.Černý M., Vítězová M., Vítěz T., Bartoš M., Kushkevych I. Variation in the distribution of hydrogen producers from the clostridiales order in biogas reactors depending on different input substrates. Energies. 2018;11:3270. [Google Scholar]
- 38.Kushkevych I., Vítězová M., Vítěz T., Bartoš M. Production of biogas: Relationship between methanogenic and sulfate-reducing microorganisms. Open Life Sci. 2017;12:82–91. [Google Scholar]
- 39.Roediger W.E.W., Duncan A., Kapaniris O., Millard S. Reducing sulfur compounds of colon impair colonocyte nutrition: implications for ulcerative colitis. Gastroenterology. 1993;104:802–809. doi: 10.1016/0016-5085(93)91016-b. [DOI] [PubMed] [Google Scholar]
- 40.Levitt M.D., Furne J., Springfield J., Suarez F., DeMaster E. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J Clin Investing. 1999;104:1107–1114. doi: 10.1172/JCI7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohge H. The effect of antibiotics and bismuth on fecal hydrogen sulfide and sulfate-reducing bacteria in the rat. In: Ohge H., Furne J.K., Springfield J., editors. Vol. 228. 2003. pp. 137–142. (FEMS Microbiol Lett). [DOI] [PubMed] [Google Scholar]
- 42.Willis Caroline L., Cummings John H., Neale Graham, Gibson Glenn R. Nutritional aspects of dissimilatory sulfate reduction in the human large intestine. Curr Microbiol. 1997;35(5):294–298. doi: 10.1007/s002849900257. [DOI] [PubMed] [Google Scholar]
- 43.Levine Jimmy, Ellis Carol J, Furne Julie K, Springfield John, Levitt Michael D. Fecal hydrogen sulfide production in ulcerative colitis. Am J Gastroenterol. 1998;93(1):83–87. doi: 10.1111/j.1572-0241.1998.083_c.x. [DOI] [PubMed] [Google Scholar]
- 44.Loubinoux J., Valente F.M.A., Pereira I.A.C. Reclassification of the only species of the genus Desulfomonas, Desulfomonas pigra, as Desulfovibrio piger comb. nov. Int J Syst Evol Microbiol. 2002;52:1305–1308. doi: 10.1099/00207713-52-4-1305. [DOI] [PubMed] [Google Scholar]
- 45.Moore W.E. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species of the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium and Ruminococcus. In: Moore W.E., Johnson J.L., Holdeman L.V., editors. Vol. 26. 1976. pp. 238–252. (Int J Syst Bact). [Google Scholar]
- 46.Langendijk P.S. Isolation of Desulfomicrobium orale sp. nov. and Desulfovibrio strain NY682, oral sulfate-reducing bacteria involved in human periodontal disease. In: Langendijk P.S., Kulik E.M., Sandmeier H., editors. Vol. 51. 2001. pp. 1035–1044. (Int J Syst Evol Microbiol). [DOI] [PubMed] [Google Scholar]
- 47.Florin T.H., Neale G., Goretski S. Sulfate in food and beverages. J Food Compos Anal. 1993;6:140–151. [Google Scholar]
- 48.Kushkevych I., Vítězová M., Vítěz T., Kovac J., Kaucká P., Jesionek W. A new combination of substrates: Biogas production and diversity of the methanogenic microorganisms. Open Life Sci. 2018;13:119–128. doi: 10.1515/biol-2018-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kushkevych I., Kováč J., Vítězová M., Vítěz T., Bartoš M. The diversity of sulfate-reducing bacteria in the seven bioreactors. Arch Microbiol. 2018;200:945–950. doi: 10.1007/s00203-018-1510-6. [DOI] [PubMed] [Google Scholar]