Graphical abstract
Keywords: Breast cancer, Hydrogen sulphide, miR-155/NOS2/NO signaling pathway, PI3K/AKT signaling pathway, Nitric oxide, miR-4317, Natural killer cells, CAR T cells
Abbreviations: BC, Breast Cancer; CAR, Chimeric antigen receptor; 51Cr-release, Chromium release assay; CD80, Cluster of differentiation 80; CD86, Cluster of differentiation 86; CBS, Cystathionine β-synthase; CSE, Cystathionine γ-lyase; CTL, Cytotoxic T lymphocyte; NOS3, Endothelial nitric oxide synthase-3; HCC, Hepatocellular carcinoma; HLA-DR, Human Leukocytic antigen DR; H2S, Hydrogen sulphide; NOS2, Inducible nitric oxide synthase-2; IFN-γ, Interferon gamma; KD, Knock down; LDH, Lactate dehydrogenase Assay; MICA/B, MHC class I polypeptide-related sequence A/B; miRNA, MicroRNA; NKG2D, Natural Killer Group 2D ; NK, Natural killer; NO, Nitric oxide; ncRNAs, Non-coding RNAs; PD-L1, Programmed death-ligand 1; Scr-miRNAs, Scrambled microRNAs; Scr-siRNAs, Scrambled siRNAs; siRNAs, Small interfering RNAs; TNBC, Triple negative breast cancer; TNF-α, Tumor necrosis factor-α; ULBP2/5/6, UL16 binding protein 2/5/6; 41BBL, 41BB Ligand
Abstract
Introduction
Hydrogen sulphide (H2S) has been established as a key member of the gasotransmitters family that recently showed a pivotal role in various pathological conditions including cancer.
Objectives
This study investigated the role of H2S in breast cancer (BC) pathogenesis, on BC immune recognition capacity and the consequence of targeting H2S using non-coding RNAs.
Methods
Eighty BC patients have been recruited for the study. BC cell lines were cultured and transfected using validated oligonucleotide delivery system. Gene and protein expression analysis was performed using qRT-PCR, western blot and flow-cytometry. In-vitro analysis for BC hallmarks was performed using MTT, BrdU, Modified Boyden chamber, migration and colony forming assays. H2S and nitric oxide (NO) levels were measured spectrophotometrically. Primary natural killer cells (NK cells) and T cell isolation and chimeric antigen receptor transduction (CAR T cells) were performed using appropriate kits. NK and T cells cytotoxicity was measured. Finally, computational target prediction analysis and binding confirmation analyses were performed using different software and dual luciferase assay kit, respectively.
Results
The H2S synthesizing enzymes, cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE), exhibited elevated levels in the clinical samples that correlated with tumor proliferation index. Knock-down of CBS and CSE in the HER2+ BC and triple negative BC (TNBC) cells resulted in significant attenuation of BC malignancy. In addition to increased susceptibility of HER2+ BC and TNBC to the cytotoxic activity of HER2 targeting CAR T cells and NK cells, respectively. Transcriptomic and phosphoprotein analysis revealed that H2S signaling is mediated through Akt in MCF7, STAT3 in MDA-MB-231 and miR-155/ NOS2/NO signaling in both cell lines. Lastly, miR-4317 was found to function as an upstream regulator of CBS and CSE synergistically abrogates the malignancy of BC cells.
Conclusion
These findings demonstrate the potential role of H2S signaling in BC pathogenesis and the potential of its targeting for disease mitigation.
Introduction
Breast cancer (BC) is the most common malignancy in females and the leading cause of cancer-related mortailty [1], [2]. BC incidence and mortality rates widely vary in countries with different socio-economic levels, with estimated incidence rates of 54.4% and 31.3% in developed and developing countries, respectively [2]. BC presents as diverse BC subtypes, which differ in prognosis and treatment options, reflecting disease heterogeneity. Hormone receptor (HR) positive tumors show high response rates to hormonal therapies. Human epidermal growth factor receptor 2 (HER2) positive tumors are amenable for treatment using HER2 targeting antibodies and HER2 specific tyrosine kinase inhibitors [3]. For triple negative breast cancer (TNBC), targeted treatment options are limited and outcomes have been substantially worse [4], [5], [6]. Collectively, this underscores BC, and particularly TNBC, as a significant worldwide problem in dire need for more efficacious and innovative therapies [7], [8].
Hydrogen sulfide (H2S) is a recently identified member of a well-characterized family of gaseous biological mediators, known as gasotransmitters [9]. Since its discovery, endogenous H2S has been proven to play pivotal roles in various physiological and pathological conditions [10]. In cancer, we and others have recently reported that H2S acts as a key player in modulating several canonical and non-canonical oncogenic signaling pathways such as PI3K/AKT/mTOR [11], JAK/STAT [12], [13], Ras/Raf/MEK/ERK [11], and nitric oxide (NO) [14] signaling cascades. Our group also highlighted the regulatory effects of H2S on the non-coding RNA (ncRNA) machinery in BC cells [14], [15].
The H2S synthesizing enzymes, cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE), have been reported to have an organ- and tissue- specific expression pattern. High expression profiles of CBS and CSE have been linked to the aggressiveness of several solid tumors, but are generally expressed at lower levels in hematological malignancies [16], [17]. For instance, upregulation of CBS was reported in androgen-dependent prostate, colon and ovarian cancer cells [18], [19]. The latter two showed increased H2S production when compared to non-malignant epithelial cells. In contrary, the expression level of CBS has been described to be significantly low in hepatocellular carcinoma (HCC) [18], [20]. Although CBS expression tends to be higher in most of the solid tumors, variability between patients with different histopathologic malignancies suggests the need for further molecular characterization of tumors. CSE was also found to enhance cellular proliferation and migration of colon cancer cell lines [21], [22], [23]. Similarly, in HCC, prostate cancer and glioma cell lines, endogenous H2S produced by CSE has been involved in the survival and proliferation of these malignant cell lines [24], [25], [26]. Collectively, CBS and CSE expression patterns and functional correlations have been reported in different solid tumors, but their role in BC is yet to be clarified.
In the present study, we aim to evaluate the expression patterns of H2S synthesizing enzymes in a cohort of BC patients, to unravel the mechanisms by which H2S modulates the oncogenic and the immunogenic properties of BC cells, and lastly to explore the therapeutic potential of targeting H2S synthesizing enzymes using ncRNAs.
Materials and methods
Study patients
BC tissues and normal breast tissues were surgically resected from 80 female patients with BC. Tumor and normal tissues were morphologically proven by a pathologist. Classification of the BC patients according to clinical data is provided in Table 1. Detailed immuno-histochemical and pathological profiles of patients are provided in (Supplementary Table S1). Fifty healthy donors were also recruited in this study. All performed experiments are compliant with the guidelines of the Institutional Review Board of Kasr El Aini Medical School, Cairo University and German University in Cairo and with the ethical standards of the declaration of Helsinki (Ethical Approval Number: N-70-2016). All participants were included after giving written informed consents.
Table 1.
Clinical data summary of the recruited female BC patients.
| BC patients | Percentage (Count) | |
|---|---|---|
| Age (Years) | ≥40 <40 |
73.75% (59/80) 26.25% (21/80) |
| Grade | I II III |
3.75% (3/80) 77.5% (62/80) 18.75% (15/80) |
| Histological type | Ductal Lobular Both |
96.25% (77/80) 1.25% (1/80) 2.5% (2/80) |
| Molecular Subtype | Luminal A Luminal B HER-2 enriched TNBC |
22.5% (18/80) 43.75% (35/80) 2.5% (2/80) 31.25% (25/80) |
| ER/PR status | Positive Negative |
66.25% (53/80) 33.75% (27/80) |
| HER2 status | Positive Negative |
26.25% (21/80) 73.75% (59/80) |
| Lymphatic involvement | Yes No |
66.25% (53/80) 33.75% (27/80) |
| Proliferation Index (Ki-67) | High (≥14%) Low (<14%) |
70% (56/80) 30% (24/80) |
ER: estrogen receptor; PR: progesterone receptor; HER2: human epidermal growth factor receptor 2.
Cell culture and treatment
MDA-MB-231 and MCF-7 BC cell lines were obtained from ATCC and Vacsera, Egypt. BC cells were cultured in DMEM (Lonza, Switzerland) media as previously described [14], [27], [28], [29].
Oligonucleotides transfection
Breast cancer cells were transfected using different oligonucleotides (such as: CBS siRNAs, CSE siRNAs, miR-4317 mimics, scrambled siRNAs (Scr-siRNAs), scrambled miRNAs (Scr-miRNAs)) (Qiagen, Germany). All transfection experiments were executed in triplicates employing HiPerfect Transfection Reagent (Qiagen, Germany). All Experiments were repeated 3 times or more [14], [28], [30], [31].
Cellular viability and proliferation experiments
3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent was used for the cellular viability experiments. BC cells (10,000 cells) were seeded in 200 µL media in a 96-well plate. Post-transfection, 48 h later media in each well was replaced by 20 µL working solution. After 6 h, the absorbance of the formazan crystals, solubilized in 200 µL buffer, was measured [14], [27], [28], [30]. Bromo-deoxyuridine (BrdU) incorporation assay was used for the proliferation experiments. BC cells were seeded into black 96-well plates at a cell density of 5 × 104 cells/well. In accordance with the Cell Proliferation ELISA kit (Roche Applied Science, Penzberg, Germany), BC cells were incubated with BrdU for 4 h, then fixed using Fix-Denate for 30 min and incubated with Anti-BrdU POD for 90 min [14], [27], [28], [30]. All experiments were performed in triplicates and repeated 3 times or more.
Cellular migration and invasion
Wound-healing assay was done to assess BC cellular migration. Transfected cells were left to grow to a confluency of 90–95% in 24-well plates. Post-transfection, 3 scratches were made in each well using a 10- µL pipette tip. Cells were washed using PBS and replenished with new low-serum media (1% FBS). Twenty four hours later, the surface areas of the scratches were measured and wound closure was quantified with Zen2012 software [14], [27], [28], [30]. The Modified Boyden chamber assay (BD Bioscience, Bedford, USA) was done to assess the in-vitro invasion capacity. In 24-well plates, BC cells were transfected with different oligonucleotides. After transfection, 6 × 104 cells, re-suspended in 200 μL low-serum media (1% FBS), were seeded in the upper well, while the lower well contained high-serum media (20% FBS). Cells were washed from the upper surface using a cotton swab 8 h later. The trans-well migrant cells were fixed and stained using 1% crystal violet (Sigma Chemical Co., California, USA) and then counted under an inverted light microscope. All experiments were performed in triplicate and repeated 3 times or more [6], [14], [27].
Colony-forming assay
For the colony forming assay, 48 h post-transfection, cells were harvested and seeded in 6-well plate at a count of 900 cells/well. Cells were incubated in full DMEM under normal conditions (37 °C and 5% CO2) for 2–3 weeks. Colonies were fixed using 6% glutaraldehyde, stained by 0.05% crystal violet and then manually counted [14], [27], [32].
Total RNA and miRNAs extraction
Biazol reagent was used for total RNA and miRNAs extraction from primary tissues and cell lines. RNA integrity was examined by 18 s rRNA bands detection on 1% agarose gel electrophoresis. RNA samples were examined for possible contamination with other molecules such as proteins, cellular fragments, or organic compounds using spectrophotometric analysis. The harvested RNA samples were spectrophotometrically assessed at two different wavelengths (260 and 280 nm) to assess its purity. The 260/280 ratio is used to assess the amount of protein contamination which could be present in trace amount during the process of RNA isolation. A ratio of ~ 2.0 is generally accepted as “pure” for RNA. If the ratio is appreciably lower, it may indicate the presence of protein, phenol or other contaminants that absorb strongly at or near 280 nm [14], [27], [28], [30], [33].
Quantitative real-time PCR analysis
Reverse transcription of CBS, CSE, NOS2, NOS3, MICA, ULBP2, TNF-α, IFN-γ, β-actin and 18srRNA mRNAs into cDNA was done using the High-Capacity cDNA Reverse Transcription Kit (ABI, California, USA) according to the manufacturer’s instruction. The extracted miRNAs were reverse transcribed into single-stranded complementary DNA (cDNA) using TaqMan MicroRNA Reverse Transcription Kit (ABI, California, USA) and specific primers for hsa-miR-155, hsa-miR-146, hsa-miR-4317 and RNU6B. Relative expression of CBS, CSE, NOS2, NOS3, MICA, ULBP2, TNF-α, IFN-γ, β-actin and 18 s rRNA (for normalization), miR-155, miR-146a, miR-4317 and RNU6B (for normalization) was quantified using TaqMan Real-Time q-PCR on StepOne™ Systems (ABI, California, USA). Relative expression was calculated using the 2-ΔΔCt method. All PCR reactions were done in triplicates and repeated 3 times or more [14], [27], [28], [30], [34].
Western blot analysis
Post oligonucleotides transfection, BC cells were lysed using RIPA lysis buffer (Pierce Biotechnology, Massachusetts, United States) containing Protease Inhibitor Cocktail Set III and Phosphatase Inhibitor Cocktail Set II (EMD Biosciences, California, USA). BCA protein assay kit (Pierce Biotechnology, Massachusetts, USA) was adopted to determine the concentration of proteins. Protein lysates (25–35 μg) were diluted with 4x laemmli buffers and incubated for 10 min at 95 °C. Proteins were then separated on 12% SDS-PAGE gels in mini electrophoresis system (Bio-Rad Laboratories, California, USA). Amersham™ ECL™ Rainbow™ Marker - Full range (GE Healthcare, California, USA) was applied as a protein size detection marker. Separated proteins were then transferred to iBlot™ 2 Transfer Stack nitrocellulose membranes (Life Technologies, California, USA) using an iBlot™ 2 dry transfer system (Life Technologies, California, USA). Membranes were blocked for 1 h using 5% fat-free dry milk in TBS (Bio-Rad Laboratories, California, USA) supplied by 0.1% tween 20 (Sigma-Aldrich, California, USA) then incubated over-night with CBS (ab140600, Abcam, California, USA), CSE (ab54573, Abcam, California, USA), Akt (pan) (Cell Signaling Technology), Phospho-Akt (s473) (Cell Signaling Technology), Phospho-STAT3 (y705) (BD Biosciences/ Cell Signaling Technology) and GAPDH (Cell Signaling Technology, California, USA) antibodies. Fluorophore labeled IRDye® (LI-COR Biosciences, California, USA) secondary antibodies were used for protein detection. Fluorescence visualization was done using an Odyssey CLx membrane imaging system (LI-COR Biosciences, California, USA). All western blot experiments were done in triplicates and repeated at least 3 times [6].
Quantification of H2S production
A microplate colorimetric H2S detection assay designed for H2S measurement was used as previously described [35], [36]. Briefly, poly vinyl pyrrolidone (PVP, 5% w/v) solution was added to Nafion® perfluorinated resin solution in a ratio of 9:1 (v/v). Thirty μL of 0.1 M silver nitrate (AgNO3) was then combined with the Nafion/PVP mixture. The Ag/Nafion/PVP solution was vortexed, and then 20 μL of solution was dropped per carefully on the lower side of the cover. The cover was dried for at least 3 h to form a membranous coating. To obtain calibration curve, 300 μL of the freshly prepared Na2S solution, at concentrations ranging from 6.25 to 50 μM, was added to a 96-well plate. The plate was covered with the fabricated Ag/Nafion/PVP coated cover. H2S was allowed to form Na2S for 4 h at 37 °C in a 5% CO2 incubator. The absorbance at 310 nm was measured using a Synergy HTX multi-mode reader (BioTek, Winooski, VT, United States). Next, we analyzed the absorbance of the Ag/Nafion/PVP membrane for H2S produced from BC cells. Finally, H2S concentration was calculated against the calibration curve (sensitivity: 0.0049 Abs/μM H2S). AgNO3, PVP (K90), Nafion® perfluorinated resin solution, Na2S, GSH, DTT, H-Cys and Cys were purchased from Sigma Aldrich (California, USA).
Quantification of NO production
NO production was measured using Griess reagent assay (Promega, USA) [14]. Briefly, 50 µL of cell culture supernatant mixed with 50 μL of the sulfanilamide solution were incubated for 10 min. Fifty µL of N-1-napthylethylenediamine dihydrochloride (NED) solution were then added and absorbance was measured at 540 nm using Wallac 1420 Victor 2 Multilabel Counter (Perkin Elmer, USA). Experiments were performed in triplicates and repeated 3 times or more [14].
Detection of cell surface markers using flow cytometry
After oligonucleotides transfection according to the manufacturing protocol, tumor cells were trypsinized then washed with cold PBS. Cells were immunostained for 45 min − 1 h with fluorophore-conjugated primary antibodies. Antibodies used included: anti-HER2_FITC, anti-HLA-DR_FITC, anti-CD80_PE, anti-CD86_APC (BD Biosciences, San Jose, California, USA), and PD-L1_PE-Cy5 (BioLegend, California, United States). CAR expression was detected by immunostaining with a recombinant human ErbB2/Her2 Fc chimeric protein (R&D Systems) at 4 °C for 1 h followed by a PE-conjugated goat anti-human IgG (Fc gamma-specific) (eBioscience). Cells were analyzed on Accuri C6 or BD Canto II flok6w cytometers (BD bioscience, California, USA). Data analysis was done using FlowJo™ 10 (BD bioscience, California, USA) [31].
NK cell isolation
Peripheral blood mononuclear cells (PBMCs) were separated from peripheral blood of healthy donors within 4 h of collection using Ficoll–Hypaque centrifugation (Axis-Shield PoC AS, Norway). NK cells were then enriched by negative selection using a MACS NK cell isolation kit (Miltenyi Biotec, Cologne, Germany). Enriched NK cell populations were 97.4% CD56/CD3 and 1.2% CD3 positive, assessed by flow cytometry [14], [28], [37].
NK cell-mediated cytolytic assay
MDA-MB-231 cells transfected with different oligonucleotides were seeded in a U-shaped 96-well plate at a cell density of 15,000 cells/well. After 2 h, primary NK cells were added to the target MDA-MB-231 cells at a 5:1 effector to target ratio (E:T) and incubated for 6–8 h. Later, the lactate dehydrogenase (LDH) activity assay kit (MAK066-1 K1-Sigma-Aldrich, St. Louis, MO, USA) was used to measure the in-vitro cytotoxicity following the manufacturer’s instructions. The percentage of lysis was calculated according to the following equation: % cytotoxicity = (target maximum release – experimental release)/ (target maximum release) × 100. The experiment was done in triplicate and repeated at least 3 times [14], [28], [37].
T cell isolation and chimeric antigen receptor (CAR) transduction
Chimeric antigen receptor (CAR) encoding γ-retroviruses were produced by co-transfecting 293 T cells with an FRP5.CD28.ζ containing SFG retroviral vector plasmid, a Peq-Pam plasmid, encoding Moloney GagPol, and RD114 envelope containing plasmid using Gene Juice transfection reagent (EMD Biosciences). Supernatants having the pseudo-retroviruses were harvested 72 h later. Briefly, PBMCs were separated from fresh blood of healthy donors, consented by a protocol approved by the IRB of Baylor College of Medicine, using Lymphoprep density gradient media (STEM CELL technologies). PBMCs were activated with CD3 (OKT3 –OrthoBiotech) / CD28 antibodies (BD Biosciences) to stimulate T cell proliferation. Twenty-four hours later, recombinant human interleukin 7 and 15 (IL-7/IL-15) were added. On day 3, cells were harvested for retroviral transduction. CAR encoding γ-retroviruses were centrifuged for 1 h at 3000 g on non-tissue culture–treated 24-well plate coated with recombinant fibronectin (RetroNectin - Takara Bio, USA). Subsequently, 1 × 105 T cells per well were transduced with retroviruses in the presence of IL-7/IL-15, by centrifugation for 5 min at 1000 g. After 48 h, cells were removed.
T cell cytotoxicity assay
Radioactive chromium (51Cr)-release assays were performed. Briefly, MCF-7 cells transfected with different oligonucleotides for 72 h were incubated with 51Cr for 1 h. In v-shaped 96 well plates T cells and BC cells were co-cultured at different concentrations. After 6 h, the supernatant was collected and analyzed for 51Cr. The average cytolysis of triplicate wells = (test release-spontaneous release) / (max release-spontaneous release) × 100.
Bioinformatic analysis
The computational software: miRDB (www.mirdb.org/miRDB), miRanda (www.microrna.org), DIANA Lab (www.diana.cslab.ece.ntua.gr), and Target Scan (www.targetscan.org) were adopted to predict novel miRNAs that could dually target CBS and CSE simultaneously with good binding scores as previously described [28], [31], [34], [37], [38]. This software use special algorithms for prediction and the results are based on multiple criteria including miRNA-mRNA complementarity strength, binding score, hybridization energy, and number of predicted binding sites within the transcripts’ 3′-UTR and the candidate miRNA. It is important to note that according to miRanda software, the more negative the hybridization energy is, the more favorable the binding between the miRNA and its target transcript. Moreover, complementarity percentage was evaluated manually by calculating how many nucleotides out of the 22 nucleotide-length miRNA were occupied by base pairing with its complementary target sequence on the 3′-UTR of the target genes mRNA.
Reporter constructs and luciferase assay
To confirm the binding of miR-4317 to CBS and CSE 3′-UTR, a dual firefly luciferase reporter construct was utilized (pmirGLO) (Promega, Madison, WI). pmirGLO Dual Luciferase microRNA target expression vector was co-digested using SacI and XbaI restriction enzymes (Thermoscientific, Boston, MA, USA). For wild-type constructs (WT), the miR-4317 binding region in 3′-UTRs of CBS and CSE were inserted in the pmirGLO vector separately. Moreover, mutant constructs (MUT) were designed (Supplementary Table S2). The sequences in bold refer to the binding region nucleotides that were deleted in the mutant constructs. MDA-MB-231 cells were transfected with the constructs or empty pmirGLO vector using SuperFect Tranfection Reagent (Qiagen, Germany) then co-transfected 24 h later with miR-4317 mimics or Scr-miR using HiPerFect Tranfection Reagent (Qiagen, Germany). After 48 h, relative luciferase activity was measured using SteadyGlo luciferase reporter assay kit (Promega, Germany) [14], [37].
Statistical analysis
Data is presented as mean ± standard error of the mean (SEM) for at least 3 different experiments, unless otherwise indicated in figure legends. Non-parametric unpaired student-t-test was executed to compare between every two independent groups (e.g. screening results). One-way analysis of variance with post hoc analysis was adopted for multiple comparisons (e.g. tranfection experiments). Correlation analysis was performed by Spearman analysis. P-value of < 0.05 was considered statistically significant, and the threshold of significance is denoted by *= p < 0.05, **p < 0.01, ***= p < 0.001, ****=p < 0.0001. Data was analyzed using GraphPad Prism 8.2.1 software.
Results
Breast cancer patients express high levels of CBS, CSE and H2S
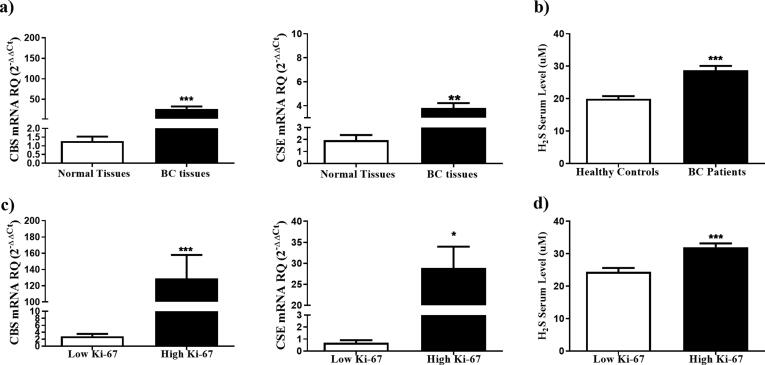
First, we assessed the levels of CBS and CSE in tumor biopsies from a cohort of 80 women, diagnosed with BC (Table 1). Tumors from BC patients showed significantly higher expression levels of CBS (P = 0.0005) and CSE (P = 0.0019) transcripts relative to histologically normal breast tissues isolated from the same mastectomy sample (Fig. 1a). All BC subtypes (Luminal A, luminal B, HER2 enriched and TNBC) showed elevated levels of CBS and CSE with no significant differences in the expression levels of those enzymes between the different subtypes. Moreover, serum H2S levels were significantly higher in BC patients (28.74 ± 1.33 µM) when compared to healthy gender and age matched controls (19.94 ± 0.85 µM; p < 0.0001) (Fig. 1b). The elevation of serum H2S and its synthesizing enzymes in BC patients, independent of the BC subtype, prompted us to investigate the correlation between the expression pattern of CBS and CSE with the proliferation index of BC tissues, an established prognostic marker of the disease [39]. Interestingly, tumors with a high proliferation marker Ki-67 expression (≥14%) showed significantly elevated levels of CBS (P = 0.0002), CSE (P = 0.0103) (Fig. 1c) and H2S (P = 0.0008) (Fig. 1d) compared to tumors with low Ki-67 (<14%).
Fig. 1.
Overexpression of CBS and CSE enzymes in BC tissues. (a) CBS and CSE expression profiles were analyzed in breast tissues isolated from 80 BC patients using qRT- PCR, normalized to 18 s rRNA as an internal control, and compared to normal BC tissues from the same patients (b) Serum H2S was compared between BC patients and healthy volunteers. (c) CBS and CSE expression was compared according to the tumor proliferation index ki-67. (d) Serum H2S levels were compared according to the tumor proliferative index ki-67.
Inhibition of endogenous H2S production attenuates the malignant properties of BC cells in vitro
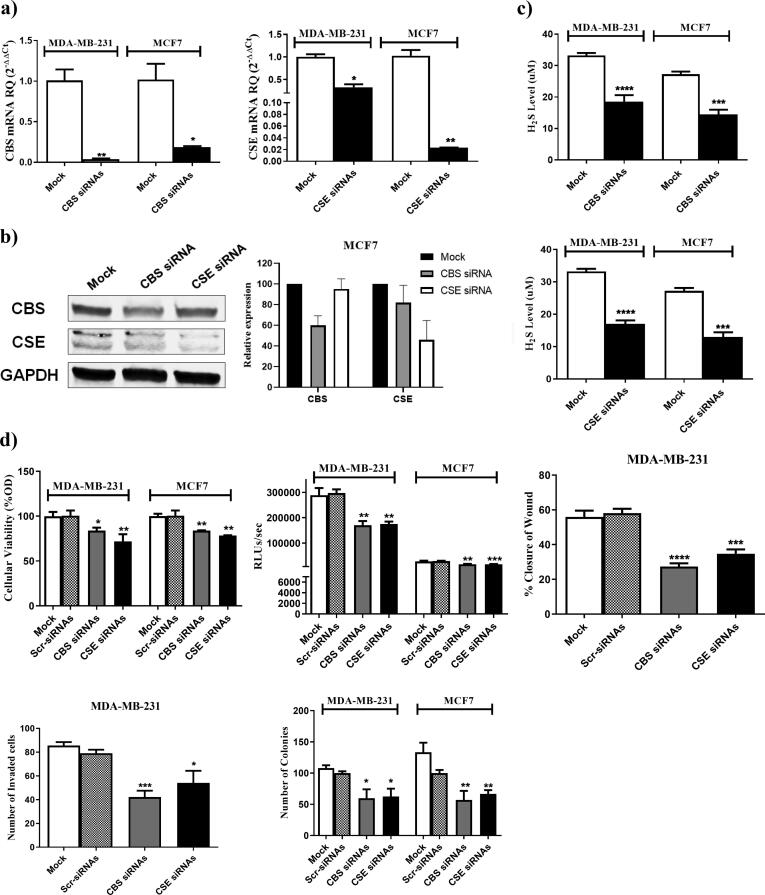
Since CBS and CSE were found to be elevated in various subtypes of BC, MCF7, a HR+/HER2+ BC cell line, and MDA-MB-231, a TNBC cell line, were used to analyze the impact of CBS and CSE silencing on the malignant phenotype of BC cells. Knocking down (KD) of CBS or CSE using siRNAs resulted in efficient reduction of CBS and CSE transcript and protein levels as evaluated via qRT-PCR (Fig. 2a) and western blot analysis (Fig. 2b, Supplementary Fig. S1), respectively. This was associated with a significant decrease in the H2S levels produced by both BC cell lines (Fig. 2c). KD of CBS or CSE led to a marked reduction in BC cellular viability, proliferation, migration, invasion, and colony forming ability (Fig. 2d).
Fig. 2.
CBS and CSE sequence specific siRNAs downregulate RNA, protein and H2S production levels and impact BC malignant properties. The expression of CBS and CSE transcripts and protein levels in mock and siRNAs transfected MDA-MB-231 and MCF7 cells were determined using (a) qRT-PCR 48 h post-transfection and (b) western blot analysis 72 h post-transfection (c) H2S levels were determined in the supernatant of MDA-MB-231 and MCF7 after silencing of CBS and CSE (d) Knocking down of CBS and CSE using CBS and CSE siRNAs resulted in a marked repression of cellular viability assessed by MTT assay (top left), cellular proliferation assessed using BrdU incorporation assay (top middle), migration capacity measured by wound healing assay (top right), invasion capacity measured by Boyder chamber assay (bottom left) and colony formation ability (bottom right).
Inhibition of endogenous H2S production modulates PI3K/Akt, JAK/STAT3 and miR-155/NOS2/NO signaling pathways in BC cells
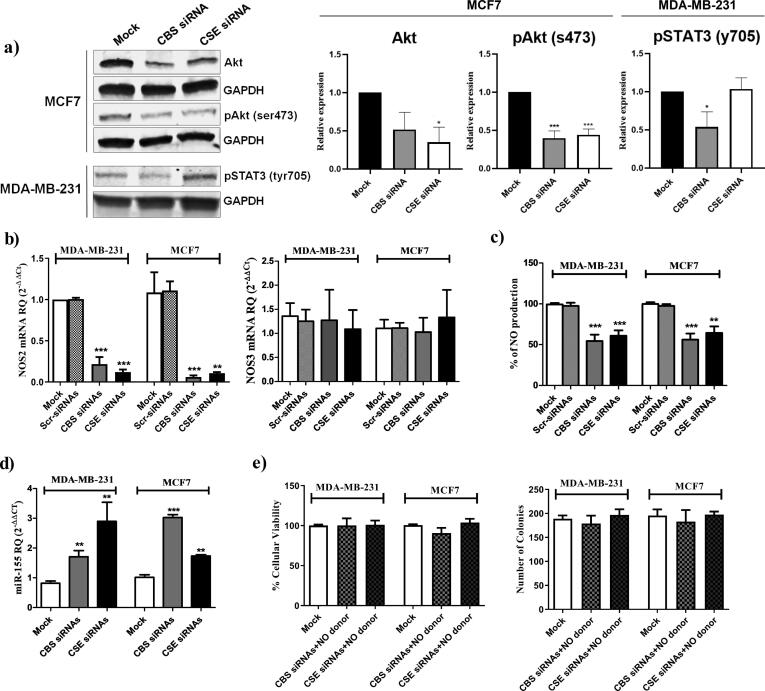
HER2 signaling is a key mediator of the malignant phenotype of HER2 expressing BC cell lines. In order to study the underlying mechanisms for the altered tumorigenicity/malignant potential of the HR+/HER2+ MCF7 cell line, we evaluated the effect of CBS/CSE KD/silencing on the PI3K/Akt pathway which is a key HER2 signaling pathway. We found that total Akt and active phosphorylated Akt levels significantly decreased upon repression of endogenous H2S production (Akt: CBS siRNA p = 0.092952, CSE siRNA p = 0.042246; pAkt: CBS siRNA p = 0.000366, CSE siRNA p = 0. 0.000779) (Fig. 3a, Supplementary Fig. S1). Of note, no effect on Akt and phosphorylated Akt levels was observed in the TNBC MDA-MB-231 cell line (data not shown). In TNBC, the JAK/STAT3 pathway has been reported to be an important mediator of tumorgenicity [40]. Accordingly, the effect of CBS/CSE KD on STAT3 was investigated in the TNBC cell line, MDA-MB-231, which resulted in a significant decrease in the activated/phosphorylated STAT3 levels in CBS silenced cells (Fig. 3a, Supplementary Fig. S1).
Fig. 3.
CBS/CSE silencing modulates PI3K/Akt, JAK/STAT3 and miR-155/NOS2/NO signaling pathways. (a) The protein expression of Akt, phosho-Akt (serine 473) in mock or siRNA transfected MCF7 and phosphor-STAT3 (tyrosine 705) in mock or siRNA transfected MDA-MB-231 was determined using western blot 72 h post transfection. Relative expression plots were normalized to the internal control GAPDH, then compared to the mock ± standard deviation from 3 separate experiments (b) the RNA trnascripts of NOS2 and NOS3 in mock and siRNAs transfected MDA-MB-231 and MCF7 cells determined by qRT-PCR 48 h post-transfection. (c) NO levels were determined in the supernatant of MDA-MB-231 and MCF7 after silencing of CBS and CSE. (d) The expression pattern of miR-155 normalized to RNU6B as an internal control in mock, CBS siRNAs and CSE siRNAs MDA-MB-231 and MCF7 cells were determined by qRT-PCR 48 h post-transfection. (e) Cellular viability and colony forming ability of MDA-MB-231 and MCF-7 cells co-treated with a NO donor (NaNO2) and CBS or CSE siRNA.
NO signaling has been associated with several malignancies including BC. We previously reported on the cross-talk between exogenous H2S and NO pathway in TNBC [14]. We, therefore, investigated the effect of CBS/CSE silencing on NO production. Interestingly, CBS and CSE KD resulted in a marked repression of NOS2 transcript levels but had no significant effect on the NOS3 transcript levels (Fig. 3b). Additionally, this significantly attenuated NO production levels (Fig. 3c). The microRNA molecules: miR-146a and miR-155 were previously found to regulate the NOS2 transcript levels [41], [42]. Accordingly, the expression levels of miR-146a and miR-155 were evaluated in CBS or CSE siRNA transfected BC cells. The expression of miR-155 was significantly up-regulated in CBS or CSE KD BC cells (Fig. 3d), whereas miR-146a expression did not change significantly (Supplementary Fig. S2). Furthermore, co-treatment of CBS or CSE siRNA-transfected MDA-MB-231 and MCF7 cells with a NO donor (NaNO2) abrogated the anti-neoplastic effects of CBS and CSE siRNAs (Fig. 3e). Collectively, these results propose an interaction between the endogenous H2S and NO pathways in BC cells whereby the pro-carcinogenic effects of H2S may be, at least in part, mediated through its effects on NO production via the miR-155/NOS2/NO signaling pathway.
Inhibition of endogenous H2S production enhances the susceptibility of BC cells to immune-mediated killing
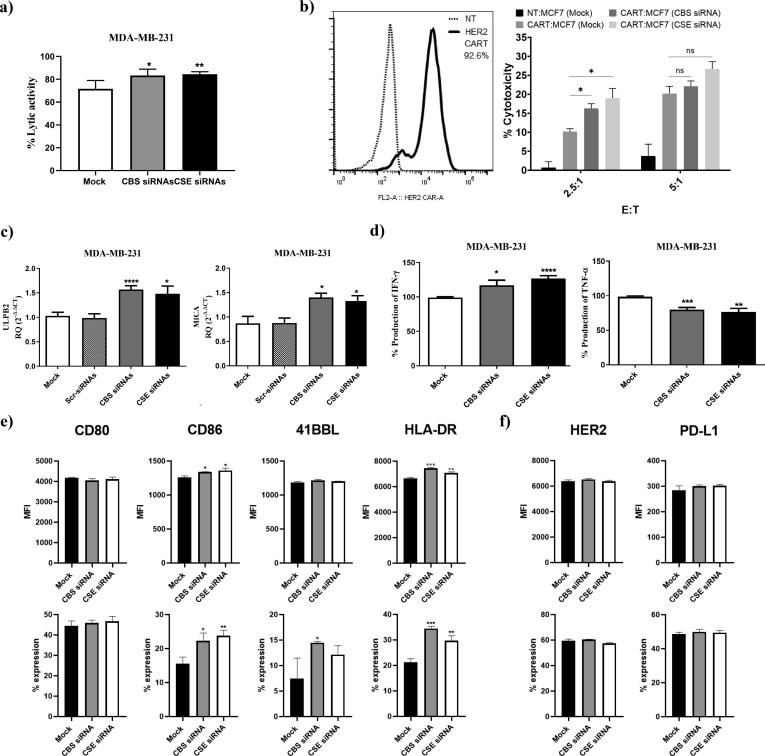
Tumor cells utilize various mechanisms to evade immune responses mediated by effector cells including T cells and NK cells [43], [44]. The results showed that interruption of H2S signaling via CBS/CSE silencing lead to an improvement in NK cell-mediated cytotoxicity against MDA-MB-231 (Fig. 4a) and HER2-targeting Chimeric Antigen Receptor (CAR) T cell-mediated cytotoxicity against HR+/HER2+ MCF7 cells (Fig. 4b). To explore the possible underlying mechanisms that may have contributed to the increased susceptibility to immune-mediated killing, we evaluated the effect of CBS/CSE silencing on NKG2D ligands including ULBP2 and MICA in the MDA-MB-231 cells. We observed that ULBP2 and MICA mRNA transcripts were significantly increased upon repressing the endogenous production of H2S (Fig. 4c). Moreover, there was also a significant reduction in the level of TNF-α and an elevation in the level of interferon-γ production by MDA-MB-231 cells, which may promote a more inflammatory tumor microenvironment (Fig. 4d). In MCF7 cells, H2S silencing led to increased expression of the T cell co-stimulatory ligands: CD80 (B7-1) / CD86 (B7-2) and 41BB ligand that activate T cell through interacting with CD28 and CD137 (41BB), respectively (Fig. 4e). Nonetheless, repression of endogenous H2S increased expression of MHC II (HLA-DR), a molecule that has been described to mediate antigenicity to adaptive immune responses (Fig. 4e and Supplementary Fig. S3). Importantly, no significant changes were found in the expression of HER2 or the T cell inhibitory ligand, PD-L1, upon CBS or CSE silencing (Fig. 4f and Supplementary Fig. S3). Collectively, inhibition of endogenous H2S production led to increased expression of activating/co-stimulatory ligands on BC cells and increased their susceptibility to NK cell- and T cell-mediated immune responses.
Fig. 4.
Impact of silencing CBS and CSE on immune cell mediated cytotoxicity, immune ligand expression and cytokines release. (a) NK cell mediated cytotoxicity was tested via co-culturing experiments that were done between mock, CBS/CSE KDMDA-MB-231 and primary NK cells using LDH assay. (b) T cell mediated cytotoxicity was tested using HER2 targeting CAR T cells. CAR transduction rate was tested using a recombinant human ErbB2/Her2 Fc chimeric protein + PE-conjugated goat anti-human IgG (Fc gamma-specific) andcompared to non-transduced (NT) T cells. CAR T cell mediated cytotoxicity was tested by a chromium (51Cr) release assay. CAR T cells were incubated with MCF7 cells at effector to target ratios of 2.5:1 and 5:1 for 6 h. Cytotoxicity is presented as percentage normalized to triton-X induced cytotoxicity.(c) ULBP2 and MICA were analyzed on the mRNA transcripts level in mock, CBS or CSE KD MDA-MB-231 using qRT-PCR 48 h post-transfection. (d) The supernatant of mock, CBS/CSE KD MDA-MB-231 was screened for IFN-γ and TNF-α production using ELISA. (e) CD80, CD86, 41BBL and HLA-DR surface expression on mock, CBS or CSE KD MCF7, represented as percentage expression and median fluorescence intensity, were analyzed using flow cytometry 72 h post-transfection (f) HER2 and PD-L1 surface expression on mock, CBS/CSE KD MCF7, represented as percentage expression and median fluorescence intensity, was analyzed using flow cytometry 72 h post-transfection.
Identification of miR-4317 as a potential regulator of CBS and CSE in BC
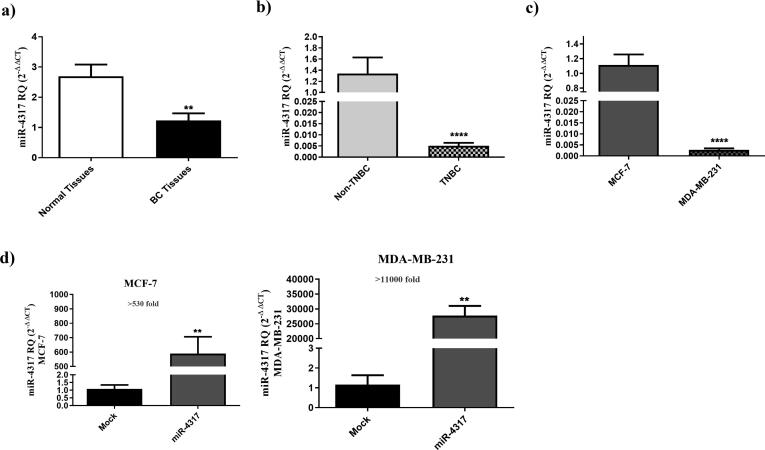
Having demonstrated a possible role of CBS and CSE in modulating BC oncogenic and immunogenic profiles, we sought to explore the effect of dual targeting the CBS and CSE in BC cells. Since miRNAs possess the potential to bind and down-regulate multiple mRNA targets, we used in-silico prediction models to identify miRNA candidates that can simultaneously target CBS and CSE transcripts. These models identified miR-4317 as a candidate that can potentially bind to the CBS 3′-UTR sequence at two different binding regions and the CSE 3′-UTR sequence at 1 binding region with high binding scores (Supplementary Table S3 and Supplementary Fig. S4). We first examined the expression pattern of miR-4317 in primary BC specimens. miR-4317 expression level was found to be significantly lower in BC tissues as compared to normal breast tissues (P = 0.0017) (Fig. 5a). Additionally, it was significantly lower in TNBC specimens when compared to other BC subtypes (P < 0.0001) (Fig. 5b). In a similar pattern, the TNBC cell line, MDA-MB-231, exhibited levels of miR-4317 lower than the HR+/HER2+ MCF7 cell line (P < 0.0001) (Fig. 5c), a phenotype that could be reversed in both MCF7 and MDA-MB-231 by ectopic expression of miR-4317 using miR-4317 mimics (Fig. 5d).
Fig. 5.
miR-4317 is downregulated in TNBC clinical samples and cell line. (a) miR-4317 expression profile was analyzed in tumor tissues from 80 BC patients and compared to patient matched normal tissues using qRT-PCR, and normalized to RNU6B as an internal control. (b) Repressed expression levels of miR-4317 in TNBC patients compared to non-TNBC patients, (c) Reduced expression levels of miR-4317 in MDA-MB-231 cell line when compared to MCF7 cells. The expression of miR-4317 in MDA-MB-231 and MCF7 cells was determined by qRT-PCR 48 h post-transfection. (d) Levels of miR-4317 showed more than 530 folds increase in miR-4317 plasmid transfected MCF7 cells compared to mock cells. miR-4317 plasmid transfected MDA-MB-231 cells showed more than 11,000 folds increase in the expression level of miR-4317 when compared to mock cells, determined by qRT-PCR 48 h post-transfection.
miR-4317 directly co-targets CBS and CSE in BC and inhibits its malignant phenotype
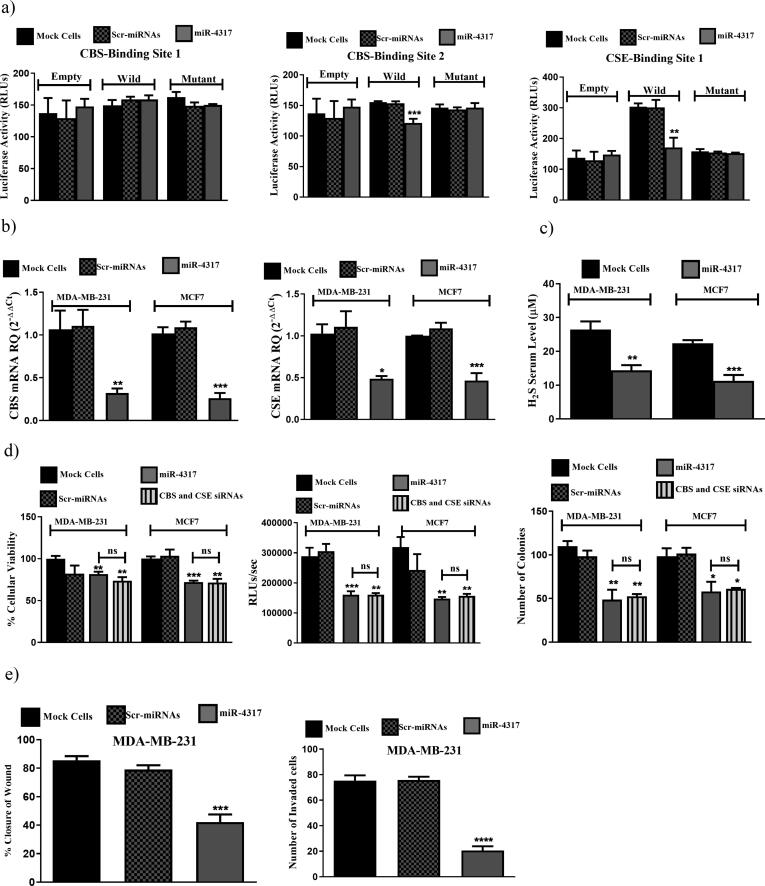
To assess whether miR-4317 directly targets the 3′-UTR of CBS and CSE, a dual luciferase assay was used. Luciferase-expressing constructs were prepared and incorporated in it the predicted miR-4317-CBS binding sites 1or 2, or the miR-4317-CSE binding site as indicated in Supplementary Table S2. These constructs were either transfected alone or with the miR-4317-expressing construct. In the presence of miR-4317, the luciferase-expressing constructs containing the miR-4317-CBS binding site 2 and the miR-4317-CSE binding site showed markedly lower levels of luciferase activity compared to cells transfected with the luciferase-expressing constructs alone. No significant reduction in the luciferase activity was detected for the construct containing the miR-4317-CBS binding site 1 when co-transfected with miR-4317 (Fig. 6a). Ectopic expression of miR-4317 in the two BC cell lines resulted in a marked repression of CBS and CSE transcript levels (Fig. 6b), alongside a reduction in H2S production levels (Fig. 6c). This also led to a significant reduction in BC cellular viability, proliferation and colony formation in both BC cell lines in a manner comparable to CBS/CSE siRNAs co-transfection (Fig. 6d). Moreover, a decrease in the cellular migration and invasion of the aggressive TNBC cell line, MDA-MB-231, was noted following miR-4317 induced expression (Fig. 6e).
Fig. 6.
miR-4317 co-targets CBS and CSE simultaneously mitigating BC malignant properties. (a) Luciferase reporter assay for binding confirmation of miR-4317 on 3′UTR of CBS and CSE. miR-4317-CBS first binding site construct showed no inhibition of luciferase activity. miR-4317-CBS second binding site construct showed significant reduction of luciferase activity in cells co-transfected with miR-4317 mimics plus WT construct compared to mock and scr-miRNAs transfected cells transfected with the WT construct alone. Likewise, for the miR-4317-CSE binding site construct, luciferase activity was also significantly repressed in cells co-transfected with miR-4317 mimics plus WT construct compared to mock and scr-miRNAs transfected cells transfected with the WT construct alone. (b) Down-regulation of CBS and CSE expression by miR-4317 mimics in MDA-MB-231 and MCF7 cells. (c) Reduced H2S levels produced from BC cells upon transfection with miR-4317 mimics. (d) miR-4317 mimics reduced cellular viability (left), cellular proliferation (middle), and colony formation ability (right) in MDA-MB-231 and MCF7. (e) miR-4317 mimics resulted in reduction in migration (left) and invasion (right) capacities in MDA-MB-231.
Discussion
Gasotransmitters, including H2S, are emerging as potential mediators in different pathological conditions and as possible diagnostic and prognostic markers in cancer [45], [46]. The synthesizing enzymes of those gasotransmitters were recently discovered to be significantly altered in tumor tissues, suggesting a potential role in the process of carcinogenesis [45]. Published studies in colon and ovarian cancer demonstrated the up-regulation of CBS in primary tumors and that CBS seems to be the predominant source of endogenous H2S. In the present study we observed that both CBS and CSE were up-regulated in clinical samples from primary BC patients and that both contribute to H2S production in BC cells. Similarly, others have shown elevated levels of CBS and CSE in hepatoma cell lines [47] and bladder urothelial cell carcinoma [48]. In this study, we demonstrate a marked attenuation of key measures of aggressiveness of BC cell lines upon CBS and/or CSE knock-down supporting a possible role of H2S synthesis in mediating the malignant phenotype of BC cells. Other studies have also shown that silencing CBS and CSE can alter the progression of several malignancies [18], [19], [48].
Silencing of the H2S synthetic pathway impacted the tumorgenicity of MDA-MB-231 (TNBC) and MCF-7 (HR+/HER2+) cell lines via distinct mechanisms. In MCF7 cells, CBS/CSE silencing resulted in a marked down-regulation of Akt and phosphorylated Akt. The importance of the PI3K/Akt pathway in the MCF7 cell line is further supported by a previous report showing that inhibition of this pathway in MCF7 had a negative impact on cell survival, but did not significantly affect the survival of the TNBC cell line, MDA-MB-231 [49]. On the other hand, MDA-MB-231 cells oncogenicity was reported to be more dependent on the cytokine receptor-mediated JAK/STAT signaling [40]. Therefore, decreased phosphorylated STAT3 levels in MDA-MB-231 cells following CBS/CSE silencing in this study may partly explain the observed attenuation of MDA-MB-231 malignant phenotype.
Recently, our group has reported a tangled crosstalk between “exogenous” H2S and endogenous NO production in TNBC cells through the sONE/NOS3 axis [14]. Of note, in this study we investigated the possible relationship between the CBS/CSE induced H2S production in BC cells and the NO pathway. It was evident that the biological effects of endogenous H2S in both BC cell lines are, in part, mediated by NO. However, compared to exogenous H2S, the intrinsic mechanism underlying this crosstalk seems to be different. While exogenous H2S mediates its effect through the sONE/NOS3/NO axis [14], the results of this study showed that endogenous H2S mediates its effects mainly through the miR-155/NOS2/NO axis. This is concordant with the recently reported role of several miRNAs in transducing endogenous H2S signaling [50], [51]. Altogether, this study demonstrated that CBS/CSE induced H2S production has been implicated with PI3k/Akt/mTOR, JAK/STAT and miR-155/NOS2/NO signaling pathways.
Tumor immunogenicity and the role of NK cells and cytotoxic T lymphocytes in mediating anti-tumor immune responses have been extensively investigated [52], [53], [54]. Targeting NKG2D ligands and/or blocking PD-1/PD-L1 interactions (checkpoint inhibitors) have been the subject of clinical research and application [55], [56]. Since alterations in the miR-155/NOS2/NO signaling pathway have been reported to attenuate the immune suppressive tumor microenvironment [31], [57], [58], [59], we hypothesized that endogenous H2S may modulate the immunogenic profile of BC cells. Tumor cells tend to down-regulate NKG2D ligands through intracellular retention and/or extracellular shedding to evade NKG2D receptor-mediated NK cell killing. Interestingly, CBS and CSE silencing induced the expression of the NKG2D ligands, MICA and ULBP2, in the MDA-MD-231 TNBC cell line. This was associated with improved NK cell cytotoxicity against H2S-depleted tumor targets. These findings are concordant with the CBS/CSE KD associated attenuation of STAT3, which was described as a transcriptional repressor of the NKG2D ligands, MICA/B and ULBP2, in colon cancer, liver cancer and multiple myeloma [28], [60]. Furthermore, CBS and CSE siRNAs led to a reduced production of TNF-α, a cytokine that may contribute to immune cell apoptosis within the tumor microenvironment. Interestingly, a recent report highlighted the role of H2S in tuning the levels of several pro-inflammatory cytokines including TNF-α in diabetic rat model [61].
On the other hand, silencing of CBS and CSE led to improved killing of the HR+/HER2+ MCF7 cells by anti-HER2 CAR T cells. This may have been partly mediated by the up-regulation of co-stimulatory ligands, including CD86 (B7-2) and 4-1BB ligand (41BBL), on MCF7 cells since the binding of these ligands to their cognate receptors (CD28 and 4-1BB) on T cells is critical for effective T cell activation and function [62], [63]. Of note, the increase in the expression of HLA-DR (class II MHC), which may expose mutant epitopes render tumor cell more immunogenic [43], [64], and co-stimulatory ligands [65] on MCF7 cells as well as the changes in NK ligands and cytokine secretion by MDA-MD-231 cells following CBS/CSE KD may be conducive of more effective immune responses against BC cells.
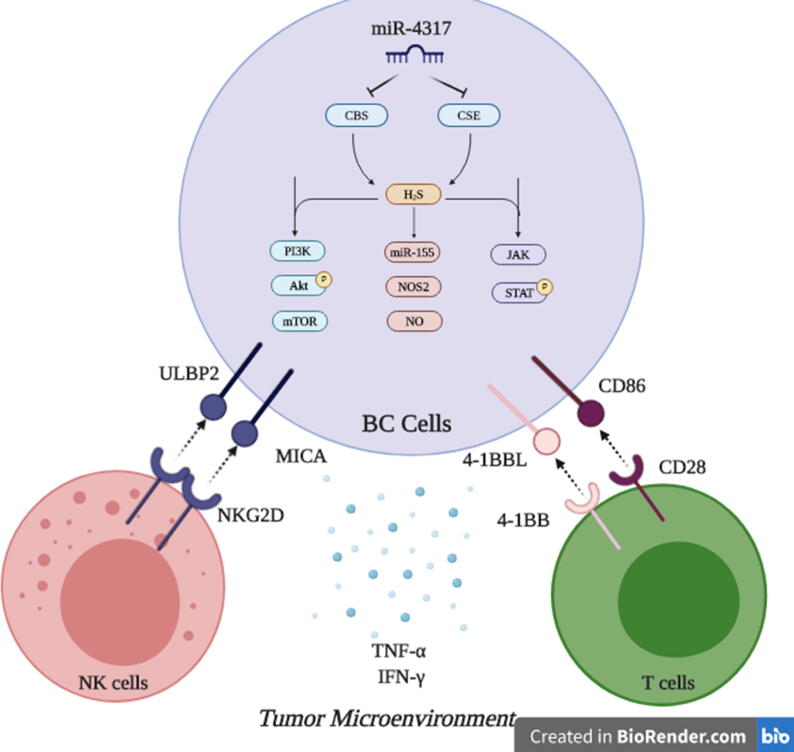
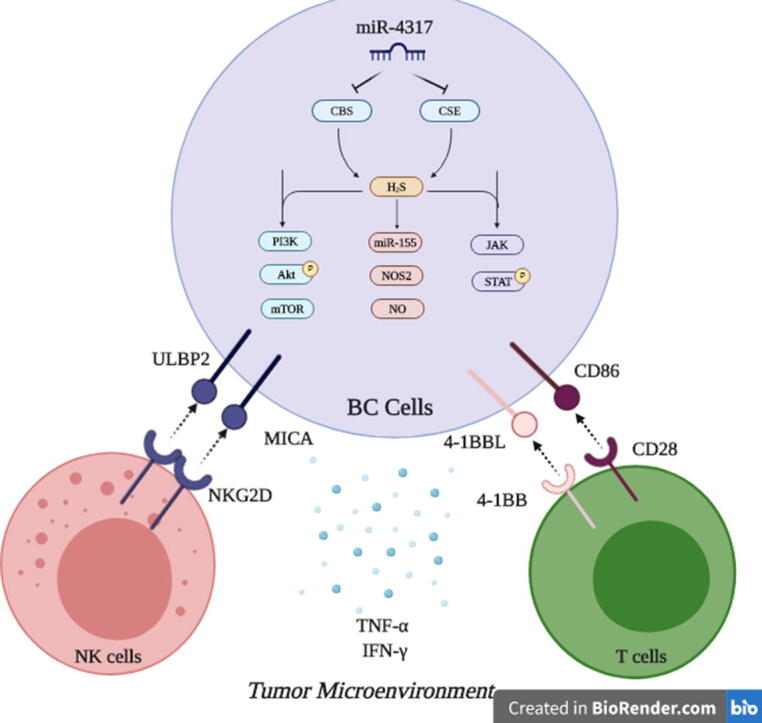
Finally, in an attempt to simultaneously target CBS and CSE using a novel miRNA to halt BC progression, we found that miR-4317 can potentially target both CBS and CSE transcripts through in silico analysis and this was functionally confirmed using a luciferase-based assay. This adds miR-4317 to the list of validated miRNAs that modulate the endogenous production of H2S along with miR-21 [66], miR-22 [67] and miR-30 [68]. Yet, all the previously reported H2S-modulating miRNAs directly target CSE. However, to our knowledge, CBS has not been described as a direct target for miRNAs. Thus, miR-4317 is the first validated miRNA to co-target CBS and CSE simultaneously. While miR-4317 tumor suppressor activity has only been reported in gastric carcinoma [69], this is the first study to describe the tumor suppressor activity of miR-4317 in BC. Noteworthy, results confirmed that miR-4317 was significantly down-regulated in TNBC patients compared to other BC subtypes. Therefore, the consideration of miR-4317 as a possible diagnostic or prognostic marker in TNBC warrants further validation. Additionally, targeted therapies aimed at up-regulating tumor suppressor miRNAs [70] such as miR-4317 expression may have important anti-tumor activities and may be particularly appealing considering the lack of targeted therapies and the dissatisfactory outcomes in TNBC. In conclusion, this study revealed that tissue levels of CBS and CSE were associated with high proliferation index in clinical samples, suggesting a prognostic value of these enzymes. Inhibition of the H2S synthesizing enzymes suppresses BC oncogenicity, as evident by its effect on cell proliferation, survival, invasion, and migration, and increases their susceptibility to both innate and adaptive cellular immune responses. Finally, miR-4317 was portrayed as a novel tumor suppressor miRNA in BC that acts through simultaneous repression of CBS and CSE (Fig. 7).
Fig. 7.
. Targeting H2S synthesizing machinery in BC cells. This is a conclusive descriptive figure highlighting the major roles of endogenously produced H2S in BC cells. Mechanistically, PI3K/Akt/mTOR, miR-155/NOS2/NO, JAK/STAT can be drawn downstream CBS and CSE induced H2S in BC cells. Functionally, this results in abrogation of BC hallmarks, potentiating NK cells and cytotoxic T cells activity and alleviating the immune suppressive microenvironment in BC tumors. Dual Targeting of CBS and CSE in BC cells was achieved using the novel tumor suppressor miR-4317.
Compliance with ethics requirements
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study.
Ethics declarations
Ethics approval and consent to participate
Ethical approvals were obtained from the ethical committees of German University in Cairo, Cairo University and written informed consent was obtained from each patient and normal individual.
Availability of supporting data
All supporting data are available with their detailed descriptions.
Consent for publication
The authors asked for full waiver for publication fees due to funds limitations.
Authors’ contributions
The research conception and design was done by RAY and MZG.
RAY, AZG, and KS completed data acquisition, and analysis. Interpretation of data was performed by RAY, AZG, KS, NA, AAM, and MZG. Clinical data of BC patients was provided by EK an HMH. The H2S detection plates were designed/supplied by YJA and GJL. Writing, review, and/or revision of the manuscript were done by RAY, AZG, KS, NA, AAM, and MZG. NA, AAM, and MZG provided study supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Stand Up to Cancer–St Baldrick’s Pediatric Dream Team Translational Research Grant (SU2C-AACR-DT1113). Stand Up to Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.07.006.
Contributor Information
Rana Ahmed Youness, Email: rana.ahmed-youness@guc.edu.eg.
Mohamed Zakaria Gad, Email: mohamed.gad@guc.edu.eg.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Waks A.G., Winer E.P. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 2.Bray F. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen D.L. Efficacy of HER2-targeted therapy in metastatic breast cancer. Monoclonal antibodies and tyrosine kinase inhibitors. Breast. 2013;22(1):1–12. doi: 10.1016/j.breast.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Denkert C. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389(10087):2430–2442. doi: 10.1016/S0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]
- 5.Youness R.A., Gad M.Z. Long non-coding RNAs: Functional regulatory players in breast cancer. Noncoding RNA Res. 2019;4(1):36–44. doi: 10.1016/j.ncrna.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youness R.A. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J Cell Physiol. 2019;234(11):20286–20297. doi: 10.1002/jcp.28629. [DOI] [PubMed] [Google Scholar]
- 7.Ibrahim A.S. Cancer incidence in egypt: results of the national population-based cancer registry program. J Cancer Epidemiol. 2014;2014 doi: 10.1155/2014/437971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferlay J. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 9.Ataollahi F. Evaluation of copper concentration in subclinical cases of white muscle disease and its relationship with cardiac troponin I. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0056163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu D.D. Hydrogen sulfide as a novel regulatory factor in liver health and disease. Oxid Med Cell Longev. 2019;2019:3831713. doi: 10.1155/2019/3831713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Q. A novel hydrogen sulfide-releasing donor, HA-ADT, suppresses the growth of human breast cancer cells through inhibiting the PI3K/AKT/mTOR and Ras/Raf/MEK/ERK signaling pathways. Cancer Lett. 2019;455:60–72. doi: 10.1016/j.canlet.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Wang L. I157172, a novel inhibitor of cystathionine gamma-lyase, inhibits growth and migration of breast cancer cells via SIRT1-mediated deacetylation of STAT3. Oncol Rep. 2019;41(1):427–436. doi: 10.3892/or.2018.6798. [DOI] [PubMed] [Google Scholar]
- 13.You J. Cystathionine- gamma-lyase promotes process of breast cancer in association with STAT3 signaling pathway. Oncotarget. 2017;8(39):65677–65686. doi: 10.18632/oncotarget.20057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Youness R.A. A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric Oxide. 2018;80:12–23. doi: 10.1016/j.niox.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Youness R.A. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J Cell Physiol. 2019 doi: 10.1002/jcp.28629. [DOI] [PubMed] [Google Scholar]
- 16.Brahmer J.R. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H. Cystathionine beta-synthase in physiology and cancer. Biomed Res Int. 2018;2018:3205125. doi: 10.1155/2018/3205125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharyya S. Cystathionine beta-synthase (CBS) contributes to advanced ovarian cancer progression and drug resistance. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szabo C. Tumor-derived hydrogen sulfide, produced by cystathionine-beta-synthase, stimulates bioenergetics, cell proliferation, and angiogenesis in colon cancer. Proc Natl Acad Sci U S A. 2013;110(30):12474–12479. doi: 10.1073/pnas.1306241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J. Expression of cystathionine beta-synthase is downregulated in hepatocellular carcinoma and associated with poor prognosis. Oncol Rep. 2009;21(6):1449–1454. doi: 10.3892/or_00000373. [DOI] [PubMed] [Google Scholar]
- 21.Fan K. Wnt/beta-catenin signaling induces the transcription of cystathionine-gamma-lyase, a stimulator of tumor in colon cancer. Cell Signal. 2014;26(12):2801–2808. doi: 10.1016/j.cellsig.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 22.Cai W.J. Hydrogen sulfide induces human colon cancer cell proliferation: role of Akt, ERK and p21. Cell Biol Int. 2010;34(6):565–572. doi: 10.1042/CBI20090368. [DOI] [PubMed] [Google Scholar]
- 23.Cao Q. Butyrate-stimulated H2S production in colon cancer cells. Antioxid Redox Signal. 2010;12(9):1101–1109. doi: 10.1089/ars.2009.2915. [DOI] [PubMed] [Google Scholar]
- 24.Pan Y. Hydrogen sulfide (H2S)/cystathionine gamma-lyase (CSE) pathway contributes to the proliferation of hepatoma cells. Mutat Res. 2014;763–764:10–18. doi: 10.1016/j.mrfmmm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Yin P. Sp1 is involved in regulation of cystathionine gamma-lyase gene expression and biological function by PI3K/Akt pathway in human hepatocellular carcinoma cell lines. Cell Signal. 2012;24(6):1229–1240. doi: 10.1016/j.cellsig.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Pei Y. Hydrogen sulfide mediates the anti-survival effect of sulforaphane on human prostate cancer cells. Toxicol Appl Pharmacol. 2011;257(3):420–428. doi: 10.1016/j.taap.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 27.Youness R.A. A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Nat Prod Res. 2018:1–6. doi: 10.1080/14786419.2018.1509326. [DOI] [PubMed] [Google Scholar]
- 28.Youness R.A. Contradicting interplay between insulin-like growth factor-1 and miR-486-5p in primary NK cells and hepatoma cell lines with a contemporary inhibitory impact on HCC tumor progression. Growth Factors. 2016;34(3–4):128–140. doi: 10.1080/08977194.2016.1200571. [DOI] [PubMed] [Google Scholar]
- 29.Awad A.R. An acetylated derivative of vitexin halts MDA-MB-231 cellular progression and improves its immunogenic profile through tuning miR- 20a-MICA/B axis. Nat Prod Res. 2019:1–5. doi: 10.1080/14786419.2019.1686372. [DOI] [PubMed] [Google Scholar]
- 30.Youness R.A. MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncol Lett. 2016;12(4):2567–2573. doi: 10.3892/ol.2016.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aboelenein H.R. Reduction of CD19 autoimmunity marker on B cells of paediatric SLE patients through repressing PU.1/TNF-alpha/BAFF axis pathway by miR-155. Growth Factors. 2017;35(2–3):49–60. doi: 10.1080/08977194.2017.1345900. [DOI] [PubMed] [Google Scholar]
- 32.Ahmed Youness R. A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Nat Prod Res. 2020;34(10):1475–1480. doi: 10.1080/14786419.2018.1509326. [DOI] [PubMed] [Google Scholar]
- 33.Youssef S.S. PNPLA3 and IL 28B signature for predicting susceptibility to chronic hepatitis C infection and fibrosis progression. Arch Physiol Biochem. 2019:1–7. doi: 10.1080/13813455.2019.1694039. [DOI] [PubMed] [Google Scholar]
- 34.Mekky R.Y. Epigallocatechin gallate (EGCG) and miR-548m reduce HCV entry through repression of CD81 receptor in HCV cell models. Arch Virol. 2019;164(6):1587–1595. doi: 10.1007/s00705-019-04232-x. [DOI] [PubMed] [Google Scholar]
- 35.Kim T.J. Characterization of H2S releasing properties of various H2S donors utilizing microplate cover-based colorimetric assay. Anal Biochem. 2019;574:57–65. doi: 10.1016/j.ab.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Ahn Y.J. Colorimetric detection of endogenous hydrogen sulfide production in living cells. Spectrochim Acta A Mol Biomol Spectrosc. 2017;177:118–124. doi: 10.1016/j.saa.2017.01.040. [DOI] [PubMed] [Google Scholar]
- 37.Rahmoon M.A. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors. 2017;35(2–3):76–87. doi: 10.1080/08977194.2017.1354859. [DOI] [PubMed] [Google Scholar]
- 38.Shaalan Y.M. Destabilizing the interplay between miR-1275 and IGF2BPs by Tamarix articulata and quercetin in hepatocellular carcinoma. Nat Prod Res. 2018;32(18):2217–2220. doi: 10.1080/14786419.2017.1366478. [DOI] [PubMed] [Google Scholar]
- 39.Chen X. The predictive value of Ki-67 before neoadjuvant chemotherapy for breast cancer: a systematic review and meta-analysis. Future Oncol. 2017;13(9):843–857. doi: 10.2217/fon-2016-0420. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y. Ganoderic acid A exerts antitumor activity against MDA-MB-231 human breast cancer cells by inhibiting the Janus kinase 2/signal transducer and activator of transcription 3 signaling pathway. Oncol Lett. 2018;16(5):6515–6521. doi: 10.3892/ol.2018.9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xin H. Hydrogen sulfide attenuates inflammatory hepcidin by reducing IL-6 secretion and promoting SIRT1-mediated STAT3 deacetylation. Antioxid Redox Signal. 2016;24(2):70–83. doi: 10.1089/ars.2015.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Y. Inflammatory response of macrophages cultured with Helicobacter pylori strains was regulated by miR-155. Int J Clin Exp Pathol. 2015;8(5):4545–4554. [PMC free article] [PubMed] [Google Scholar]
- 43.Axelrod M.L. Biological consequences of MHC-II expression by tumor cells in cancer. Clin Cancer Res. 2019;25(8):2392–2402. doi: 10.1158/1078-0432.CCR-18-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Driessens G., Kline J., Gajewski T.F. Costimulatory and coinhibitory receptors in anti-tumor immunity. Immunol Rev. 2009;229(1):126–144. doi: 10.1111/j.1600-065X.2009.00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szabo C. Gasotransmitters in cancer: from pathophysiology to experimental therapy. Nat Rev Drug Discov. 2016;15(3):185–203. doi: 10.1038/nrd.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu D. Hydrogen sulfide in cancer: Friend or foe? Nitric Oxide. 2015;50:38–45. doi: 10.1016/j.niox.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Zhen Y. Exogenous hydrogen sulfide exerts proliferation/anti-apoptosis/angiogenesis/migration effects via amplifying the activation of NF-kappaB pathway in PLC/PRF/5 hepatoma cells. Int J Oncol. 2015;46(5):2194–2204. doi: 10.3892/ijo.2015.2914. [DOI] [PubMed] [Google Scholar]
- 48.Gai JW, et al. Expression profile of hydrogen sulfide and its synthases correlates with tumor stage and grade in urothelial cell carcinoma of bladder. Urol Oncol, 2016. 34(4): p. 166 e15-20. [DOI] [PubMed]
- 49.Marina M., Wang L., Conrad S.E. The scaffold protein MEK Partner 1 is required for the survival of estrogen receptor positive breast cancer cells. Cell Commun Signal. 2012;10(1):18. doi: 10.1186/1478-811X-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu T.X. MiR 20a,-20b and -200c are involved in hydrogen sulfide stimulation of VEGF production in human placental trophoblasts. Placenta. 2016;39:101–110. doi: 10.1016/j.placenta.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Y. Hydrogen sulfide promotes angiogenesis by downregulating miR-640 via the VEGFR2/mTOR pathway. Am J Physiol Cell Physiol. 2016;310(4):C305–C317. doi: 10.1152/ajpcell.00230.2015. [DOI] [PubMed] [Google Scholar]
- 52.Sun H., Sun C. The rise of NK cell checkpoints as promising therapeutic targets in cancer immunotherapy. Front Immunol. 2019;10:2354. doi: 10.3389/fimmu.2019.02354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ihara F. Regulatory T cells induce CD4(-) NKT cell anergy and suppress NKT cell cytotoxic function. Cancer Immunol Immunother. 2019 doi: 10.1007/s00262-019-02417-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang J., Natarajan K., Margulies D.H. MHC molecules, T cell receptors, natural killer cell receptors, and viral immunoevasins-key elements of adaptive and innate immunity. Adv Exp Med Biol. 2019;1172:21–62. doi: 10.1007/978-981-13-9367-9_2. [DOI] [PubMed] [Google Scholar]
- 55.Garrido F. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol. 2016;39:44–51. doi: 10.1016/j.coi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spear P. NKG2D ligands as therapeutic targets. Cancer Immun. 2013;13:8. [PMC free article] [PubMed] [Google Scholar]
- 57.Fahim Golestaneh A. Large scale in vivo micro-RNA loss of function screen identified miR-29a, miR-100 and miR-155 as modulators of radioresistance and tumor-stroma communication. Int J Cancer. 2019;144(11):2774–2781. doi: 10.1002/ijc.32019. [DOI] [PubMed] [Google Scholar]
- 58.Huffaker T.B. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J Biol Chem. 2017;292(45):18530–18541. doi: 10.1074/jbc.M117.808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poles W.A. Targeting the polarization of tumor-associated macrophages and modulating mir-155 expression might be a new approach to treat diffuse large B-cell lymphoma of the elderly. Cancer Immunol Immunother. 2019;68(2):269–282. doi: 10.1007/s00262-018-2273-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zingoni A. NKG2D and Its Ligands: “One for All, All for One”. Front Immunol. 2018;9:476. doi: 10.3389/fimmu.2018.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu M. Hydrogen sulfide attenuates myocardial fibrosis in diabetic rats through the JAK/STAT signaling pathway. Int J Mol Med. 2018;41(4):1867–1876. doi: 10.3892/ijmm.2018.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawalekar O.U. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity. 2016;44(2):380–390. doi: 10.1016/j.immuni.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 63.Velasquez M.P. CD28 and 41BB costimulation enhances the effector function of CD19-specific engager T cells. Cancer Immunol Res. 2017;5(10):860–870. doi: 10.1158/2326-6066.CIR-17-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kreiter S. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma R.K. Tumor cells engineered to codisplay on their surface 4–1BBL and LIGHT costimulatory proteins as a novel vaccine approach for cancer immunotherapy. Cancer Gene Ther. 2010;17(10):730–741. doi: 10.1038/cgt.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cindrova-Davies T. Reduced cystathionine gamma-lyase and increased miR-21 expression are associated with increased vascular resistance in growth-restricted pregnancies: hydrogen sulfide as a placental vasodilator. Am J Pathol. 2013;182(4):1448–1458. doi: 10.1016/j.ajpath.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L. MiR-22/Sp-1 links estrogens with the up-regulation of cystathionine gamma-lyase in myocardium, which contributes to estrogenic cardioprotection against oxidative stress. Endocrinology. 2015;156(6):2124–2137. doi: 10.1210/en.2014-1362. [DOI] [PubMed] [Google Scholar]
- 68.Shen Y. miRNA-30 family inhibition protects against cardiac ischemic injury by regulating cystathionine-gamma-lyase expression. Antioxid Redox Signal. 2015;22(3):224–240. doi: 10.1089/ars.2014.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu X. miRNA-4317 suppresses human gastric cancer cell proliferation by targeting ZNF322. Cell Biol Int. 2017 doi: 10.1002/cbin.10870. [DOI] [PubMed] [Google Scholar]
- 70.ElKhouly A.M., Youness R.A., Gad M.Z. MicroRNA-486-5p and microRNA-486-3p: Multifaceted pleiotropic mediators in oncological and non-oncological conditions. Noncoding RNA Res. 2020;5(1):11–21. doi: 10.1016/j.ncrna.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.