Abstract
Studies on induced pluripotent stem (iPS) cells highly rely on the investigation of their gene expression which requires normalization by housekeeping genes. Whether the housekeeping genes are stable during the iPS reprogramming, a transition of cell state known to be associated with profound changes, has been overlooked. In this study we analyzed the expression patterns of the most comprehensive list to date of housekeeping genes during iPS reprogramming of a mouse neural stem cell line N31. Our results show that housekeeping genes’ expression fluctuates significantly during the iPS reprogramming. Clustering analysis shows that ribosomal genes’ expression is rising, while the expression of cell-specific genes, such as vimentin (Vim) or elastin (Eln), is decreasing. To ensure the robustness of the obtained data, we performed a correlative analysis of the genes. Overall, all 70 genes analyzed changed the expression more than two-fold during the reprogramming. The scale of this analysis, that takes into account 70 previously known and newly suggested genes, allowed us to choose the most stable of all genes. We highlight the fact of fluctuation of housekeeping genes during iPS reprogramming, and propose that, to ensure robustness of qPCR experiments in iPS cells, housekeeping genes should be used together in combination, and with a prior testing in a specific line used in each study. We suggest that the longest splice variants of Rpl13a, Rplp1 and Rps18 can be used as a starting point for such initial testing as the most stable candidates.
Subject terms: Cell biology, Computational biology and bioinformatics, Stem cells
Introduction
Induced pluripotent stem cells (iPSCs)1 are a promising technology that is becoming increasingly important for medical treatment development2 and basic research. iPSCs are generated by forced expression of pluripotent genes, such as Oct4, Klf4, Sox2 and Myc, and this process is associated with profound changes in cell metabolism3, gene expression, and epigenetics4. In the great majority of previous studies, the analysis of gene or protein expression rely on the use of housekeeping genes used as reference for normalization of the data5. Typical housekeeping genes relate to the basic functioning of cells, such as cytoskeletal genes (Actb (actin), Tubb5 (tubulin), Vim (vimentin) etc.), ribosomal genes (multiple genes for large and small subunits of the ribosome, such as Rpl7, Rps9 etc.), ATP generation-related genes (such as Gapdh, Pgk1 etc.), various essential enzyme genes (such as fumarate hydratase Fh1, DNA mismatch repair gene Mlh3, Peptidylprolyl Isomerase A (Ppia) etc.), and these genes are generally thought to be stable regardless of the cell state.
The question of stability of housekeeping genes has previously been investigated in mouse embryonic stem cells (mESCs). In 2006, E. Willems and colleagues have identified Actb and Gapdh as the most stable genes in mouse embryos and in differentiating mouse and human ES cells6. A subsequent study by S. Mamo in 2007 focused on reference genes in mouse oocytes and embryos, and pointed out the instability of housekeeping genes, while suggesting that Ppia, H2az1 and Hprt were the most stable genes in the embryos7. The same team has later published a 2008 study in rabbit oocytes and preimplantation stage embryos. The team identified H2az1, Hprt and Ywhaz as the most stable reference genes, while indicating that Ubc, Tbp and B2m were the least stable and unsuitable for normalization in qPCR experiments in pluripotent stem cells8. Another study found that Sdha, Tbp and Ywhaz were the most stable genes during the differentiation of mESCs in vitro9. A study on human ES cells conducted in 2013, on the other hand, identified B2M and RPL13A as the most stable genes during differentiation10. In 2015, another study on differentiating human ES cells expanded the set of analyzed genes because there were concerns about the variability of the housekeeping genes’ expression, and used large-scale datasets to perform global transcriptional analysis using a technique less trusted today, namely microarrays. This study included 9 different datasets and 144 microarrays to identify a set of non-varying genes, while highlighting the fact that commonly used ACTB, GAPDH, HPRT1, PPIA, SDHA and B2M varied substantially during human ESC differentiation. This study put HPRT1 and B2M in the group of highly varied genes11. The most notable study conducted in stem cells is the 2007 study by Synnergren and colleagues12. This study used microarrays to highlight that conventionally used housekeeping genes such as HPRT1, ACTB and GAPDH fluctuate in differentiating human ESCs. The study has identified a special set of housekeeping genes for use as a reference in pluripotent stem cell experiment but failed to validate the genes by qPCR.
The aforementioned studies have focused on ES cells with the exception of one study, conducted in differentiating iPS cells, where the team investigated the stability of commonly used housekeeping genes13. In this study, ACTB, C1orf43, PSMB4, GAPDH and HMBS were identified as the most stable genes out of 16 genes during iPS differentiation. Therefore, currently, there exist no studies on housekeeping genes during the iPS reprogramming process except for a limited study by our team in 2018 that analyzed 13 housekeeping genes14. Thus, the stability of common housekeeping genes and the newly suggested housekeeping genes12, as well as their performance compared to conventional housekeeping genes, still needs to be investigated in iPS systems, especially during the reprogramming process.
The goal of this work was to check the stability of the majority of all housekeeping genes used in the aforementioned stem cell studies, including the newly suggested genes discovered by Holmgren et.al11, so as to provide the most comprehensive list of housekeeping genes as of today. In this work, we have used the most precise method of gene expression quantification15, namely RT-qPCR, combined with newly developed high-precision data analysis method, Pairwise Efficiency16, to uncover gene expression patterns in 70 housekeeping genes during iPS reprogramming. This work includes all of the most commonly used housekeeping genes as well as recently suggested pluripotency-related genes12, and takes into account possible splice variants of the genes. This work uncovers the fluctuations in housekeeping genes during iPS reprogramming and underlines important considerations for the researchers who are using iPS cells in their research to avoid normalization to housekeeping genes without prior check.
Materials and methods
Cell culture
The iPS reprogramming was carried out in a reprogrammable cell system previously described in Hikichi et. al., 201217, and applied by Panina and colleagues14. The system consists of a mouse neural progenitor cell line named N31 and does not express pluripotency markers nor possesses pluripotent phenotype prior to reprogramming. The “reprogrammability” of this system has been shown in the publications mentioned above. This system enables reprogramming with a doxycycline-inducible cassette with Oct4, Sox2, Klf4 and c-Myc. Addition of doxycycline activates the four factors and initiates reprogramming. The cells were maintained on plastic gelatin-coated dishes (Corning) in RHB neural stem cell media (Takara) supplemented with Ndiff and 1 mg/ml FGF and 1 mg/ml EGF. For reprogramming initiation, the medium was changed to Essential 8 iPS reprogramming medium (home-made) and 1 µg/ml doxycycline was added to the dish. From that point on, the media were changed every day to avoid pH fluctuations. The reprogramming process continued until day 15, and the cell samples were collected on days 0, 5, 10, and 15.
Gene selection and primer design
Full information about genes, primers and amplicons is available as supplementary data. All primers in this work have been carefully designed to include the longest splice variants of the genes (in contrast to other studies which did not take splicing into account), and have been tailored to work in the same melting temperature using free NCBI tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) in order to allow screening of different genes on the same qPCR plate.
RNA isolation and cDNA synthesis
Total RNA was isolated with RNeasy kit (Invitrogen) according to the manufacturer's instructions, and RNA absorbance ratios (A260/280 and A260/230) were assessed by Nanodrop 2000 Spectrophotometer (NanoDrop Technologies). 300 ng of RNA from each sample was converted to cDNA using SuperScript First-Strand Synthesis System (Thermo Fisher Scientific). The DNA was assessed by the Nanodrop spectrophotometer and diluted to 100 ng/µl. The reprogramming process (day 0–day 15) was repeated 2 times, thus 2 biological replicates were obtained for each time point, amounting to 18 cell material samples per primer tested. In total, each time point had 2 biological and 4 technical replicas.
Experiment design and PCR dataset generation
To assess the stability of chosen 70 genes, we gathered total RNA at four time points throughout the process, namely at day 0, day 5, day 10 and day 15. We then performed qPCR of these genes at all four time points using Pairwise Efficiency approach16. This approach takes into account the efficiency of qPCR reaction, and is approximately two times more precise compared to conventional Delta Ct method. 4 time points * 70 housekeeping genes resulted in 280 independent DNA-primer combinations. Six replicas were run for each combination, as required by Pairwise Efficiency method, and a total of 1680 amplification curves were obtained. The Baseline Subtracted PCR datasets were generated from each PCR run and processed using Bio-Rad CFX Manager 2.0 (2.0.885.0923). These datasets were imported for analysis into the Pairwise Efficiency software (unpublished).
Quantitative real-time PCR
qPCR was performed on a CFX96 Connect (BioRad). SYBR Green PCR supermix (BioRad) was used as per the manufacturer's instructions. Each reaction contained 5 ng of cDNA in a final volume of 10 μL, and the primers’ concentration was 300 nM. Thermocycler program was as following: 1) hot start cycle at 95 °C for 3 min, 2) 40 cycles at 95 °C for 10 s, and 3) 60 °C for 30 s. For product specificity confirmation, a melting curve analysis was performed.
Statistical analyses according to most commonly used algorithms
For selecting the most stably expressed genes we have used the following software freely available, namely, RefFinder18 which calculates a comprehensive score from DeltaCt, BestKeeper19, NormFinder20 and Genorm21.
Statistical analyses and data processing
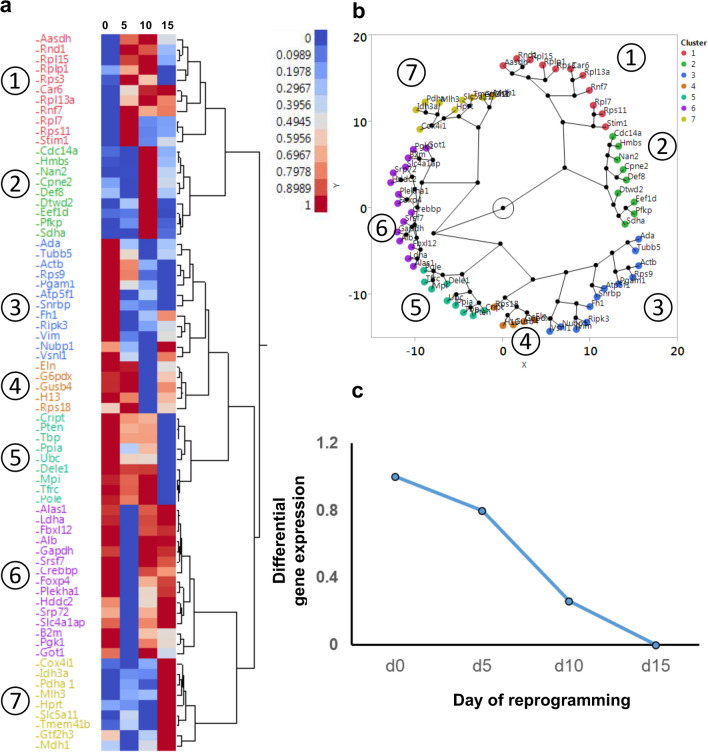
The data processing was carried out in Microsoft Excel, R, and JMP Version 11 (https://www.jmp.com). Clustering was used to visualize the patterns of variation of gene expression during reprogramming, such as tendencies toward decrease, increase or fluctuation during reprogramming. To cluster the groups of genes that displayed similar tendencies (such as, increasing during reprogramming, or decreasing during reprogramming, or fluctuating), we performed a hierarchical clustering on the gene expression value normalized from 0 to 1 (for each gene). The distances were calculated using the Ward method, which is a well known metric for clustering. Ward’s method joins clusters to maximize the likelihood at each level of the hierarchy. The output of this clustering is presented using two visualizations, a dendrogram in Fig. 2a and a constellation plot in Fig. 2b. In the constellation plot, each housekeeping gene is represented by an endpoint and each cluster join is represented by a new node. The axis scaling, orientation of points, and angles of the lines are arbitrary. However, the lengths of the lines represent the distance between clusters and the lengths are meaningful with respect to each other. The constellation plot is an alternative visualization of the hierarchical clustering dendrogram shown in Fig. 2b and is but another way to look at the statistical distance between clusters. It notably highlights that the first two subgroups (red and green), according to the statistical ward distance calculated between clusters, are classified relatively far apart from the other 5 subgroups, showing that these genes behave significantly differently than the genes of other clusters.
Figure 2.
Expression change of 70 housekeeping genes throughout the iPS reprogramming process. (a) Clustering analysis of the dynamics of gene expression levels during reprogramming. Gene expression levels relative to Day 0 were considered and scaled from 0 to 1 for each gene. The color scale attributes red to a value of 1, indicating the highest gene expression during reprogramming. Four time points during the reprogramming are indicated over the heatmap (Days zero, five, ten and fifteen), and the gene names are shown on the left. JMP Version 11 software was used to create the figure (https://www.jmp.com). (b) Alternative visualization of the clustering analysis of (a) using a constellation plot. Genes are grouped by their similarity of expression dynamics. Lengths of the lines represent the statistical distance between clusters. (c) Relative dynamic of Actb gene expression by comparison to Day 0 during reprogramming.
Results
The choice of housekeeping genes for the study
In this study, we investigated 70 genes to provide a comprehensive overview of the most of housekeeping genes used in literature (Table 1). The list of 70 genes was assembled using previously published articles for expression patterns in differentiating pluripotent stem cells. The list contains a portion of very commonly used genes as well as genes identified by microarray analysis as stable and suitable for qPCR normalization in differentiating human ES cells12. The Supplementary Table S1, available in the supplementary materials, lists gene names, associated symbols and accession numbers, general protein information and splice variants, the functions of the gene products by Gene Ontology, and the mRNA expression in RPKM units obtained in ENCODE transcriptome project for the CNS of mouse embryo (E11.5). The Supplementary Table S2 available in the same excel file lists all the information about primers. All primers were checked for splice variants and were carefully designed to include the longest splice variants to ensure the robustness of the study.
Table 1.
Summary of 70 housekeeping genes evaluated in this study. Gene symbol, accession number and general protein information are shown.
| Symbol | Accession number | General protein information |
|---|---|---|
| Aasdh | NM_173765.3 | Aminoadipate-semialdehyde dehydrogenase |
| Actb | NM_007393.5 | Actin, beta |
| Ada | NM_001272052.1 | Adenosine deaminase |
| Alas1 | NM_001291835.1 | Aminolevulinic acid synthase 1 |
| Alb | NM_009654.4 | albumin |
| Atp5f1 | NM_009725.4 | ATP synthase, H + transporting, mitochondrial F0 complex, subunit B1 |
| B2m | NM_009735.3 | Beta-2 microglobulin |
| Car6 | NM_009802.2 | Carbonic anhydrase 6 |
| Cdc14a | NM_001080818.2 | CDC14 cell division cycle 14A |
| Cox4i1 | NM_009941.3 | Cytochrome c oxidase subunit 4I1 |
| Cpne2 | NM_153507.2 | Copine II |
| Crebbp | NM_001025432.1 | CREB binding protein |
| Cript | NM_019936.3 | Cysteine-rich PDZ-binding protein |
| Def8 | NM_001253783.1 | Differentially expressed in FDCP 8 |
| Dele1 | NM_024179.5 | DAP3 binding cell death enhancer 1 |
| Dtwd2 | NM_026854.3 | DTW domain containing 2 |
| Eef1d | NM_029663.2 | Eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| Eln | NM_007925.4 | Elastin |
| Fbxl12 | NM_013911.3 | F-box and leucine-rich repeat protein 12 |
| Fh1 | NM_010209.2 | Fumarate hydratase 1 |
| Foxp4 | NM_001110824.1 | Forkhead box P4 |
| G6pdx | NM_008062.2 | Glucose-6-phosphate dehydrogenase X-linked |
| Gapdh | NM_001289726.1 | Glyceraldehyde-3-phosphate dehydrogenase |
| Got1 | NM_010324.2 | Glutamic-oxaloacetic transaminase 1, soluble |
| Gtf2h3 | NM_181410.3 | General transcription factor IIH, polypeptide 3 |
| Gusb | NM_010368.2 | Glucuronidase, beta |
| H13 | NM_001159551.1 | Histocompatibility 13 |
| Hddc2 | NM_027168.2 | HD domain containing 2 |
| Hmbs | NM_001110251.1 | Hydroxymethylbilane synthase |
| Hprt | NM_013556.2 | Hypoxanthine guanine phosphoribosyl transferase |
| Idh3a | NM_029573.2 | Isocitrate dehydrogenase 3 (NAD +) alpha |
| Ldha | NM_001136069.2 | Lactate dehydrogenase A |
| Mdh1 | NM_001316675.1 | Malate dehydrogenase 1, NAD (soluble) |
| Mlh3 | NM_001304475.1 | mutL homolog 3 |
| Mpi | NM_025837.2 | Mannose phosphate isomerase |
| Nubp1 | NM_011955.2 | Nucleotide binding protein 1 |
| Pdha1 | NM_008810.3 | Pyruvate dehydrogenase E1 alpha 1 |
| Pfkp | NM_001291071.1 | Phosphofructokinase, platelet |
| Pgam1 | NM_023418.2 | Phosphoglycerate mutase 1 |
| Pgk1 | NM_008828.3 | Phosphoglycerate kinase 1 |
| Plekha1 | NM_001346515.1 | Pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| Pole | NM_011132.2 | Polymerase (DNA directed), epsilon |
| Ppia | NM_008907.2 | Peptidylprolyl isomerase A |
| Pten | NM_008960.2 | Phosphatase and tensin homolog |
| Ripk3 | NM_019955.2 | Receptor-interacting serine-threonine kinase 3 |
| Rnd1 | NM_172612.3 | Rho family GTPase 1 |
| Rnf7 | NM_011279.3 | Ring finger protein 7 |
| Rpl13a | NM_009438.5 | Ribosomal protein L13A |
| Rpl15 | NM_001359897.1 | Ribosomal protein L15 |
| Rpl7 | NM_011291.5 | Ribosomal protein L7 |
| Rplp1 | NM_018853.3 | Ribosomal protein, large, P1 |
| Rps11 | NM_013725.4 | Ribosomal protein S11 |
| Rps18 | NM_011296.2 | Ribosomal protein S18 |
| Rps3 | NM_012052.2 | Ribosomal protein S3 |
| Rps9 | NM_029767.2 | Ribosomal protein S9 |
| Sdha | NM_023281.1 | Succinate dehydrogenase complex, subunit A, flavoprotein |
| Slc4a1ap | NM_001347328.1 | Solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| Slc5a11 | NM_146198.2 | Solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| Snrpb | NM_009225.2 | Small nuclear ribonucleoprotein B |
| Srp72 | NM_025691.1 | Signal recognition particle 72 |
| Srsf7 | NM_146083.2 | Serine/arginine-rich splicing factor 7 |
| Stim1 | NM_009287.5 | Stromal interaction molecule 1 |
| Tbp | NM_013684.3 | TATA box binding protein |
| Tfrc | NM_011638.4 | Transferrin receptor |
| Tmem41b | NM_153525.5 | Transmembrane protein 41B |
| Tubb5 | NM_011655.5 | Tubulin, beta 5 class I |
| Ubc | NM_019639.4 | Ubiquitin C |
| Vim | NM_011701.4 | Vimentin |
| Vsnl1 | NM_012038.4 | Visinin-like 1 |
| Ywhaz | NM_001253805.1 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
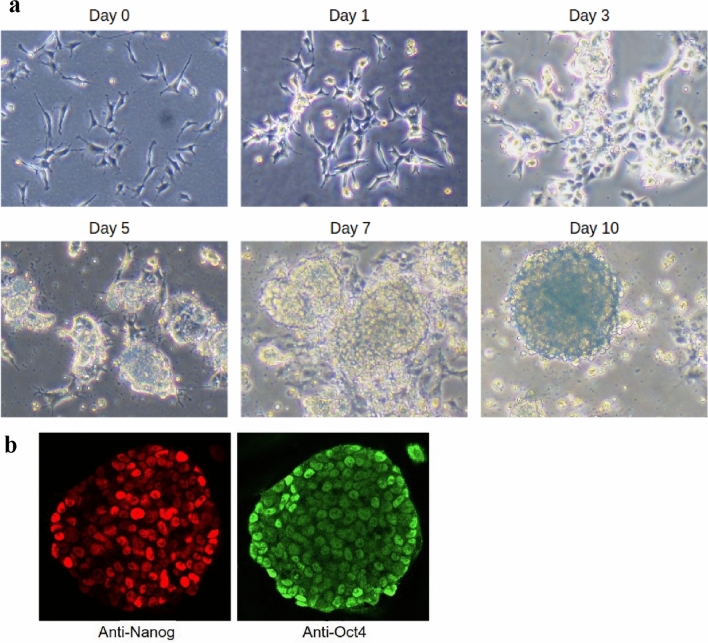
Reprogramming of mouse neural progenitors into iPS cells
To investigate housekeeping genes’ expression changes throughout the reprogramming process, we performed reprogramming of mouse neural progenitors using previously described system14,17, and monitored them for standard markers of pluripotency (shape changes and pluripotency markers Nanog and Oct4). The cells began to grow in lumps starting from day 5, and by day 10 they formed characteristic round-shaped colonies with tight edges. Immunostaining for pluripotency markers Nanog and Oct 4 showed that the colonies fully expressed these genes at day 10 (Fig. 1) (the markers were absent in the parental neural progenitors, data not shown).
Figure 1.
Reprogramming of neural progenitors into the pluripotent iPS cells. (a) Colony formation during the iPS reprogramming of neural progenitors. The truly round-shaped colonies with a characteristic “glow” indicative of reprogrammed cells appear between days 7 and 10. (b) The results of the immunofluorescent analysis of nascent colonies for pluripotency markers Nanog and Oct4 (Day 10).
Housekeeping genes fluctuate during iPS reprogramming
The stability of the 70 candidate genes was monitored at four time points throughout the reprogramming process, and gene expression values were calculated by Pairwise Efficiency approach. To reveal patterns in the expression fluctuation of the genes, the expression was scaled from 0 to 1 across the four time points. Data were then submitted to a hierarchical clustering to visualize the group of genes that behaved similarly during reprogramming (Fig. 2). The analysis revealed three major types of patterns, and a total of 7 subgroups (clusters). This analysis (Fig. 2a,b) clearly placed together most of the ribosomal genes such as Rpl15, Rplp1, Rpl13a, Rpl7, Rps3 and Rps11 (cluster 1), and they displayed a rising pattern, especially on Day 5 and Day 10 of the reprogramming. Cluster 2 contained some of the biosynthetic genes (such as Hmbs), cell cycle genes (such as Cdc14a), and other genes with unclear function (such as Def8), showing a fluctuating pattern. Clusters 3 and 4 were separated by a small statistical distance (represented by the length of the branches on Fig. 2b) and contained cytoskeleton-related genes, such as Actin (Actb), Tubulin (Tubb5), Elastin (Eln) and Vimentin (Vim), as well as two small ribosomal subunits Rps9 and Rsp 18. The mRNA expression of these genes tended to fall throughout the process of iPS reprogramming. Clusters 6 and 7 contained genes related to growth (specifically, ATP production) and cell cycle, such as Gapdh, Ldha, Mdh1, Pgk1, Got1, Foxp4, Pdha, Idh3a, Mlh3 and Cox4i1. Most of the genes in this group are involved in the steps of glucose breakdown and ATP generation (e.g. Gapdh is a glyceraldehyde-3-phosphate dehydrogenase which turns Glyceraldehyde 3-phosphate into 1,3-bisphosphoglyceric acid, and Pgk1 is the next step in the chain, converting 1,3-bisphosphoglyceric acid into 3-Phosphoglyceric acid). The genes in this group were found to fluctuate (rise and fall) during the reprogramming.
Assessing the degree of change in individual genes
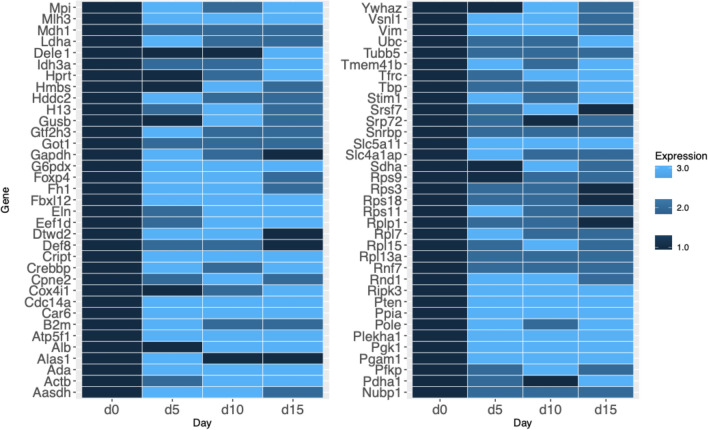
To assess the degree of change of the individual genes and reveal which genes are the most stable overall during the iPS reprogramming, we calculated the relative gene expression by comparison to Day 0. Then, we associated 3 colors to the gene levels to reflect the following: (1) The expression level equal to that of Day 0 (i.e., no change compared to non-reprogrammed cells), dark blue, (2) the expression level at least 1.5 times greater but less than threefold (transcripts showing a |FC|≥ 1.5 and a False Discovery Rate (FDR) < 0.05), medium blue, and (3) gene expression change of more than threefold, light blue. We found that all genes showed significant changes in the expression levels, and there was not a single gene that would show constant (dark blue only) expression during the whole process of reprogramming (Fig. 3). Moreover, 80% of all genes displayed changes in gene expression levels that were more than three-times fold at least on one of the days of reprogramming (light blue tiles). Only 20% of all genes stayed within the three-times fold change in expression levels (no light blue tiles), among them Tubulin (Tubb5), ribosomal subunits Rsp3, Rpl13a and Rsp18 and less known housekeeping genes Snrpb and Srp72. Importantly, this visualization showed that the most popular reference genes, Actb, Gapdh, Hprt, Ppia and B2m belonged to the group which fluctuated more than threefold. Since for the purposes of normalization it is necessary that the reference gene does not change its expression, we compiled a list of genes that stayed relatively constant, that is, within the category 1 and category 2 (dark blue and medium blue tiles) at all four time points during the iPS reprogramming process (Table 2).
Figure 3.
A heatmap representing fold changes in the housekeeping genes’ expressions throughout the iPS reprogramming process. The genes are grouped by fold-expression change into one of three groups, represented by dark blue, medium blue and light blue. Four time points during the reprogramming are indicated under the heatmaps, and the gene names are written on the left. The dark blue color represents no expression change, and Day 0 is taken as the “time point zero” before initiation of reprogramming. The medium blue color represents the expression change of more than statistically significant 1.5-fold, but less than threefold. Light blue represents gene expression change greater than threefold, compared to time point zero (Day 0).
Table 2.
Selected genes that do not change their expression more than three-fold during the iPS reprogramming.
| Mouse gene symbol | Human gene symbol | Gene name |
|---|---|---|
| Tubb5 | TUBB | Tubulin beta class I |
| Rps3 | RPS3 | Ribosomal protein S3 |
| Rsp18 | RPS18 | Ribosomal protein S18 |
| Rplp1 | RPLP1 | Ribosomal protein lateral stalk subunit P1 |
| Rpl13a | RPL13A | Ribosomal protein L13a |
| Mdh1 | MDH1 | Malate dehydrogenase 1 |
| Got1 | GOT1 | Glutamic-oxaloacetic transaminase 1 |
| Srp72 | SRP72 | Signal recognition particle 72 |
| Snrpb | SNRPB | Small nuclear ribonucleoprotein polypeptides B and B1 |
| Rnf7 | RNF7 | Ring finger protein 7 |
| Nubp1 | NUBP1 | Nucleotide binding protein 1 |
| Def8 | DEF8 | Differentially expressed in FDCP 8 |
The most stably expressed genes during iPS reprogramming
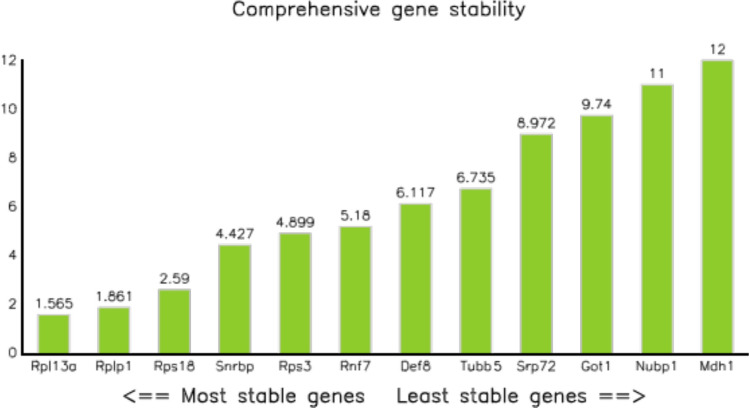
Finally, we subjected the selected twelve genes (Table 2) to the comprehensive ranking analysis that takes into account most of existing stability analyses software available, namely, RefFinder which calculates a comprehensive score from DeltaCt, BestKeeper, NormFinder and Genorm (see “Materials and methods”), and determined the most stable gene overall using the software (Fig. 4). Rpl13a was ranked the most stable overall, following by Rplp1 and Rps18.
Figure 4.
The most stable genes out of 70 selected for this study. The most stable genes are shown on the left, and the least stable genes are shown on the right. The small ribosomal subunit 13a is ranked as the most stable by all algorithms, with the large ribosomal subunit Rplp1 following it. Overall, the ribosomal subunits Rpl13a, Rplp1 and Rps18 were selected as the most stable genes throughout the iPS reprogramming.
Discussion
iPS cells are increasingly used for basic research, medical applications and drug testing. Since the research using these cells progresses rapidly, and more and more works include RNA sequencing data or qPCR data that rely on housekeeping genes for normalization, it is very important to know whether such normalization is suitable in iPS cells and specifically during the reprogramming process. In this work, we have analyzed the stability of 70 housekeeping genes throughout the reprogramming of mouse neural progenitors. Genes were selected from all known literature, to our best knowledge, making this study the first comprehensive comparison of stability in housekeeping genes. Importantly, we found that all housekeeping genes changed their expression more than two-fold during the iPS reprogramming. Moreover, 80% of the genes changed more than three-fold (Fig. 3). While these results were obtained in only one cell line, namely, mouse neural progenitors, it illustrates several important points: (1) housekeeping genes should not be assumed to be stable until actually tested, (2) iPS reprogramming affects even the cell maintenance-related genes, (3) therefore, more studies in other cell lines may uncover differences between species and cell lines.
It was of interest to us to see how the pattern of gene expression changed across different genes, for which we employed clustering analysis. Clustering approach revealed patterns of gene expression dynamics (Fig. 2). Ribosomal genes, which appeared in the same cluster, had a tendency to increase their expression throughout the reprogramming. This result is in agreement with the concept of iPS reprogramming, where the change in pluripotency state from less pluripotent to more pluripotent is associated with the increase in cell cycle progression and speed22–24. The increase in cell cycle speed would logically be associated with the increased need for ribosomal RNA synthesis. Interestingly, the analyses revealed that all ribosomal genes were grouped together, suggesting their temporal dynamics is similar during reprogramming. To validate this hypothesis, further investigations (for example, the measurements of nucleoli size or modern RNA staining techniques) would be needed. It is also worth noting that, while most ribosomal genes were found to increase during the iPS reprogramming process, some ribosomal genes displayed a fluctuating pattern (e.g. Rps18 and Rps9). Both of these genes belong to the small ribosomal subunit. Hypothetically, these two genes might play a distinctive role in regulating the production of ribosomes and by that, regulate the cell growth and division state. Indeed, recent research has also pointed at such a possibility, simulating the behavior of ribosomal genes and identifying them as a major speed-regulating hub for cell cycle progression25.
It is important to note that, in the current work, we have used a different strategy for primer design compared to our previous publication (Panina et al., 2018). In the present study, we conducted an analysis of splice variants of each gene (Sup. Table S1), and found that several genes possessed different splice variants. In the present work, we designed all primers so as to include the longest splice variant (Sup. Table S2). For example, the Rps18 gene in the present work includes the Exon 1 at the beginning of the gene sequence. This could be one explanation why the Rps18 gene in the current work was among the most stable ones, while in the previous publication (Panina et.al., 2018), where we used primer pairs that corresponded to the central exon of Rps18 gene (Sup. Figure), Rps18 was among the least stable ones. We thus caution the reader to use the same primers when attempting to reproduce results from other publications.
Our clustering analysis also grouped several ATP-generating genes into two categories: the genes whose expression decreases following a similar pattern (decreasing genes), and the genes whose expression fluctuates throughout the reprogramming. For example, such genes as Pgk1 and Pgam1 were grouped together as decreasing. Interestingly, Pgk1 and Pgam1 represent consequent steps in glycolysis. Pgk1 is a phosphoglycerate kinase, a glycolytic enzyme involved in step 2 of the pathway and catalyzes the conversion of 1,3-diphosphoglycerate to 3-phosphoglycerate, and Pgam1 catalyzes the reaction of 3-phosphoglycerate to 2-phosphoglycerate. On the other hand, Pfkp, Idh3a and Pdha1 were clustered next to each other on the other, “increasing” part of the clustering spectrum. Pfkp is the platelet-specific isoform of phosphofructokinase and plays a key role in glycolysis regulation, Idh3a is catalytic subunit of isocitrate dehydrogenase and catalyze the allosterically regulated rate-limiting step of the tricarboxylic acid (TCA) cycle. Pdha is a subunit of pyruvate dehydrogenase E1, a mitochondrial multienzyme complex that provides the primary link between glycolysis and the TCA cycle. TCA cycle is a process happening in mitochondria and is related to oxidative phosphorylation (OXPHOS). Since iPS reprogramming is well-known to involve profound changes in the balance between glycolysis and OXPHOS3, it would be interesting to research glycolytic genes further to shed more light on the control of these events.
It should also be noted that the current study focuses on the mouse neural stem cells and cannot claim that the results would be the same in other cell lines. Different cell lines may differ in the expression of the possible variants of housekeeping genes. As a case in point, MiniSox9 is a shorter splice variant of the Sox9 gene, and acts as a silencer of the full‐size (the longest) splice variant of Sox9, which can lead to drastic differences in the actual Sox9 expression depending on cell line and primers26. We thus caution the reader to investigate the actual performance of their primers of choice in their particular cell line while keeping in mind which splice variants would be amplified. This study should serve as a general guideline for the possible genes that can be tested.
In the end, we have evaluated all selected genes for their relative stability during the iPS reprogramming. Our results showed that the longest splice variants of Rpl13a, Rplp1 and Rps18 were the most stable during the process, which makes them the best candidates for normalization during RNA experiments. Overall, our analysis has shown that all currently known housekeeping genes fluctuate during the iPS reprogramming, and that the best strategy would be to combine several genes. We have suggested candidates for that.
Supplementary Information
Author contributions
Y.P. conceived the study, performed experiments, performed data analysis and wrote the manuscript, A.G. contributed to data analysis and wrote the manuscript, T.W. contributed to data analysis, checked the manuscript and supervised the project.
Funding
This research was supported by Japan Agency for Medical Research and Development under grant number 17bm0804008.
Data availability
All data concerning genes and primers are available in the Supplementary Material.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-78863-5.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Ebben JD, Zorniak M, Clark PA, Kuo JS. Introduction to induced pluripotent stem cells: Advancing the potential for personalized medicine. World Neurosurg. 2011;76:270–275. doi: 10.1016/j.wneu.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panopoulos AD, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagci H, Fisher AG. DNA demethylation in pluripotency and reprogramming: The role of tet proteins and cell division. Cell Stem Cell. 2013;13:265–269. doi: 10.1016/j.stem.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Evans C, Hardin J, Stoebel DM. Selecting between-sample RNA-Seq normalization methods from the perspective of their assumptions. Brief Bioinform. 2018;19:776–792. doi: 10.1093/bib/bbx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willems E, et al. Selection of reference genes in mouse embryos and in differentiating human and mouse ES cells. Int. J. Dev. Biol. 2006;50:627–635. doi: 10.1387/ijdb.052130ew. [DOI] [PubMed] [Google Scholar]
- 7.Mamo S, Gal AB, Bodo S, Dinnyes A. Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev. Biol. 2007;7:14. doi: 10.1186/1471-213X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mamo S, Gal AB, Polgar Z, Dinnyes A. Expression profiles of the pluripotency marker gene POU5F1 and validation of reference genes in rabbit oocytes and preimplantation stage embryos. BMC Mol. Biol. 2008;9:67. doi: 10.1186/1471-2199-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veazey KJ, Golding MC. Selection of stable reference genes for quantitative rt-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLoS ONE. 2011;6:e27592. doi: 10.1371/journal.pone.0027592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vossaert L, et al. Reference loci for RT-qPCR analysis of differentiating human embryonic stem cells. BMC Mol. Biol. 2013;14:21. doi: 10.1186/1471-2199-14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmgren G, et al. Identification of stable reference genes in differentiating human pluripotent stem cells. Physiol. Genomics. 2015;47:232–239. doi: 10.1152/physiolgenomics.00130.2014. [DOI] [PubMed] [Google Scholar]
- 12.Synnergren J, et al. Differentiating human embryonic stem cells express a unique housekeeping gene signature. Stem Cells. 2007;25:473–480. doi: 10.1634/stemcells.2006-0247. [DOI] [PubMed] [Google Scholar]
- 13.Artyukhov AS, et al. New genes for accurate normalization of qRT-PCR results in study of iPS and iPS-derived cells. Gene. 2017;626:234–240. doi: 10.1016/j.gene.2017.05.045. [DOI] [PubMed] [Google Scholar]
- 14.Panina Y, Germond A, Masui S, Watanabe TM. Validation of common housekeeping genes as reference for qPCR gene expression analysis during iPS reprogramming process. Sci. Rep. 2018;8:8716. doi: 10.1038/s41598-018-26707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaffl, M. W. in Polymerase Chain Reaction: Theory and Technology. 10.21775/9781912530243.05 (Caister Academic Press, 2019).
- 16.Panina Y, Germond A, David BG, Watanabe TM. Pairwise efficiency: A new mathematical approach to qPCR data analysis increases the precision of the calibration curve assay. BMC Bioinform. 2019;20:295. doi: 10.1186/s12859-019-2911-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hikichi T, et al. Transcription factors interfering with dedifferentiation induce cell type-specific transcriptional profiles. Proc. Natl. Acad. Sci. USA. 2013;110:6412–6417. doi: 10.1073/pnas.1220200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012 doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 19.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 20.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 21.Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol.3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed]
- 22.Ghule PN, et al. Reprogramming the pluripotent cell cycle: Restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. J. Cell Physiol. 2011;226:1149–1156. doi: 10.1002/jcp.22440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz S, et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapinas K, et al. The abbreviated pluripotent cell cycle. J. Cell Physiol. 2013;228:9–20. doi: 10.1002/jcp.24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J, Amir A. Homeostasis of protein and mRNA concentrations in growing cells. Nat. Commun. 2018;9:4496. doi: 10.1038/s41467-018-06714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdel-Samad R, et al. MiniSOX9, a dominant-negative variant in colon cancer cells. Oncogene. 2011;30:2493–2503. doi: 10.1038/onc.2010.621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data concerning genes and primers are available in the Supplementary Material.