Abstract
Several Lactobacillus plantarum strains are marketed as probiotics for their potential health benefits. Prebiotics, e.g., galacto-oligosaccharides (GOS), have the potential to selectively stimulate the growth of L. plantarum probiotic strains based on their phenotypic diversity in carbohydrate utilization, and thereby enhance their health promoting effects in the host in a strain-specific manner. Previously, we have shown that GOS variably promotes the strain-specific growth of L. plantarum. In this study we investigated this variation by molecular analysis of GOS utilization by L. plantarum. HPAEC-PAD analysis revealed two distinct GOS utilization phenotypes in L. plantarum. Linking these phenotypes to the strain-specific genotypes led to the identification of a lac operon encoding a β-galactosidase (lacA), a permease (lacS), and a divergently oriented regulator (lacR), that are predicted to be involved in the utilization of higher degree of polymerization (DP) constituents present in GOS (specifically DP of 3–4). Mutation of lacA and lacS in L. plantarum NC8 resulted in reduced growth on GOS, and HPAEC analysis confirmed the role of these genes in the import and utilization of higher-DP GOS constituents. Overall, the results enable the design of highly-selective synbiotic combinations of L. plantarum strain-specific probiotics and specific GOS-prebiotic fractions.
Subject terms: Food microbiology, Bacterial genes, Dietary carbohydrates
Introduction
Immediately after birth the gut of infants is rapidly colonized by microbes, which marks the start of a dynamic and interactive development during the first years of human life1. Amongst other factors, diet can have a profound effect on the modulation of the early gut microbiota. Human milk oligosaccharides (HMO) are a structurally diverse group of carbohydrates present in human milk that are strong drivers of infant gut microbiota development, and it has been shown that in particular Bifidobacterium species are HMO-utilization specialists2. In situations where breastfeeding is not feasible, infant formula provides an alternative to mimic the nutritional composition of breastmilk3. Although bovine milk is mostly devoid of oligosaccharides, lactose based oligosaccharides can be produced in large quantities by incubation of various microbial β-galactosidases with high concentrations of lactose to catalyze transglycosylation reactions to produce galacto-oligosaccharides (GOS)4. The chemical composition of GOS components (i.e., linkage type, and degree of branching and degree of polymerization) is highly dependent of the reaction conditions, as well as the specific β-galactosidase enzyme used during production, which results in GOS preparations that are commonly a composite of multiple different oligosaccharides5. GOS, together with fructo-oligosaccharides (FOS), can support the early gut development in a similar fashion as HMO6–8, and are a continued subject of research to improve infant formula.
The application of GOS also extends to health benefits associated with its dietary-supplementation in human adults and animals that are mainly related to its bifidogenic effects9,10 and the stimulation of intestinal lactobacilli (and enterococci) in lactose-intolerant individuals and healthy elderly11,12. GOS and HMO compounds can facilitate the growth of several lactobacilli13,14, and various strains of different Lactobacillus species are marketed as probiotics, including strains of L. rhamnosus, L. reuteri, L. casei, L. acidophilus and L. plantarum15. Previously, we have analyzed the growth of 77 L. plantarum strains on a specific GOS substrate (Vivinal GOS), revealing that all strains could grow on this substrate but to a significantly different extent, indicating strain-specific variation of GOS-utilization within the L. plantarum species16. L. plantarum is found in a wide range of ecological habitats including silage, fermented and non-fermented food products and the gastro-intestinal tract of humans and animals, and displays a notable phenotypic and genotypic diversity, in particular in its carbohydrate metabolism repertoire17,18. In lactobacilli in general, it is thought that GOS is imported by lactose permeases (LacS), which was verified in L. acidophilus NCFM to be the sole importer for GOS and lactose19. Once imported, lactose is hydrolyzed by β-galactosidases of Glycoside Hydrolase family 220 (LacZ/LacLM), whereas GOS is proposed to be digested by GH family 42 (LacA-type) β-galactosidases21,22. Not much is known about the mechanism of import and subsequent hydrolysis of GOS in L. plantarum, although a recent comparative transcription analysis of L. plantarum STIII revealed the GOS-growth induced upregulation of two gene clusters harboring various galactose- and lactose utilization genes, including lacS, lacA, lacL and lacM23. Similar genes were also identified to be involved in the utilization of GOS in L. reuteri ATCC PTA-647522.
In this study we further explored the variable capacity of 21 L. plantarum strains to utilize GOS for growth and determined the strain-specific utilization pattern of specific components of GOS using high performance anion exchange with pulsed amperometric detection (HPAEC-PAD), revealing two distinct GOS utilization phenotypes. Particularly the constituents of GOS with a higher degree of polymerization (i.e., DP of 3–4) (HDP-GOS) were differentially used by a subset of strains. This distinctive phenotype could be linked to the presence of a specific gene cluster (lacAS, lacR) by comparative genomic analysis and the subsequent construction of lacA and lacS deficient derivatives allowed the confirmation of the involvement of these genes in HDP-GOS utilization in L. plantarum.
Results
Differential GOS utilization in L. plantarum strains
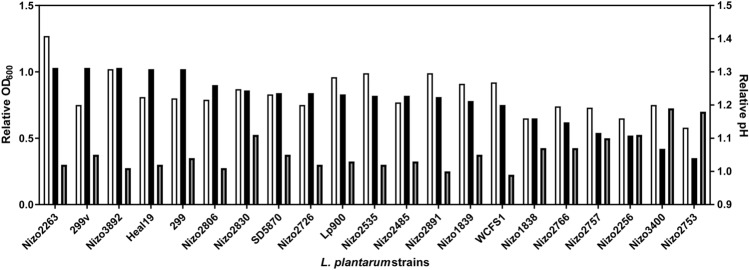
A panel of 21 L. plantarum strains with known genome sequences (Table 1) was selected from a larger panel of strains that was employed in an in vitro screening of strain-specific growth on Vivinal GOS containing media, to appropriately reflect the range of growth capacities observed in strains of this species16. These previously reported results illustrate that despite the significant variation in strain-specific growth on GOS, all L. plantarum strains grow relatively well on GOS-supplemented media, which was further confirmed in the present study by batch-growth experiments using optical density (OD600) and pH as growth reflecting read-outs (Fig. 1). The obtained strain-specific relative growth (OD600) of the 21 strains in GOS-supplemented medium, was significantly correlated (Pearson correlation r = 0.5967; p = 0.0043) with previously reported growth observed in the screening model (Supplemental Figure S1A). Moreover, the final pH of the same cultures was significantly correlated (Spearman correlation ρ = − 0.5381, p = 0.0119) with growth (OD600) (Supplemental Figure S1B). These results establish that despite the relatively moderate differences in GOS-dependent growth efficiency observed for this panel of L. plantarum strains these differences are reproducibly observed using different culturing conditions (96-well plate screening, versus more controlled batch cultures) and are reflected by the final pH after overnight cultivation.
Table 1.
Strains used in this study.
| Strain | Relevant feature | Reference | Source of isolation |
|---|---|---|---|
| E. coli NEB 5-alpha | Phage T1 resistant (fuhA2) DH5α™ derivative | 24 | n.a. |
| L. plantarum NC8 | Plasmid free strain of L. plantarum | 25 | Silage |
| L. plantarum NC8ΔlacS | CmR; EmR; NC8 derivative; chromosomal disruption of lacS by pNZ5319-lacS-KO integration | This study | n.a. |
| L. plantarum NC8ΔlacA | CmR; EmR; NC8 derivative; chromosomal disruption of lacA by pNZ5319-lacA-KO integration | This study | n.a. |
| L. plantarum WCFS1a | Single-colony isolate of L. plantarum NCIMB8826 | 26 | Human saliva |
| L. plantarum Nizo1838a | 18 | Human stool | |
| L. plantarum Nizo1839a | 18 | Sour cassava | |
| L. plantarum Nizo2256a | 18 | Human stool | |
| L. plantarum Nizo2263a | 18 | Silage | |
| L. plantarum Nizo2485a | 18 | Pork pickled sour Vietnam sausage | |
| L. plantarum Nizo2535a | 18 | Fermented orange | |
| L. plantarum Nizo2726a | 18 | Maize silage | |
| L. plantarum Nizo2753a | 27 | Fermented sourdough | |
| L. plantarum Nizo2757a | 18 | Fermented sourdough | |
| L. plantarum Nizo2766a | 18 | Fermented sourdough | |
| L. plantarum Nizo2806a | 28 | Sauerkraut | |
| L. plantarum Nizo2830a | 18 | n.a. | |
| L. plantarum Nizo2891a | 18 | Pickled radish | |
| L. plantarum Nizo3400a | 18 | Milk | |
| L. plantarum Nizo3892a | 17 | Human spinal fluid | |
| L. plantarum Lp900b | 16 | Ogi (red sorghum) | |
| L. plantarum Heal19b | 29 | Human GI tract | |
| L. plantarum 299vb | 30 | Human colon | |
| L. plantarum 299b | 30 | Human intestine | |
| L. plantarum SD5870b | 16 | Human GI tract |
aNizo strain collection, bProbi A/B strain collection. n.a. not available.
Figure 1.
L. plantarum strain-specific relative growth on GOS. The relative growth (OD600) after 24 h of L. plantarum strains on ½ MRS-C supplemented with 0.5% GOS in comparison to their growth on ½ MRS-C supplemented with 0.5% glucose in batch growth experiments (OD600; black bars, n = 1) and pH (grey bars, n = 1) and its congruency with the relative optical density (OD600; white bars, n = 3) obtained in a previous reported in vitro screening16.
Refinement of strain-specific utilization of GOS in L. plantarum
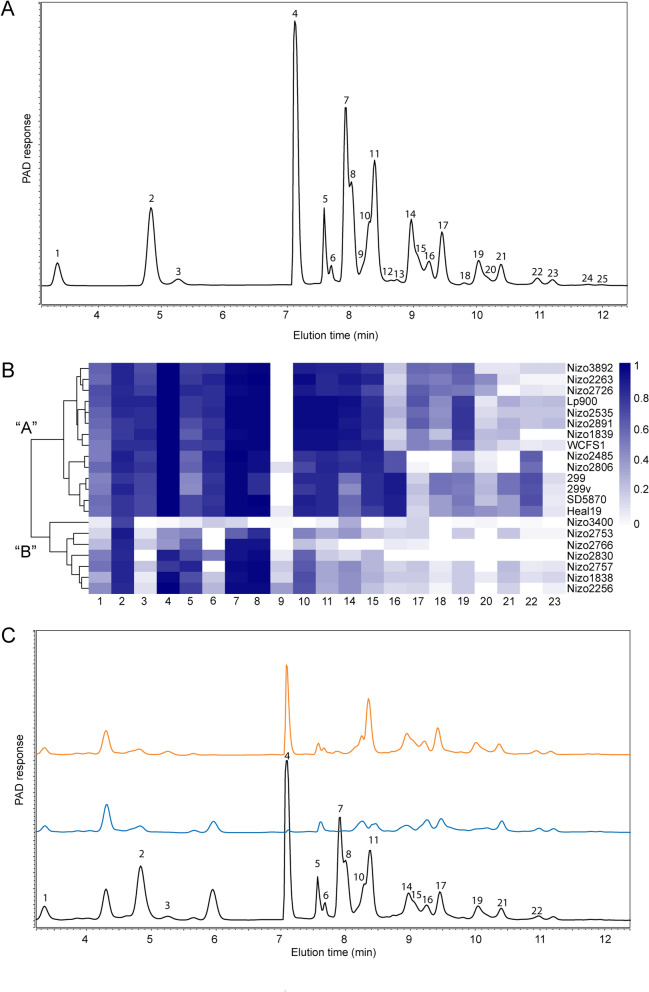
To further explain the observed differences in relative growth performance on GOS, substrate utilization was assessed by HPAEC-PAD. The GOS used in this study is a diverse mixture of linear and branched oligosaccharides with degrees of polymerization (DP) between 2 and 8 and a multitude of β-linked glycosidic linkages, although most of the information available is limited to lower DP fractions5. HPAEC-PAD analysis of GOS (Fig. 2A) revealed 25 distinct peaks in an elution profile that strongly resembles a previously reported similar analysis of the same GOS product in which the chemical identity of several individual peaks was further characterized5,31. The similarity of the elution patterns enables the tentative identification of the majority of the separately eluting compounds (Supplemental Table S1). HPAEC-PAD elution patterns of L. plantarum spent culture supernatants revealed a clear distinction of two major GOS-utilization phenotypes (Fig. 2B). The majority of the strains was able to utilize the majority of the GOS substrate (14 out of 21 strains), including the oligomers of higher DP (HDP-GOS utilizers, designated phenotype “A”), whereas the remaining strains (designated phenotype “B”) only utilized lactose with variable efficiency (peak 4) and utilized a few subsequent eluting peaks that are presumed to represent β-1,2 and β-1,3 linked galactose-glucose disaccharides (peaks 7 and 8) but failed to utilize the tentative β-1,4 and β-1,6 linked galactose disaccharides (peaks 3 and 6) (Fig. 2). Additionally, a subset of strains within the “A” phenotype (i.e., strains Nizo2485, Nizo2806, 299, 299v, SD5870 and Heal19) could utilize oligosaccharides corresponding to peak 16 and 22 more efficiently than the rest of the strains displaying the “A” phenotype, but appeared to have a reduced utilization of the oligosaccharides corresponding to peak 17 and 19. Strains with the “B” phenotype displayed a relatively slow adaptation to growth on lactose, since subsequent passaging of these cultures on GOS-containing media showed accelerating rates of growth and lactose utilization but did not lead to any utilization of HDP-GOS (data not shown). To verify that the increased overall GOS utilization in “A” phenotype strains corresponded with increased growth on this substrate, the relative reduction of the cumulative surface area of all peaks in the HPAEC-PAD elution patterns per strain in comparison to the original GOS substrate was significantly correlated with the relative optical density (OD600) (Spearman correlation ρ: − 0.4497, p = 0.0408) and the final pH (Spearman correlation ρ: 0.6643, p = 0.0010) reached by the strains (Supplemental Figure S2). These analyses also suggested that the final pH (i.e., as a proxy for overall lactate production) provides a more accurate reflection of GOS utilization in L. plantarum as compared to the final optical density reached.
Figure 2.
Refinement of GOS utilization by L. plantarum strains. (A) HPAEC-PAD elution pattern of the GOS substrate, in which the identified compounds include glucose and galactose (peak 2) and lactose (peak 4). See Table S1 in the supplementary material for tentative identification of the GOS peaks. (B) Heatmap and hierarchical clustering of relative peak surface area decrease in HPAEC-PAD response of spent culture supernatants of strains grown on 0.5% GOS compared to the original 0.5% GOS substrate after overnight incubation, with peak numbers corresponding to those presented in panel (A). Designated GOS utilization phenotypes “A” and “B” are represented at the first branching point of hierarchical clustering. (C) Exemplary HPAEC-PAD elution patterns of “A” phenotype (WCFS1, blue) and “B” phenotype (Nizo2830, orange) and the original MRS-GOS medium (black), with relevant peaks corresponding to panel (B).
Candidate L. plantarum genes involved in the utilization of GOS
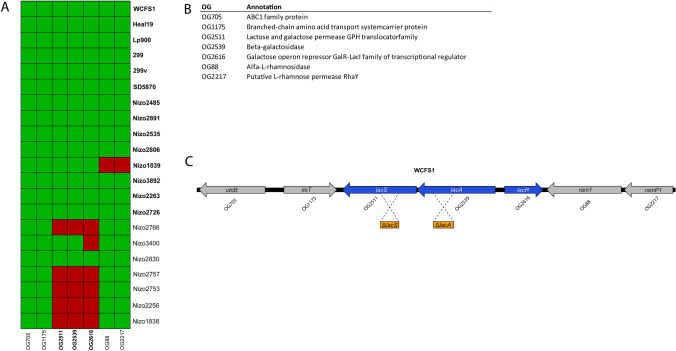
Comparative genomics to predict orthology relations between translated protein sequences of the publicly available L. plantarum genomes were performed to assemble an orthologous gene (OG) matrix for the subset of 21 strains using the orthAgogue tool32. The OG matrix was used to correlate the GOS utilization phenotypes “A” and “B” to the corresponding genotype of each strain, also known as “gene trait matching” (GTM)33–35, with the objective to identify candidate genes involved in the utilization of GOS and/or its breakdown products. GTM failed to identify genetic determinants that displayed a perfect presence-absence pattern corresponding with the two phenotypes within the L. plantarum strain panel used. However, all phenotype “A” (HDP-GOS utilizers; WCFS1, Heal19, Lp900, 299, 299v, SD5870, Nizo2485, Nizo2891, Nizo2535, Nizo206, Nizo1839, Nizo3892, Nizo2263 and Nizo2726) contain a lac operon consisting of OG2539 and OG2511 that are annotated as lacA and lacS, respectively, in the genome of the reference strain WCFS1. In contrast, most of the “B” phenotype strains lack this operon, albeit with the exception of the L. plantarum Nizo3400 and Nizo2830 strains that appeared to encode both OGs (Fig. 3). The identified genes are predicted to encode an intracellular β-galactosidase that contains a glycoside hydrolase (GH) family 42 domain (LacA), and a glycoside-pentoside-hexuronide (GPH) family lactose and galactose permease (LacS). In addition, upstream of the lacA gene a divergently oriented gene is located that encodes a transcriptional regulator (OG2616, annotated as lacR in the WCFS1 genome) that is putatively involved in the regulation of the lacAS operon. This divergently oriented lacR gene is present adjacent to the lacAS operon of all strains displaying the “A” phenotype, but is strikingly absent in the lacAS locus of the “B” phenotype strain L. plantarum Nizo3400. Therefore, it is tempting to speculate that lacR deficiency could lead to inappropriate regulation or complete lack of lacAS expression in this strain that underlies its failure to utilize HDP-GOS. Moreover, further analysis of the lacAS operon of L. plantarum Nizo2830 revealed that the lacS gene contains a disruptive mutation, rendering it a pseudogene, which explains the lack of HDP-GOS utilization in this strain. These findings provide credible explanations for the apparent genotype–phenotype discrepancy in the Nizo3400 and Nizo2830 strains, further supporting the postulated role of the lacAS operon in utilization of HDP-GOS compounds. Notably, gene-trait-matching analysis of the subtle difference detected between the utilization capacities observed for specific substrates (e.g., enhanced use of peak 16 and 22, and reduced use of peak 17 and 19 in strains Heal19, 299, 299v, SD570, Nizo2485 and Nizo2806) did not lead to any additional gene relationships that were considered worthwhile to pursue. Taken together, the lacAS operon is the prime locus associated with the difference in “A” and “B” utilization phenotypes.
Figure 3.
Gene cluster, identified in L. plantarum strains that readily utilize GOS. (A) Heatmap displaying the in silico gene-trait matching results based on the association between presence (green) or absence (red) of specific OGs and GOS utilization phenotypes “A” (strain names in bold) and “B” (strain names in normal font). (B) Predicted functions of OGs encoded in the identified operon and flanking genes in reference strain WCFS1. (C) Gene map of the lac operon and flanking genes in reference strain WCFS1 with OGs identified by gene-trait matching (blue), and genes flanking the operon (grey). The lac operon and flanking regions of WCFS1 are conserved in L. plantarum strain NC8, and homologous regions cloned in suicide vector pNZ5319 for disruption of candidate genes by single cross-over events in NC8 are presented in orange boxes for lacS (ΔlacS) and lacA (ΔlacA).
Inactivation of lacAS results in loss of HDP-GOS utilization in L. plantarum NC8
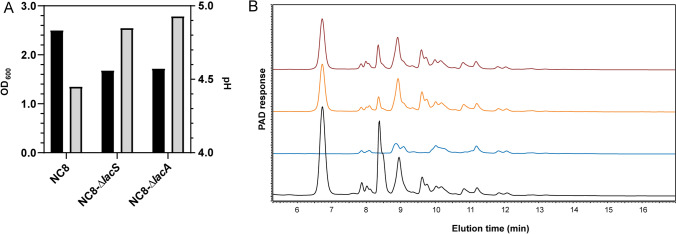
The role of the lacA and lacS orthologues in GOS utilization was investigated in L. plantarum strain NC8, a plasmid free L. plantarum strain (Table 1). Although this strain was not part of the original set of 21 strains, based on its high efficiency in transformation it was selected as a suitable host for mutagenesis, and its chromosome encodes the lacAS operon. NC8 can efficiently grow on GOS as carbon substrate (OD600) and displays the typical HDP-GOS utilizing, “A” phenotype (Fig. 4). To construct lacA and lacS mutagenesis vectors, internal fragments of either lacA or lacS of L. plantarum NC8 (Fig. 3) were cloned into suicide vector pNZ5319, which contains a P32-cat cassette that confers chloramphenicol resistance when integrated into the chromosome36. The resulting mutagenesis vectors, designated pNZ5319-lacA-KO and pNZ5319-lacS-KO (Table 2) were transformed to L. plantarum NC8 and single-cross-over plasmid integration derivatives were selected and designated NC8ΔlacA and NC8ΔlacS (Table 1). The two mutant strains and their parental strain NC8, were grown overnight on GOS supplemented media and final optical densities (OD600) and pH were determined, demonstrating that both gene knockout strains displayed a reduction of growth and acidification as compared to the L. plantarum NC8 parent strain (Fig. 4). Longer incubation times (i.e., 24 h) did not lead to increased utilization. Subsequently, spent culture supernatants of these overnight cultures were analyzed by HPAEC to investigate differential substrate utilization, showing that in contrast to their parental strain both knockout strains failed to utilize the HDP-GOS constituents and displayed the typical “B” phenotype (Fig. 4). Notably, with the chosen mutation strategy for the construction of NC8ΔlacA it cannot be excluded that this mutant also lacks expression of the downstream lacS gene due to polar effects. Nevertheless, these results demonstrate that at least the LacS transport function is required for the import of HDP-GOS in L. plantarum NC8, and suggests that once internalized, the oligosaccharides are likely to be hydrolyzed by LacA. Finally, both NC8ΔlacA and NC8ΔlacS displayed the reduced efficiency in lactose utilization as well as the utilization of the additional GOS constituents that are typically utilized by strains of the “B” phenotype, which is apparently depends on an alternative import and intracellular hydrolysis system in L. plantarum (Fig. 2).
Figure 4.
Growth of L. plantarum strain NC8 lacS- and lacA knockouts on GOS. (A) Exemplary growth characteristics of overnight cultures of L. plantarum NC8, NC8ΔlacS or NC8ΔlacA cultured on ½ MRS-C supplemented with 0.5% GOS with optical densities (black bars) and final pH (grey bars). (B) HPAEC-PAD elution patterns of MRS-C + 0.5% GOS without inoculum (black) and spent culture supernatants of NC8 (blue), NC8-ΔlacS (orange) and NC8-ΔlacA (red) grown on ½ MRS-C + 0.5% GOS.
Table 2.
Plasmids and primers used in this study.
| Plasmid or primer | Relevant feature | References |
|---|---|---|
| Plasmids | ||
| pNZ5319 | CmR, EmR; pNZ5318 derivative for multiple gene replacements in gram-positive bacteria | 36 |
| pNZ5319-lacA-KO | CmR, EmR; pNZ5319 derivative containing a 612 bp region homologous to lacA | This study |
| pNZ5319-lacS-KO | CmR, EmR; pNZ5319 derivative containing a 522 bp region homologous to lacS | This study |
| Primersa | ||
| S1 | 5′-CGTATCTCGAGAAGCTTAGTCGCAAACTGGG-3′ | This study |
| S2 | 5′-CGTATGGTACCGTTACGAAAGTACGGCCCAC-3′ | This study |
| S3 | 5′-CGTATGGTACCTTCACGGCTCTTGGCAGTTT-3′ | This study |
| S4 | 5′-CGTATCTCGAGATTGAGCATTGACCGAGTGC-3′ | This study |
| S5 | 5′-CGTATGGTACCATGCAGTTTAAATT-3′ | This study |
| S6 | 5′-CGTATCTCGAGCGCGTTATCGGTCC-3′ | This study |
| S7 | 5′-ATAAGGGCGGGAGCAGAATG-3′ | This study |
| S8 | 5′-AATAATACGCCGCTCCCTGG-3′ | This study |
| S9 | 5′-ATTCGGTCCTCGGGATATGA-3′ | This study |
| S10 | 5′-CTAAAGCAATGGCACCGACG-3′ | This study |
| S11 | 5′-TTGGATTTTTGTGAGCTTGGACT-3′ | This study |
CmR chloramphenicol resistant, EmR erythromycin resistant. aUnderlined sequences indicate enzymatic restriction sites.
Discussion
Various studies have demonstrated the prebiotic effect of dietary GOS to influence the host microbiome9–12. Especially the stimulation of endogenous Bifidobacterium has extensively been studied, while there are also reports on stimulation of endogenous lactobacilli11–14. In synbiotic applications, GOS may also selectively stimulate the in situ growth of the accompanying probiotic strain(s). Several strains of L. plantarum have demonstrated probiotic properties37–40, and their intrinsic genetic and phenotypic diversity in carbohydrate metabolism17,18,41 offers opportunities for selective and strain-specific stimulation by prebiotics. The GOS preparation employed here supports the growth of a large amount of L. plantarum strains, although this growth stimulation displays a substantial degree of variability that is reflected by the strain-specific growth stimulation detected16. Notably, the GOS preparation employed here contains approximately 30% mono- and di- saccharides, which tend to reduce the selectivity of growth stimulation because most bacteria can utilize these sugars. The remaining oligomeric proportion of the GOS substrate, is a complex substrate that contains a variety of oligo-saccharides that differ in glycosidic linkages and degrees of polymerization, that can be branched or linear5. The chemical complexity of GOS can obscure the selective growth stimulation by its specific constituents, and the analysis of the strain-specific utilization pattern of GOS that we used in this study can provide avenues towards the identification of specific GOS-constituents that allow strain-specific stimulation.
The variation in growth on GOS within individual strains of L. plantarum combined with HPAEC-PAD analysis of the spent culture supernatants, revealed two distinct GOS utilization phenotypes in this species. The majority of the strains, that displayed the most effective growth on GOS (i.e., “A”-phenotype) utilized the majority of the substrate, including the HDP-GOS constituents that could not be used by strains that grew less well on the GOS substrate and only appeared to utilize the lower DP constituents of the substrate (i.e., “B”-phenotype). Due to the structural complexity of GOS, it is likely that several of the peaks identified in HPAEC-PAD correspond to a combination of GOS constituents rather than a single chemical entity5,42. This agrees with the observation that specific compounds remain after L. plantarum growth that overlap with peaks in the original GOS substrate, reflecting their partial utilization. Tentative comparative analysis of the GOS elution pattern with previously reported GOS-structure analysis studies5 suggests that strains displaying the “A”-phenotype can utilize readily lactose as well as linear GOS up to a DP of 4, while the “B”-phenotype strains utilize only lactose and other galactose-glucose disaccharides. Moreover, peaks that are tentatively corresponding to branched GOS constituents appear not to be utilized by the “B” phenotype strains, suggesting that besides DP also saccharide structure constrains import in these strains. Interestingly, the “B”-phenotype strains also do not utilize the tentative galactose-galactose disaccharides and utilize lactose with reduced and variable efficiency relative to the “A” phenotype strains, suggesting that in these strains the pathway involved in HDP-GOS utilization also contributes to a more preferred lactose utilization. There is no correlation between the source of isolation of these strains and their capacity to utilize GOS, as both phenotypes harbor strains derived from human origin, fermented food or plant material (Table 1), confirming that the nomadic lifestyle of L. plantarum has consequences for the prediction of a strain’s genotype and corresponding phenotype based on the reported niche of isolation17. The distinction between the utilization of GOS-derived disaccharides and HDP-GOS constituents has also been reported for a panel of lactic acid bacteria strains of various species, showing that L. acidophilus W37 could utilize HDP-GOS, whereas L. salivarius W57, L. paracasei W20, L. casei W56, as well as Pediococcus acidilactici W143 and Enterococcus faecium W54 utilized only the disaccharide fraction of GOS43. Intriguingly, these observations are comparable to the “A” and “B” phenotype distinction observed for L. plantarum, illustrating that screening for prebiotic utilization should be performed at strain level, especially for species with high genetic and phenotypic diversity like L. plantarum.
Gene trait matching enabled the identification of the lacAS operon that was predicted to be involved in HDP-GOS utilization (“A” phenotype), which was confirmed in lacA and lacS deficient derivatives of L. plantarum NC8, which effectively led to the switching of the GOS-utilization “A” phenotype of this strain to the typical “B” phenotype. Although possible polar effects of lacA mutation on the downstream encoded lacS prevent the unambiguous confirmation of the role of the lacA encoded β-galactosidase in GOS utilization, the role of LacS-mediated import of HDP-GOS in L. plantarum is convincingly evidenced. Interestingly, inactivation of a LacS homologue in Lactobacillus acidophilus NCFM (~ 60% protein identity with the lacS of WCFS1), resulted in the complete loss of lactose, GOS and lactitol utilization in this species19. In contrast, the LacS deficient derivative of L. plantarum NC8 (NC8ΔlacS) still utilized lactose and several other (disaccharide) constituents of GOS, albeit with reduced efficiency. This implies that these GOS constituents are utilized by L. plantarum via a redundant, alternative pathway that is apparently absent in L. acidophilus NCFM. This alternative pathway in L. plantarum is likely to involve a lactose and galactose permease (OG2178) that is encoded in the core genome of this species (annotated as rafP in WCFS1), and displays 39% protein-sequence identity with both the LacS homologues of L. plantarum and L. acidophilus. Domain prediction tools suggest that OG2178 exhibits similar function as LacS, but our results indicate that its import capacity is restricted to glucose- and galactose disaccharides and that it imports lactose with reduced efficiency compared to the LacS function identified in this study. OG2178 is encoded within the core-genome encoded gal cluster, that encompasses three polycistronic transcriptional units associated with galactose metabolism (i.e., import, hydrolysis, Leloir pathway and transcriptional regulation)23. This cluster also encodes the large- (lacL) and small (lacM) β-galactosidase subunit (OG1755 and OG816, respectively) that form the heterodimer LacLM that belongs to the GH family-2 β-galactosidases, and is homologous to the L. acidophilus NFCM LacLM (64 and 61% protein sequence identity, respectively). Comparative transcriptome analysis in strain L. plantarum STIII, revealed that both the gal cluster and the lacAS operon identified in this study, were upregulated when this strain was grown on GOS as a substrate, supporting the role of both clusters in the utilization of this prebiotic substrate23. Interestingly, heterologous expression of the L. plantarum FUA3112 derived LacLM in Lactococcus lactis established its lactose hydrolysis activity21, while experiments using the purified L. plantarum LacLM revealed that the enzyme also has high transglycosylation activity and could be used for the synthesis of GOS di, tri- and tetra-saccharides with a high preference of β-1,6 linkages44. It remains to be established what the precise roles of LacA and LacLM in GOS hydrolysis are in L. plantarum (i.e., DP and branched or linear structure). Intriguingly, a recent investigation of GOS metabolism in L. reuteri ATCC PTA-6475 established a role of its LacA homologue (~ 37% identity with LacA in L. plantarum) in hydrolysis of galactose-galactose linkages in GOS, but also showed that this enzyme could not hydrolyze lactose, which was dependent on the LacLM homologue in this species (~ 64% identity with L. plantarum)22. Moreover, a LacS deficient L. reuteri derivative (~ 58% identity with L. plantarum LacS) displayed reduced growth efficiency on GOS as well as lactose, which is similar to the LacS mutant of L. plantarum in this study. Finally, analogous to L. plantarum, L. reuteri ATCC PTA-6475 encodes a redundant galactose- and lactose permease (~ 56% identity with L. plantarum OG2178), suggesting that in both species LacS and LacA are involved in import and hydrolysis of HDP-GOS, while lactose and other galactose-glucose disaccharides can also be imported either by LacS or the alternative permease (i.e., OG2178 in L. plantarum, annotated as RafP in strain WCFS1) followed by hydrolysis by LacLM.
In conclusion, our study establishes that the L. plantarum capacity to utilize HDP-GOS depends on the strain-specific capacity to import these substrates via LacS, while it also strongly suggests a co-dependence on the genetically linked LacA. Thereby, the presence of these genes in combination with their putative regulator LacR can accurately predict the growth capacity of L. plantarum strains on HDP-GOS substrates. Since commercially available GOS prebiotics commonly contain a varying fraction of mono- and di-saccharides, the distinctive phenotype among L. plantarum strains may be obscured in GOS-growth studies. Our study also exemplifies that much more pronounced differences in growth, and more selective and strain-specific growth stimulation may be achieved by further purification of HDP-GOS constituents, specifically those of DP3-4 since import of higher DP GOS appears to be restricted in L. plantarum. Moreover, detailed chemical characterization of the constituents of GOS5,43 may further enhance their selective application for stimulatory of growth and or activity of specific (intestinal) bacteria dependent on the substrate-specificities of their import and hydrolysis machinery. The approach taken in this study would be suitable to explore such future applications of specific fractions of GOS. In L. plantarum we detect only two distinctive GOS-utilization phenotypes, but this may be different in other species. In support of such possibilities, we have previously shown that the different degree of utilization of isomalto-oligosaccharides in L. plantarum coincides with multiple phenotype and corresponding genotype variations in specific strains of this species16. Finally, the results presented in this study offer opportunities for the design of strongly selective L. plantarum and HDP-GOS synbiotic products, aiming to synergistically enhance the health benefit to the consumer45,46, underpinning that deciphering the relationships between the chemical complexity of prebiotics and the selective stimulation of growth of probiotic strains by specific prebiotic constituents could enhance the health effects of synbiotics.
Materials and methods
Prebiotic substrate, bacterial strains, and growth conditions
Vivinal GOS was kindly provided by Friesland Campina (Domo-branch, Amersfoort, the Netherlands) and consists of 70% galacto-oligosaccharides (DP 3–8), 25% lactose and 5–6% monosaccharides glucose and galactose. The DP2–DP5 pools of GOS contain over 40 different structures42. The majority consists of a terminal glucose moieties elongated with β-1,6 linked galactose subunits, but also contains β-1,2, β-1,3 and β-1,6 linkages and may branched or linear. The L. plantarum strains used in this study were selected from the NIZO and Probi A/B strain collections (Table 1). The strains were selected from a previously studied larger panel of strains (77 strains), to reflect the range of relative growth capacities on GOS-supplemented media16. L. plantarum strains were routinely propagated in MRS medium (Merck KGaA, Darmstadt, Germany) at 37 °C without agitation. Media were supplemented with 10 μg/mL chloramphenicol when appropriate.
To verify and refine the previous results of L. plantarum strain-specific growth capacity on GOS as a substrate the selected strains were inoculated 1:500 from overnight cultures grown on standard MRS in 15 mL Falcon tubes (Corning, New York, USA) containing twofold diluted MRS47 without the addition of any carbon source (½ MRS-C) supplemented with 0.5% (w/v) GOS or glucose and grown for 24 h at 37 °C. The strain panel was also grown on ½ MRS-C without carbon source supplementation, to account for background growth on carbohydrates that are naturally present in MRS constituents. All media and GOS suspensions were autoclaved at 100 °C for 10 min. Final culture densities (OD600, Microplate Spectrophotometer SpectraMax M5) and final pH were determined as proxies for growth on both substrates. The relative growth capacities of the tested strains were expressed as the optical density (OD600) or pH of strains grown on medium supplemented with GOS compared to those obtained when grown on medium supplemented with glucose. Cells were removed by centrifugation (4000×g, 10 min, 4 °C) and spent culture supernatants were stored at − 20 °C for further analysis (see below).
HPAEC-PAD peak profile characterization
Spent culture supernatants or GOS (5 mg/ml) were diluted 1:20 in MilliQ and eventual debris was removed by centrifugation (16,000×g, 15 min, room temperature), and subsequently analyzed by High-Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) essentially as described previously16. Relative peak utilization per strain was estimated by determination of the area under the curve in the spent culture supernatant relative to the corresponding peaks in the original GOS substrate and was visualized in heatmaps using the pheatmap package 1.0.748 in R version 3.4.0.
Gene-trait matching
The genomes of the 21 L. plantarum strains for which the GOS utilization was analyzed with HPAEC-PAD, were obtained from the National Center for Biotechnology Information (NCBI) database49. Protein sequences were aligned with an all-versus-all bi-directional BLASTP (cut-off: E-value < 1e−05)50 and the resulting output was processed by OrthAgogue32 to cluster orthologue genes into orthologue groups (OGs). Orthologue gene families were respectively appointed as either part of the core genome or variome based on their presence in the entire- or subset of the 21 strains. An OG matrix was constructed from the assigned OGs where each OG is linked to the corresponding genome-locus tag for each of the strains, which was subsequently converted to a binary gene-presence (1) and -absence (0) matrix (Supplemental Table S2). An in silico genotype/phenotype gene-trait matching (GTM) was performed based on the generated binary matrix of orthologous genes, correlating the presence and absence of genes with the observed constituent-specific GOS utilization phenotypes (i.e., HDP-GOS utilizers [phenotype “A”] and non-utilizers [phenotype “B”]) of each strain visualized by HPAEC-PAD. Candidate OGs were ranked based on their importance (predictive value), and presence in phenotype “A” and absence in phenotype “B” to predict their correlation with the phenotype as previously described33. Visualization of genomic regions containing the phenotype-related candidate OGs, were performed in Artemis version 16.0.051, and the gene map was created using Easyfig 2.2.252, while the presence or absence of candidate OGs and their corresponding operons within the strain panel were visualized in a heatmap using pheatmap 1.0.748 in R version 3.4.0. Function-annotation of candidate genes were investigated with InterProScan 553 and the CazY database20. To predict the cellular localization of candidate OGs, the encoded proteins were analyzed with SignalP-5.054, and TMHMM server v.2.055.
LacA and lacS knock-outs by homologous recombination in L. plantarum strain NC8
Internal fragments of the coding regions of lacA (612 bp, locus tag: nc8_2941) and lacS (522 bp; locus tag: nc8_2940) were amplified by colony PCR using a single colony of L. plantarum strain NC8 and primers S1-2 and S3-4, respectively (Table 2). The vector fragment of the pACYC184-derived pNZ5319 that contains a P32-cat cassette that enables selection on basis of chloramphenicol resistance36, was amplified by PCR using primers S5-6 (Table 2). All PCR-amplicons were purified using MSB Spin PCRapace (STRATEC Molecular, Germany), and digested with XhoI and KpnI (Thermo Fisher Scientific). The internal fragments of lacA and lacS were separately ligated with the cloning-vector fragment using T4 ligase for 90′ at room temperature (Thermo Fisher Scientific) and subsequently transformed to NEB 5-alpha competent Escherichia coli (New England Biolabs, Massachusetts, USA) according to the manufacturers protocol. Transformants were selected on Luria–Bertani (LB) agar plates supplemented with 10 μg/mL chloramphenicol, and colonies harboring the correct lacA and lacS mutation constructs were identified by colony PCR using primers S6 and S2 or S6 and S4 for the pNZ5319-lacA-KO and pNZ5319-lacS-KO constructs, respectively (Table 2). The selected E. coli clones that harbored pNZ5319-lacA-KO and pNZ5319-lacS-KO were grown overnight at 37 °C in LB media supplemented with 10 μg/mL chloramphenicol. The plasmids pNZ5319-lacA-KO and pNZ5319-lacS-KO were isolated from these overnight cultures using QIAGEN Plasmid Plus Midi Kit (Qiagen, Hilden, Germany) and 4 μg of the plasmids was transformed to electrocompetent L. plantarum NC8 using a Gene Pulser Xcell Electroporation System (Biorad, Hercules, California, USA) using a voltage of 1500 V, capacity of 25 μF and a resistance of 400 Ω. Electrocompetent L. plantarum NC8 cells were prepared as previously described36, by culturing the cells in glycine-containing media followed by several washing steps using ice-cold solution of 30% (w/v) polyethylene glycol 1450, finally using 100-fold concentrated cell-suspensions for electroporation. Electroporated L. plantarum NC8 cells were resuspended in MRS supplemented with 0.5 M sucrose and 0.1 M MgCl2 and incubated at 37 °C for 90 min, followed by plating on MRS plates containing 10 μg/mL chloramphenicol. Notably, pNZ5319-derived plasmids can replicate extrachromosomally in E. coli, but not in L. plantarum, whereby selection of chloramphenicol resistant L. plantarum transformants selects for a single-cross over plasmid-integration resulting from homologous recombination of the plasmid encoded internal fragment of lacA or lacS and the corresponding homologous region in the NC8 chromosome, effectively disrupting the lacA or lacS gene, respectively. Chloramphenicol resistant plasmid-integrant colonies were selected after 3 days of incubation at 37 °C and the anticipated chromosomal integration of the plasmid was verified by PCR using primers S8-9 and S10-11 for lacA and lacS integrant colonies, respectively (Table 2). Selected single colony isolates were inoculated in MRS supplemented with 10 μg/mL chloramphenicol and grown overnight at 37 °C. Overnight cultures were subcultured (1:500) in ½ MRS-C supplemented with 0.5% GOS or glucose and grown overnight at 37 °C. Optical densities of the overnight cultures were determined (OD600) using a Microplate Spectrophotometer SpectraMax M5 and final pH of the cultures was measured.
Statistical analysis
To determine whether data sets were normally distributed, Shapiro–Wilk normality tests were performed with a confidence interval of 95%. Spearman and Pearson correlations were performed two-tailed with a confidence interval of 95%. All analyses were performed using GraphPad Prism version 8.00 for Windows (GraphPad Software, San Diego, California, USA).
Supplementary Information
Acknowledgements
This work was financially supported by Probi A/B, Lund, Sweden, without having a direct involvement in study design, data acquisition, data interpretation and the decision to submit this work for publication. However, we would like to thank Kerstin Holmgren, Gunilla Önning and Niklas Larson from Probi A/B for the fruitful discussions about the results obtained. We want to thank Friesland Campina for providing the Vivinal GOS tested in this study. We also want to thank Probi A/B and NIZO food research for providing the L. plantarum strains that were used in this study.
Author contributions
J.F., H.A.S. and M.K. conceived the study. J.F. performed the primary growth experiments. J.F., L.P.M. and M.K. performed the gene-trait matching. J.F., M.S. and M.K. performed the genetic knock-outs. J.F. and C.R. performed HPAEC-PAD analysis of spent culture supernatants. J.F., C.R., H.A.S. and M.K. contributed to the interpretation of HPAEC-PAD results. J.F. and M.K. contributed drafting the manuscript. All authors contributed to editing the final manuscript. All authors read and approved the final manuscript.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-78721-4.
References
- 1.Milani C, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017;81:1–67. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakanaka M, et al. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: Prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. 2020;12:1–21. doi: 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin CR, Ling P-R, Backburn GL. Review of infant feeding: key features of breast milk and infant formula. Nutrients. 2016;8:1–11. doi: 10.3390/nu8050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer C, Kleinschmidt T. Synthesis of galactooligosaccharides in milk and whey: a review. Compr. Rev. Food Sci. Food Saf. 2018;17:678–697. doi: 10.1111/1541-4337.12344. [DOI] [PubMed] [Google Scholar]
- 5.Van Leeuwen SS, Kuipers BJH, Dijkhuizen L, Kamerling JP. Comparative structural characterization of 7 commercial galacto-oligosaccharide (GOS) products. Carbohydr. Res. 2016;425:48–58. doi: 10.1016/j.carres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Moro G, et al. Dosage-related bifidogenic effects of galacto- and fructooligosaccharides in formula-fed term infants. J. Pediatr. Gastroenterol. Nutr. 2002;34:291–295. doi: 10.1097/00005176-200203000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 2006;72:2359–2365. doi: 10.1128/AEM.72.4.2359-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakker-Zierikzee AM, et al. Effects of infant formula containing a mixture of galacto- and fructo-oligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br. J. Nutr. 2005;94:783–790. doi: 10.1079/BJN20051451. [DOI] [PubMed] [Google Scholar]
- 9.Davis LMG, Martínez I, Walter J, Hutkins R. A dose dependent impact of prebiotic galactooligosaccharides on the intestinal microbiota of healthy adults. Int. J. Food Microbiol. 2010;144:285–292. doi: 10.1016/j.ijfoodmicro.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Bouhnik Y, et al. The capacity of nondigestible carbohydrates to stimulate fecal bifidobacteria in healthy humans: a double-blind, randomized, placebo-controlled, parallel-group, dose-response relation study. Am. J. Clin. Nutr. 2004;80:1658–1664. doi: 10.1093/ajcn/80.6.1658. [DOI] [PubMed] [Google Scholar]
- 11.Azcarate-Peril MA, et al. Impact of short-chain galactooligosaccharides on the gut microbiome of lactose-intolerant individuals. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E367–E375. doi: 10.1073/pnas.1606722113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vulevic J, Drakoularakou A, Yaqoob P, Tzortzis G, Gibson GR. Modulation of the fecal microflora profile and immune function by a novel trans-galactooligosaccharide mixture (B-GOS) in healthy elderly volunteers. Am. J. Clin. Nutr. 2008;88:1438–1446. doi: 10.3945/ajcn.2008.26242. [DOI] [PubMed] [Google Scholar]
- 13.Thongaram T, Hoeflinger JL, Chow JM, Miller MJ. Human milk oligosaccharide consumption by probiotic and human-associated bifidobacteria and lactobacilli. J. Dairy Sci. 2017;100:7825–7833. doi: 10.3168/jds.2017-12753. [DOI] [PubMed] [Google Scholar]
- 14.Schwab C, Michael G. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiol. Lett. 2010;315:141–148. doi: 10.1111/j.1574-6968.2010.02185.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Lv J, Pan L, Zhang Y. Roles and applications of probiotic Lactobacillus strains. Appl. Microbiol. Biotechnol. 2018;102:8135–8143. doi: 10.1007/s00253-018-9217-9. [DOI] [PubMed] [Google Scholar]
- 16.Fuhren J, Rösch C, ten Napel M, Schols HA, Kleerebezem M. Synbiotic matchmaking in Lactobacillus plantarum; substrate screening and gene-trait matching to characterize strain-specific carbohydrate utilization. Appl. Environ. Microbiol. 2020;86:e01081–e1120. doi: 10.1128/AEM.01081-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martino ME, et al. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016;18:4974–4989. doi: 10.1111/1462-2920.13455. [DOI] [PubMed] [Google Scholar]
- 18.Siezen RJ, et al. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 2010;12:758–773. doi: 10.1111/j.1462-2920.2009.02119.x. [DOI] [PubMed] [Google Scholar]
- 19.Andersen JM, et al. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17785–17790. doi: 10.1073/pnas.1114152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwab C, Sørensen KI, Gänzle MG. Heterologous expression of glycoside hydrolase family 2 and 42 β-galactosidases of lactic acid bacteria in Lactococcus lactis. Syst. Appl. Microbiol. 2010;33:300–307. doi: 10.1016/j.syapm.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Rattanapprasert M, et al. Genes involved in galactooligosaccharide metabolism in Lactobacillus reuteri and their ecological role in the gastrointestinal tract. Appl. Environ. Microbiol. 2019;85:1–15. doi: 10.1128/AEM.01788-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Wang L, Lu Y, Yu H, Tian H. Comparative transcriptional analysis of Lactobacillus plantarum and its ccpA-knockout mutant under galactooligosaccharides and glucose conditions. Front. Microbiol. 2019;10:1–12. doi: 10.3389/fmicb.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anton BP, Raleigh EA. Complete genome sequence of NEB 5-alpha, a derivative of. Genome Announc. 2016;4:e01245–e1316. doi: 10.1128/genomeA.01245-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axelsson L, et al. Genome sequence of the naturally plasmid-free Lactobacillus plantarum strain NC8 (CCUG 61730) J. Bacteriol. 2012;194:2391–2392. doi: 10.1128/JB.00141-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleerebezem M, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pepe O, et al. Technological and molecular diversity of Lactobacillus plantarum strains isolated from naturally fermented sourdoughs. Syst. Appl. Microbiol. 2004;27:443–453. doi: 10.1078/0723202041438446. [DOI] [PubMed] [Google Scholar]
- 28.Vermeiren L, Devlieghere F, Debevere J. Evaluation of meat born lactic acid bacteria as protective cultures for the biopreservation of cooked meat products. Int. J. Food Microbiol. 2004;96:149–164. doi: 10.1016/j.ijfoodmicro.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Vásquez A, Ahrné S, Jeppsson B, Molin G. Oral administration of Lactobacillus and Bifidobacterium strains of intestinal and vaginal origin to healthy human females: Re-isolation from faeces and vagina. Microb. Ecol. Health Dis. 2005;17:15–20. [Google Scholar]
- 30.Johansson ML, et al. Administration of different Lactobacillus strains in fermented oatmeal soup: In vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl. Environ. Microbiol. 1993;59:15–20. doi: 10.1128/AEM.59.1.15-20.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladirat SE, et al. In vitro fermentation of galacto-oligosaccharides and its specific size-fractions using non-treated and amoxicillin-treated human inoculum. Bioact. Carbohydr. Diet. Fibre. 2014;3:59–70. doi: 10.1016/j.bcdf.2014.02.002. [DOI] [Google Scholar]
- 32.Ekseth OK, Kuiper M, Mironov V. OrthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics. 2014;30:734–736. doi: 10.1093/bioinformatics/btt582. [DOI] [PubMed] [Google Scholar]
- 33.Bayjanov JR, Molenaar D, Tzeneva V, Siezen RJ, van Hijum SAFT. PhenoLink—a web-tool for linking phenotype to ~omics data for bacteria: Application to gene-trait matching for Lactobacillus plantarum strains. BMC Genomics. 2012;13:170. doi: 10.1186/1471-2164-13-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pretzer G, et al. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 2005;187:6128–6136. doi: 10.1128/JB.187.17.6128-6136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siezen RJ, van Hylckama Vlieg JET. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb. Cell Fact. 2011;10:S3. doi: 10.1186/1475-2859-10-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert JM, Bongers RS, Kleerebezem M. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 2007;73:1126–1135. doi: 10.1128/AEM.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panigrahi P, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017;548:407–412. doi: 10.1038/nature23480. [DOI] [PubMed] [Google Scholar]
- 38.Van Baarlen P, Troost FJ, Van Hemert S, Van Der Meer C, De Vos WM. Differential NF-κB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc. Natl. Acad. Sci. 2009;106:2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoppe M, Önning G, Berggren A, Hulthén L. Probiotic strain Lactobacillus plantarum 299v increases iron absorption from an iron-supplemented fruit drink: a double-isotope cross-over single-blind study in women of reproductive age. Br. J. Nutr. 2015;114:1195–1202. doi: 10.1017/S000711451500241X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klarin B, et al. Lactobacillus plantarum 299v reduces colonisation of Clostridium difficile in critically ill patients treated with antibiotics. Acta Anaesthesiol. Scand. 2008;52:1096–1102. doi: 10.1111/j.1399-6576.2008.01748.x. [DOI] [PubMed] [Google Scholar]
- 41.Molenaar D, et al. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 2005;187:6119–6127. doi: 10.1128/JB.187.17.6119-6127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Leeuwen SS, Kuipers BJH, Dijkhuizen L, Kamerling JP. 1H NMR analysis of the lactose/β-galactosidase-derived galacto-oligosaccharide components of Vivinal® GOS up to DP5. Carbohydr. Res. 2014;400:59–73. doi: 10.1016/j.carres.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 43.Böger M, Van Leeuwen SS, Lammerts Van Bueren A, Dijkhuizen L. Structural identity of galactooligosaccharide molecules selectively utilized by single cultures of probiotic bacterial strains. J. Agric. Food Chem. 2019;67:13969–13977. doi: 10.1021/acs.jafc.9b05968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iqbal S, Nguyen T, Thanh T, Maischberger T, Haltrich D. β-Galactosidase from Lactobacillus plantarum WCFS1: biochemical characterization and formation of prebiotic galacto-oligosaccharides. Carbohydr. Res. 2010;345:1408–1416. doi: 10.1016/j.carres.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 45.Pandey KR, Naik SR, Vakil BV. Probiotics, prebiotics and synbiotics—a review. J. Food Sci. Technol. 2015;52:7577–7587. doi: 10.1007/s13197-015-1921-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 47.de Man J, Rogosa M, Elisabeth Sharpe M. A medium for the cultivation of lactobaclli. J. Appl. Bacteriol. Bacteriol. 1960;23:130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 48.Raivo K. Pheatmap: pretty heatmaps. R Package Version. 2012;61:915. [Google Scholar]
- 49.Geer LY, et al. The NCBI BioSystems database. Nucleic Acids Res. 2009;38:492–496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschup SF, Gish W, Pennsylvania T, Park U. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 51.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almagro Armenteros JJ, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 55.Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.