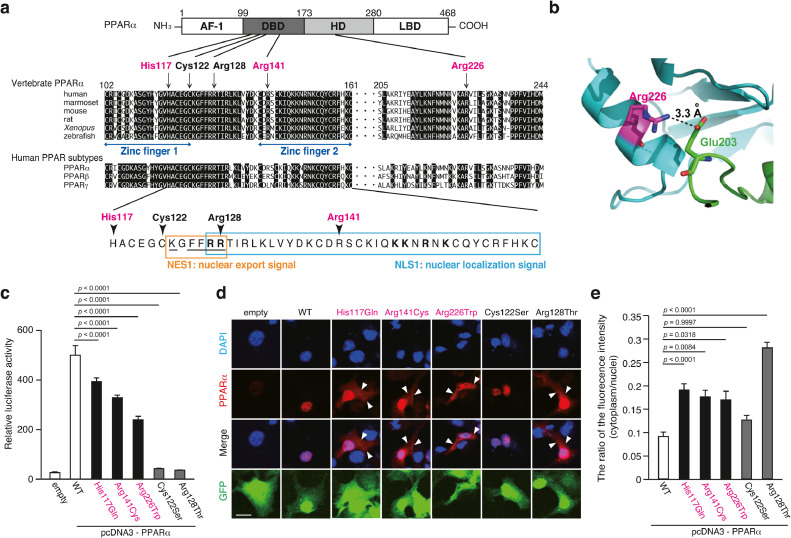
Fig. 3.
Three missense variants in the PPARA gene
(a) Schematic representation of the PPARα protein. (upper) Amino acid sequence alignment among vertebrates and human subtypes of PPARα. Magenta characters and arrows indicate the position of the missense mutations found in patients with schizophrenia. Black characters and arrows indicate the position of the functional mutations reported in the previous studies [66, 67]. (lower) Amino acid sequences of NLS1 (blue square) and NES1 (orange square) of human PPARα. Bold letters in NLS1 designate a potential bipartite basic motif similar to the consensus binding signal for importin α. The underline in NES1 designates a potential KXFF(K/R)R motif similar to the consensus binding signal for CRT (calreticulin) and a putative hydrophobic NES motif for CRM1.
(b) A salt bridge between Arg226 in PPARα and Glu203 in RXRα.
(c) Activity of PPARα mutants as a transcription factor in the CV-1 cells.
The values represent the mean ± standard error (n = 3/group). The data were analyzed using one-way ANOVA (p < 0.001), followed by Dunnett's multiple comparison tests. These results were replicated in three independent experiments (data not shown).
(d) Intracellular localization of PPARα in the COS-7 cells was examined using the anti-PPARα antibody under a confocal microscope. Scale bar = 10 µm.
(e) The ratio of PPARα fluorescent intensity in the cytoplasm to that in the nuclei. The values represent the mean ± standard error (n > 97 group). The data were analyzed using a one-way ANOVA (p < 0.001), followed by Dunnett's multiple comparison tests.