Abstract
Glaucoma is a progressive and deteriorating optic neuropathy that leads to visual field defects. The damage occurs as glaucoma is irreversible, so early and timely diagnosis is of significant importance. The proposed system employs the convolution neural network (CNN) for automatic segmentation of the retinal layers. The inner limiting membrane (ILM) and retinal pigmented epithelium (RPE) are used to calculate cup-to-disc ratio (CDR) for glaucoma diagnosis. The proposed system uses structure tensors to extract candidate layer pixels, and a patch across each candidate layer pixel is extracted, which is classified using CNN. The proposed framework is based upon VGG-16 architecture for feature extraction and classification of retinal layer pixels. The output feature map is merged into SoftMax layer for classification and produces probability map for central pixel of each patch and decides whether it is ILM, RPE, or background pixels. Graph search theory refines the extracted layers by interpolating the missing points, and these extracted ILM and RPE are finally used to compute CDR value and diagnose glaucoma. The proposed system is validated using a local dataset of optical coherence tomography images from 196 patients, including normal and glaucoma subjects. The dataset contains manually annotated ILM and RPE layers; manually extracted patches for ILM, RPE, and background pixels; CDR values; and eventually final finding related to glaucoma. The proposed system is able to extract ILM and RPE with a small absolute mean error of 6.03 and 5.56, respectively, and it finds CDR value within average range of ± 0.09 as compared with glaucoma expert. The proposed system achieves average sensitivity, specificity, and accuracies of 94.6, 94.07, and 94.68, respectively.
Keywords: Convolution neural network (CNN), Inner limiting membrane (ILM), Retinal pigmented epithelium (RPE), Cup-to-disc ratio (CDR), Glaucoma
Introduction
Glaucoma is a progressive and degenerative optic neuropathy that causes visual field defects, which further leads to permanent blindness [1]. Glaucoma is an eye condition that occurs due to increase in intraocular pressure (IOP). In early stages of glaucoma, peripheral vision loss occurs, which is difficult for even patient to diagnose. It has been witnessed that significant retinal nerve fiber layer (RNFL) loss occurs approximately 6 years prior to any detectable visual field defects [2, 3]. As the vision loss due to glaucoma is permanent and unobtrusive until the evident visual field defect appears, its timely and accurate diagnosis is always a concern for ophthalmologist [4]. Tonometer, gonioscopy, and fundoscopy are the tests widely used for initial screening of glaucoma. Tonometer is used by ophthalmologists to measure the IOP. Gonioscopy is used to measure the angle between iris and cornea, which amenities an ophthalmologist to determine the glaucoma type. Fundoscopy has been used for interior eye examination; optic nerve head, macula, and peripheral and central retinal regions are being evaluated. Funds image has been employed to investigate initial finding of the ocular disease but does not provide quantitative assessments of retinal layers. Imaging techniques used for the quantitative assessment of retinal layers include confocal scanning laser ophthalmoscopy (CSLO), scanning laser polarimetry (SLP), and optical coherence tomography (OCT). However, SLP measures peripapillary RNFL thickness, CSLO only provides the analysis of optic nerve head (ONH), whereas OCT provides assessments of both RNFL and ONH.
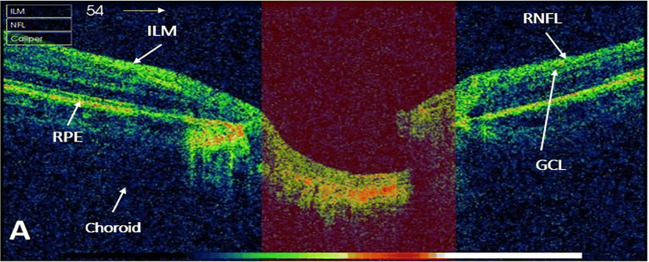
OCT is a noninvasive imaging technology that uses low-coherence light to have a cross-sectional image with higher resolution [5, 6]. OCT technique has a number of clinical applications including cardiology, dermatology, oncology, and gastroenterology [7], but classically has been used in field of ophthalmology. A study [8] presented a systematic review of OCT technology and evaluated its diagnostic accuracy for the glaucoma detection. Glaucoma affects the inner three retinal layers: inner limiting membrane layer (ILM), ganglion cell layer (GCL), and RNFL, as shown in Fig. 1. The loss of ganglion cell resulted into thinning of RNFL, which ultimately raises the cup-to-disc ratio (CDR). Clinical assessment of macula, ONH, GCL, and RNFL has been significant areas for diagnosis of glaucoma [8, 9]. To calculate CDR, firstly cup and disc boundaries have to be found by extracting ILM and RPE layers, respectively [10].
Fig. 1.
An OCT image ONH centered. Highlighted retinal layers which are affected by glaucoma (ILM, RNFL, and GCL)
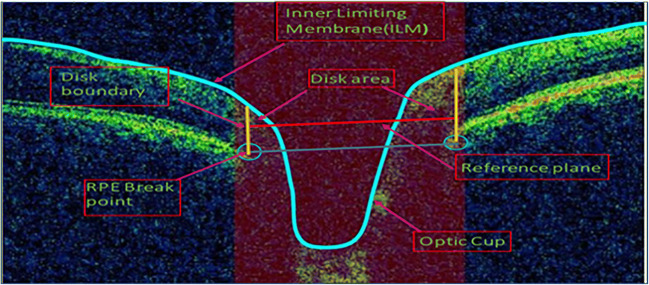
The boundary of the optic disc is defined as the endpoint of the retinal pigment epithelium (RPE) layer, and the line joining these points is the reference plane. Intersection point of ILM layer and a reference plane, which is moved 150 μm above the level of RPE, denotes the cup margins. After identifying the cup and disc borders, their diameters are measured. OCT image of ONH is shown in Fig. 2. ILM and RPE endpoints are marked as blue line and blue circle, respectively. Reference plane at 150 μm is highlighted by red line, and disc boundary is fitted by yellow line in Fig. 2. Finally, using the cup and disc diameter, CDR computation was performed.
Fig. 2.
OCT image ONH centered and showing how to find the CDR by locating the cup and disc border. ILM layer and RPE layer endpoints are marked with blue line and blue circle, respectively. Reference plane (blue line) has been constructed by joining the endpoint of RPE layer; disc boundary was highlighted (yellow line); thus, the length of plane is referred as disc diameter. Cup edges are the intersection point of ILM layer and reference plane (moved 150 μm upward shown in red line); red portion represents the rim area [10]
Convolutional neural network (CNN) has been gaining popularity, thus customized and optimized for various medical imaging applications [9, 11–14]. There are various CNN architectures presented in literature, including LeNet, AlexNet, VGG, ResNet, and Inspection models. In 2014, VGG architecture was proposed and gained popularity as they achieved increased network depth by applying very small convolution filters of dimensions 3 × 3 [15]. Best-performing models presented was VGG-16 and VGG-19 having depth of 16 and 19 layers, respectively. Here, we used the VGG-16 architecture as it has achieved substantial high accuracy on ILSVRC classification. However, this model is also applicable to other image classification tasks. The limitation of the VGG model is the large number of parameters, thus making it hard to train the network on normal machine specifications.
The paper is organized as follows: “Related Work” presents the related work. Materials used in this study are discussed in “Materials.” “Proposed Methodology” presents the proposed methodology for extraction of retinal layer and calculation of cup-to-disc ratio. “Results” articulates the results, and discussion is presented in “Discussion.” Finally, in “Conclusion,” we conclude our study and provide the future directions.
Related Work
A number of automated algorithms have been proposed in literature for segmenting various retinal layers. Automated segmentation of retinal layers helps ophthalmologists in accurate diagnosis of several eye diseases. However, accurate retinal layer extraction is of significant importance for the calculation of various diagnostic parameters such as horizontal rim area, vertical rim area, RNFL thickness, and cup-to-disc ratio.
Literature presented here is divided into three categories based upon the techniques used for the extraction of retinal layers through an OCT image. First category includes studies that used image processing techniques such as intensity variation, edge detection, thresholding, texture analysis [16–24], and graph theory [25–27] for segmenting the layers. Second set covers the related work of classic machine learning algorithms including support vector machine, linear discriminant analysis, and principal component analysis [28–31]. The last group encapsulates the deep learning algorithms based upon CNN for the extraction of retinal layers [32–35].
R. Kromer et al. [21] segmented peripapillary retinal layers through SD-OCT images and compared with manual delineation of layers done by two spectators. Median filter, gray level equalization, and thresholding had been performed for feature extraction. Euclidean distance was used to measure the efficiency of algorithm and was discovered that accuracy of segmented retinal layer highly dependent on image quality and machine time. Fernández et al. [23] employed complex diffusion filtering combined with texture analysis to segment the retinal layers. RPE layer extraction was performed by finding the maximum intensity level, and following peaks correspond to next retinal layers. The spurious edges and discontinuities in the retinal layers were removed by linear interpolation. Tehmina et al. [17] proposed a system that segmented ILM and RPE layers and then computed CDR. A novel technique was proposed to remove outliers and to interpolate missing points. Aneeqa et al. [16] proposed an automated algorithm for the extraction of the ILM layer. They introduced algorithm based on a novel quality valuation technique that takes different parameters for denoising. In addition, a unique time-effective linear interpolation technique was proposed to refine the extracted layers. Hina et al. [18] presented an automated algorithm for the extraction of inner limiting membrane layer especially in noisy OCT images. The wiener filter was applied, followed by thresholding and combination of morphological operation to remove the noise from images.
The drawback of the techniques discussed above is their reliance on specific “ad hoc” rules; thus, they result into poor performance in the various artifacts. Another constraint to these is techniques that their design highly depends upon expert’s analysis. Recent studies have endeavored to address these concerns by exploiting machine learning techniques for segmentation retinal layers in an OCT image. The study [36] reviewed different machine learning algorithms for the glaucoma diagnosis and compared their performance accuracy. The study presented an automated algorithm for delineation of retinal layers in an SD-OCT image by using graph theory dynamic programing and support vector machine; the proposed technique accurately extracted the seven to ten retinal layers [30]. A neural network was proposed, which was used to identify points for each retinal layer that further aids to boundaries estimation of layers [29]. The study applied the recurrent neural network along with the patch-based image classifier and graph search for the boundary extraction of retinal layers through an OCT image [31]. The study [37] presented the fully automated algorithm for dry age-related macular degeneration (AMD) and diabetic macular edema through an OCT image. The multiscale histograms have been used as a feature for support vector machine. Another review-based study [38] presented and compared the diagnostic accuracy of machine learning techniques for the detection of glaucoma.
The problem with the classic machine learning algorithms is that they have to model a lot of constraints such as feature extraction, cost functions, and model parameters. It has been always difficult to design automated algorithm using classic machine learning methods because they need human expertise. Recently, deep learning computational models have achieved tremendous better results in computer vision, speech recognition, genomics, drug discovery, and various other fields [9, 11, 12]. Deep learning learns complicated structure in large data sets even in millions of statistics, and system can train itself, so feature extraction has been performed without explicit instructions. Deep learning approaches are getting attention from various imaging methods in ophthalmology; the study highlighted deep learning techniques and its current applications in the field of optical coherence tomography [38].
The study [33] presented a deep learning technique to segment the surface in volumetric OCT images and AMD diagnosis. Features and transformation were cultured from the training data, and regional approach was employed to learn the local surface profiles. A single CNN was used to learn the surfaces for both normal and diseased images. The method was validated on data set containing 40 OCT volumes from 20 normal and 20 age-related macular degeneration (AMD) subjects. The proposed approach achieved significant improved accuracy compared with graph-based optimal surface segmentation with convex priors (G-OSC). X. Sui et al. proposed multiscale and end-to-end CNN architecture to segment the choroidal boundaries [32] . The approach worked on multiscale and combined the global and local information. The optimal graph-edge weight was updated directly from raw pixels, and method was validated on 912 OCT B-scans and shown that learned graph-edge weights outperformed. L. Fang et al. [34] presented a framework which combines graph search method and convolutional neural networks and for the automatic extraction of nine retinal layer in an OCT images. Algorithms termed as CNN-GS first employed a CNN to extract features for each retinal layer and train a corresponding classifier to have an estimated delineation of eight retinal layers. Furthermore, final boundaries demarcation was done by applying graph search method that used probability maps. The dataset considered the 2915 B-scans (60 volumes) from twenty human eyes of non-exudative AMD patients. The study [35] segmented the multiple retinal surface to detect intermediate AMD using convolution neural network. Single CNN was trained to segment the three retinal layers in one pass for each B-scan and classify them into healthy and intermediate AMD images. The algorithm was validated on 3000 B-scans (50 OCT volumes) acquired from 50 patients including 25 normal and 25 intermediate AMD images. The proposed method was compared with optimal surface segmentation method with convex priors (OSCS) and two deep learning convolution neural networks UNET-1 and UNET-2.
The study [39] developed an algorithm for the glaucoma diagnosis based upon transfer learning of convolution neural network through fundus and OCT images. They utilized findings like disc RNFL thickness map, macular ganglion cell complex (GCC) thickness map, disc RNFL deviation map, and macular GCC deviation map, which were given by commercialized software. Another study presented for glaucoma detection, which directly classify unsegmented OCT volumes into normal and glaucomatous eye using a 3D CNN [40]. They employed 3D convolution in order to have depth information, thus identified the significant region for the detection of glaucoma. The architecture was composed of five 3D convolutional layers with ReLU activation function. Class activation maps were used to extract the features from raw data. The approach had limitations to detect glaucoma through OCT images of older subjects and extreme cases.
The studies presented above are mostly not targeting glaucoma disease using OCT images due to unavailability of benchmarked OCT dataset for glaucoma. The studies [16, 17, 36, 41–44] proposed automated glaucoma detection based upon traditional image processing technique. To our knowledge, very few state-of-the-art studies have been proposed for the glaucoma detection through an OCT image using CNN. Studies like [39, 40] utilized CNN but in a limited scenario as already explained. Furthermore, to the best of our knowledge, no studies have been found which have detected glaucoma by automated retinal layer segmentation using CNN and have utilized CDR value for glaucoma detection.
In this paper, we have proposed an automated system to diagnose glaucoma from OCT images. The proposed system utilizes CNN to extract ILM and RPE. Based on these ILM and RPE, CDR is calculated, which is further used to detect glaucoma using OCT images. One of major contribution of proposed system is that it compliments CNN with structure tensor-based input to have a faster, real-time analysis of test images and then further uses graph search theory [25] to smooth layers generated through CNN. The study also presents the comparison of our work with the algorithms proposed in the literature, which extracted retinal layers using traditional image processing methods [16–18]. We have also compared the CDR diagnostic accuracy of our proposed algorithm to Tehmina et al. [17], which has been simulated on the same data set.
Contributions
Major contributions that we have made in this article are as follows:
We have presented a novel automated framework, designed to detect glaucoma based upon cup-to-disc ratio (CDR) through an OCT image using CNN.
In order to reduce the computational complexity, structure tensor flow has been employed to find the candidate pixel in an OCT image.
To the best of our knowledge, no work has been found that detect glaucoma by automated retinal layer segmentation using CNN and utilized CDR.
The proposed algorithm is robust and able to handle the scan artifacts that are present in OCT scans.
In addition, we have generated a dataset for the detection of glaucoma, which is comprised of 196 OCT image. The dataset will be available for researchers to develop the automated algorithm and find out other structure abnormalities related to glaucoma diagnosis.
Materials
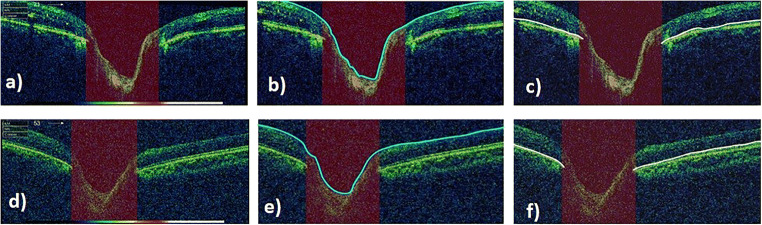
The data used in this work was obtained from Armed Forces Institute of Ophthalmology (AFIO). A subset of dataset, DATA1, is publicly available [45]. In this study, OCT images of human eye is under investigation; hence, OCT images are collected from year 2016 to June 2019. TOPCON’S 3D OCT-1000 camera was employed to acquire OCT images. Frequency to acquire B-scan (1024 A-scans) acquisition is 5 Hz, thus plunging the eye movement effects. Scanning speed was 27,000 to 50,000 A-scans per second with the depth of 2.3 mm. The data set includes both healthy and glaucomatous images with resolution of 951 × 456. Examination is done through dilated pupil with Ø4.0 mm (45°) diameter. The data is divided into two groups, DATA1 and DATA2. DATA1 consists of 50 OCT images from 26 subjects, having 18 healthy and 32 glaucomatous images, whereas DATA2 encompasses a total of 146 OCT images from 75 subjects, including 32 healthy and 114 glaucoma affected images. The dataset includes B-scan OCT images with ONH centered. Original healthy and glaucomatous images from DATA1 are shown in Fig. 3 a and d, respectively. Manual annotation of ILM and RPE layers is performed with the help of ophthalmologist using Illustrator CS6, which can be visualized in Fig. 3b, c, e, f. The dataset is available with original B-scans and also annotated ILM and RPE layers. Other than these, CDR values and final grading with respect to healthy and glaucoma are also given along with original B-scans. Table 1 summarizes the specifications of data collected and then used for this research.
Fig. 3.
OCT images centered at ONH from DATA1. a An image of a health subject. b ILM layer annotation performed by ophthalmologist. c RPE layer annotation done by ophthalmologist. d Glaucoma-affected image and its corresponding ILM and RPE demarcation in e and f
Table 1.
Dataset specifications
| DATA1 | DATA2 | |
|---|---|---|
| Imaging device | TOPCON’S 3D OCT-1000 | TOPCON’S 3D OCT-1000 |
| Pathology | Healthy (H) | Healthy (H) |
| Glaucoma (G) | Glaucoma (G) | |
| Scan reference | Optic nerve head (ONH) centered | Optic nerve head (ONH) centered |
| Scan type | B-scan | B-scan |
| Examination | Dilated pupil with Ø4.0 mm (45°) diameter | Dilated pupil with Ø4.0 mm (45°) diameter |
| Images | 50 OCT images | 146 OCT images |
| Resolution | 951 × 456 | 951 × 456 |
| Subjects | 26 | 75 |
| Categories | 18 H, 32G | 32 H, 114G |
Proposed Methodology
The automated algorithm is proposed to detect glaucoma through an ONH-centered B-scan OCT image. The techniques are designed for the accurate extraction of retinal layers, which will help in computing cup-to-disc ratio. First, patches of ILM, RPE layers, and background are extracted from an OCT image. The extracted patches are fed into CNN to train the network. In testing phase, the structure tensor is applied on OCT image to find out the candidate pixels. Afterwards these candidate pixels help in extraction of patches for retinal layers, which are then passed to pre-trained CNN. The trained CNN classify them into three classes ILM, RPE, and other. Class labels trace the ILM layer and RPE layer, which are further refined by graph search. Finally, cup-to-disc ratio is computed, which is used for glaucoma diagnosis. The block diagram of proposed methodology is shown in Fig. 4.
Fig. 4.
Block diagram of proposed framework for the diagnosis of glaucoma using CNN
Training Phase
Patch Extraction
First step of training is to extract patches from images in each class. The proposed technique employs the extracted patches for training the VGG-16 network. We have designed an illustrating tool in Matlab for extracting the patches from a B-scan OCT image. The tool extracts the patches of size 224 × 224 × 3, which are input dimensions of VGG-16 network architecture. Moving the center of cursor along the image, we can go to the desired point of patch extraction (Fig. 5a). Place the center of cursor on point of interest and a single click will extract the patch. We have extracted the patches for three classes; ILM, RPE layers, and others. Other class includes the patches from background and layers other than ILM and RPE layers. Figure 5 shows the extracted patches for all the classes. Figure 5 b shows ILM layer. RPE layer patch is displayed in Fig. 5d, and patch from other class is seen in Fig. 5c.
Fig. 5.
Patch extraction tool. a Pointer shown, which moves over OCT image to select the point of interest. b ILM layer patch. c Extracted patch for third class (pixels from background and other layers). d Patch extracted from RPE layer
Training Convolution Neural Network: VVG-16
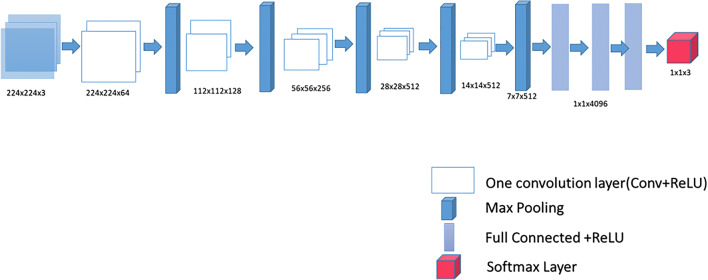
Patch extraction is performed for three classes: ILM layer, RPE layer, and other. The extracted patches for each class are assigned with their corresponding class labels. The proposed methodology utilizes the VGG-16 architecture, which has small convolution filters of dimensions 3 × 3. The convolution filter with stride of one and same padding is applied to all the layers. The number of channels in convolution filters increases as they move deeper into the network. Max pooling applied with stride of two thus reduces the spatial dimensions. Convolutional layers have been followed by three fully connected layers. The first fully connected (FC) layer becomes convolution layer with 4096 filters of size 7 × 7 × 512. The next tailed FC layers have 4096 and 3 filters of size 1 × 1 × 4096. Finally, the layer responsible for classification is SoftMax layer. Table 2 shows the hyperparameters for all layers of VGG-16. A nonlinear activation function ReLU is applied after every convolution and fully connected layers, thus making decision function more discriminating. Figure 6 illustrates the VGG-16 model with 13 convolution layers followed by three fully connected layers (FC). To train the network, the extracted patches for each class along with their class labels are fed into VGG-16. The network trains itself and extracts the features without any explicit information.
Table 2.
Hyperparameters for VGG-16 architecture
| S. No | Layers | Transformation | Filter/stride | Feature map | Activation | Size | Parameters |
|---|---|---|---|---|---|---|---|
| Input | Image | – | – | 1 | – | 224 × 224 × 3 | – |
| 1. | Conv1 | Convolution | 3 × 3/1 | 64 | ReLU | 224 × 224 × 64 | 1792 |
| 2. | Conv2 | Convolution | 3 × 3/1 | 64 | ReLU | 224 × 224 × 64 | 36,928 |
| Pool 1 | Max pooling | 3 × 3/2 | 64 | 112 × 112 × 64 | 0 | ||
| 3. | Conv3 | Convolution | 3 × 3/1 | 128 | ReLU | 112 × 112 × 128 | 73,856 |
| 4. | Conv4 | Convolution | 3 × 3/1 | 128 | ReLU | 112 × 112 × 128 | 147,584 |
| Pool 2 | Max pooling | 3 × 3/2 | 128 | 56 × 56 × 128 | 0 | ||
| 5. | Conv5 | Convolution | 3 × 3/1 | 256 | ReLU | 56 × 56 × 256 | 295,168 |
| 6. | Conv6 | Convolution | 3 × 3/1 | 256 | ReLU | 56 × 56 × 256 | 590,080 |
| 7. | Conv7 | Convolution | 3 × 3/1 | 256 | ReLU | 56 × 56 × 256 | 590,080 |
| Pool 3 | Max pooling | 3 × 3/2 | 256 | 28 × 28 × 256 | 0 | ||
| 8. | Conv8 | Convolution | 3 × 3/1 | 512 | ReLU | 28 × 28 × 512 | 1,180,160 |
| 9. | Conv9 | Convolution | 3 × 3/1 | 512 | ReLU | 28 × 28 × 512 | 2,359,808 |
| 10. | Conv10 | Convolution | 3 × 3/1 | 512 | ReLU | 28 × 28 × 512 | 2,359,808 |
| Pool 4 | Max pooling | 3 × 3/2 | 512 | 14 × 14 × 512 | 0 | ||
| 11. | Conv11 | Convolution | 3 × 3/1 | 512 | ReLU | 14 × 14 × 512 | 2,359,808 |
| 12. | Conv12 | Convolution | 3 × 3/1 | 512 | ReLU | 14 × 14 × 512 | 2,359,808 |
| 13. | Conv13 | Convolution | 3 × 3/1 | 512 | ReLU | 14 × 14 × 512 | 2,359,808 |
| Pool 5 | Max pooling | 3 × 3/2 | 512 | 7 × 7 × 512 | 0 | ||
| 14. | FC (flatten) | Fully connected | – | ReLU | 25,088 | 0 | |
| 15. | FC (dense) | Fully connected | – | ReLU | 4096 | 102,764,544 | |
| 16. | FC (dense) | Fully connected | – | ReLU | 4096 | 16,781,312 | |
| Output | FC | Fully connected | – | SoftMax | 3 | 4,097,000 |
Fig. 6.
Illustration of convolutional neural networks architecture VGG-16 with sixteen convolution layers, five Max pooling transformations, three fully connected layers, and one SoftMax layer
Testing Phase
In testing phase, the aim is to extract all ILM and RPE layer pixels to construct these layers. The proposed testing phase is fully automated, and in order to support already trained CNN and to reduce computational time, we introduce structure tensors to extract candidate layer pixels. As shown in Fig. 4, in testing phase, candidate pixels of test OCT image are extracted by 2D structure tensor. The tensor with highest degree of coherence is determined to obtain the maximum information of retinal layers. Afterwards, patches are extracted for each candidate pixel from an OCT image, and these patches are fed into pretrained CNN model for classification into any of three proposed classes, which are ILM, RPE, and background. The CNN model assigns the class labels along with class probabilities to each patch. The ILM and RPE layers extracted from CNN are not refined, having some missing and false points. So, the interpolation of missing points and removal of outliers are performed by graph search. After the demarcation of ILM and RPE layer, cup and disc are located. Finally, CDR is calculated to classify the OCT image into healthy or glaucoma-affected subject.
Candidate Pixels Extraction Through Structure Tensor
In testing phase, two-dimensional discrete structure tensor is applied to find the candidate pixels of an OCT image [46]. The purpose of introducing this phase in proposed scheme is to reduce overall layer extraction time during testing phase. This module extracts all possible candidate layer pixels that will eventually be passed to pretrained CNN module. In the proposed structure tensor, the degrees of coherency within candidate matrix are measured to get more information about the image. The structure tensor calculates non-overlapping orthogonal tensors by using Eqs. (1-4).
| 1 |
| 2 |
| 3 |
| 4 |
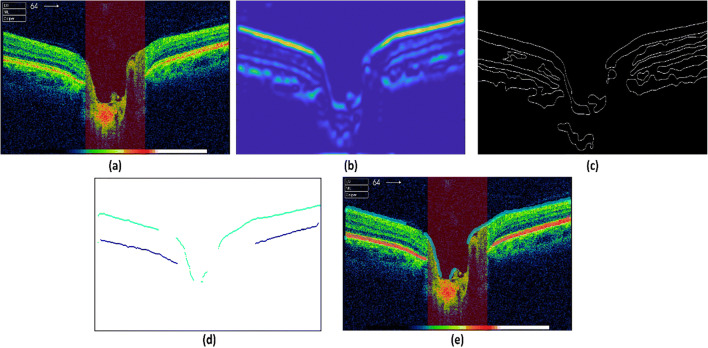
where δ is the 2nd order tensor and w(xi, yj) represents the localized Gaussian filter. and represent the square of horizontal image and vertical gradients, respectively. The product of vertical and horizontal gradients is represented by TXY and TYX; directional derivatives of the candidate are donated as TϕX, TϕY, and TϕXY. Each tensor describes the respective foremost orientation of the retinal layers in an OCT image. Tensor with maximum coherency encompasses most of the information in an image. For each tensor, degree of coherency is measured through eigen values, and the tensor with maximum degree is selected. These provide us with candidate layer pixels for next stage. Figure 7 a, b, and c show original OCT scan, layers highlighted using most coherent structure tensors, and candidate layer pixels, respectively.
Fig. 7.
Extraction of ILM and RPE. a Original OCT B scan. b Enhanced layers using most coherent structure tensor. c Candidate layer pixels after applying adaptive thresholding on structure tensor layers. d ILM and RPE pixels extracted after applying trained CNN models on all candidate layer pixels. e Original OCT B scan with super imposed ILM and RPE layers
Patch Classification by CNN: VGG-16 Model
The proposed framework extracts patches of size 224 × 224 × 3 for each pixel in an OCT image. As we have already mentioned, instead of extraction patches from the entire image, we have extracted patches for candidate pixels that are highlighted by the application of structure tensors. Further, the extracted patches are fed into pretrained VGG-16 network to classify them into three categories: ILM layer (0), RPE layer (1), and others (2). Figure 7 d and e show ILM and RPE layers extracted after applying CNN and then super imposing them on original OCT, respectively.
Graph Search
The layers extracted from structure tensor and CNN do have some missing pixels, and sometimes they do have outliers as well (Fig. 7d). To cater these things, the graph search is applied to refine retinal layer boundaries. The ILM and RPE layer pixels are decomposed into an undirected graph node for tracing the ILM and RPE layers. Interpolation of missing points is performed by the graph search and dynamic programing as shown in Fig. 7.
A directed graph is constructed for each layer, where graph vertices correspond to the pixels in layer. Each vertex is contacted to its neighbors, and weights to these connections are updated by Eq. 5.
| 5 |
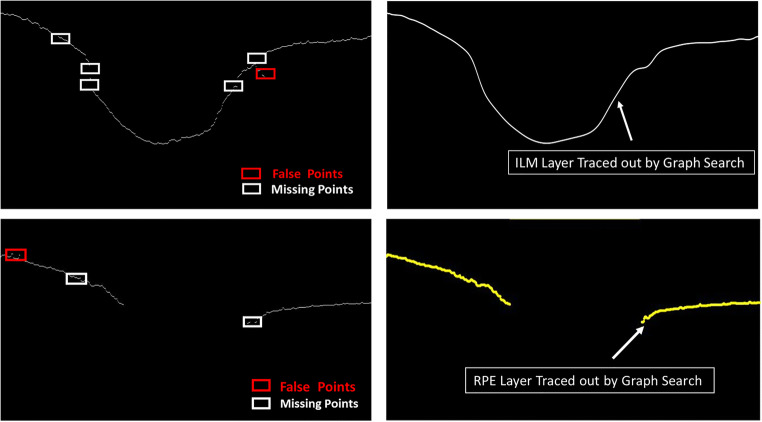
where wmin is minimum possible weight, and PS and PD are source and destination vertice values. A column with maximum intensity is appended to right and left of an image to automate initialization. Each column vertex is vertically connected from top to bottom. The Dijkstra’s shortest path algorithm is used to find the optimal path and that will be the predicted location of retinal boundary. Figure 8 demonstrates the graph search algorithm; the ILM layer, RPE layer, and missing points are denoted by yellow, red, and black boxes, respectively. The confused pixel is represented by a blue box and the foreground pixel as purple box. The foreground or confused pixel connects to the layer with shortest distance found. The shortest distance (i.e., acceptable distance) is marked in the green color arrows and unacceptable distance by the peach color arrows, in Fig. 8b. Interpolated ILM and RPE layers after applying the graph search are displayed in Fig. 8c. Figure 9 shows the extracted ILM and RPE layers through an OCT image from DATA1. Graph search applied on extracted retinal layers, interpolated ILM and RPE layers, is shown in Fig. 9b, d.
Fig. 8.
Interpolating of missing points of retinal layers by graph search. ILM and RPE layers are shown by yellow and red boxes, respectively. Black boxes are referred as missing pixels in ILM and RPE layers. Dijkstra’s shortest path algorithm has been used to find the optimal distance for particular pixels; acceptable distance is represented as green color, and unacceptable distance is indicated by peach color
Fig. 9.
Extracted retinal layers through an OCT image from DATA1. White boxes represent the missing point, and red boxes identified the outliers on ILM layer (a) and RPE layer (c). ILM and RPE interpolated by graph search algorithm are shown in b and d, respectively
Calculation of CDR
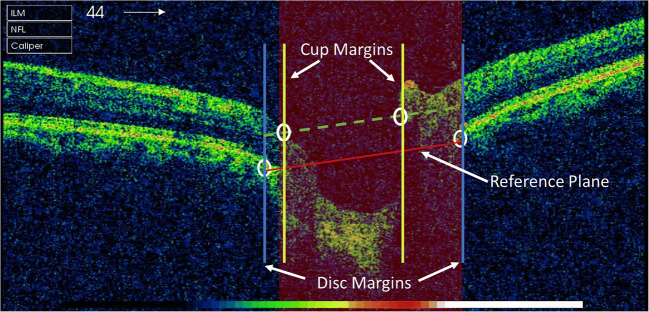
The cup-to-disc ratio is a significant investigating factor for diagnosis of glaucoma. After the extraction of retinal layers, next step is to locate the cup and disc margins. The inner limiting membrane and retinal pigmented epithelium layer correspond to the cup and disc boundaries, respectively. First, we have to find the endpoints of RPE layer that will demarcate the disc margins. The distance between the endpoints of RPE will determine the disc diameter (DS_d). To find the cup boundaries, there is a need of reference plane. As the RPE layer is not affected by glaucoma progression, the reference plane positioned on RPE point would be stable. The line joining the endpoints of RPE acts as the reference plane. The intersection points of ILM layer and the reference plane, which is moved 150 μm above the RPE level, are considered the cup margins. The distance between the cup margins is cup diameter (CP_d). To calculate cup-to-disc ratio, we have used Eq. (6). Figure 10 shows the reference plane marked with red line with cup and disc margins demarcated with yellow and blue lines. By following clinical rules, the CDR value < 0.5 is considered healthy, whereas ≥ 0.5 is classified as glaucomatous image [47].
| 6 |
Fig. 10.
An OCT image centered on optic nerve head; the reference plane (red line) is constructed by joining the endpoints of RPE layer, and disc margins are represented by blue vertical line. As we move reference plane 150 μm above RPE layer, cup edges are located at intersection point of plane, and ILM layer and cup margins are marked as yellow vertical lines
Results
To validate the effectiveness of our proposed frame work, we have used 196 B-scans OCT images of 101 subjects as explained in “Materials.” The data was obtained from Armed Forces Institute of Ophthalmology (AFIO). Manual annotation of ILM and RPE layers was performed with the help of ophthalmologist using Illustrator CS6. The proposed extraction tool has been used to extract the patches of size 224 × 224 from each OCT image. We extracted 600 patches per B-scan i.e. 200, 150, and 250 patches of ILM, RPE, and background, respectively. A total of 117,600 patches are extracted, including 39,200 patches of ILM layer, 29,400 of RPE layer, and 49,000 patches for others. From the dataset, we randomly selected 80% of data that is 94,080 patches for training and remaining 20% of data containing 23,520 patches for testing of our proposed methodology. In training phase, average number of patches in one batch is 940, and VGG-16 network is trained for 50 epochs. The learning and weight decay rates and other hyperparameters are kept at their default values of VGG-16. The evaluation of proposed system is carried out on a system with Core i7-8700 with3.2GHz, 32GB RAM, and 1080Ti Nvidia GPU with 8GB and 256GB SSD.
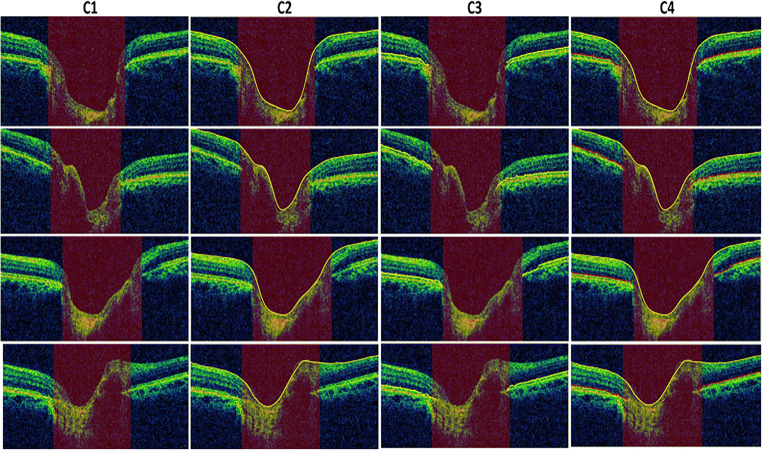
The structure tensor is applied to extract the candidate pixels that have maximum amount of information regarding the retinal layers. The next step in proposed technique is to extract patch of the candidate pixels from OCT image. The pertained CNN model is fed with extracted patches, and it assigns class labels and class probabilities to each input patch of an image. The boundaries traced by proposed techniques are as shown in Fig. 11. Graph search algorithm interpolated the missing points and also removed false portions on the retinal layers; extracted ILM and RPE layers are traced out in Fig. 11 (C2) and (C3). It is evident from Fig. 11 that the proposed technique accurately extracted the ILM and RPE layers.
Fig. 11.
Retinal layer segmentation. Column C1 showing original B-scan OCT images. Extracted ILM and RPE layers (yellow color) are shown in columns C2 and C3. Both ILM (marked as yellow color) and RPE (highlighted as red color) layers are traced out on original image
We have performed evaluation of proposed system at different stages i.e. layer extraction, CDR calculation, and finally glaucoma detection. To evaluate the performance of our proposed method, difference has been calculated between manually annotated and automatically labeled ILM and RPE layers. The errors are measured by Euclidean distance formula (Eq. 7) for each patch, and then their mean is calculated for each OCT image. Next, absolute mean error for all the images has been determined for both datasets DATA1 and DATA2, presented in Table 3.
| 7 |
where i and j are the x and y coordinates of extracted retinal layer (s) and the manually annotated layer (m), respectively.
Table 3.
Evaluation of extracted retinal layers
| Dataset | Layers | Mean difference |
|---|---|---|
| DATA1 | Inner limiting membrane (ILM) | 6.056842 ± 2.03 |
| Retinal pigmented epithelium (RPE) | 5.775647 ± 3.03 | |
| DATA2 | Inner limiting membrane (ILM) | 6.002345 ± 2.45 |
| Retinal pigmented epithelium (RPE) | 5.345677 ± 3.23 |
The proposed model diagnosed glaucoma based upon the value of CDR. The system was able to correctly classify 17 out of 18 normal cases and 30 out of 32 glaucomatous cases in DATA1. In DATA2, proposed algorithm correctly detects 31 out of 32 normal images and 109 out of 114 glaucomatous images. The confusion matrix has been filled up for both the datasets and is shown in Table 4. We have also measured the accuracy, specificity, sensitivity, precision, and F-score of the presented work; all the performance metrics are seen in Table 5.
Table 4.
Confusion matrices for data1 and data2
| DATA1 | DATA2 | ||||
|---|---|---|---|---|---|
| Healthy (H) | Glaucoma (G) | Healthy (H) | Glaucoma (G) | ||
| Healthy (H) | 17 | 1 | Healthy (H) | 30 | 2 |
| Glaucoma (G) | 2 | 30 | Glaucoma (G) | 5 | 109 |
Table 5.
Performance metrics for methodology
| Dataset | Images | CDR mean variation | CDR standard deviation | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F-score |
|---|---|---|---|---|---|---|---|---|
| DATA1 | 50 B-scan OCT | 0.094 | 0.081 | 94 | 94.4 | 93.75 | 94.44 | 0.9188 |
| DATA2 | 146 B-scan OCT | 0.089 | 0.094 | 95.20 | 93.75 | 95.61 | 93.75 | 0.8954 |
The proposed system fails in some cases where layers are not present properly due to noise and its almost hard to find candidate layer pixels. Figure 12 shows a couple of failure cases, where proposed algorithm is unable to extract the retinal layers. It is clear from these cases that layers are not properly visible, which makes it difficult for algorithm to extract layer pixels.
Fig. 12.
Showing the failure cases of proposed methodology, unable to extract retinal layer in images having missing structural information due to high speckle noise. a OCT image from DATA2. b, c Extracted layer by proposed algorithm. d OCT image from DATA2. e, f Extracted layer by proposed algorithm
Discussion
As there is no publicly available dataset for glaucoma and the published works related to automatic detection of glaucoma worked on their own privately collected datasets, it is difficult for fair comparison of different automated algorithms proposed to detect glaucoma. So, here we are comparing this proposed technique with pervious published works [16–18]. The used data in previous studies is a subset of dataset used in this work; therefore, we have divided our data into two sets DATA1 and DATA2. DATA1 is the same dataset that has been used in previous work containing 50 images, whereas DATA2 have different 146 images. Tables 6 and 7 present the comparison of our proposed methodology with other studies, and our work showed improved results.
Table 6.
Performance evaluation of extracted ILM and RPE layers
Table 7.
Comparison of algorithms to detect glaucoma on the basis of CDR
| Dataset | Images | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| Tehmina [17] | 50 B-scan OCT | 86 | 81 | 94 |
| Proposed method | 50 B-scan OCT DATA1 | 94 | 94.4 | 93.75 |
The studies [12, 13, 20] are based upon traditional image processing techniques including histogram equalization, thresholding, and morphological operations. However, these studies are highly dependent on image quality and other artifacts. There are a number of parameters and threshold that need to be adjusted before application of these methods. Whereas, the proposed methodology resolves this problem by employing deep convolution neural network that learns from each OCT image in training set. A CNN organizes series of transforming layers to learn and classify the retinal layers from an image without any explicit information. The proposed technique reduces the dependence on image quality and effect of other artifacts thus increasing the algorithm robustness. Structure tensors are applied to find the candidate pixel to get the region of interest, thus reducing the image size of further processing. Employing structure tensor proposed framework able to reduce the computational complexity and processing time. The training time is 5 h and 45 min, whereas test OCT image takes 30 s to extract the retinal layers and classify it into heathy or glaucomatous image. The proposed framework has achieved accuracy of 94% and 95.20% for datasets DATA1 and DATA2, respectively. As the dataset is unbalanced, other performance metrics have been measured to evaluate the proposed algorithm. The proposed framework obtained high precision for both datasets, 94.44% for DATA1 and 93.75% for DATA2. Furthermore, algorithm has high F-score for datasets DATA1 and DATA2. The proposed method has achieved significantly improved results in terms of retinal layers extraction and classification as compared with state-of-the art studies reported in literature. It is observed from Tables 6 and 7 that proposed framework has better performance in terms of mean error, accuracy, specificity, and sensitivity as compared with existing techniques.
Conclusion
In this paper, we have presented a method for glaucoma diagnosis through a B-scan OCT image. The diagnostic parameter considered for detection of glaucoma was cup-to-disc ratio. To calculate CDR, first ILM and RPE layers are extracted. A single CNN is used to train the model for both normal and glaucoma-affected data. Inner limiting membrane and retinal pigmented epithelium layers extracted by proposed method are very close to manual annotation performed by the ophthalmologist. The proposed techniques achieved robustness by reducing the dependence on image quality and other artifacts’ effects. The results of the proposed technique showed significant increase in performance as compared with previous state-of-the-art approaches. Hence, our proposed system will help make accurate diagnosis of glaucoma from an OCT image.
The methodology presented is not limited to the modality of glaucoma, rather this work can be applicable for diagnosis of other ocular abnormalities especially in optic nerve head region. This methodology can work for other ocular diseases by training using that particular diseased data and slight modifications. The proposed model to segment the retinal layers can be extended to higher dimensional data such as OCT volumes.
Acknowledgments
The data was provided by the Armed Forces Institute of Ophthalmology (AFIO), Pakistan, and the research was carried out under Biomedical Image/Signal Analysis (BIOMISA) Research Lab in NUST, Islamabad, Pakistan.
Funding
Higher Education Commission (HEC) Pakistan funded this researcher. This research was co-funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Approval was obtained from the ethics committee of NUST College of Electrical and Mechanical Engineering, Pakistan. The procedures used in this study adhere to the tenets of the Declaration of Helsinki.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent to Publish
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/25/2021
A Correction to this paper has been published: 10.1007/s10278-021-00468-9
Contributor Information
Hina Raja, Email: hinaraja65@gmail.com.
M. Usman Akram, Email: usman.akram@ceme.nust.edu.pk.
Arslan Shaukat, Email: arslan.asp@gmail.com.
Shoab Ahmed Khan, Email: drshoabakhan@gmail.com.
Norah Alghamdi, Email: dr.norah.alghamdi@hotmail.com.
Sajid Gul Khawaja, Email: sajid.gul@ceme.nust.edu.pk.
Noman Nazir, Email: noman.nazir@gmail.com.
References
- 1.Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: a review. JAMA. 2014;311(18):1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeoung JW, Park KH. OCT on the ability to detect localized retinal nerve fiber layer defects in preperimetric glaucoma. Invest Ophthalmol Vis Sci. 2010;51:938–945. doi: 10.1167/iovs.08-3335. [DOI] [PubMed] [Google Scholar]
- 3.Sommer A, Miller NR, Pollack I, Maumenee AE, George T. The nerve fiber layer in the diagnosis of glaucoma. Arch Ophthalmol. 1977;95(12):2149–56. doi: 10.1001/archopht.1977.04450120055003. [DOI] [PubMed] [Google Scholar]
- 4.Khan KB, Khaliq AA, Jalii A, Iftikhar MA, Ullah N, Aziz MW, Ullah K, Shahid M. A review of retinal blood vessels extraction techniques: challenges, taxonomy, and future trends. Pattern Anal Applic. 2019;22(3):767–802. doi: 10.1007/s10044-018-0754-8. [DOI] [Google Scholar]
- 5.Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, Chang W, Hee MR, Flotte T, Gregory K, Puliafito CA, Fujimoto JG. Optical coherence tomography. Science. 1991;254(5035):1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D. W and F. JG. State-of-the-art retinal optical coherence tomography. Prog Retin Eye Res. 2008;27(1):45–88. doi: 10.1016/j.preteyeres.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Zysk AM, Nguyen FT, Oldenburg AL, Marks DL, Boppart SA. Optical coherence tomography: a review of clinical development from bench to bedside. J Biomed Opt. 2007;12(5):051403. doi: 10.1117/1.2793736. [DOI] [PubMed] [Google Scholar]
- 8.Wollstein G, Ishikawa H, Wang J, Beaton SA, Schuman JS. Comparison of three optical coherence tomography scanning areas for detection of glaucomatous damage. Am J Ophthalmol. 2005;139:39–43. doi: 10.1016/j.ajo.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 9.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 10.Wang YP, Chen Q, Lu ST: Quantitative assessments of cup-to-disk ratios in spectral domain optical coherence tomography images for glaucoma diagnosis. In 2013 6th International Conference on Biomedical Engineering and Informatics, China, 2013.
- 11.Mohammadimanesh F, Salehi B, Mahdianpari M, Gill E, Molinier M. A new fully convolutional neural network for semantic segmentation of polarimetric SAR imagery in complex land cover ecosystem. ISPRS J Photogramm Remote Sens. 2019;151:223–236. doi: 10.1016/j.isprsjprs.2019.03.015. [DOI] [Google Scholar]
- 12.Staar B, Lütjen M, Freitag M. Anomaly detection with convolutional neural networks for industrial surface inspection. Procedia CIRP. 2019;79:484–489. doi: 10.1016/j.procir.2019.02.123. [DOI] [Google Scholar]
- 13.Singadkar G, Mahajan A, Thakur M, Talbar S. Deep deconvolutional residual network based automatic lung nodule segmentation. J Digit Imaging. 2020;3:678–684. doi: 10.1007/s10278-019-00301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. Zhao, C. Zhang, D. Li and J. Niu, "Combining multi-scale feature fusion with multi-attribute grading, a CNN model for benign and malignant classification of pulmonary nodules," J Digit Imaging, 2020. [DOI] [PMC free article] [PubMed]
- 15.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In International Conference on Learning Representations, Banff, Canada, 2015
- 16.Ramzan A, Akram MU, Salam A, Ramzan J, Mubarak Q, Yasin AUU: Automated inner limiting membrane segmentation in OCT retinal images for glaucoma detection. In: IEEE 2018 Computing Conference, London, UK, 2018
- 17.Khalil T, Akram MU, Raja H, Jameel A, Basit I. Detection of glaucoma using cup to disc ratio from spectral domain optical coherence tomography images. IEEE Access. 2018;6:4560–2576. doi: 10.1109/ACCESS.2018.2791427. [DOI] [Google Scholar]
- 18.Raja H, Akram MU, Ramzan A, Khalil T, Aziz AHR: A framework for extraction of inner limiting membrane in high speckle noisy images. In IEEE 6th International Conference on Control, Decision and Information Technologies, Paris, France, 2019
- 19.Ishikawa H, Stein D, Wollstein G, Beaton S, Fujimoto J, Schuman J. Macular segmentation with optical coherence tomography. Invest Ophthalmol Vis Sci. 2005;46(6):2012–7. doi: 10.1167/iovs.04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahidi M, Wang Z, Zelkha R. Quantitative thickness measurement of retinal layers imaged by optical coherence tomography. Am J Ophthalmol. 2005;139:1056–61. doi: 10.1016/j.ajo.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Kromer R, Rahman S, Filev F, Klemm M. An approach for automated segmentation of retinal layers in peripapillary spectralis SD-OCT images using curve regularisation. Insights Ophthalmol. 2017;1(7):1–6. [Google Scholar]
- 22.George A, Dillenseger J, Weber A, Pechereau A. Optical coherence tomography image processing. Invest Ophthalmol Vis Sci. 2000;41:165–73. [Google Scholar]
- 23.Fernández DC, Salinas HM, Puliafito CA. Automated detection of retinal layer structures on optical coherence tomography images. Opt Express. 2005;13(25):10200–10216. doi: 10.1364/OPEX.13.010200. [DOI] [PubMed] [Google Scholar]
- 24.A.González-López, J. Moura, J.Novo, M.Ortega and M.G.Penedo, "Robust segmentation of retinal layers in optical coherence tomography images based on a multistage active contour model," Heliyon, vol. 5, no. 2, p. e01271, 2019. [DOI] [PMC free article] [PubMed]
- 25.Chiu SJ, Li XT, Nicholas P, Toth CA, Izatt JA, Farsiu S. Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation. Opt Express. 2010;18(18):19413–19428. doi: 10.1364/OE.18.019413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garvin MK, Abramoff MD, Kardon R, Russell SR, Wu X, Sonka M. Intraretinal layer segmentation of macular optical coherence tomography images using optimal 3-D graph search. IEEE Trans Med Imaging. 2008;27(10):1495–1505. doi: 10.1109/TMI.2008.923966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garvin MK, Abramoff MD, Wu X, Russell SR, Burns TL, Sonka M. Automated 3-D intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE Trans Med Imaging. 2009;28(9):1436–1447. doi: 10.1109/TMI.2009.2016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang A, Carass A, Hauser M, Sotirchos ES, Calabresi PA, Ying HS, Prince JL. Retinal layer segmentation of macular OCT images using boundary classification using boundary classification. Biomed Opt Express. 2013;4(7):1133–1152. doi: 10.1364/BOE.4.001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonough K, Kolmanovsky I, Glybina IV: A neural network approach to retinal layer boundary identification from optical coherence tomography images. In 2015 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Niagara Falls, ON, Canada, 2015
- 30.Srinivasan PP, Heflin SJ, Izatt JA, Arshavsky VY, Farsiu S. Automatic segmentation of up to ten layer boundaries in SD-OCT images of the mouse retina with and without missing layers due to pathology. Biomed Opt Express. 2014;5(2):348–365. doi: 10.1364/BOE.5.000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kugelman J, Alonso-Caneiro D, Read SA, Vincent SJ, Collins MJ. Automatic segmentation of OCT retinal boundaries using recurrent neural networks and graph search. Biomed Opt Express. 2018;9(11):5759–5777. doi: 10.1364/BOE.9.005759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sui X, Zheng Y, Bi BWH, Wu JF, Pan X, Yin Y, Zhang S. Choroid segmentation from Optical Coherence Tomography with graph-edge weights learned from deep convolutional neural networks. Neurocomputing. 2017;237:332–341. doi: 10.1016/j.neucom.2017.01.023. [DOI] [Google Scholar]
- 33.Shah A, Abramoff MD, Wu X. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. 2017. Simultaneous multiple surface segmentation using deep learning; pp. 3–11. [Google Scholar]
- 34.Fang L, David Cunefare CW, Guymer RH, Li S, Farsiu S. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed Opt Express. 2017;8(5):2732–2744. doi: 10.1364/BOE.8.002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah A, Zhou L, Abrámoff MD, Wu X. Multiple surface segmentation using convolution neural nets: application to retinal layer segmentation in OCT images. Biomed Opt Express. 2018;9(9):4509–4526. doi: 10.1364/BOE.9.004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M. Usman, M. M. Fraz and S. A. Barman, "Computer vision techniques applied for diagnostic analysis," Arch Comput Methods Eng, pp. 1-17, 2016.
- 37.Srinivasan PP, Kim LA, Mettu PS, Cousins SW, Comer GM, Izatt JA, Farsiu S. Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images. Biomed Opt Express. 2014;5(10):3568–3577. doi: 10.1364/BOE.5.003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Treder M, Eter N. Deep learning and neuronal networks in ophthalmology: applications in the field of optical coherence. Ophthalmologe. 2018;115(9):714–721. doi: 10.1007/s00347-018-0706-0. [DOI] [PubMed] [Google Scholar]
- 39.G. An, K. Omodaka, K. Hashimoto, S. Tsuda, Y. Shiga, N. Takada, T. Kikawa, H. Yokota, M. Akiba and T. Nakazawa, "Glaucoma diagnosis with machine learning based on optical coherence tomography and color fundus images," J Healthc Eng, vol. 2019, pp. ID 4061313, 9, 2019. [DOI] [PMC free article] [PubMed]
- 40.S. Maetschke, B. Antony, H. Ishikawa, G. Wollstein, J. Schuman and R. Garnavi, "A feature agnostic approach for glaucoma detection in OCT volumes," PLoS ONE 1, 2019. [DOI] [PMC free article] [PubMed]
- 41.Babu T, Devi SS, Venkatesh R. Optic nerve head segmentation using fundus images and optical coherence tomography images for glaucoma detection. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2015;159(4):607–615. doi: 10.5507/bp.2015.053. [DOI] [PubMed] [Google Scholar]
- 42.Nithya R, Venkateswaran N. Analysis of segmentation algorithms in colour fundus and OCT images for glaucoma detection. Indian J Sci Technol. 2015;8(24):1–6. doi: 10.17485/ijst/2015/v8i24/80151. [DOI] [Google Scholar]
- 43.Babu T, Devi SS, Venkatesh R. Automatic detection of glaucoma using optical coherence tomography image. J Appl Sci. 2012;12(20):2128–2138. doi: 10.3923/jas.2012.2128.2138. [DOI] [Google Scholar]
- 44.Rajan A, Ramesh G. Automated early detection of glaucoma in wavelet domain using optical coherence tomography images. Biosci Biotechnol Res Asia. 2015;12(3):2821–2828. doi: 10.13005/bbra/1966. [DOI] [Google Scholar]
- 45.Raja H, Akrama MU, Khawaja SG, Arslan M, Ramzan A. Data on OCT and fundus images for the detection of glaucoma. Data Brief. 2020;29:105342. doi: 10.1016/j.dib.2020.105342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hassan B, Raja G, Hassan T, Akram MU. Structure tensor based automated detection of macular edema and central serous retinopathy using optical coherence tomography images. J Opt Soc Am A USA. 2016;33(4):455–63. doi: 10.1364/JOSAA.33.000455. [DOI] [PubMed] [Google Scholar]
- 47.Garway-Heath DF, Ruben ST, Viswanathan A, Hitchings RA. Vertical cup/disc ratio in relation to optic disc size: its value in the assessment of the glaucoma suspect. Br J Ophthalmol. 1998;82(10):1118–1124. doi: 10.1136/bjo.82.10.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]