Abstract
Microvascular invasion (mVI) is the most significant independent predictor of recurrence for hepatocellular carcinoma (HCC), but its pre-operative assessment is challenging. In this study, we investigate the use of multi-parametric MRI radiomics to predict mVI status before surgery. We retrospectively collected pre-operative multi-parametric liver MRI scans for 99 patients who were diagnosed with HCC. These patients received surgery and pathology-confirmed diagnosis of mVI. We extracted radiomics features from manually segmented HCC regions and built machine learning classifiers to predict mVI status. We compared the performance of such classifiers when built on five MRI sequences used both individually and combined. We investigated the effects of using features extracted from the tumor region only, the peritumoral marginal region only, and the combination of the two. We used the area under the receiver operating characteristic curve (AUC) and accuracy as performance metrics. By combining features extracted from multiple MRI sequences, AUCs are 86.69%, 84.62%, and 84.19% when features are extracted from the tumor only, the peritumoral region only, and the combination of the two, respectively. For tumor-extracted features, the T2 sequence (AUC = 80.84%) and portal venous sequence (AUC = 79.22%) outperform other MRI sequences in single-sequence-based models and their combination yields the highest AUC of 86.69% for mVI status prediction. Our results show promise in predicting mVI from pre-operative liver MRI scans and indicate that information from multi-parametric MRI sequences is complementary in identifying mVI.
Keywords: Hepatocellular carcinoma, Microvascular invasion, Radiomics, Machine learning, MRI
Introduction
Liver cancer was predicted to be the sixth most diagnosed cancer and the fourth major cause of death due to cancer worldwide in 2018 [1]. Hepatocellular carcinoma (HCC) is the most common primary liver cancer, accounting for 75–85% of total cases [1, 2], and its incidence rates have also been steadily increasing since 2000 and are projected to increase further until 2030 [3]. In the United States, 1-year survival for HCC is lower than 50% [4], and median survival was estimated to be 11 months in a German population [5]. Treatment of HCC can be surgical (i.e., liver resection or transplantation), locoregional (e.g., chemoembolization), or systemic (e.g., immunotherapy) [6] and the Barcelona Clinic Liver Cancer (BCLC) system [7] offers a way to assign a treatment option to each HCC case. The treatment of choice for patients without cirrhosis is resection [6, 8], while liver transplantation can treat both tumor and the underlying hepatic condition. Survival rates after liver transplantation can be as high as 75% (at 4 years) with recurrence below 15% in patients meeting the Milan criteria [9] (i.e., one HCC of 5 cm or smaller, or up to three nodules smaller than 3 cm), while recurrence after resection can be as high as 70% at 5 years [10].
Predicting recurrence can inform treatment selection as well as liver allocation, and many independent predictors of recurrence have been analyzed in the literature. For example, the biological markers neutrophil-lymphocyte ratio (NLR) and alpha-fetoprotein (AFP) and tumor size were found superior to the Milan criteria in predicting recurrence [11]. In addition, bad tumor differentiation and previous hepatectomy were found to be risk factors for recurrence [12]. Nonetheless, studies show that the most important predictor of recurrence is microvascular invasion (mVI) [13].
Unfortunately, pre-operative assessment of mVI is difficult since mVI cannot be reliably diagnosed by needle biopsy [14]. Serum markers like AFP are currently used to predict mVI [8], but AFP lacks sensitivity and specificity in detecting HCCs [15] and assessing mVI [16]. Tumor size and nodule number can be determined pre-operatively and are also useful to predict mVI [17], but size alone cannot predict survival; recurrence-free survival rates can be greater than 80% for tumors larger than 5 cm if no vascular invasion is found [18]; tumors larger than 10 cm can be resected achieving 64.8% 1-year survival if no vascular invasion is present (vs. 28% if vascular invasion is found) [18].
Liver imaging such as MRI and CT (especially with the use of contrast agents) and its visual assessment by radiologists revealed that some image features are correlated with mVI [19]. For example, non-smooth tumor margins [20–22] and an incomplete tumor capsule [22, 23] are predictive of mVI, together with tumoral and peritumoral enhancement in the arterial phase of contrast-enhanced (CE) MRI [23, 24] or CT [20] and peritumoral hypointensity in the hepatobiliary phase of CE-MRI [24, 25]. Recently, a few studies showed that a set of pre-defined radiomics/texture features extracted from the liver tumor region are also associated with mVI. These features may come from CT [26] or gadoxetic acid-enhanced MRI [27–29], and the results appear to be preliminary but encouraging.
In this work, we investigated a radiomics-based approach for pre-operative mVI detection from gadopentetic acid (Gd-DTPA)-enhanced MRI. Different from previous work, we studied five MRI sequences (i.e., diffusion-weighted imaging, T1, T2, late arterial phase, and portal venous phase) and compared their effects when used individually or combined; furthermore, we investigated the use of imaging features extracted from not only the tumor region but also its peripheral region. Our results show that combining different MRI sequences is beneficial for mVI prediction.
Materials and Methods
Study Cohort and Dataset
With institutional review board approval and waiver of the written informed consent from patients, we retrospectively identified 99 patients who underwent surgical treatment for primary HCC at our institution. All patients had pre-operative multi-parametric MRI scans (described below) taken within a week before surgery and mVI was assessed by pathologic inspection after removal of the tumor, according to the criteria in Sumie et al. [30] (i.e., three mVI classes: “no mVI”, “mild mVI”, and “severe mVI”).
Imaging Protocol
All liver MRI scans were obtained using a 1.5T MR scanner (Sonata, Siemens Healthcare, Erlangen, Germany) and the following protocols for the five sequences:
T2: HASTE acquisition protocol [31] with TR = 900 msec, TE = 90 msec, slice thickness = 8 mm, spatial resolution = 1.32 × 1.15 mm, matrix = 256 × 256
T1 (pre-contrast): FLASH acquisition protocol [32] with TR = 118 msec, TE = 2.49 msec, slice thickness = 8 mm, spatial resolution = 1.32 × 1.15 mm, matrix = 256 × 256
DWI: EPI-DWI acquisition protocol with TR = 2800 msec, TE = 84 msec, slice thickness = 8 mm, spatial resolution = 2.65 × 2.30 mm, matrix = 128 × 128
T1 post-contrast arterial phase: VIBE acquisition protocol [33] with TR = 4.78 msec, TE = 2.26 msec, slice thickness = 5 mm, resolution = 1.32 × 1.00 mm, matrix = 256 × 256
T1 post-contrast portal phase: FLASH acquisition protocol with TR = 118 msec, TE = 2.49 msec, slice thickness = 8 mm, spatial resolution = 1.32 × 1.15 mm, matrix = 256 × 256
For dynamic T1-weighted images, a three-dimensional, volumetric interpolated breath-hold examination (VIBE) sequence (TR = 4.78 ms, TE = 90 ms, slice thickness = 5 mm, matrix = 256 × 256) was used and 0.1 mmol/kg of Gd-DTPA was injected at a rate of 2 mL/s.
Tumor and Margin Segmentation
Since previous studies indicate that tumor margins may contain useful information in classifying mVI [20, 23–25, 29], we studied both the tumor and its margin for feature extraction and machine learning modeling. To this end, we assessed the following three experimental setups:
Tumor-only features
Margin-only features
Tumor and margin features
The purpose of the three setups was to assess the amount of information stored in the tumor itself and in its marginal region, as well as to investigate the potential added value of combining the two. In the third setup, we combined features separately extracted from the tumor only and the margin only. We also compared the effects of extracting features directly from the combined tumor and margin area.
Regions of interest (ROIs) were manually drawn outlining the tumor region (single tumor or the biggest lesion if there were more than one) in all five MRI sequences for each patient. When analyzing patients with more than one lesion, our physicians suggested we consider the biggest lesion only. Two experienced radiologists worked on tumor segmentation, where X. X. (3 years of experience) performed the initial segmentation and X. X. (18 years of experience) reviewed, adjusted, and confirmed the segmentation. Table 1 reports information on the number of slices per tumor for each sequence.
Table 1.
Number of tumor slices (for each sequence, the minimum number of slices per tumor is 1)
| Sequence | Average number slices | Max number slices |
|---|---|---|
| Arterial | 12 | 31 |
| DWI | 6.5 | 18 |
| T1 | 6 | 18 |
| T2 | 6 | 24 |
| Venous | 7 | 29 |
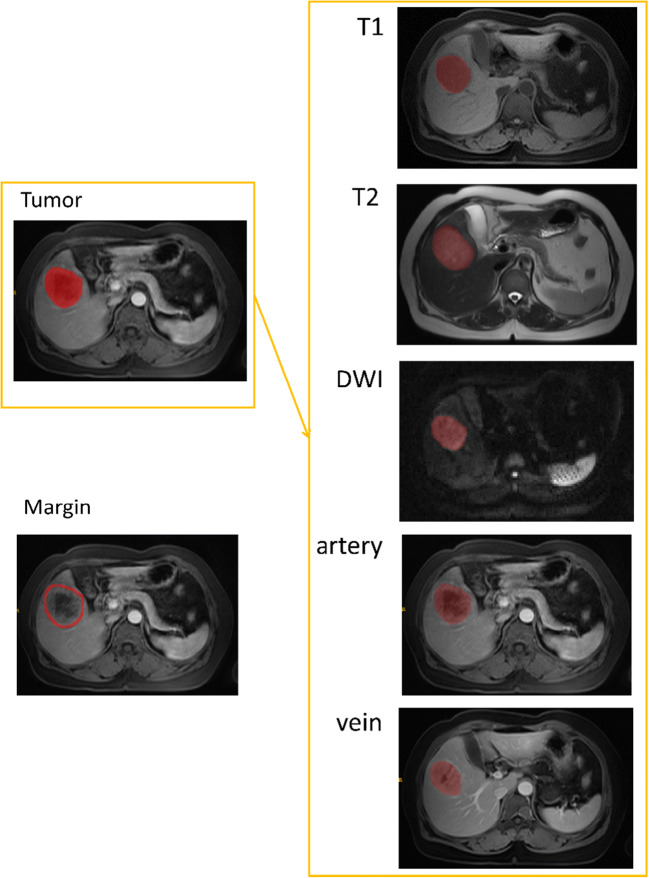
For each segmented tumor, we automatically obtained a tumor margin for the peritumoral region. To that end, we applied a dilation operation to the tumor’s ROI using a squared kernel of size 11 × 11. This enlarged the ROI by about 10 pixels around the tumor. From the dilated segmentation, we subtracted the original tumor’s ROI, thus isolating the marginal region. A 10-pixel margin corresponds to approximately 1 cm around the tumor (spatial resolution: 1.32 × 1.15 mm for all sequences but the late arterial sequence, whose resolution is 1.32 × 1.00 mm). The choice of a 10-pixel (or 1 cm) margin is based on previous studies [29, 34] as well as on our expert physician’s suggestions with the consideration of allowing the inclusion of effective information in the margin while limiting the potential influence of extra-hepatic tissue. Figure 1 shows examples of tumor and margin segmentation.
Fig. 1.
Examples of tumor and margin segmentation in five MRI sequences
Evaluation of Single and Combined MRI Sequences
In order to evaluate the effect of each individual MRI sequence and whether combining different sequences may provide additional information to improve mVI prediction, we trained models on features extracted from individual sequences and on combinations of such features. To understand how each sequence’s features contribute in a multi-sequence model, we checked which features in the multi-sequence models were also selected in the corresponding single-sequence models. The idea is that, if two sequences capture different aspects relevant to the mVI prediction task, this should be reflected when combining the two sequences. It is important to acknowledge that a feature may not be relevant when considered in a single-sequence model, but become relevant when combined with features from a different sequence.
Radiomics Feature Extraction and Modeling
We employed Pyradiomics [35] to extract 100 3D radiomics features, including shape features, first-order features, and various texture features,1 on each of the five MRI sequences (namely, T1, T2, DWI, late arterial, and portal venous). The 100 features were extracted separately from the volumetric tumor segmentation and the tumor margin. Since marginal features and tumor features may be complementary, we also combined them to obtain 200 features (100 from the tumor and 100 from the margins) per sequence.
We performed feature selection by Least Absolute Shrinkage and Selection Operation (LASSO) [36]. When using features from tumor or margin only, if a multi-sequence combination is considered and feature selection is run, only feature sets that include features from all the sequences in the combination are kept. This is because we tested all possible combinations of the five sequences and we did not want to learn the same model when considering different sequence combinations. Similarly, when using combined features from tumor and margin, we discarded all LASSO-selected feature sets that did not include both tumor and margin features. This means that all single-sequence models must include features from margin and tumor for that sequence, and that all multi-sequence models must include features from margin (and tumor) from at least one sequence. This is to avoid learning models from feature sets found when using tumor or margin features only.
We built binary machine learning classifiers to predict mVI status: “no mVI” vs. “mild or severe mVI.” The machine learning models we employed are support vector machines (SVM), decision trees, k-nearest neighbor (k = 5), and Naïve-Bayes. The implementation we used is that provided by the MATLAB R2018a Statistics and Machine Learning Toolbox.
Considering our sample size, we used stratified 5-fold cross-validation to evaluate the radiomics models. In each fold, the ratio of positive and negative samples is kept constant. The same folds are used to train all models to ensure results are directly comparable. We used the area under the receiver operating characteristic curve (AUC) [37] as a primary performance metric, reporting accuracies as well for additional information.
We performed feature stability analysis to better understand the learned models. During 5-fold cross-validation, we looked at how many features are shared among models built on the five folds. The idea is that, if a feature is selected in many cross-validation rounds, it can be considered “stable” and it is likely to be meaningful/essential for the classification task. Conversely, if a feature is selected only once, it may be capturing differences in the data that were introduced by chance during the cross-validation split. We considered a feature to be stable if it was chosen in at least three cross-validation folds.
Statistical Analysis
When comparing the demographic characteristics between the two groups (“no mVI” vs. “mVI [mild or severe mVI]”), the Wilcoxon signed-rank test (age, max diameter, and KI67) or the Chi-squared or Fisher exact test (all others) were used as appropriate. We did not use t tests for continuous variables because normality in the mVI and no mVI group could not be verified using a Kolmogorov-Smirnov test.
After identifying which stable features are shared by the best model and the corresponding single-sequence models, we analyzed the relationship between such features and factors that are currently used to make clinical decisions about HCC (e.g., tumor size and number in the Milan criteria). We report correlation between the shared radiomics features and the tumors’ maximum diameter, and p values for Wilcoxon tests on median values (given our limited sample size, normality cannot be assumed and a t test cannot be performed) for the radiomics features grouped by AFP value (dichotomized at a threshold of 400 ng/mL), by histopathological grade (dichotomized as {1,2}, {3,4}), and by number of lesion (dichotomized as one lesion only vs. more than one lesion). All tests were performed using R version 3.5.1.
Results
Patient Characteristics
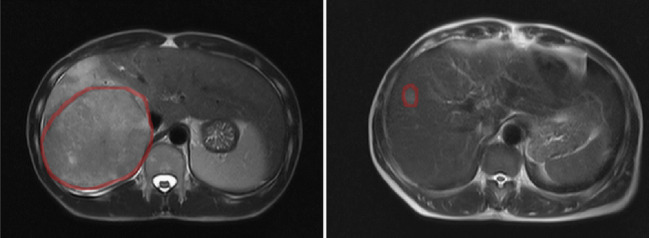
Table 2 reports the demographic information for the patient cohort. Sixty-one patients out of 99 presented histological evidence of mVI; most patients in our cohort are male (83) and the mean age is 51 years in the mVI group and 54 years in the no mVI group. Most of our patients have hepatitis B virus (HBV), hepatitis C virus (HCV), or both. It is worth noticing that the tumor size in our cohort is large with average size of more than 6.5 cm in both groups. In addition, only 33 patients have their biggest lesion smaller than 5 cm and 14 have their biggest lesion smaller than 3 cm. Figure 2 shows two examples of different sizes of tumor.
Table 2.
Patient demographic characteristics (notice that AFP values are not available for six patients). Continuous variables are reported as mean (standard deviation)
| No mVI | mVI | Number of patients | p value | |
|---|---|---|---|---|
| Age | 54.34 (9.27) | 51.42 (12.27) | 99 | 0.301 |
| Number of lesions | 99 | 0.0005 | ||
| 1 | 36 | 39 | ||
| > 1 | 2 | 22 | ||
| Sex | 99 | 0.273 | ||
| M | 34 | 49 | ||
| F | 4 | 12 | ||
| Histopathologic grade [according to Edmondson-Steiner [38]] | 99 | 0.031 | ||
| 1 or 2 | 10 | 5 | ||
| 2—3 | 7 | 9 | ||
| 3 or 4 | 21 | 47 | ||
| KI67 | 28.08 (21.85) | 30.49 (19.23) | 99 | 0.578 |
| Max diameter (mm) | 67.18 (38.43) | 71.22 (34.51) | 99 | 0.485 |
| Alpha FetoProtein (ng/mL) | 93 | 0.1 | ||
| ≤ 400 | 31 | 37 | ||
| > 400 | 6 | 19 | ||
| Hepatitis | 99 | 0.145 | ||
| HBV | 28 | 52 | ||
| HBC | 2 | 0 | ||
| Both | 1 | 3 | ||
| No | 7 | 6 | ||
Fig. 2.
Margins we automatically identified for two different tumors of 111 mm of diameter (left) and of 17 mm of diameter (right)
The two sub-populations (mVI vs. no mVI) were not statistically different for all characteristics except for histopathologic grade (p value 0.03) and number of lesions (p value 0.0074). These findings are compatible with previously reported study populations [28, 39].
Effects of mVI Prediction Models
The best-performing machine learning model we tested is SVM, which is used as the classifier for all experiment results reported in this article. Table 3 shows the performance for single-sequence models and Table 4 shows the summarized results when using different combinations of features (tumor and margin) and multiple MRI sequences. Note that we focus on the results based on the combination of features separately extracted from the tumor region only and the margin region only, while the results from the features directly extracted from the combined tumor and margin area show a consistently inferior performance (results not shown).
Table 3.
Single-sequence models’ performance (means and standard deviations)
| Sequence | Tumor | Tumor margin | Tumor + margin | |||
|---|---|---|---|---|---|---|
| AUC mean and SD (%) | Accuracy mean and SD (%) | AUC mean and SD (%) | Accuracy mean and SD (%) | AUC mean and SD (%) | Accuracy mean and SD (%) | |
| DWI | 62.21 (12.28) | 59.79 (11.28) | 60.18 (11.11) | 64.63 (3.63) | 63.97 (11.70) | 58.68 (11.04) |
| T1 | 72.88 (6.08) | 65.79 (11.63) | 71.08 (7.05) | 60.58 (5.65) | 71.11 (5.65) | 67.68 (7.51) |
| T2 | 80.84 (12.5) | 68.74 (12.80) | 81.82 (5.95) | 72.84 (8.83) | 84.19 (11.29) | 70.74 (12.36) |
| Late arterial | 68.74 (12.03) | 57.47 (10.34) | 60.61 (10.84) | 63.63 (8.93) | 74.34 (14.41) | 66.68 (2.37) |
| Portal venous | 79.22 (3.68) | 71.74 (4.29) | 70.43 (10.26) | 65.68 (8.05) | 84.18 (4.48) | 78.84 (6.34) |
Table 4.
Summary of the performance for the best models in each experimental setup (means and standard deviations)
| Features | Best sequence combination | AUC mean (%) | AUC SD (%) | Accuracy mean (%) | Accuracy SD (%) |
|---|---|---|---|---|---|
| Tumor | T2 + venous | 86.69 | 8.09 | 79.68 | 10.41 |
| Margin | DWI + T2 + arterial + venous | 84.62 | 4.49 | 75.79 | 6.37 |
| Tumor + margin | T2 | 84.19 | 11.29 | 70.74 | 12.36 |
We observe that the two best-performing single-sequence models are trained on the T2 sequence (AUC = 80.84%) and on the portal venous sequence (AUC = 79.22%), whose combination also achieves the highest performance (AUC = 86.69%). We can also notice that T2 and portal venous remain the highest performing sequences in the tumor + margin setup (AUCs = 84.19% and 84.18%, respectively) and that adding the margin slightly increases the performance of these two single-sequence models, although both performance measures are within about one standard deviation between the tumor only setup and the tumor + margin setup (for T2, AUC = 80.84 ± 12.5% vs. 84.19 ± 11.29%, for portal venous, AUC = 79.22 ± 3.68% vs. 84.18 ± 4.48% between tumor and tumor + margin).
Radiomics Feature Stability Analysis
Table 4 shows AUCs and accuracies for the highest-performing SVM models in the three experimental setups, and Table 5 reports the feature stability analysis for those models (with feature percentage being calculated over the number of features found in at least one fold). We observe that the percentage of features consistently selected across folds decreases as we increase the stability threshold. This pattern was expected and is common to all three experimental setups although the decrease is faster in the margin-only setup and in the tumor + margin setup. In fact, of the 105 (over 200 total features; 100 from T2 and 100 from portal venous) features selected in at least one fold in the tumor-only setup, 17 (16%) are shared by at least three folds. This percentage decreases to 10% in the other two experimental setups, where 40 features and 15 features are shared by at least three folds.
Table 5.
Feature stability analysis results
| Number of folds | Tumor (T2 + venous) | Margin (T2 + DWI + arterial + venous) | Tumor + margin (T2) | |||
|---|---|---|---|---|---|---|
| Feature no. | Feature % | Feature no. | Feature % | Feature no. | Feature % | |
| ≥ 1 | 105 | 100 | 400 | 100 | 147 | 100 |
| ≥ 2 | 48 | 45.7 | 106 | 26.5 | 46 | 31.3 |
| ≥ 3 | 17 | 16.2 | 40 | 10 | 15 | 10.0 |
| ≥ 4 | 4 | 3.8 | 12 | 3 | 3 | 2 |
| 5 | 2 | 1.9 | 8 | 2 | 1 | 0.7 |
For models built on tumor features, our results show that most of the stable predictors used in the best-performing multi-sequence model (i.e., T2 and portal venous) are also stable predictors in the single-sequence ones (14 out of 17). This indicates that features that are strong predictors in single-sequence models work synergistically when considered together.
Correlation of Radiomics Features with Other Factors
Table 6 reports the 14 features that are in common between the single-sequence models and the best-performing model for the tumor-only setup, together with their relations with some demographic variables (shown as either correlations or p values). We can observe moderate correlation between feature #7 (gray-level size zone matrix-gray level non-uniformity) and maximum tumor diameter and that the means (medians) of feature #13 (shape–elongation) are significantly different between patients when grouped by AFP value and histopathologic grade.
Table 6.
Features common to the single-sequence models and the best model for the tumor feature experiment and their relation to demographic variables. For each grouping (i.e., column in the table), we only report p values that are < 0.05. If no test in that grouping reaches this significance level, we show p values < 0.1
| Features | Sequence | Diameter (correlation) | AFP (threshold = 400 ng/mL) | Histopathologic grade | Number of lesions | |
|---|---|---|---|---|---|---|
| 1 | 1st order—kurtosis | T2 | 0.24 | |||
| 2 | 1st order—skewness | T2 | 0.31 | |||
| 3 | Glcm—Imc2 | T2 | 0.69 | 0.087 | ||
| 4 | Gldm—dependence variance | T2 | 0.66 | 0.014 | ||
| 5 | Gldm—low gray level emphasis | T2 | − 0.49 | |||
| 6 | Gldm—small dependence emphasis | T2 | − 0.57 | |||
| 7 | Glszm—gray level non-uniformity | T2 | 0.80 | |||
| 8 | Glszm—low gray level zone emphasis | T2 | − 0.60 | |||
| 9 | Shape—flatness | T2 | 0.22 | 0.055 | ||
| 10 | Shape—sphericity | T2 | − 0.37 | |||
| 11 | Glcm—difference variance | Portal venous | 0.19 | |||
| 12 | Glszm—size zone non-uniformity normalized | Portal venous | − 0.53 | |||
| 13 | Shape—elongation | Portal venous | 0.22 | 0.023 | 0.056 | |
| 14 | Shape—sphericity | Portal venous | − 0.31 |
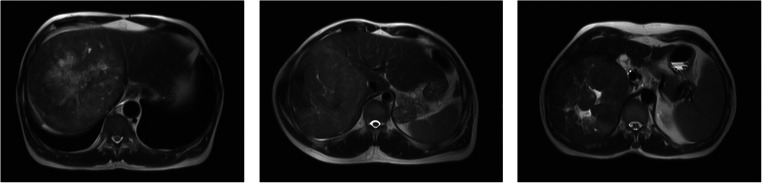
Table 6 reveals a noticeable correlation between tumor diameter and feature #7 (Glszm–gray level non-uniformity). This feature measures how heterogeneous the gray-level intensities are in the ROI. In order to better understand its relationship with the tumor’s diameter, Fig. 3 shows the three patients with the highest value for feature #7.
Fig. 3.
Sample T2 scans for patients with the highest value of feature #7. Left: diameter: 130 mm, center: diameter: 138 mm, right: diameter: 138 mm
In all the three cases, the inside of the tumor is heterogeneous with respect to gray-level intensities. In particular, we can see “patches” of relatively uniform gray-level intensity. The size and distribution of such patches are captured by the gray-level size zone matrix (Glszm) features, which may be the reason why the diameter and feature #7 are correlated. In addition, very small tumors cannot exhibit a heterogeneous distribution of gray-level intensities, so their value for feature #7 is reasonably low.
Comparison with Previous Work
Here, we put the results of our study in context with several previously published related studies addressing mVI prediction from pre-operative images. Our study using 1.5T MRI scanners and Gd-DTPA as a contrast agent is different from the study using triphasic CT scans [26] or other MRI-based studies in field strength (e.g., 3T [27, 29] or both 1.5T and 3T [28]), contrast agent (e.g., gadoxetic acid [27–29]), MRI sequence protocol, and the approach of incorporating information for radiomics modeling [29]. Results from these related previous studies are overall comparable to ours. For example, a study including 28 HCC patients obtained an AUC of 0.76 by using radiomics features characterizing tumors and their boundaries [26]. Similarly, another study achieved an AUC of 88.2% on an independent validation cohort of 50 patients but using only a single sequence of the Gd-EOB-DTPA-enhanced MRI [29]. Using DWI sequence on 41 patients, another study reported a sensitivity of 94.4% and a specificity of 63.6% and highlighted D-value and irregular shape of the tumor as important predictors [27]. Similarly, we found that shape features are important factors too, but, in our case, these features were extracted from the T2 and portal venous sequences. Including MRI sequences for 179 patients with single HCC, an additional study showed features such as the presence of satellite nodules, peritumoral hypointensity, tumor sphericity, and washout were mVI predictors [28]. In our study, Table 6 shows that tumor sphericity also plays a similarly important role in predicting mVI.
Discussion
The goal of this study was to analyze whether it is possible to pre-operatively assess microvascular invasion for hepatocellular carcinoma from multi-parametric liver MRI scans. In addition, we tested whether the peritumoral region provides valuable information for this task. Our results show that radiomics features extracted from the tumor region on pre-operative MRI scans capture information relevant to the mVI prediction task. At the same time, radiomics information from tumor margin also shows an association with mVI but with a slightly lower AUC in comparison to tumor radiomics. When combining the tumor and margin radiomics features together, however, the performance of the model drops, indicating that information from the margin is not complementary to (and may even be in conflict with) information from the tumor.
Our results from the tumor radiomics setup show that different sequences (i.e., T2 and portal venous) can capture complementary information, thus achieving increased performance when combined. Likewise, when it comes to margin radiomics, a similar behavior is seen because the combination of DWI, T2, late arterial, and portal venous exhibits substantially improved performance over individual sequences alone. These findings show that a multi-sequence approach to mVI prediction is advisable.
The feature stability analysis results help better understand the potential of the proposed radiomics approach. In all three experimental setups, the number of features shared across folds decreases as we require them to be shared by more folds. This pattern is expected, although it seems to also reveal an underlying heterogeneity in the 5-fold split, which likely causes different features to be relevant when observed on different datasets.
Tumor Margin
When correlating radiologic assessment with mVI, previous studies report that peritumoral enhancement patterns are good predictors. For this reason, we assessed models built on features extracted from an automatically generated marginal region around the tumor. While previous studies have included peritumoral information by, for example, extracting features from an ROI around the tumor [28], we precisely defined a peritumoral margin and extracted features from this region. In addition, all our features are automatically extracted and do not require visual assessment by radiologists [26]. Our results (Table 4) show that marginal information alone can reach a close discrimination performance as tumor-extracted features, achieving an AUC of 84.62% (vs. 86.69% achieved by tumor features). Also, single-sequence models using margin radiomics perform generally worse than their tumor radiomics counterparts. When tumor features and margin features are jointly modeled, we generally see an increase in average prediction performance across the five folds, but also an increased variability. This indicates a notable effect of the features extracted from peritumoral areas for mVI prediction, meriting further evaluation in larger cohorts. Note that in our study the automatically extracted peritumoral region may include extra-hepatic areas. This is partly due to the relatively large size of the tumors in our cohort, as exemplified in Fig. 2. The margin of the tumor of diameter 111 mm includes extra-hepatic regions, while that of the tumor of diameter 17 mm only includes liver parenchyma. The extra-hepatic areas can possibly be responsible for the heterogeneity in the marginal radiomics feature extraction. This observation has to do with our choice of the margin size (i.e., 10 pixels). Choosing a wider margin would result in the inclusion of larger portions of extra-hepatic tissue, thus exacerbating the heterogeneity issue, while too narrow a margin may not contain enough information. In this study, our choice of 10 pixels (1 cm wide) margin per suggestions of our experienced radiologists is concordant with previous work in the literature [29, 34], but optimal margin size will need further investigation.
The issue with the margin features is also present when we combine tumor features and margin features. In fact, the best performing model in the third experiment is a single-sequence model, which seems to contradict the observation drawn from the tumor features experiment that different sequences capture different pieces of information that can work synergistically toward the prediction goal. As mentioned in the “Materials and Methods” section, we forced models learned in the third experiment to include features from both margin and tumor region, so the best model for the first experiment (i.e., T2 and portal venous, tumor features only) cannot be learned in the third experimental setup. Once again, we suspect that the heterogeneity in the marginal features leads to the lack of improvement in mVI prediction when margin and tumor features are combined.
Limitations
Despite our promising results, our study has some limitations. First, the sample size is limited to 99 patients. Nevertheless, we performed extensive analysis investigating five different MRI sequences per patient and extracting radiomics features from both tumor and margin region. The sample is slightly unbalanced because there are 61 mVI positive cases. Second, our cohort mostly includes large tumors (with average diameter > 6.5 cm) and patients with either HBV, HCV, or both. The fact that we only have 14 cases with tumor size < 3 cm prevents us from doing a subgroup analysis on these smaller tumors. A larger study including a broader span of patients’ characteristics is needed for results to be generalizable. Regarding the inclusion of large tumors, tumor size and number of nodules are commonly used to gauge how aggressive a tumor is and, consequently, whether to surgically treat it (e.g., the Milan criteria [9]). This notwithstanding, some studies reported that the number (and size) of nodules was not predictive of survival when corrected by mVI status [13]. Also related to generalizability, all our patients came from at the same hospital and were scanned with the same MRI scanner. While this data consistency reduces potential confounding effects, external cohorts and different MRI scanners would increase the independent validity and generalizability of our results. In addition, because tumor ROIs were manually drawn, there could be some differences between the tumor ROI outlines in different sequences, and as a result, the “tumor region” in one sequence may have been a peritumoral region in another sequence. This could be a potential factor that had influenced the model’s evaluation. More accurate and automated tumor segmentation is needed for an in-depth analysis in future work. Finally, considering our sample size, our interpretation of the results related to the multi-sequence and tumor margin as well as the feature analysis needs further evaluation. Our study has important differences with related work and our results represent an additional contribution on examining how radiomics features may relate to possibly observable characteristics of a tumor and its surrounding region in liver MRI for mVI prediction.
Conclusion
In summary, our study provides encouraging results on pre-operative assessment of mVI from multi-parametric liver MRI scans. Our radiomics-based machine learning algorithm reaches an AUC of 86.69% and shows different MRI sequences (in our case, T2 and portal venous) provide complementary information for mVI prediction. Further evaluation on larger datasets is needed.
Funding Information
This work was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) grants (#1R01CA193603, #3R01CA193603-03S1, and #1R01CA218405), a Radiological Society of North America (RSNA) Research Scholar Grant (#RSCH1530), an Amazon AWS Machine Learning Research Award, and a University of Pittsburgh Physicians (UPP) Academic Foundation Award.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
https://pyradiomics.readthedocs.io/en/latest/features.html, last accessed Jan. 7th, 2019
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Giacomo Nebbia and Qian Zhang contributed equally to this work.
Contributor Information
Xinxiang Zhao, Email: zhaoxinxiang06@126.com.
Shandong Wu, Email: wus3@upmc.edu.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. 10.3322/caac.21492 [DOI] [PubMed]
- 2.Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, Mcglynn KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978-2007. UICC Int J Cancer IJC. 2016;139:1534–1545. doi: 10.1002/ijc.30211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrick JL, Kelly SP, Altekruse SF, McGlynn KA, Rosenberg PS. Future of hepatocellular carcinoma incidence in the United States forecast through 2030. J Clin Oncol. 2016;34(15):1787–1794. doi: 10.1200/JCO.2015.64.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greten TF, Papendorf F, Bleck JS, et al. Survival rate in patients with hepatocellular carcinoma: a retrospective analysis of 389 patients. Br J Cancer. 2005;92(10):1862–1868. doi: 10.1038/sj.bjc.6602590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 7.Llovet J, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis. 1999;19(03):329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 8.Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68(2):723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 9.Mazzaferro V, Regalia E, Doci R, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334(11):693–700. doi: 10.1056/NEJM199603143341104. [DOI] [PubMed] [Google Scholar]
- 10.Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S. Recurrence of hepatocellular cancer after resection: patterns, treatments, and prognosis. Ann Surg. 2015;261(5):947–955. doi: 10.1097/SLA.0000000000000710. [DOI] [PubMed] [Google Scholar]
- 11.Halazun KJ, Najjar M, Abdelmessih RM, et al. Recurrence after liver transplantation for hepatocellular carcinoma. Ann Surg. 2017;265(3):557–564. doi: 10.1097/SLA.0000000000001966. [DOI] [PubMed] [Google Scholar]
- 12.Escartin A, Sapisochin G, Bilbao I, et al. Recurrence of hepatocellular carcinoma after liver transplantation. Transplant Proc. 2007;39(7):2308–2310. doi: 10.1016/J.TRANSPROCEED.2007.06.042. [DOI] [PubMed] [Google Scholar]
- 13.Mazzaferro V, Llovet JM, Miceli R, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 2009;10(1):35–43. doi: 10.1016/S1470-2045(08)70284-5. [DOI] [PubMed] [Google Scholar]
- 14.Pawlik TM, Gleisner AL, Anders RA, Assumpcao L, Maley W, Choti MA. Preoperative assessment of hepatocellular carcinoma tumor grade using needle biopsy: implications for transplant eligibility. Ann Surg. 2007;245(3):435–442. doi: 10.1097/01.sla.0000250420.73854.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sterling RK, Wright EC, Morgan TR, et al. Frequency of elevated hepatocellular carcinoma (HCC) biomarkers in patients with advanced hepatitis C. Am J Gastroenterol. 2012;107(1):64–74. doi: 10.1038/ajg.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta S, Bent S, Kohlwes J. Test characteristics of α-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C: a systematic review and critical analysis. Ann Intern Med. 2003;139(1):46–50. doi: 10.7326/0003-4819-139-1-200307010-00012. [DOI] [PubMed] [Google Scholar]
- 17.Pawlik TM, Delman KA, Vauthey JN, et al. Tumor size predicts vascular invasion and histologic grade: implications for selection of surgical treatment for hepatocellular carcinoma. Liver Transplant. 2005;11(9):1086–1092. doi: 10.1002/lt.20472. [DOI] [PubMed] [Google Scholar]
- 18.Roayaie S, Frischer JS, Emre SH, et al. Long-term results with multimodal adjuvant therapy and liver transplantation for the treatment of hepatocellular carcinomas larger than 5 centimeters. Ann Surg. 2002;235(4):533–539. doi: 10.1097/00000658-200204000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ünal E, İdilman İS, Akata D, Özmen MN, Karçaaltıncaba M. Microvascular invasion in hepatocellular carcinoma. Diagnostic Interv Radiol. 2016;22(2):125–132. doi: 10.5152/dir.2015.15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renzulli M, Brocchi S, Cucchetti A, et al. Can current preoperative imaging be used to detect microvascular invasion of hepatocellular carcinoma? Radiology. 2016;279(2):432–442. doi: 10.1148/radiol.2015150998. [DOI] [PubMed] [Google Scholar]
- 21.Chou C-T, Chen R-C, Lin W-C, Ko C-J, Chen C-B, Chen Y-L. Prediction of microvascular invasion of hepatocellular carcinoma: preoperative CT and histopathologic correlation. Am J Roentgenol. 2014;203(3):W253–W259. doi: 10.2214/AJR.13.10595. [DOI] [PubMed] [Google Scholar]
- 22.Yang C, Wang H, Sheng R, Ji Y, Rao S, Zeng M. Microvascular invasion in hepatocellular carcinoma: is it predictable with a new, preoperative application of diffusion-weighted imaging? Clin Imaging. 2017;41(2017):101–105. doi: 10.1016/j.clinimag.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Lei Z, Li J, Wu D, et al. Nomogram for preoperative estimation of microvascular invasion risk in hepatitis B virus–related hepatocellular carcinoma within the Milan criteria. JAMA Surg. 2016;151(4):356. doi: 10.1001/jamasurg.2015.4257. [DOI] [PubMed] [Google Scholar]
- 24.Kim H, Park MS, Choi JY, et al. Can microvessel invasion of hepatocellular carcinoma be predicted by pre-operative MRI? Eur Radiol. 2009;19(7):1744–1751. doi: 10.1007/s00330-009-1331-8. [DOI] [PubMed] [Google Scholar]
- 25.Kim KA, Kim M-J, Jeon HM, et al. Prediction of microvascular invasion of hepatocellular carcinoma: usefulness of peritumoral hypointensity seen on gadoxetate disodium-enhanced hepatobiliary phase images. J Magn Reson Imaging. 2012;35:629–634. doi: 10.1002/jmri.22876. [DOI] [PubMed] [Google Scholar]
- 26.Bakr S, Echegaray S, Shah R, Kamaya A, Louie J. Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. J Med Imaging. 2017;4(04):1. doi: 10.1117/1.JMI.4.4.041303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Zhang J, Zheng Z, et al. Preoperative histogram analysis of intravoxel incoherent motion (IVIM) for predicting microvascular invasion in patients with single hepatocellular carcinoma. Eur J Radiol. 2018;105(January):65–71. doi: 10.1016/j.ejrad.2018.05.032. [DOI] [PubMed] [Google Scholar]
- 28.Ahn SJ, Kim JH, Park SJ, Kim ST, Han JK. Hepatocellular carcinoma: preoperative gadoxetic acid–enhanced MR imaging can predict early recurrence after curative resection using image features and texture analysis. Abdominal Radiology. http://link.springer.com/10.1007/s00261-018-1768-9. Published September 18, 2018. Accessed Oct 18, 2018. [DOI] [PubMed]
- 29.Feng ST, Jia Y, Liao B, et al. Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol. 2019;29(9):4648–4659. doi: 10.1007/s00330-018-5935-8. [DOI] [PubMed] [Google Scholar]
- 30.Sumie S, Nakashima O, Okuda K, et al. The significance of classifying microvascular invasion in patients with hepatocellular carcinoma. Ann Surg Oncol. 2014;21(3):1002–1009. doi: 10.1245/s10434-013-3376-9. [DOI] [PubMed] [Google Scholar]
- 31.Semelka RC, Kelekis NL, Thomasson D, Brown MA, Laub GA. HASTE MR imaging: description of technique and preliminary results in the abdomen. Vol 6.; 1996. 10.1002/jmri.1880060420 [DOI] [PubMed]
- 32.Runge VM, Wood ML, Kaufman DM, Kevin Nelson ML, Traill MR. FLASH: clinical three-dimensional magnetic resonance imaging. Vol 8.; 1988. https://pubs.rsna.org/doi/pdf/10.1148/radiographics.8.5.3227132. Accessed Jan 4, 2019. [DOI] [PubMed]
- 33.Rofsky NM, Lee VS, Laub G, et al. Abdominal MR imaging with a volumetric interpolated breath-hold examination. Radiology. 1999;212(3):876–884. doi: 10.1148/radiology.212.3.r99se34876. [DOI] [PubMed] [Google Scholar]
- 34.Roayaie S, Blume IN, Thung SN, et al. A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology. 2009;137(3):850–855. doi: 10.1053/j.gastro.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. J R Stat Soc Ser B (Statistical Methodol) 2011;73(3):273–282. doi: 10.1111/j.1467-9868.2011.00771.x. [DOI] [Google Scholar]
- 37.Tahmassebi A, Wengert GJ, Helbich TH, et al. Impact of machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy and survival outcomes in breast cancer patients. Invest Radiol. 2019;54(2):110–117. doi: 10.1097/RLI.0000000000000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edmondson HA, Steiner PE. Primary carcinoma of the liver.A study of 100 cases among 48,900 necropsies. Cancer. 1954;7(3):462–503. doi: 10.1002/1097-0142(195405)7:3<462::AID-CNCR2820070308>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 39.Chandarana H, Robinson E, Hajdu CH, Drozhinin L, Babb JS, Taouli B. Microvascular invasion in hepatocellular carcinoma: is it predictable with pretransplant MRI? Am J Roentgenol. 2011;196(5):1083–1089. doi: 10.2214/AJR.10.4720. [DOI] [PubMed] [Google Scholar]