Abstract
Infertility affects approximately 15–20% of individuals of reproductive age worldwide. Over the last 40 years, assisted reproductive technology (ART) has helped millions of childless couples. However, ART is limited by a low success rate and risk of multiple gestations. Devising methods for selecting the best gamete or embryo that increases the ART success rate and prevention of multiple gestation has become one of the key goals in ART today. Special emphasis has been placed on the development of non-invasive approaches, which do not require perturbing the embryonic cells, as the current morphology-based embryo selection approach has shortcomings in predicting the implantation potential of embryos. An observed association between embryo metabolism and viability has prompted researchers to develop metabolomics-based biomarkers. Nuclear magnetic resonance (NMR) spectroscopy provides a non-invasive approach for the metabolic profiling of tissues, gametes and embryos, with the key advantage of having a minimal sample preparation procedure. Using NMR spectroscopy, biologically important molecules can be identified and quantified in intact cells, extracts or secretomes. This, in turn, helps to map out the active metabolic pathways in a system. The present review covers the contribution of NMR spectroscopy in assisted reproduction at various stages of the process.
Keywords: assisted reproductive technology, nuclear magnetic resonance spectroscopy, metabolomics, systems biology, in vitro fertilization
1. Introduction
Infertility is a highly prevalent global health condition affecting approximately 15–20% of individuals of reproductive age worldwide. The major causes of infertility in women include anovulation, anatomical problems and endometriosis, whereas sperm disorders are the important causes of male infertility. Several lifestyle factors such as stress, obesity, urbanization-based work pressure and sexually transmitted infections can impair the fertility of the individuals [1,2]. Assisted reproductive technology (ART) has played an important role to partially overcome the low natural reproductive ratio [3]. Over the past 4 decades, ART has revolutionized the treatment of infertility, and more than 8 million live births have taken place since the first successful implementation of in vitro fertilization (IVF) in 1978 [4,5]. The demand for ART has increased significantly in recent years; using which a large number of successful pregnancies have been achieved [3,6].
The ART fertility clinic success rate report of the USA showed that a total of 284 385 cycles of ART were performed in 2017 [6]. Age-dependent variation is seen in the type of ART cycles (non-donor or donor oocyte cycle) [3,6,7] and the live-birth rate is inversely related to the patient age [3,7]. As per the 2016 registry of the European Society of Human Reproduction and Embryology (ESHRE), after treatment with IVF and intracytoplasmic sperm injection (ICSI), the clinical pregnancy rates per transfer were 34.8% and 33.2%, respectively [8]. Multiple pregnancies pose a potential health risk for the mother, including pregnancy-induced hypertension and postpartum haemorrhage [9]. Hence, several countries (such as Finland, Sweden, Belgium, Australia and New Zealand) have employed elective single embryo transfer to reduce the possibility of multiple gestation and eventual complications [10,11]. However, many countries are yet to adopt the strict regulations in ART practice, presumably due to the expense involved coupled with the patient's hope to achieve pregnancy in each cycle with better chances. In general, the transfer of multiple embryos is primarily driven by the number of available embryos and overall embryo quality. Thus, the selection of an ideal gamete or embryo that results in a higher success rate together with reduced multiple gestations is the prime focus of ART today, and any improvements in the existing methods for preimplantation embryo assessment would be beneficial for this purpose.
Morphological evaluation is the conventional approach used to select a healthy embryo for transfer as it is considered to be safe, specific and simple [12]. However, this static mode of evaluation has drawbacks, such as subjectivity of the evaluator, frequent assessment during embryo development and culture stability [13]. Evaluation efficacy has been enhanced with the recent development of time-lapse imaging systems allowing embryologists to perform a non-invasive and dynamic evaluation of embryo quality in the steady culture environment [14,15]. In addition, an important observation has been made about the uptake and release of metabolites by sibling human embryos, having identical development and morphology showing significant differences, which could be attributed to genetic and functional heterogeneity in gametes [16,17]. The metabolic activity of the embryo has been found to be an indicator of its viability [18]. Hence, it is considered important to assimilate the metabolic profile of the embryo in the selection process to predict its implantation potential. It provides specific metabolic markers to identify competent embryos. Thus, understanding the metabolism of embryos is central to ART.
In recent years, different analytical techniques have been used in ART for studying embryo metabolism. These include near-infrared spectroscopy [19], Raman spectroscopy [20], mass spectrometry (MS) [21] and nuclear magnetic resonance (NMR) spectroscopy [22,23]. NMR spectroscopy is one of the widely used methods for identification and quantification of molecular species in a sample and is an alternative to MS, which is another popular technique used in metabolomics [24–28]. NMR is quantitative and does not require special sample preparation approaches like separation or derivatization that is needed for MS. However, compared with liquid chromatography-coupled mass spectrometry (LC-MS) or gas chromatography-coupled mass spectrometry (GC-MS), NMR is often 10–100 times less sensitive, and NMR-based metabolomics study usually provides reliable information on metabolites with concentrations greater than 10 µM [25].
The present review covers the contribution of NMR spectroscopy in assisted reproduction at various stages of the process. The general principles of NMR in metabolomics are first introduced, followed by its application to various procedures in ART. Apart from this, NMR plays a significant role in understanding the pathophysiology of polycystic ovarian syndrome (PCOS) and endometriosis, which are known to impair fertility in reproductive-age women [29,30]. NMR studies performed on animal models are also included as they have helped in probing the embryo directly and provided translational knowledge, which is not possible in human embryos due to ethical restrictions [31].
2. NMR spectroscopy in metabolomics
NMR has emerged as one of the important analytical techniques used in metabolomics. Despite its lower sensitivity, NMR spectroscopy offers many unparalleled advantages over other analytical techniques for studying the metabolic profile of a system [32–35]: (i) the NMR spectra obtained are highly reproducible [36]; (ii) the non-destructive nature of NMR analysis allows repeating experimentation and the sample can be recovered and stored for a long time and used for other studies [37]; (iii) NMR spectroscopy is inherently quantitative as the signal intensity is directly proportional to the metabolite concentrations and the number of particular nuclei in the molecule [38,39]; (iv) NMR can be used for both targeted and untargeted analyses of metabolic flux both in vitro and in vivo [40]; (v) its inherently quantitative nature enables precise quantification of precursors and products [41]; and (vi) it is well suited for studying intact tissues, organs, and other solid or semisolid samples through solid-state NMR (ssNMR) and magic-angle spinning (MAS-NMR) [42]. However, NMR also has its disadvantages, with the most significant challenge being its lack of sensitivity. In recent years, the development of ultra-high-field NMR spectrometers of 1 GHz and above [43], and advancements made in NMR probes, have greatly improved the application of NMR in metabolomics. Cryogenically cooled probes can increase signal sensitivity by a factor of 3–4. In addition, microprobes or microcoil probes (1 mm TXI and 1.7 mm TXI probes) not only enhance sensitivity but also reduce the required sample size down to a few microlitres [44–47].
ART in humans is a multistep process involving the superovulation of the ovaries through gonadotropin administration to the female partner followed by the surgical aspiration of oocytes which are suspended in the follicular fluid (FF), within the ovary. Oocytes surrounded by the granulosa and cumulus cells are assessed for their maturity and prepared for fertilization. Parallelly, healthy spermatozoa are separated from the seminal plasma of the ejaculate, and fertilization is achieved. Resulting embryos are developed in the laboratory using appropriate culture media, under a controlled environment for 3–5 days. Morphologically normal embryos that followed a developmental timeline [48] are selected for transfer to the patient's womb.
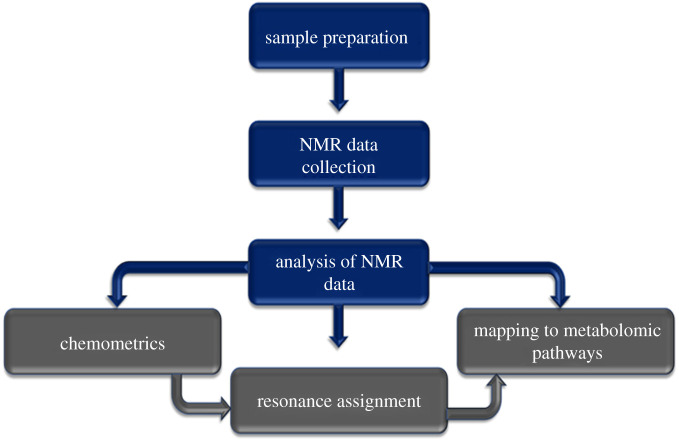
NMR-based metabolomics study generally has three major steps as depicted in figure 1: (i) sample preparation, (ii) recording NMR spectra from the sample, and (iii) analysis of these data to identify metabolites, biomarkers and biological pathways.
Figure 1.
Schematic illustration representing the key steps involved in an NMR-based metabolomics study.
The first step in the NMR-based metabolomics involves the collection and preparation of the sample. The volume of the sample needed for NMR data collection is 10–600 µl depending on the probe and the sample tube used. With the advent of low volume and high-sensitivity NMR probes, it is possible to reduce the sample volume even to 5 µl as mentioned above [44–47]. In most of the studies, the sample collected from the source can be directly used for NMR analysis after adding a small amount of deuterated solvent [35]. However, in cases where the sample contains macromolecules such as proteins and lipids, the large difference in the size of the macromolecule and small molecules of interest hamper the analysis due to significantly broadened NMR peaks, resulting in low accuracy of quantification. It is therefore necessary at times to remove high-molecular-weight molecules from the samples. There are two approaches to address this problem. One is to physically separate the macromolecules from the small molecule metabolites using ultra-filtration, solid-phase extraction or protein precipitation using organic solvents such as methanol, acetonitrile, acetone, perchloric acid and trichloroacetic acid [32]. An alternative is to allow the macromolecules to be present in the sample but suppress their signals in the spectrum by special NMR techniques. This is described below while discussing the NMR methods.
The second step is NMR data collection, where the primary goal is to identify and assign the metabolites present in the sample; the metabolites are identified based on their signature pattern in the NMR spectrum. This is usually accomplished using the one-dimensional proton NMR experiment and a set of homonuclear and heteronuclear two-dimensional (2D) NMR experiments. A detailed description of the theory of NMR experiments is beyond the scope of this review. The reader is encouraged to refer to articles on the basics of NMR [49–51]. In certain cases, it is not necessary to remove the protein component from the sample (as alluded to above). The signals from the proteins can be suppressed using specific NMR experiments. Typically, one-dimensional (1D) NMR spectra are recorded using a ‘T2 filter’ procedure, which attenuates the signal from macromolecules such as proteins and lipids [52,53]. One potential drawback of not removing macromolecules from the sample is that it can affect the quantification of small molecules that interact with the macromolecules [54].
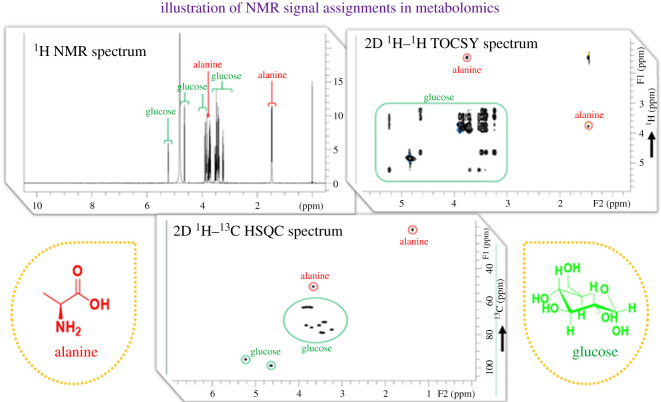
The third step is the analysis of NMR spectra, which involves the identification/assignment of metabolites and their quantification using 1D and 2D NMR. The 2D NMR techniques are preferred for obtaining assignments (identification) of the metabolites observed in the NMR spectrum and generally, it is acquired in the beginning phase of the study. The most popular 2D NMR techniques are 1H–1H total correlation spectroscopy (TOCSY) and 1H–13C heteronuclear single quantum coherence spectroscopy (HSQC) [36,55]. Figure 2 illustrates the three types of NMR spectra required for peak assignments (1D and 2D NMR). The spectra show the signatures of a mixture of glucose and alanine prepared in an aqueous solvent. Peak assignments are highlighted. The identification of metabolites by NMR is accomplished by the comparison of observed spectral parameters such as the observed chemical shifts, peak intensities and/or coupling patterns with those observed in the reference data of the molecules available in various databases. Ideally, these reference spectra should be available for all measured nuclei (1H, 13C, 15N and 31P), and for all types of NMR spectra and for all detected metabolites (about 1000 compounds), at the commonly used spectrometer frequencies (500–800 MHz) [35]. Through collaborative efforts of several metabolomics-based laboratories from around the world, these data are now available in several high-quality, web-based NMR spectral databases containing reference NMR spectra for hundreds of metabolites collected over a wide range of spectrometer frequencies and for a diverse range of nuclei. Most of these databases are freely accessible, including the Human Metabolome Database (HMDB), the Biological Magnetic Resonance Data Bank (BMRB), the Madison-Qingdao Metabolomics Consortium Database (MMCD), the NMRShiftDB2 database and the AIST spectral database in Japan [56–60].
Figure 2.
Illustration of assignment of peaks and identification of metabolites from NMR. The 1D and 2D NMR spectra recorded on a mixture of glucose and alanine sample is shown.
Software such as Chenomx NMRSuite [35] can be used for identifying compounds based on the 1D NMR spectrum. Some open-source programs have been designed to identify compounds by deconvoluting 1D 1H NMR spectra [61,62]. Several programs such as complex mixture analysis by NMR (COLMAR), MetaboMiner, PROMEB and ChemSMP are also available to identify compounds from 2D NMR spectra [63–66].
The assignment process is hindered if the spectrum is crowded with overlapping cross-peaks. The conventional approach is to use higher dimensional (2D) experiments. However, higher dimensional experiments require prohibitively long experimental acquisition time (in the order of days). Fast NMR data acquisition is, therefore, important in metabolomics in general. Towards this end, several methods have been developed to shorten the data collection time [67–76]. Our group has developed methods for rapid NMR data acquisition and assignments of molecules in molecules mixtures [65,66,72,73,75,77–79]. These methods provide time gain of a significant magnitude when compared with conventional NMR methods.
3. NMR spectroscopy in assisted reproduction
NMR spectroscopy has contributed significantly to identify and quantify biomarkers in the field of ART during the last 3 decades. As discussed above, in NMR, the level of metabolites in each ART sample type can be quantified accurately. Any statistical change (increase or decrease) in the metabolite levels, when compared with an appropriate control, helps to identify specific molecules or biomarkers that are responsible for the condition being studied. However, in a clinical scenario, NMR-based evaluation of various samples from ART is consumptive as NMR tested samples like spermatozoa, testicular tissues, embryos, etc. cannot be used in ART procedures.
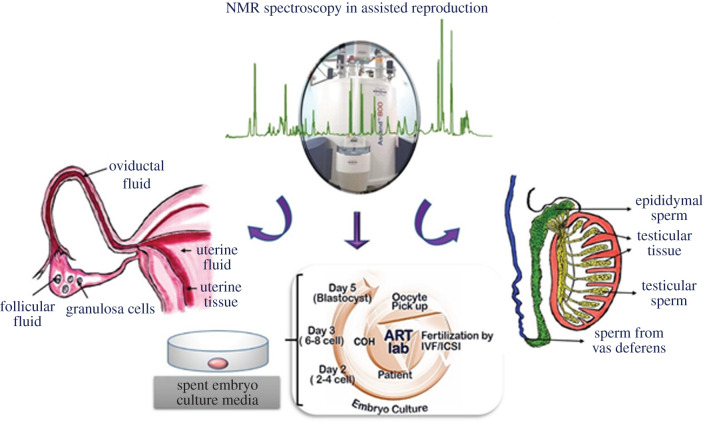
Initial studies on oocytes, spermatozoa and embryos using NMR spectroscopy focused on using 31P NMR to understand energy metabolism and related physiological conditions like intracellular pH, motility and growth [80–83]. The application of NMR spectroscopy in assisted reproduction on a variety of experimental samples collected during various stages of ART procedures is schematically represented in figure 3. All the samples are prepared using different methods, and relevant preparation methods are explained below. Following this, the NMR-based studies that have been carried out till date are discussed for each sample type.
Figure 3.
Schematic of the utility of NMR spectroscopy in assisted reproduction and a variety of sample types that have been studied using NMR spectroscopy.
3.1. Studies on the embryo spent culture medium
3.1.1. Composition of the different embryo culture media
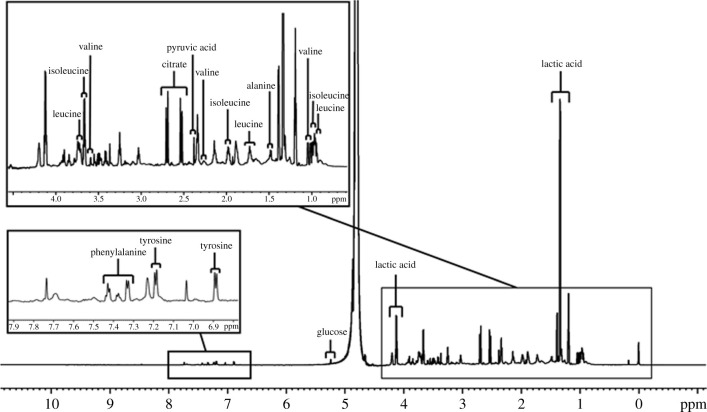
In clinical IVF, single embryo transfer is gaining popularity to prevent multiple gestations. The blastocyst is preferred over cleavage stage embryos due to a higher implantation rate of single blastocyst transfer [84,85]. A widely used approach is to investigate the metabolome of the culture media in which the embryo is grown, termed as the embryo spent culture media (SCM) as it is a clear reflection of an embryo's physiological or pathological status. Embryo SCM is altered by the developing embryo. A range of metabolic parameters of embryo SCM has been studied using a variety of non-invasive techniques [86–88]. Studies have focused on addressing the association between amino acid uptake, embryo viability and ART outcome [89–91]. NMR can detect the metabolic changes non-invasively and correlate with the embryo growth or implantation [23,92,93]. The preparation of SCM for NMR studies has been described earlier [23]. A representative 1D NMR spectrum of an embryo culture medium is shown in figure 4. The identity/assignment of metabolites is indicated on the peaks.
Figure 4.
One-dimensional proton NMR spectrum of a one-step embryo culture medium (Vitromed) with peak assignments shown for metabolites.
Human embryo culture was first introduced using a physiological salt solution containing glucose, lactate and pyruvate, supplemented with the patient's serum [94]. Later, the addition of amino acids to the culture medium both in humans and animals demonstrated a positive impact on embryo development and viability [95]. The quantitative analysis was done by Houghton [96] to estimate the amino acid turnover of individual human embryos during the early period of embryo development. Presently, most of the human embryo culture media are enriched with amino acids, energy substrates, growth factors and vitamins.
Currently, two types of media are used in human ART, one-step/single step (monoculture), and sequential culture systems [97]. One-step systems use a single medium component to support the zygote up to the blastocyst stage development. On the other hand, sequential media are used in two stages, first from zygote to the compaction stage and second from the compaction stage to the blastocyst stage. Various factors such as type of incubator, temperature, humidity and air quality result in some changes in the culture medium [98], and collectively, these alterations are known to affect (epi) genetic integrity of the embryo [99]. It has been shown that Sequential ISM1 (ORIGIO) medium resulted in a better embryo quality than that of a one-step Universal IVF (ORIGIO) medium [100]. Another study compared four one-step culture media compositions that varied notably in pyruvate, lactate and amino acids and concluded that blastocyst development was affected by culture media and its interaction with reduced (5%) and ambient (20%) oxygen [101].
Tarahomi et al. [101] compared 15 commercial embryo culture media to understand the composition and stability during storage. This study revealed that no two-culture media had an identical composition. Further, significant variation in the stability of the components is observed [102]. These observations suggest that there is no consensus on the media composition for human embryo development. The variation in media components is the primary cause of the conflicting observations in NMR studies on SCM. Sunde et al. [98] communicated a strong case for demanding full transparency and the scientific rationale concerning the compositions of embryo culture media as the composition can have a significant influence on the metabolic activity of the embryo.
Initially, SCM metabolites were studied by the modified ultra-microfluorescence method [18]. The global metabolomics approach using NIR and Raman spectra was used for the first time to assess the human embryo viability which could differentiate the embryos that resulted in successful or failed implantation [19]. Subsequently, NMR spectroscopy was applied to profile the human embryo SCM metabolites which revealed a high level of glutamate and alanine/lactate ratio in the embryos that were implanted and delivered successfully [22]. An attempt was made to combine non-invasive metabolomics and chemometric approach for the selection of best quality ART-derived embryos [103]. Subsequently, several groups have used NMR for profiling the metabolites in the SCM. The outcome of such studies is summarized in table 1 and discussed in detail below.
Table 1.
NMR studies of SCM of human embryos. LAH, laser-assisted hatching; Glc, glucose; Ala, alanine; Lac, lactate; Ile, isoleucine; Gly, glycine; Cit, citrate; Acac, acetoacetate; Thy, thymine; OAc, acetate; Trp, tryptophan; Lys, lysine; Pro, proline; Glu, glutamate; n.a., not available.
| study type |
study | study population |
outcome parameters | metabolic changes observed |
|||
|---|---|---|---|---|---|---|---|
| embryos transferred | day, post-fertilization | patients (N) | spent media (N) | upregulated | downregulated | ||
| multiple | 3 | Seli et al. [22] | 18 | 34 | implantation and pregnancy in ICSI patients | Glu and Ala/Lac | n.a. |
| multiple | 3 | Marhuenda-Egea et al. [103] | 23 | 46 | implantation and pregnancy for IVF patients | triglycerides, cholesterol compounds and phospholipids were compared | |
| single and multiple | 3 | Rinaudo et al. [104] | 108 | 228 | implantation, ICSI patients | NMR profiling of SCM cannot predict implantation | |
| single | 3 | Sánchez-Ribas et al. [105] | 85 | 171 | trisomy 21 versus monosomy 21 and euploid | caproate and androsterone sulfate | Ile |
| multiple | 2 and 3 | Pudakalakatti et al. [23] | 48 | 127 | implantation for ICSI patients | n.a. | Pyr/Ala |
| single | 3 and 4 | Nadal et al. [106] | 12 | 39 | embryo developmental arrest | (model with poor predictability) NMR profiles alone were not predictive of developmental arrest | |
| single, double and triple | 2 | Wallace et al. [107] | 37 | 58 | IVF/ICSI pregnancy outcome |
3-formate/Gly, formate/3-aminoisobutyrate, formate/Ac, formate/Glu and formate/Try | Cit/Ala, aminoisobutyrate/AcAc, 3-aminoisobutyrate/Ala, Cit/formate |
| single | 3 and 5 | Kirkegaard et al. [84] | 161 | 278 | pregnancy, IVF/ICSI patients, positively selected by good prognosis factors | NMR profiles were not predictive of pregnancy | |
| multiple | 3 | Uppangala et al. [108] | 25 | 50 | influence of LAH on embryo developmental potential | Glc, Pyr, Lac Ala, Val, Lys, Pro, Ile, and they were comparable between the groups | |
| multiple | 3 | Uppangala et al. [109] | 34 | 4 | sperm-mediated influence on the embryo developmental potential | Pyr and Pyr/Ala | Ala and Glu |
Pudakalakatti et al. [23] showed that an embryo with high-implantation potential consumed more pyruvate from the media for their energy requirements, converting it to into lactate by lactate dehydrogenase or into alanine by transamination reaction to avoid the accumulation of ammonia, which causes cell toxicity. The excess alanine thus generated resulted in the increase in alanine in the spent media of embryos with high-implantation potential. Hence, low pyruvate to alanine ratio in the SCM was proposed as a potential biomarker for the selection of embryo with high-implantation potential [23]. On the other hand, a study conducted by Nadal-Desbarats et al. [107] characterized the metabolites from individually cultured human embryos on days 3 and 4 [106]. However, this study failed to demonstrate any statistical significance between successful and unsuccessful implantation groups. Wallace et al. characterized the SCM metabolites from patients undergoing single, double and triple embryo transfers to predict the pregnancy outcome. This study identified a wide variety of ratios, among which formate/glycine ratio was 17-fold high in day 2 embryo resulting in successful implantation, whereas the citrate/alanine ratio was 22-fold less in the same group. Further, a significant increase in the ratios of formate/3-amino butyrate, formate/acetate, formate/glutamate and formate/tryptophan were characterized for embryos that resulted in successful implantation [107].
While the above studies found the value of NMR analysis in predicting ART outcome based on the level of the metabolites in SCM, Rinaudo et al. [104] reported that NMR-based metabolomics profiling cannot be used as a tool for the prediction of embryo implantation potential. Similarly, Kirkegaard et al. [84] found no correlation between the NMR metabolic profile of SCM and pregnancy outcome, patient or treatment characteristics.
Notably, the NMR-based profiling of metabolites from SCM has also been used for the differentiation of euploid and aneuploid embryos [105]. Sánchez-Ribas et al. [105] investigated the metabolomic signatures of trisomy 21 (T21)/monosomy 21 (M21) and the euploid embryos using NMR and were able to differentiate between normal and aneuploid embryos; they also suggested that isoleucine levels could be used as a differentiator between T21/M21 and normal embryos. Similarly, embryo derived from sperm carrying high DNA damage had reduced pyruvate uptake from the media, reduced alanine released into the media and glutamine intensity was significantly low in the SCM [109]. These approaches suggest the possibilities of using SCM metabolites as non-invasive biomarkers in screening out the genetically abnormal embryos. However, there needs to be more research to understand the association between the turnover of metabolites and the type of genetic lesions and the extent of genetic abnormalities in the embryos.
Laser-assisted hatching (LAH) is one of the commonly used techniques in ART to enhance the implantation process. Since hyperthermia induced by the laser can negatively affect the embryo viability, Uppangala et al. [108] conducted a study to understand the impact of LAH on embryo quality. LAH was done on day 2 of the embryonic development and SCM collected 24 h later was subjected to NMR analysis. Metabolites were compared between LAH embryos and the sibling unhatched embryos and results suggested that the LAH does not affect the metabolism of embryos [108].
The metabolic profiling of SCM has also been carried out in mouse and bovine embryo culture systems. Unlike human embryos, murine models allow the selective manipulation of embryo growth conditions and to monitor the effectiveness of such changes on the embryo metabolism. Historically, bovine embryos have been used to study early embryo developmental stages as the early developmental mechanism in cattle and humans are very similar. Such studies have always contributed towards the advancement in human IVF procedures. The details of these studies are summarized in table 2.
Table 2.
NMR studies of animal embryo SCM. CL, cleavage stage; BL, blastocyst stage; IVF, in vitro fertilization; PA, parthenogenetically activated; Cit, citrate; Pyr, pyruvate; Lys, lysine; Ac, acetate; Phe, phenylalanine; His, histidine; Trp, tryptophan; Val, valine; Glc, glucose; Lac, lactate; Thy, thymine; Pro, proline; Ala, alanine, Ile, isoleucine; n.a., not available.
| study model | study type | study | study population | analysis carried out | metabolic changes observed |
|
|---|---|---|---|---|---|---|
| upregulated | downregulated | |||||
| bovine | 2-day culture, IVF-derived and parthenogenetic-activated embryos | Rubessa et al. [110] | CL 20, BL 20, | blastocyst development | Cit, Pyr and Lys | Myo-inositol |
| IVF 20, PA 20 | IVF versus PA | Phe | ↓OAc | |||
| individual culture and group culture (30/drop) | Perkel et al. [111] | 113 | fast-growing and slow-growing embryos | 4 cell-Pyr | 4 cell: n.a. | |
| 8 cell and 16 cell: His, Trp, Val | 8 cell and 16 cell: n.a. | |||||
| 2 cell and 16 cell: Ile, Leu | 2 cell and 16 cell: n.a. | |||||
| group culture system | Rubessa et al. [83] | male 20, female 20 | sex determination of embryo at early (1–3 days) and late development stage (5–7 days) | 1–2 days: Val | 1–2 days: n.a. | |
| 5–7 days: n.a. | 5–7 days: Pyr | |||||
| mouse | group culture (10/drop) | D'Souza et al. [112] | 185 | embryo carrying induced DNA lesion on day 3.5 | Glc, Lac, Pyr, Thy, Pro, Val, Ala, Ile and Lys | n.a. |
| individual culture | D'Souza et al. [93] | 118 | blastocyst in vitro outgrowth | Pyr and Lac | Pyr/Ala | |
| individual culture | D'Souza et al. [113] | 90 | blastocyst | Pyr | Ala, Pyr/Ala and Pyr/Lac | |
Perkel & Madan [111] carried out the first NMR-based metabolomics study of individually cultured bovine embryos. The study revealed a difference in the requirement of metabolites between slow-growing (SG) and fast-growing (FG) bovine embryos [111]. Global metabolome analysis of spent culture medium can also be used for gender differentiation [83]. A study conducted by Rubessa et al. [83] in bovine embryos showed that gender-dependent metabolite differences can be seen in the global metabolome analysis. While the bovine model is extensively used to understand the pathophysiology of human reproductive function, cattle are known ruminants; hence, one can expect significant differences in the metabolism between bovine and humans. Since the present review is focused on biomarkers for human IVF, there is a potential pitfall in extrapolating bovine data to the clinical IVF.
D'Souza et al. [93] used mouse embryo SCM to predict the implantation ability of the embryos which showed the increased requirement of pyruvate and lactate accompanied by a significant reduction of pyruvate/alanine ratio in the SCM. Further, sperm-induced genetic lesions in the embryos are associated with a significant increase in the uptake of pyruvate and reduced pyruvate/alanine ratio in SCM [113]. Data from animal studies provide only fundamental knowledge but cannot be directly extrapolated to human ART due to species-specific differences in the metabolic requirements, chromatin architecture and developmental timelines.
Despite the promising observations by several researchers, embryo SCM metabolomics by NMR has not been implemented clinically, possibly due to technical limitations in establishing collaboration between ART clinics and NMR facilities, the time requirement for NMR analysis and the need of experts to interpret the NMR results. Further, the discrepancies observed so far could be attributed to the great heterogeneity in the variables used in each study, such as the composition of embryo culture media, culture conditions, collection technique, handling, and transportation of SCM, and measurement technique. Thus, the standardization of variables is essential before using NMR metabolomics as a complementary or independent tool for embryo selection in human ART.
3.2. NMR studies of follicular fluid
The accomplishment of oocyte developmental competence is crucial for the formation of viable, good quality embryos. The oocyte growth and development depend primarily on the nurturing microenvironment of the follicle, including the FF. The FF consists of exudates of the circulating plasma, secretion from the follicular and granulosa cells and essential substances like growth factors, cytokines, amino acid, energy substrates, steroids, lipids and cholesterol that are necessary for the oocyte maturation and fertilization in vivo [114,115]. Several NMR studies have been performed in search of a biomarker from FF for assessing the embryo quality (table 3). The preparation of FF for NMR analysis has been described [118].
Table 3.
NMR studies of the follicular fluid. Lac, lactate; Glc, glucose; HDL, high-density lipoprotein; Pro, proline; Leu, leucine; Ile, isoleucine; Chol, choline; ChoP, phosphocholine; GPC, glycerophosphocoline; Glyc, glycerol; TMA, trimethylamine; Pyr, pyruvate; Ac, acetate; Gly, glycine; Gln, glutamine; Ala, alanine; Asp, aspartate; Asn, asparagine; Cit, citrate; Cr, creatinine; 3-HB, 3-hydroxybutyrate; Lys, lysine; Met, methionine; Phe, phenylalanine; n.a., not available; FF, follicular fluid.
| study model | study type | study | study population |
metabolic changes observed |
|||
|---|---|---|---|---|---|---|---|
| no. of patients | no. of FF | analysis carried out | upregulated | downregulated | |||
| human | oocyte donors women undergoing IVF | Piñero et al. [114] Wallace et al. [116] | 30 | 30 | oocyte quality | 42 metabolites identified | |
| 58 | 108 | developmental and implantation potential of the embryo | 2 cell: Lac and Chol/ ChoP | 2 cell: Glc and HDL | |||
| implantation: Pro, Lac and Leu+ Ile | implantation: Glc | ||||||
| women undergoing natural cycle versus IVF/ICSI | McRae et al. [117] | 10 | 10 | effect of exogenous hCG administration during pre- and mid-ovulatory period | FF: Lac, Pyr | FF: Glc | |
| plasma: Glc and Ac | plasma: glycoprotein, Gly and trimethylamine | ||||||
| PCOS patients | Zhang et al. [118] | 51 | 51 | metabolic profile and oocyte quality | glycoprotein, Ac and cholesterol | Lac, Gln, Pyr and Ala | |
| PCOS patients | Nunzia et al. [119] | 41 | 41 | AMH versus FF metabolites | Glc | Lac, Pyr and Ala | |
| different stages of endometriosis | Marianna et al. [120] | 30 | 60 | metabolic profile and oocyte quality | Lac | Lys, Asp, Chol, Pro, Ala, Leu, Val and ChoP | |
| benign and malignant | Morelli et al. [121] | 20 | 20 | metabolic profile and fertility preservation | Cit, Cr, Glyc, GPC and Glc | Asn, Asp, Pro, cholesterol, Chol, Lac and lipids | |
| endometriosis | Karaer et al. [122] | 24 | 24 | metabolic profile | Lac, β-Glc, Pyr and Val | n.a. | |
| different female factor infertility | Morelli et al. [123] | 53 | 53 | metabolic profile | PCOS: Glc, Glycerol, Cr | Ac, Leu, Thr, β-HB | |
| endometriosis: Glc, Lac | Ac, β-HB, Cit and Val | ||||||
| sheep, pig and cow | FF, ovarian venous blood | Gosden et al. [124] | sheep 3, pigs 5, cows 4 | metabolic profile | Ac, Ala, Cr/Creatine, Gly, D-3-HB, Lac and Val identified | ||
| mare | FF and serum | Gérard et al. [125] | FF 20 and serum 20 | early, late and pre-ovulatory stage dormant follicle | Ala and lipoprotein | TMA, glyco-conjugates, Ac and Glc | |
| pig | FF | Bertoldo et al. [126] | summer small 8 and large 15, winter small 9 and large 15 | metabolic profile of good and poor follicular environments | follicular size alone: Glc | follicular size alone: Lac, hypoxythine, Ac, inositol, TMA, Ala, Leu, Lys, Met and Phe | |
| follicular size and season: Glu, Gly, N-acetyl group and uridine identified | |||||||
| season alone: succinate | n.a. | ||||||
| mare, sow and cow | FF | Gérard et al. [115] | small 5 and large 5 per animal group | metabolic profile of small and large follicle from three species | α and β Glc | n.a. | |
In general, the NMR-based metabolomics characterization of FF reveal variations in the composition in relation to species [124], oestrous cycle stage [117,125], seasonal influence [126], underlying pathology such as PCOS [118,119,123], endometriosis [120,122,123], benign and malignant ovarian cancers [114], other infertility conditions [123] and the size and quality of the developing oocyte [114,115,126]. Association has also been established between FF composition and the reproductive potential of the developing oocyte and embryo [116]. Gosden et al. [124] carried out the first NMR-based study to characterize the FF metabolites from cow, sheep and pig and showed a species-specific difference in the composition of metabolites.
Few studies have investigated the clinical value of FF analysis by NMR in human ART. Wallace et al. [116] for the very first time studied FF samples from women undergoing IVF and observed a high level of lactate and choline/phosphocholine, and a low level of glucose and high-density lipoprotein (HDL) in the FF collected from the oocyte that fertilized and formed good embryos, compared with the FF from the oocyte which fertilized but failed to develop further. Similarly, metabolomic profiling of FF was found useful in predicting the developmental competence of the human oocyte, thereby facilitating embryo selection [117].
Factors such as the day of follicular aspiration, size of the follicle, synchrony of the follicular cohort within the ovary and patients' pathology do influence the follicular microenvironment. Gonadotrophin stimulation in ART normally results in the recruitment of multiple follicles where time-dependent increase in the follicle size and their synchronous development play a crucial role in producing mature and competent oocytes. Follicle size-dependent variation in metabolic profile was observed in the FF composition [115]. Gérard et al. studied the FF of the small and large follicles collected from three different species; cow, sow and mare. Even though there exists a species-specific difference in the FF composition, in general, all species were found to have a higher concentration of α and β glucose in the large follicles (possibly mature) compared with the small follicles (possibly immature) [115]. However, it has been reported that not only the follicle size but also the season and the quality of the follicle have an influence on the porcine FF composition [126]. Nonetheless, at this juncture, there is no conclusive evidence to apply FF metabolites as biomarkers in human ART.
PCOS is one of the major causes of female factor infertility, affecting 5–10% of women of reproductive age [123]. The major metabolic dysregulation among PCOS patients is glucose intolerance due to insulin resistance, dyslipidaemia and hyper-androgenization which eventually influences the follicular environment and oocyte quality. Biochemical studies of the FF found that carbohydrate/glucose metabolism is impaired due to hyperinsulinemia in PCOS patients, whereas NMR-based metabolic profiles have suggested that glucose is positively correlated with the serum level of anti-Müllerian hormone (AMH) which is a marker for ovarian reserve. On the other hand, the level of lactate, pyruvate and alanine were negatively correlated with AMH. From the metabolomics point of view, glucose, lactate and pyruvate are directly involved in the glycolytic pathway and cumulus and granulosa cells that are responsible for the transfer of energy intermediate of the glycolytic pathway to the developing oocyte are unable to do so due to the insulin resistance, leading to an elevated level of glucose and overuse of the alternative pathways [119]. A significant difference in the FF metabolites between PCOS and non-PCOS patients and their impact on oocyte quality and fertilization outcome was demonstrated by NMR [123]. Hence, the alterations in pyruvate metabolism, glycolysis, and amino acid metabolism in FF help in explaining the PCOS pathogenesis and its impact on the oocyte environment within the follicle [118].
Endometriosis is another major cause of female infertility which compromises the oocyte quality and implantation potential. The metabolomics-based analysis of FF collected from the patient with endometriosis showed elevated concentration of glucose, lactate and pyruvate indicating more demand for glycolysis and anaerobic metabolism compared with the healthy control [120,122,123]. On the other hand, the concentration of acetate, citrate and β-hydroxybutyrate were reduced in the FF of endometriosis patients [123]. Valine is an essential branched chain amino acid attributed to the inflammatory response in endometriosis patients. Studies conducted by Castiglione Morelli et al. [123] and Marianna et al. [120] reported a low level of FF valine, whereas Karaer et al. [122] showed an elevated level in the endometriosis patients compared with the healthy control. Further, the concentration of few fatty acids, lysine, choline, glucose, aspartate, alanine, leucine, valine, proline, phosphocholine, total LDH as well its LDH-3 isoform were found to be reduced in endometriosis patients compared with the healthy control [120].
These observations can aid-in understanding the impact of specific pathological conditions such as PCOS and endometriosis on the follicular microenvironment and how altered metabolites can further affect the functional ability of the oocytes in ART.
Restoration of fertility in cancer survivors is a challenging task and still experimental. It is well known that cancer affects several metabolic pathways in general. Castiglione Morelli et al. [121] looked at the metabolite profile of FF from cancer patients which showed a reduction in the concentration of asparagine, aspartate, proline, cholesterol, choline, lactate, lipids compared with the healthy group. Also, an upregulation of the citrate, creatine, glycerol, glycerophosphocholine and glucose concentration was observed in cancer patients compared with the healthy control [121]. Hence, NMR-based metabolomics could serve as a prognostic tool for identifying and selecting the oocytes in relation to FF metabolites, and thereby help in predicting the ART outcome when cancer survivors are undergoing fertility restoration treatment.
3.3. NMR studies of blood serum/plasma
Examination of plasma composition during the follicular and periovulatory phases of the menstrual cycle in patients undergoing fertility treatment provides insights into the underlying pathology. PCOS is known to impair oocyte quality, endometrial receptivity and embryo implantation potential, which can be captured through metabolites from several body fluids including blood serum or plasma. Standard protocols are followed for the collection and preparation of the serum or plasma samples used in NMR [29,30]. A study conducted by Zhao et al. showed that the difference in serum metabolomic profile in PCOS and control patients was characterized by elevated glycolysis, reduced tricarboxylic acid cycle (TAC), reduced ratio of branched chain amino acid/aromatic amino acid in PCOS. The results also suggested upregulated concentration of serine, threonine, phenylalanine, tyrosine and ornithine in PCOS patients could be the main reason for ovulatory dysfunction [29].
RoyChoudhury et al. explored the value of serum metabolites to understand the pathophysiology of recurrent implantation failures. The concentration of eight metabolites (valine, adipic acid, lysine, creatine, ornithine, glycerol, d-glucose, urea) were elevated in the serum of patients with recurrent implantation failure compared with control [127]. Details of these studies are stated in table 4.
Table 4.
NMR studies of the human blood serum/plasma. Ser, serine; Thr, threonine; Phe, phenylalanine; Val, valine; Leu, leucine; Gly, glycine; Orn, ornithine; Glc, glucose; Lys, lysine; n.a., not available.
| study type | study | n | analysis carried out | metabolic changes observed |
|
|---|---|---|---|---|---|
| upregulated | downregulated | ||||
| PCOS | Zhao et al. [29] | 265 | potential metabolic profiles for different phenotypes of PCOS | Ser, Thr, Phe, Tyr, Val, Leu and Orn | Gly |
| repeated implantation failures (RIF) versus repeated implantation success (RIS) | RoyChoudhury et al. [127] | 52 | metabolic dysfunction associated with RIF | Vali, adipic acid, Lys, creatine, Orn, glycerol, d-Glc and urea | n.a. |
These studies have significance not only in identifying novel metabolic biomarkers that predict specific pathological conditions but also in better understanding of the metabolic pathways directly affected during these diseases, which may provide a rationale for the development of novel therapeutics.
3.4. NMR studies of oocytes and embryos
The oocyte contributes 70–80% towards the successful fertilization and further embryonic development. Hence, knowing the biochemical status of the oocyte helps to enhance the reproductive outcome. One of the rate-limiting steps in ART is the selection of a single embryo with the highest implantation potential. Understanding the embryo kinetics and metabolism by NMR may help to tackle this problem. Towards this end, a few studies have been performed, the details of which are summarized in table 5. However, it is important to note that in-cell analysis of oocytes/embryos is restricted to non-human oocytes and embryos due to ethical restriction in exposing them to invasive preparation such as centrifugation and NMR as described earlier [129]. Rubessa et al. [110] used NMR to study the metabolic requirements of bovine gametes and embryos during the in vitro production and thereby made modification in the culture media composition. This study analysed the pyruvate, lactate and alanine concentration of SCM during of gametes co-incubation. The concentration of pyruvate was significantly reduced during the culture of oocyte–cumulus complex (OCC) alone and OCC co-incubated with sperm. This observation helped investigators to modify the embryo culture media composition by reducing the concentration of pyruvate and lactate which improved the bovine embryonic development [110].
Table 5.
NMR studies of the oocytes and embryos. Pyr, pyruvate; Lac, lactate; n.a., not available.
| study type | study | study population |
analysis carried out | metabolic changes observed |
||
|---|---|---|---|---|---|---|
| no. of animals | n | upregulated | downregulated | |||
| bovine oocyte | Rubessa et al. [110] | n.a. (911 oocytes) | 35+ | evaluation of energy consumption during gamete co-incubation and reformulation of IVF media | n.a. | Pyr and Lac |
| mouse embryo | Lyman et al. [128] | n.a. | 402 | embryo viability in real time using NMR | n.a. | n.a. |
In-cell NMR analysis of the mouse embryos was conducted to understand the embryo viability and the study found that in-cell NMR analysis has an adverse effect on the in vitro development of the mouse embryos. However, the observed effect was mainly because of the medium used for the analysis. Hence, modification in the analysing medium may provide better information on embryo viability [128]. However, it is important to note that exposing embryos to NMR may have potential long-term detrimental effects, hence practically not possible to apply this method in a clinical setting.
3.5. NMR studies of testicular tissue
The biochemical status of the testicular tissue has a direct influence on the process of spermatogenesis. NMR-based metabolomics helps to understand the hidden cause of male infertility. In the experimental set-up, making good use of NMR spectroscopy, characterization of metabolites could help in understanding bio-dysfunction of gonads and aid in developing therapeutic options [130]. The details of such studies are summarized in table 6. Sample preparation for NMR analysis has been described earlier [133]. Human testis was initially studied by Chew et al. [131] using 31P NMR and found that that phosphomonoester (PM)/β adenosine triphosphate (ATP) and PM/phospho-diester ratios were low in infertile patients compared with the healthy control, whereas the ratio of phosphate/PM was high in patients with primary testicular failure and chronic tubular obstruction compared with the control. Rat testicular tissue had a high concentration of lactate, alanine, phosphatidylcholine and creatinine, which help to meet the energy requirements and to maintain the cell viability [132]. Jarak et al. [133] addressed the association between rat testicular senescence and the related decline of fertility using 31P NMR which revealed an age-dependent decline in the antioxidant metabolites like betaine, creatine and glutathione; the elevated level of phenylalanine and tyrosine; decrease in nucleotide synthesis (IMP, CMP, ATP) and increase in testicular content of phospholipid (choline, ethanolamine, myo-inositol, glycerol) precursors. At present, results are inconclusive to suggest the utility of testicular tissue NMR analysis in the diagnosis and clinical management of infertility.
Table 6.
NMR studies of the testicular tissue. PM, phosphomonoester; ATP, adenosine triphosphate; Pi, inorganic phosphate; n.a., not available; Cr, creatine; Lac, lactate; Ala, alanine; Phe, phenylalanine; Tyr, tyrosine.
| model | study | study population |
analysis carried out | metabolic changes observed |
||
|---|---|---|---|---|---|---|
| patients/animals | samples | upregulated | downregulated | |||
| human | Chew et al. [131] | 23 | 23 | testicular metabolic integrity and differentiation of normal testicles | Pi/PM | PM/β-ATP and PM/phospho-diester |
| rat | Griffin et al. [132] | 6 | 1 | differential metabolomics profiling | Cr, phosphatidylcholine, Lac and Ala | n.a. |
| Jarak et al. [133] | 40 | 5 | senescence and declining reproductive potential | choline, ethanolamine, myo-inositol, glycerol, Phe and Tyr | betaine, Cr and glutathione | |
3.6. NMR studies of spermatozoa
The structural and functional integrity of spermatozoan can seriously affect its fertilizing ability and the developmental competence of embryos. NMR has been used extensively to address the biochemical environment of spermatozoa (table 7). 31P NMR analysis of male gametes in the sea urchin, turbot, Xenopus laevis, etc. have addressed the structure and differences in functional characteristics [80,139–142]. Using NMR, Smith et al. [143] found that bovine spermatozoa use an unknown energy source for the phosphorylation of ATP and thereby maintains acidic cytosolic pH. This study also found that prolonged incubation of sperm with media promotes the accumulation of lactate in the surrounding media and leads to an exchange of internal K+ for H+, thereby reducing the intracellular sperm pH.
Table 7.
NMR studies of the spermatozoa. Lac, lactate; GPC, glycerophosphorylcholine; Chol, choline; n.a., not available.
| model | study | study population |
analysis carried out | metabolic changes observed |
||
|---|---|---|---|---|---|---|
| animals/subjects | samples | upregulated | downregulated | |||
| rhesus macaque | Lin et al. [134] | 6 | n.a. | season-dependent energy metabolism | formate, carnitine and acetyl carnitine | Lac |
| human | Paiva et al. [135] | 9 | 3 | endogenous metabolism | 42 metabolites identified | |
| Reynolds et al. [136] | n.a. | n.a. | progressive motility versus oxidative phosphorylation and lactate fermentation | Lac and bicarbonates | n.a. | |
| Reynolds et al. [137] | n.a. | n.a. | metabolomics of live sperm (40% and 80%) | Lac, lipid and Chol/GPC | n.a. | |
| Calvert et al. [138] | 97 | n.a. | glycolysis and oxidative phosphorylation for viable sperm population | Lac | n.a. | |
NMR studies have proved useful for the development of metabolomic markers that signal sperm metabolic impairments. The preparation of spermatozoa for 1H NMR has been described earlier [135]. NMR profiling of monkey spermatozoa collected during different seasons (i.e. autumn and spring) have shown that there exists a seasonal-dependent variation in the metabolic profile characterized by a high level of formate in the autumn season and high level of carnitine and acetylcarnitine during the spring season [134]. This study also found that glycolysis plays a major role to yield ATP in the monkey's sperm. Extensive intracellular metabolomics characterization of 69 intracellular metabolites in human spermatozoa done by the combined application of 1H NMR spectroscopy and gas chromatography–mass spectrometry (GC-MS) [135] may describe their function in male gamete physiology and to help in exploring potential causes for sperm dysfunction.
Reynolds et al. profiled percoll gradient separated live human spermatozoa to demonstrate differential metabolomic signature in relatively low concentration of sperm (approx. 3 × 106 ml−1) which could open up the possibilities of developing metabolomics-based diagnostics to test poor quality ejaculates [137]. The same group used dissolution dynamic nuclear polarization (dDNP) to enhance NMR sensitivity to study the correlation between the energy metabolism and sperm motility in human which showed 13C labelled pyruvate was converted significantly into lactate and bicarbonate, indicating the active glycolytic and oxidative phosphorylation in progressively motile spermatozoa [136].
3.7. NMR studies of seminal fluid/plasma
Seminal fluid has important roles in sperm survival and overall fertilization success. Biochemical changes in seminal plasma composition may alter the fertility potential of the spermatozoa. It is, therefore, important to consider the implication of different biomarkers in seminal plasma, both in the diagnosis and treatment of male infertility. Studies conducted on seminal plasma using NMR are summarized in table 8. Seminal plasma preparation for NMR analysis has been described earlier [86]. The metabolome of human seminal plasma secretion in patients with vasal aplasia and non-obstructive infertility revealed that the ratio of citrate/choline and spermine/choline were twofold higher in these patients compared with fertile subjects [144]. Further, an attempt to use NMR profiling of seminal plasma to distinguish the obstructive and non-obstructive forms of azoospermia and different forms of spermatogenic failure has shown that glycerophosphorylcholine (GPC)/choline ratio can serve as an important parameter to differentiate between different forms of spermatogenic failure [145].
Table 8.
NMR studies of the seminal fluid/plasma. GPC, glycerophosphorylcholine; Cit, citrate; Lac, lactate; Chol, choline; Glc, glucose; Cit, citrate; Ala, alanine; His, histidine; GPE, glycerylphosphorylethanolamine; UMP, uridine 5 monophosphate; Thy, thyrosine; Phe, phenylalanine; Lys, lysine; Fra, fructose; Trp, tryptophan; Cr, creatinine; PCr, creatine phosphate; Gly, glycine; n.a., not available.
| model | study | study population |
analysis carried out | metabolic changes observed |
||
|---|---|---|---|---|---|---|
| animals/subjects | samples | upregulated | downregulated | |||
| human | Lynch et al. [144] | 19 | 19 | vas deferens obstruction and sperm antibodies | Cit/Chol and spermine/Chol | n.a. |
| Hamamah et al. [145] | 60 | 60 | differential metabolomics profiling of different infertility condition | n.a. | GPC, Cit and Lac | |
| glycerylphosphorylethanolamine (GPE), Cit/Lac, GPC/Lac and GPE/GPC are significant | ||||||
| Tomlins et al. [146] | n.a. | n.a. | time-dependent changes | phosphorylcholine to Ch and UMP to uridine identified | ||
| Sharma et al. [147] | 37 | n.a. | effect of new injectable male contraceptive | n.a. | Glu, Lac, GPC, Chol, Cit/Lac and GPC/Chol | |
| Gupta et al. [148] | 230 | 230 | effect of Mucuna pruriens | Ala, Cit, GPC and His | Phe | |
| Gupta et al. [149] | 185 | 185 | oligoazoospermia | Phe and Tyr | Ala, Cit and GPC | |
| Gupta et al. [150] | 230 | 230 | effect of Withania somnifera | Lac, Ala, Cit, GPC, His and Glu | Phe | |
| Bonechi et al. [151] | different infertility condition | GPC, Chol, Phe, Cit, Lac, His and uridine identified | ||||
| 83 | 83 | |||||
| Zhang et al. [152] | 63 | 63 | asthenozoospermia | 19 metabolites and 5α-cholesterol and 7-ketocholesterol are upregulated | ||
| Darbandi et al. [153] | 151 | 151 | ROS induced changes | TMA N-oxide | Trp and Tyr/Tyrosol | |
| Mumcu et al. [154] | 59 | 59 | oligoastheno teratozoospermia | Tyr | Cit, spermine, putrescine, α-keto glutaric acid, Val, Lac, Cr, Lys, Arg and Gln | |
| Mehrparvar et al. [155] | 29 | 29 | teratozoospermia | Cit, Ch, d-Glc, l-Tyr, l-Ala, l-Leu, l-Pro, l-Lys, myoinsitol, l-Lac, Thr, Pyr, Gln, Val and Ile | l-Glu, taurine and cholesterol | |
| rat | Ebrahimi et al. [156] | 24 | n.a. | Ala, Lac and His | n.a. | |
Time-dependent biochemical changes were analysed in the seminal fluid by co-incubation with EDTA. NMR profile revealed a rapid observable change in the conversion of phosphorylcholine to choline and uridine-5-monophosphate (UMP) to uridine [146]. This observation may help in understanding the interaction between spermatozoa and seminal plasma and to address abnormalities in the seminal liquefaction process.
Seminal plasma analysis by NMR was used to test the effects of natural products on sperm functional improvement. Mucuna pruriens therapy rectifies the hormonic balance of endogenous metabolites such as alanine, citrate, GPC, histidine and phenylalanine content in seminal plasma and improved the semen quality in infertile men [148]. The effectiveness of Withania somnifera plant extract in male infertility management showed restoration of lactate, alanine, glutamate, citrate, GPC and histidine in the human seminal plasma over a period of three months and reduced phenylalanine level [150]. Similarly, NMR helped in understating the effects of Eurycoma longifolia extracts in improving rat sperm concentration by studying the level of alanine, lactic acid and histidine in the post-treated rat seminal plasma [156].
The use of NMR for metabolic profiling of seminal plasma has rendered it as a diagnostic tool for male infertility. Lower levels of seminal plasma metabolites such as alanine, citrate and GPC and a higher level of tyrosine, and phenylalanine were found in oligozoopsermic (decreased sperm number in the ejaculate) men [149]. Asthenozoospermia (decreased sperm motility) was characterized by the upregulation or downregulation of 19 metabolites responsible for lipid metabolism, phospholipids (choline) metabolism, cholesterol metabolism, nucleoside metabolism, the Krebs cycle and energy metabolism [152]. Further, a high level of 5α-cholesterol and 7-ketocholesterol was clearly evident in asthenozoospermic patients, indicating the degree of oxidative stress in the group compared with the control [152]. The level of citric acid, choline, d-glucose, tyrosine, alanine, proline, leucine, lysine, myo-inositol, lactic acid, threonine, pyruvate, glutamine, valine and isoleucine were high and glutamic acid, cholesterol and taurine levels were low in the seminal plasma of teratozoospermia (increased number of morphologically abnormal spermatozoa) [155]. It has been proposed that NMR signature of seminal plasma along with semen analysis data can be used to categorize the different infertility condition [151]. The model showed a well discrimination between leukocytospermia (pus cells in the ejaculate) and concomitant varicocele/ex varicocele, also testicular cancer, necrozoospermia (increased number of dead spermatozoa) and azoospermia (no sperm in the ejaculate) from other conditions. The metabolites including GPC, a cluster of metabolites consist of choline, phenylalanine, citrate, lactate, and histidine and uridine played an important role in the clustering different infertility condition [151]. Oxidative stress can significantly influence functional and genetic integrity of spermatozoa. Elevated seminal plasma reactive oxygen species (ROS) can result in the downregulation of tyrosine/tyrosol and tryptophan and upregulation of the trimethylamine N-oxide levels [156].
Since seminal plasma is a complex biological fluid, NMR application in studying the metabolites so far have provided general information on the association between type of spermatogonial disorders [144–146,149,151,152,155], and effect of treatment modalities on change in the level of metabolites [148,150,156]. Though these observations may not have any direct implications on human ART, development of specific biomarkers in predicting genetic and functional integrity of male gametes non-invasively may open up a new era in the non-invasive sperm selection.
4. Emerging trends and future challenges
Non-invasive evaluation of embryos in ART clinic has advanced by multiple emerging techniques such as NMR, Raman, NIR and MS. Lately, the ability to predict embryo potential has been enhanced by introduction of time-lapse imaging systems (TLS) in ART clinics. It allows the assessment of embryo quality without physically removing them from the incubator. TLS has been used to explore the associations between the morphologic, morphometric and morpho kinetic parameters of oocytes, zygotes and embryos, and their associations with live birth [15,157].
The field of NMR is constantly evolving, and several new techniques are being developed to improve upon the limitations of existing NMR-based metabolomics. These include hyperpolarization methods [158], ultrafast 2D NMR methods [37,55], pure-shift NMR techniques [159,160] and hybrid NMR approaches [24,26,161,162]. Inhomogeneous intact tissue samples can be analysed with the use of high-resolution magic-angle spinning (HRMAS) spectroscopy [163,164]. Using HRMAS, tissue samples can be examined without the need for sample extraction or other sample preparation steps [163,164]. Hyperpolarization methods such as DNP can be used to enhance the NMR signal intensities of metabolites, hence improving the sensitivity of NMR experiments [165]. In principle, the sensitivity enhancement of three orders of magnitude can be obtained [166,167]. DNP allows for the detection of metabolites present in very low concentrations [168].
Together, with the advent of new NMR methods and stronger magnets, NMR spectroscopy is providing a complementary tool to explore various aspects of assisted reproduction. There are several isotope labelling methods in NMR-based metabolomics [78] that can be employed in ART. NMR serves as a research tool to explore and unravel the metabolic signature of embryos and to discover biomarkers. However, in the clinical IVF centre, an NMR set-up is not needed. Instead, a simple assay or kit-based approach can be developed to rapidly detect and quantify the biomarkers on-site that have been discovered by NMR. For detection of the metabolites at the IVF site, two approaches can be implemented: (i) a colorimetric assay involving enzymatic conversion of metabolites followed by detection and quantification using absorption spectroscopy, or (ii) HPLC. Both these methods can be calibrated with known standards and cross-validated with the quantitative results obtained from NMR spectroscopy. Our laboratories are currently working in this direction to develop IVF diagnostic kits.
Precision medicine, an emerging branch of medicine, functions with prospects of the customization of healthcare (i.e. medical practices, decisions and treatments are tailored to individual patients [169]). The introduction of automatized embryo morphological evaluation using time-lapsed imaging in conjunction with using artificial intelligence (AI) is an attractive possibility [15]. Integration of NMR-based metabolomics and clinical parameters into an AI model will have powerful diagnostic and prognostic values in assisted reproduction. This will help embryologists to optimize techniques and personalize the treatment to maximize the reproductive outcome in the field of ART.
Data accessibility
This article has no additional data.
Authors' contributions
The manuscript was written through contributions of all authors and all authors have given final approved for publication. G.A. has coordinated with corresponding authors for designing outline of the review and written the abstract, introduction, NMR in ART, embryo culture media and part of the last section of the review. A.C. has written major contents of all the tables and all the text explaining tables of the review. D.J. has written the NMR in metabolomics, emerging trend and future challenges section. H.S.A. and S.K.A. have designed the outline, helped in writing and revising the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by Science and Engineering Research Board (SERB), DST, MAHE and CSIR. The facilities provided by the NMR Research Centre at Indian Institute of Science (IISc) funded by Department of Science and Technology of India (DST), Manipal Assisted Reproduction Centre (MARC) and Department of Clinical Embryology, Kasturba Medical College, Manipal, are gratefully acknowledged. H.S.A. and S.K.A. acknowledge support from the Science and Engineering Research Board (SERB) research grant (EMR/2017/003181). G.A. acknowledges financial assistance given by Manipal Academy of Higher Education (MAHE) under Post-Doctoral fellowship (PDF). A.C. acknowledges Dr TMA Pai, structured PhD fellowship from MAHE. D.J. acknowledges the financial support given by the Council of Scientific and Industrial Research CSIR under Junior Research Fellowship (JRF).
References
- 1.Sharma R, Biedenharn KR, Fedor JM, Agarwal A. 2013. Lifestyle factors and reproductive health: taking control of your fertility. Reprod. Biol. Endocrinol. 11, 66 ( 10.1186/1477-7827-11-66) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ilacqua A, Izzo G, Emerenziani GP, Baldari C, Aversa A. 2018. Lifestyle and fertility: the influence of stress and quality of life on male fertility. Reprod. Biol. Endocrinol. 16, 115 ( 10.1186/s12958-018-0436-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kushnir VA, Barad DH, Albertini DF, Darmon SK, Gleicher N. 2017. Systematic review of worldwide trends in assisted reproductive technology 2004–2013. Reprod. Biol. Endocrinol. 15, 6 ( 10.1186/s12958-016-0225-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steptoe PC, Edwards RG. 1978. Birth after the reimplantation of a human embryo. Lancet 312, 366 ( 10.1016/S0140-6736(78)92957-4) [DOI] [PubMed] [Google Scholar]
- 5.ESHRE Monitoring. 2018. More than 8 million babies born from IVF since the world's first in 1978. See https://www.sciencedaily.com/releases/2018/07/180703084127.htm.
- 6.Health D of R. 2019. Assisted reproductive technology 2017: fertility clinic success rates report. Atlanta, GA: National Centre for Chronic Disease Prevention and Health Promotion.
- 7.Report NS. 2018. Assisted reproductive technology 2016: national summary report. Atlanta, GA: National Centre for Chronic Disease Prevention and Health Promotion.
- 8.Wyns C, et al. 2020. ART in Europe, 2016: results generated from European registries by ESHRE. Hum. Reprod. Open 3, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luke B, Brown MB. 2007. Contemporary risks of maternal morbidity and adverse outcomes with increasing maternal age and plurality. Fertil. Steril. 88, 283–293. ( 10.1016/j.fertnstert.2006.11.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiitinen A. 2019. Single embryo transfer: why and how to identify the embryo with the best developmental potential. Best Pract. Res. Clin. Endocrinol. Metab. 33, 77–88. ( 10.1016/j.beem.2019.04.001) [DOI] [PubMed] [Google Scholar]
- 11.Meczekalski B, Szeliga A, Podfigurna A, Miechowicz I, Adashi EY. 2020. Assisted reproductive technology outcome in United States of America and Australia with New Zealand: comparison of annual reports 2005–2016. Gynecol. Endocrinol. 36, 1–9. ( 10.1080/09513590.2020.1737006) [DOI] [PubMed] [Google Scholar]
- 12.Cummins JM, Breen TM, Harrison KL, Shaw JM, Wilson LM, Hennessey JF. 1986. A formula for scoring human embryo growth rates in in vitro fertilization: its value in predicting pregnancy and in comparison with visual estimates of embryo quality. J. Vitr. Fertil. Embryo Transf. 3, 284–295. ( 10.1007/BF01133388) [DOI] [PubMed] [Google Scholar]
- 13.Rocha JC, Passalia F, Matos FD, Maserati Júnior MP, Alves MF, de Almeida TG, Cardoso BL, Basso AC, Nogueira MFG. 2016. Methods for assessing the quality of mammalian embryos: how far we are from the gold standard? JBRA Assist. Reprod. 20, 150–158. ( 10.5935/1518-0557.20160033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conaghan J. 2014. Time-lapse imaging of preimplantation embryos. Semin. Reprod. Med. 32, 134–140. ( 10.1055/s-0033-1363555) [DOI] [PubMed] [Google Scholar]
- 15.Tran D, Cooke S, Illingworth PJ, Gardner DK. 2019. Deep learning as a predictive tool for fetal heart pregnancy following time-lapse incubation and blastocyst transfer. Hum. Reprod. 34, 1011–1018. ( 10.1093/humrep/dez064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botros L, Sakkas D, Seli E. 2008. Metabolomics and its application for non-invasive embryo assessment in IVF. Mol. Hum. Reprod. 14, 679–690. ( 10.1093/molehr/gan066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leese HJ. 2012. Metabolism of the preimplantation embryo: 40 years on. Reproduction 143, 417–427. ( 10.1530/REP-11-0484) [DOI] [PubMed] [Google Scholar]
- 18.Leese HJ, Hooper MAK, Edwards RG, Ashwood-Smith MJ. 1986. Uptake of pyruvate by early human embryos determined by a non-invasive technique. Hum. Reprod. 1, 181–182. ( 10.1093/oxfordjournals.humrep.a136376) [DOI] [PubMed] [Google Scholar]
- 19.Seli E, Sakkas D, Scott R, Kwok SC, Rosendahl SM, Burns DH. 2007. Noninvasive metabolomic profiling of embryo culture media using Raman and near-infrared spectroscopy correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil. Steril. 88, 1350–1357. ( 10.1016/j.fertnstert.2007.07.1390) [DOI] [PubMed] [Google Scholar]
- 20.Ding J, Xu T, Tan X, Jin H, Shao J, Li H. 2017. Raman spectrum: a potential biomarker for embryo assessment during in vitro fertilization. Exp. Ther. Med. 13, 1789–1792. ( 10.3892/etm.2017.4160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortezzi SS, et al. 2013. Prediction of embryo implantation potential by mass spectrometry fingerprinting of the culture medium. Reproduction 145, 453–462. ( 10.1530/REP-12-0168) [DOI] [PubMed] [Google Scholar]
- 22.Seli E, Botros L, Sakkas D, Burns DH. 2008. Noninvasive metabolomic profiling of embryo culture media using proton nuclear magnetic resonance correlates with reproductive potential of embryos in women undergoing in vitro fertilization. Fertil. Steril. 90, 2183–2189. ( 10.1016/j.fertnstert.2008.07.1739) [DOI] [PubMed] [Google Scholar]
- 23.Pudakalakatti SM, Uppangala S, D'Souza F, Kalthur G, Kumar P, Adiga SK, Atreya HS. 2013. NMR studies of preimplantation embryo metabolism in human assisted reproductive techniques: a new biomarker for assessment of embryo implantation potential. NMR Biomed. 26, 20–27. ( 10.1002/nbm.2814) [DOI] [PubMed] [Google Scholar]
- 24.Bingol K, Brüschweiler R. 2015. Two elephants in the room: new hybrid nuclear magnetic resonance and mass spectrometry approaches for metabolomics. Curr. Opin. Clin. Nutr. Metab. Care 18, 471–477. ( 10.1097/MCO.0000000000000206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emwas AM. 2015. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research . New York, NY: Springer. [DOI] [PubMed] [Google Scholar]
- 26.Bingol K, Brüschweiler R. 2017. Knowns and unknowns in metabolomics identified by multidimensional NMR and hybrid MS/NMR methods. Curr. Opin. Biotechnol. 43, 17–24. ( 10.1016/j.copbio.2016.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauli GF, Gödecke T, Jaki BU, Lankin DC. 2012. Quantitative 1H NMR. Development and potential of an analytical method: an update. J. Nat. Prod. 75, 834–851. ( 10.1021/np200993k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markley JL, Brüschweiler R, Edison AS, Eghbalnia HR, Powers R, Raftery D, Wishart DS. 2017. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 43, 34–40. ( 10.1016/j.copbio.2016.08.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y, et al. 2012. Metabolic profiles characterizing different phenotypes of polycystic ovary syndrome: plasma metabolomics analysis. BMC Med. 10, 153–175. ( 10.1186/1741-7015-10-153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.RoyChoudhury S, et al. 2016. Serum metabolomics of Indian women with polycystic ovary syndrome using 1H NMR coupled with a pattern recognition approach. Mol. Biosyst. 12, 3407–3416. ( 10.1039/c6mb00420b) [DOI] [PubMed] [Google Scholar]
- 31.Bavister BD. 2002. How animal embryo research led to the first documented human IVF. Reprod. Biomed. Online 4, 24–29. ( 10.1016/S1472-6483(12)60008-X) [DOI] [PubMed] [Google Scholar]
- 32.Nagana Gowda GA, Raftery D. 2015. Can NMR solve some significant challenges in metabolomics? J. Magn. Reson. 260, 144–160. ( 10.1016/j.jmr.2015.07.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan TWM, Lane AN. 2016. Applications of NMR spectroscopy to systems biochemistry. Prog. Nucl. Magn. Reson. Spectrosc. 92–93, 18–53. ( 10.1016/j.pnmrs.2016.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takis PG, Ghini V, Tenori L, Turano P, Luchinat C. 2019. Uniqueness of the NMR approach to metabolomics. Trends Anal. Chem. 120, 115 300–115 308. ( 10.1016/j.trac.2018.10.036) [DOI] [Google Scholar]
- 35.Emwas AH, et al. 2019. NMR spectroscopy for metabolomics research. Metabolites 9, 123–162. ( 10.3390/metabo9070123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong KC. 2014. Review of NMR spectroscopy: basic principles, concepts and applications in chemistry. J. Chem. Educ. 91, 1103–1104. ( 10.1021/ed500324w) [DOI] [Google Scholar]
- 37.Mishkovsky M, Frydman L. 2009. Principles and progress in ultrafast multidimensional nuclear magnetic resonance. Annu. Rev. Phys. Chem. 60, 429–448. ( 10.1146/annurev.physchem.040808.090420) [DOI] [PubMed] [Google Scholar]
- 38.Wishart DS. 2008. Quantitative metabolomics using NMR. Trends Anal. Chem. 27, 228–237. ( 10.1016/j.trac.2007.12.001) [DOI] [Google Scholar]
- 39.Barding GA, Salditos R, Larive CK. 2012. Quantitative NMR for bioanalysis and metabolomics. Anal. Bioanal. Chem. 404, 1165–1179. ( 10.1007/s00216-012-6188-z) [DOI] [PubMed] [Google Scholar]
- 40.Nargund S, Joffe ME, Tran D, Tugarinov V, Sriram G. 2013. Nuclear magnetic resonance methods for metabolic fluxomics. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 41.Truong QX, Yoon JM, Shanks JV. 2014. Isotopomer measurement techniques in metabolic flux analysis I: nuclear magnetic resonance ( 10.1007/978-1-62703-661-0) [DOI] [PubMed]
- 42.Serra O, Chatterjee S, Huang W, Stark RE. 2012. Mini-review: What nuclear magnetic resonance can tell us about protective tissues. Plant Sci. 195, 120–124. ( 10.1016/j.plantsci.2012.06.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moser E, Laistler E, Schmitt F, Kontaxis G. 2017. Ultra-high field NMR and MRI—the role of magnet technology to increase sensitivity and specificity. Front. Phys. 5, 1–15. ( 10.3389/fphy.2017.00033) [DOI] [Google Scholar]
- 44.Schlotterbeck G, Ross A, Hochstrasser R, Senn H, Kuhn T, Marek D, Schett O. 2002. High-resolution capillary tube NMR: a miniaturized 5-µL high-sensitivity TXI probe for mass-limited samples, off-line LC NMR, and HT. Anal. Chem. 74, 4464–4471. ( 10.1021/ac025556e) [DOI] [PubMed] [Google Scholar]
- 45.Martin GE. 2005. Small-volume and high-sensitivity NMR probes. Paris, France: Elsevier Masson SAS. [Google Scholar]
- 46.Grimes JH, Connell TMO. 2011. The application of micro-coil NMR probe technology to metabolomics of urine and serum. J. Biomol. NMR 49, 297–305. ( 10.1007/s10858-011-9488-2) [DOI] [PubMed] [Google Scholar]
- 47.Nagato EG, Lankadurai BP, Soong R, Simpson AJ, Simpson MJ. 2015. Development of an NMR microprobe procedure for high-throughput environmental metabolomics of Daphnia magna. Magn. Reson. Chem. 53, 745–753. ( 10.1002/mrc.4236) [DOI] [PubMed] [Google Scholar]
- 48.Balaban B, et al. 2011. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum. Reprod. 26, 1270–1283. ( 10.1093/humrep/der037) [DOI] [PubMed] [Google Scholar]
- 49.Friebolin H. 2011. Basic one- and two-dimensional NMR spectroscopy, 5th edn New York, NY: Wiley-VCH. [Google Scholar]
- 50.Günther H. 2013. NMR spectroscopy: basic principles, concepts and applications in chemistry. New York, NY: Wiley-VCH. [Google Scholar]
- 51.Keeler J. 2010. Understanding NMR spectroscopy. New York, NY: Wiley. [Google Scholar]
- 52.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. 2006. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 78, 4430–4442. ( 10.1021/ac060209g) [DOI] [PubMed] [Google Scholar]
- 53.Graham SF, Ruiz-Aracama A, Lommen A, Cannizzo FT, Biolatti B, Elliott CT, Mooney MH. 2012. Use of NMR metabolomic plasma profiling methodologies to identify illicit growth-promoting administrations. Anal. Bioanal. Chem. 403, 573–582. ( 10.1007/s00216-012-5815-z) [DOI] [PubMed] [Google Scholar]
- 54.Nikolaev Y, Ripin N, Soste M, Picotti P, Iber D, Allain FHT. 2019. Systems NMR: single-sample quantification of RNA, proteins and metabolites for biomolecular network analysis. Nat. Methods 16, 743–749. ( 10.1038/s41592-019-0495-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giraudeau P, Frydman L. 2014. Ultrafast 2D NMR: an emerging tool in analytical spectroscopy. Annu. Rev. Anal. Chem. 7, 129–161. ( 10.1146/annurev-anchem-071213-020208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wishart DS, et al. 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37, D603–D610. ( 10.1093/nar/gkn810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui Q, et al. 2008. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 26, 162–164. ( 10.1038/nbt0208-162) [DOI] [PubMed] [Google Scholar]
- 58.Kuhn S, Schlörer NE. 2015. Facilitating quality control for spectra assignments of small organic molecules: Nmrshiftdb2—a free in-house NMR database with integrated LIMS for academic service laboratories. Magn. Reson. Chem. 53, 582–589. ( 10.1002/mrc.4263) [DOI] [PubMed] [Google Scholar]
- 59.Ulrich EL, et al. 2007. BioMagResBank. Nucleic Acids Res. 36, D402–D408. ( 10.1093/nar/gkm957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wishart DS, et al. 2007. HMDB: the human metabolome database. Nucleic Acids Res. 35, 521–526. ( 10.1093/nar/gkl923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hao J, Astle W, De Iorio M, Ebbels TMD. 2012. BATMAN—an R package for the automated quantification of metabolites from nuclear magnetic resonance spectra using a Bayesian model. Bioinformatics 28, 2088–2090. ( 10.1093/bioinformatics/bts308) [DOI] [PubMed] [Google Scholar]
- 62.Ravanbakhsh S, et al. 2015. Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS ONE 10, e0124219–e0124234. ( 10.1371/journal.pone.0124219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xia J, Bjorndahl TC, Tang P, Wishart DS. 2008. MetaboMiner—semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinformatics 9, 507 ( 10.1186/1471-2105-9-507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson SR, Lange BM. 2015. Open-access metabolomics databases for natural product research: present capabilities and future potential. Front. Bioeng. Biotechnol. 3, 1–10. ( 10.3389/fbioe.2015.00022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dubey A, Rangarajan A, Pal D, Atreya HS. 2015. Pattern recognition-based approach for identifying metabolites in nuclear magnetic resonance-based metabolomics. Anal. Chem. 87, 7148–7155. ( 10.1021/acs.analchem.5b00990) [DOI] [PubMed] [Google Scholar]
- 66.Dubey A, Rangarajan A, Pal D, Atreya HS. 2015. Chemical shifts to metabolic pathways: identifying metabolic pathways directly from a single 2D NMR spectrum. Anal. Chem. 87, 12 197–12 205. ( 10.1021/acs.analchem.5b03082) [DOI] [PubMed] [Google Scholar]
- 67.Hyberts SG, et al. 2007. Ultrahigh-resolution 1H−13C HSQC spectra of metabolite mixtures using nonlinear sampling and forward maximum entropy reconstruction. J. Am. Chem. Soc. 129, 5108–5116. ( 10.1021/ja068541x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ludwig C, Ward DG, Martin A, Viant MR, Ismail T, Johnson PJ, Wakelam MJO, Günther UL. 2009. Fast targeted multidimensional NMR metabolomics of colorectal cancer. Magn. Reson. Chem. 47, S68–S73. ( 10.1002/mrc.2519) [DOI] [PubMed] [Google Scholar]
- 69.Giraudeau P, Massou S, Robin Y, Cahoreau E, Portais J-C, Akoka S. 2011. Ultrafast quantitative 2D NMR: an efficient tool for the measurement of specific isotopic enrichments in complex biological mixtures. Anal. Chem. 83, 3112–3119. ( 10.1021/ac200007p) [DOI] [PubMed] [Google Scholar]
- 70.Bingol K, Brüschweiler R. 2014. Multidimensional approaches to NMR-based metabolomics. Anal. Chem. 86, 47–57. ( 10.1021/ac403520j) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Le Guennec A, Giraudeau P, Caldarelli S. 2014. Evaluation of fast 2D NMR for metabolomics. Anal. Chem. 86, 5946–5954. ( 10.1021/ac500966e) [DOI] [PubMed] [Google Scholar]
- 72.Pudakalakatti SM, Dubey A, Jaipuria G, Shubhashree U, Adiga SK, Moskau D, Atreya HS. 2014. A fast NMR method for resonance assignments: application to metabolomics. J. Biomol. NMR 58, 165–173. ( 10.1007/s10858-014-9814-6) [DOI] [PubMed] [Google Scholar]
- 73.Pudakalakatti SM, Dubey A, Atreya HS. 2015. Simultaneous acquisition of three NMR spectra in a single experiment for rapid resonance assignments in metabolomics. J. Chem. Sci. 127, 1091–1097. ( 10.1007/s12039-015-0868-0) [DOI] [Google Scholar]
- 74.Gierth P, Codina A, Schumann F, Kovacs H, Kupče E. 2015. Fast experiments for structure elucidation of small molecules: Hadamard NMR with multiple receivers. Magn. Reson. Chem. 53, 940–944. ( 10.1002/mrc.4292) [DOI] [PubMed] [Google Scholar]
- 75.Singh A, Dubey A, Adiga SK, Atreya HS. 2018. Phase modulated 2D HSQC-TOCSY for unambiguous assignment of overlapping spin systems. J. Magn. Reson. 286, 10–16. ( 10.1016/j.jmr.2017.11.005) [DOI] [PubMed] [Google Scholar]
- 76.Von Schlippenbach T, Oefner PJ, Gronwald W. 2018. Systematic evaluation of non-uniform sampling parameters in the targeted analysis of urine metabolites by 1H, 1H 2D NMR spectroscopy. Sci. Rep. 8, 4249 ( 10.1038/s41598-018-22541-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barnwal RP, Rout AK, Atreya HS, Chary KVR. 2008. Identification of C-terminal neighbours of amino acid residues without an aliphatic 13Cγ as an aid to NMR assignments in proteins. J. Biomol. NMR 41, 191–197. ( 10.1007/s10858-008-9254-2) [DOI] [PubMed] [Google Scholar]
- 78.Atreya HS. 2012. Isotope labeling in biomolecular NMR, 1st edn Dordrecht, The Netherlands: Springer. [Google Scholar]
- 79.Mulleti S, Singh A, Brahmkhatri VP, Chandra K, Mukherjee SP, Seelamantula CS, Atreya HS. 2017. Super-resolved nuclear magnetic resonance spectroscopy. Sci. Rep. 7, 1–10. ( 10.1038/s41598-017-09884-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patel AB, Srivastava S, Phadke RS, Govil G. 1998. Arginine activates glycolysis of goat epididymal spermatozoa: an NMR study. Biophys. J. 75, 1522–1528. ( 10.1016/S0006-3495(98)74071-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Inoue H, Yoshioka T. 1980. Measurement of intracellular pH in sea urchin eggs by 31P NMR. J. Cell. Physiol. 105, 461–468. ( 10.1002/jcp.1041050310) [DOI] [PubMed] [Google Scholar]
- 82.Colman A, Gadian DG. 1976. 31P nuclear-magnetic-resonance studies on the developing embryos of Xenopus laevis. Eur. J. Biochem. 61, 387–396. ( 10.1111/j.1432-1033.1976.tb10032.x) [DOI] [PubMed] [Google Scholar]
- 83.Rubessa M, Ambrosi A, Gonzalez D, Kathryn P, Matthew MP. 2018. Non-invasive nuclear magnetic resonance analysis of male and female embryo metabolites during in vitro embryo culture. Metabolomics 14, 1–9. ( 10.1007/s11306-018-1414-0) [DOI] [PubMed] [Google Scholar]
- 84.Kirkegaard K Svane ASP, Nielsen JS, HindkjæR JJ, Nielsen NC, Ingerslev HJ. 2014. Nuclear magnetic resonance metabolomic profiling of Day 3 and 5 embryo culture medium does not predict pregnancy outcome in good prognosis patients: a prospective cohort study on single transferred embryos. Hum. Reprod. 29, 2413–2420. ( 10.1093/humrep/deu236) [DOI] [PubMed] [Google Scholar]
- 85.Khosravi P, et al. 2019. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. npj Digit. Med. 2, 1–9. ( 10.1038/s41746-019-0096-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hardy K, Hooper MAK, Handyside AH, Rutherford AJ, Winston RML, Leese HJ. 1989. Non-invasive measurement of glucose and pyrovate uptake by individual human oocytes and preimplantation embryos. Hum. Reprod. 4, 348 ( 10.1093/oxfordjournals.humrep.a136905) [DOI] [PubMed] [Google Scholar]
- 87.Vergouw CG, Kieslinger DC, Kostelijk EH, Botros LL, Schats R, Hompes PG, Sakkas D, Lambalk CB. 2012. Day 3 embryo selection by metabolomic profiling of culture medium with near-infrared spectroscopy as an adjunct to morphology: a randomized controlled trial. Hum. Reprod. 27, 2304–2311. ( 10.1093/humrep/des175) [DOI] [PubMed] [Google Scholar]
- 88.Liang B, et al. 2019. Raman profiling of embryo culture medium to identify aneuploid and euploid embryos. Fertil. Steril. 111, 753–762.e1. ( 10.1016/j.fertnstert.2018.11.036) [DOI] [PubMed] [Google Scholar]
- 89.Conaghan J, Hardy K, Handyside AH, Winston RML, Leese HJ. 1993. Selection criteria for human embryo transfer: a comparison of pyruvate uptake and morphology. J. Assist. Reprod. Genet. 10, 21–30. ( 10.1007/BF01204436) [DOI] [PubMed] [Google Scholar]
- 90.Gardner DK, Lane M, Stevens J, Schoolcraft WB. 2001. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil. Steril. 76, 1175–1180. ( 10.1016/S0015-0282(01)02888-6) [DOI] [PubMed] [Google Scholar]
- 91.Brison DR, Houghton FD, Falconer D, Roberts SA, Hawkhead J, Humpherson PG, Lieberman BA, Leese HJ. 2004. Identification of viable embryos in IVF by non-invasive measurement of amino acid turnover. Hum. Reprod. 19, 2319–2324. ( 10.1093/humrep/deh409) [DOI] [PubMed] [Google Scholar]
- 92.Seli E, Vergouw CG, Morita H, Botros L, Roos P, Lambalk CB, Yamashita N, Kato O, Sakkas D. 2010. Noninvasive metabolomic profiling as an adjunct to morphology for noninvasive embryo assessment in women undergoing single embryo transfer. Fertil. Steril. 94, 535–542. ( 10.1016/j.fertnstert.2009.03.078) [DOI] [PubMed] [Google Scholar]
- 93.D'Souza F, Uppangala S, Asampille G, Salian SR, Kalthur G, Talevi R, Atreya HS, Adiga SK. 2018. Spent embryo culture medium metabolites are related to the in vitro attachment ability of blastocysts. Sci. Rep. 8, 1–10. ( 10.1038/s41598-018-35342-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Steptoe PC, Edwards RG. 1978. Birth after the reimplantation of a human embryo. Arch. Pathol. Lab. Med. 116, 321 ( 10.1016/s0140-6736(78)92957-4) [DOI] [PubMed] [Google Scholar]
- 95.Lane M, Gardner DK. 2007. Embryo culture medium: which is the best? Best Pract. Res. Clin. Obstet. Gynaecol. 21, 83–100. ( 10.1016/j.bpobgyn.2006.09.009) [DOI] [PubMed] [Google Scholar]
- 96.Houghton FD. 2003. Non-invasive amino acid turnover predicts human embryo developmental capacity. Hum. Reprod. 18, 1756–1757. ( 10.1093/humrep/deg389) [DOI] [PubMed] [Google Scholar]
- 97.Cimadomo D, et al. 2018. Continuous embryo culture elicits higher blastulation but similar cumulative delivery rates than sequential: a large prospective study. J. Assist. Reprod. Genet. 35, 1329–1338. ( 10.1007/s10815-018-1195-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sunde A, Brison D, Dumoulin J, Harper J, Lundin K, Magli MC, Van Den Abbeel E, Veiga A.. 2016. Time to take human embryo culture seriously. Hum. Reprod. 31, 2174–2182. ( 10.1093/humrep/dew157) [DOI] [PubMed] [Google Scholar]
- 99.Simopoulou M, et al. 2018. Considerations regarding embryo culture conditions: from media to epigenetics. In Vivo 32, 451–460. ( 10.21873/invivo.11261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xella S, Marsella T, Tagliasacchi D, Giulini S, La Marca A, Tirelli A, Volpe A.. 2010. Embryo quality and implantation rate in two different culture media: ISM1 versus Universal IVF Medium. Fertil. Steril. 93, 1859–1863. ( 10.1016/j.fertnstert.2008.12.030) [DOI] [PubMed] [Google Scholar]
- 101.Morbeck DE, Baumann NA, Oglesbee D. 2017. Composition of single-step media used for human embryo culture. Fertil. Steril. 107, 1055–1060; e1. ( 10.1016/j.fertnstert.2017.01.007) [DOI] [PubMed] [Google Scholar]
- 102.Tarahomi M, Vaz FM, Van Straalen JP, Schrauwen FAP, van Wely M, Hamer G, Repping S, Mastenbroek S. 2019. The composition of human preimplantation embryo culture media and their stability during storage and culture. Hum. Reprod. 34, 1450–1461. ( 10.1093/humrep/dez102) [DOI] [PubMed] [Google Scholar]
- 103.Marhuenda-Egea FC, Gonsálvez-Álvarez R, Martínez-Sabater E, Lledó B, Ten J, Bernabeu R. 2011. Improving human embryos selection in IVF: non-invasive metabolomic and chemometric approach. Metabolomics 7, 247–256. ( 10.1007/s11306-010-0245-4) [DOI] [Google Scholar]
- 104.Rinaudo P, et al. 2012. 1H NMR based profiling of spent culture media cannot predict success of implantation for day 3 human embryos. J. Assist. Reprod. Genet. 29, 1435–1442. ( 10.1007/s10815-012-9877-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sánchez-Ribas I, Riqueros M, Vime P, Puchades-Carrasco L, Jönsson T, Pineda-Lucena A, Ballesteros A, Domínguez F, Simón C. 2012. Differential metabolic profiling of non-pure trisomy 21 human preimplantation embryos. Fertil. Steril. 98, 1157–1164.e2. ( 10.1016/j.fertnstert.2012.07.1145) [DOI] [PubMed] [Google Scholar]
- 106.Nadal-Desbarats L, Veau S, Blasco H, Emond P, Royere D, Andres CR, Guérif F. 2013. Is NMR metabolic profiling of spent embryo culture media useful to assist in vitro human embryo selection? Magn. Reson. Mater. Phys. Biol. Med. 26, 193–202. ( 10.1007/s10334-012-0331-x) [DOI] [PubMed] [Google Scholar]
- 107.Wallace M, Cottell E, Cullinane J, McAuliffe FM, Wingfield M, Brennan L. 2014. 1H NMR based metabolic profiling of day 2 spent embryo media correlates with implantation potential. Syst. Biol. Reprod. Med. 6368, 58–63. ( 10.3109/19396368.2013.854426) [DOI] [PubMed] [Google Scholar]
- 108.Uppangala S, D'Souza F, Pudakalakatti S, Atreya HS, Raval K, Kalthur G, Adiga SK. 2016. Laser assisted zona hatching does not lead to immediate impairment in human embryo quality and metabolism. Syst. Biol. Reprod. Med. 62, 396–403. ( 10.1080/19396368.2016.1217952) [DOI] [PubMed] [Google Scholar]
- 109.Uppangala S, Pudakalakatti S, D'souza F, Salian SR, Kalthur G, Kumar P, Atreya HS, Adiga SK. 2016. Influence of sperm DNA damage on human preimplantation embryo metabolism. Reprod. Biol. 16, 234–241. ( 10.1016/j.repbio.2016.07.004) [DOI] [PubMed] [Google Scholar]
- 110.Rubessa M, Ambrosi A, Gonzalez-Pena D, Polkoff KM, Denmark SE, Wheeler MB. 2016. Non-invasive analysis of bovine embryo metabolites during in vitro embryo culture using nuclear magnetic resonance. AIMS Bioeng. 3, 538–551. ( 10.3934/bioeng.2016.4.538) [DOI] [PubMed] [Google Scholar]
- 111.Perkel KJ, Madan P. 2017. Spent culture medium analysis from individually cultured bovine embryos demonstrates metabolomic differences. Zygote 25, 662–674. ( 10.1017/S0967199417000417) [DOI] [PubMed] [Google Scholar]
- 112.D'Souza F, et al. 2016. Unraveling the association between genetic integrity and metabolic activity in pre-implantation stage embryos. Sci. Rep. 6, 37291 ( 10.1038/srep37291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.D'Souza F, Asampille G, Uppangala S, Kalthur G, Atreya HS, Adiga SK. 2019. Sperm-mediated DNA lesions alter metabolite levels in spent embryo culture medium. Reprod. Fertil. Dev. 31, 443–450. ( 10.1071/RD18136) [DOI] [PubMed] [Google Scholar]
- 114.Piñero-Sagredo E, Nunes S, De Los Santos MJ, Celda B, Esteve V.. 2010. NMR metabolic profile of human follicular fluid. NMR Biomed. 23, 485–495. ( 10.1002/nbm.1488) [DOI] [PubMed] [Google Scholar]
- 115.Gérard N, Fahiminiya S, Grupen CG, Nadal-Desbarats L. 2015. Reproductive physiology and ovarian folliculogenesis examined via 1H-NMR metabolomics signatures: a comparative study of large and small follicles in three mammalian species (Bos taurus, Sus scrofa domesticus and Equus ferus caballus). Omi. A J. Integr. Biol. 19, 31–40. ( 10.1089/omi.2014.0097) [DOI] [PubMed] [Google Scholar]
- 116.Wallace M, Cottell E, Gibney MJ, McAuliffe FM, Wingfield M, Brennan L. 2012. An investigation into the relationship between the metabolic profile of follicular fluid, oocyte developmental potential, and implantation outcome. Fertil. Steril. 97, 1078–1084. ( 10.1016/j.fertnstert.2012.01.122) [DOI] [PubMed] [Google Scholar]
- 117.McRae C, Baskind NE, Orsi NM, Sharma V, Fisher J. 2012. Metabolic profiling of follicular fluid and plasma from natural cycle in vitro fertilization patients—a pilot study. Fertil. Steril. 98, 1449–1457.e6. ( 10.1016/j.fertnstert.2012.07.1131) [DOI] [PubMed] [Google Scholar]
- 118.Zhang Y, Liu L, Yin TL, Yang J, Xiong CL. 2017. Follicular metabolic changes and effects on oocyte quality in polycystic ovary syndrome patients. Oncotarget 8, 80 472–80 480. ( 10.18632/oncotarget.19058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jaccarino N, Amato J, Pagano B, D'Oriano L. 2019. 1H NMR-based metabolomics study on follicular fluid from patients with polycystic ovary syndrome. Biochim. Clin. 43, 256–263. ( 10.19186/BC) [DOI] [Google Scholar]
- 120.Marianna SM, et al. 2017. Metabolomic profiling and biochemical evaluation of the follicular fluid of endometriosis patients. Mol. Biosyst. 13, 1213–1222. ( 10.1039/c7mb00181a) [DOI] [PubMed] [Google Scholar]
- 121.Castiglione Morelli MA, Iuliano A, Schettini SCA, Petruzzi D, Ferri A, Colucci P, Viggiani L, Cuviello F, Ostuni A. 2018. NMR metabolomics study of follicular fluid in women with cancer resorting to fertility preservation. J. Assist. Reprod. Genet. 35, 2063–2070. ( 10.1007/s10815-018-1281-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karaer A, Tuncay G, Mumcu A, Dogan B. 2019. Metabolomics analysis of follicular fluid in women with ovarian endometriosis undergoing in vitro fertilization. Syst. Biol. Reprod. Med. 65, 39–47. ( 10.1080/19396368.2018.1478469) [DOI] [PubMed] [Google Scholar]
- 123.Castiglione Morelli MA, Iuliano A, Schettini SCA, Petruzzi D, Ferri A, Colucci P, Viggiani L, Cuviello F, Ostuni A. 2019. NMR metabolic profiling of follicular fluid for investigating the different causes of female infertility: a pilot study. Metabolomics 15, 1–10. ( 10.1007/s11306-019-1481-x) [DOI] [PubMed] [Google Scholar]
- 124.Gosden RG, Sadler IH, Reed D, Hunter RHF. 1990. Characterization of ovarian follicular fluids of sheep, pigs and cows using proton nuclear magnetic resonance spectroscopy. Experientia 46, 1012–1015. ( 10.1007/BF01940658) [DOI] [PubMed] [Google Scholar]
- 125.Gérard N, Loiseau S, Duchamp G, Seguin F. 2002. Analysis of the variations of follicular fluid composition during follicular growth and maturation in the mare using proton nuclear magnetic resonance (1H NMR). Reproduction 124, 241–248. ( 10.1530/rep.0.1240241) [DOI] [PubMed] [Google Scholar]
- 126.Bertoldo MJ, Nadal-Desbarats L, Gérard N, Dubois A, Holyoake PK, Grupen CG. 2013. Differences in the metabolomic signatures of porcine follicular fluid collected from environments associated with good and poor oocyte quality. Reproduction 146, 221–231. ( 10.1530/REP-13-0142) [DOI] [PubMed] [Google Scholar]
- 127.RoyChoudhury S, Singh A, Gupta NJ, Srivastava S, Joshi MV., Chakravarty B, Chaudhury K. 2016. Repeated implantation failure versus repeated implantation success: discrimination at a metabolomic level. Hum. Reprod. 31, 1265–1274. ( 10.1093/humrep/dew064) [DOI] [PubMed] [Google Scholar]
- 128.Lyman JT, Rubessa M, Oh DK, Ginger L, Magin RL, Wheeler MB. 2016. Investigation into the use of nuclear magnetic resonance spectroscopy for viability predictions, in real time on mouse preimplantation embryos. Int. J. New Technol. Res. 2, 14–20. [Google Scholar]
- 129.Bodart JF, Wieruszeski JM, Amniai L, Leroy A, Landrieu I, Rousseau-Lescuyer A, Vilain JP, Lippens G. 2008. NMR observation of Tau in Xenopus oocytes. J. Magn. Reson. 192, 252–257. ( 10.1016/j.jmr.2008.03.006) [DOI] [PubMed] [Google Scholar]
- 130.Zhao SJ, Tian JS, Tai G, Gao XX, Liu HL, Du GH, Liu XJ, Qin XM. 2019. 1H NMR-based metabolomics revealed the protective effects of Guilingji on the testicular dysfunction of aging rats. J. Ethnopharmacol. 238, 111839 ( 10.1016/j.jep.2019.111839) [DOI] [PubMed] [Google Scholar]
- 131.Chew WM, Hricak H, McClure RD, Wendland FM.. 1990. In vivo human testicular function assessed with P-31 MR spectroscopy. Radiology 177, 743–747. [DOI] [PubMed] [Google Scholar]
- 132.Griffin JLL, Troke J, Walker LAA, Shore RFF, Lindon JCC, Nicholson JKK. 2000. The biochemical profile of rat testicular tissue as measured by magic angle spinning 1H NMR spectroscopy. FEBS Lett. 486, 225–229. ( 10.1016/S0014-5793(00)02307-3) [DOI] [PubMed] [Google Scholar]
- 133.Jarak I, Almeida S, Carvalho RA, Sousa M, Barros A, Alves MG, Oliveira PF. 2018. Senescence and declining reproductive potential: Insight into molecular mechanisms through testicular metabolomics. BBA—Mol. Basis Dis. 1864, 3388–3396. ( 10.1016/j.bbadis.2018.07.028) [DOI] [PubMed] [Google Scholar]
- 134.Lin CY, Hung PH, VandeVoort CA, Miller MG. 2009. 1H NMR to investigate metabolism and energy supply in rhesus macaque sperm. Reprod. Toxicol. 28, 75–80. ( 10.1016/j.reprotox.2009.03.005) [DOI] [PubMed] [Google Scholar]
- 135.Paiva C, Amaral A, Rodriguez M, Canyellas N, Correig X, Ballescà JL, Ramalho-Santos J, Oliva R. 2015. Identification of endogenous metabolites in human sperm cells using proton nuclear magnetic resonance (1H-NMR) spectroscopy and gas chromatography-mass spectrometry (GC-MS). Andrology 3, 496–505. ( 10.1111/andr.12027) [DOI] [PubMed] [Google Scholar]
- 136.Reynolds S, Fadhlina Bt Ismail N, Calvert SJ, Pacey AA, Paley MNJ. 2017. Evidence for rapid oxidative phosphorylation and lactate fermentation in motile human sperm by hyperpolarized 13C magnetic resonance spectroscopy Sci. Rep. 7, 1–8. ( 10.1038/s41598-017-04146-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Reynolds S, Calvert SJ, Paley MN, Pacey AA. 2017. 1H magnetic resonance spectroscopy of live human sperm. Mol. Hum. Reprod. 23, 441–451. ( 10.1093/molehr/gax025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Calvert SJ, Reynolds S, Paley MN, Walters SJ, Pacey AA. 2018. Probing human sperm metabolism using 13C-magnetic resonance spectroscopy. Mol. Hum. Reprod. 25, 30–41. ( 10.1093/molehr/gay046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Risten RC, Schackmann RW, Dahlquist FW, Shapiro BM, Christen R, Schackmann RW, Dahlquist FW, Shapiro BM. 1983. 31P-NMR analysis of sea urchin sperm activation: reversible formation of high energy phosphate compounds by changes in intracellular pH. Exp. Cell Res. 149, 289–294. ( 10.1016/0014-4827(83)90400-7) [DOI] [PubMed] [Google Scholar]
- 140.Robitaille PA, Robitaille P-ML, Brown GG. 1986. 31P-NMR studies of Limulus polyphemus: spermatozoa at rest and after motility. J. Exp. Zool. 238, 89–98. ( 10.1002/jez.1402380111) [DOI] [Google Scholar]
- 141.Dreanno C, Seguin F, Cosson J, Suquet M, Billard R. 2000. 1H-NMR and 31P-NMR analysis of energy metabolism of quiescent and motile turbot (Psetta maxima) spermatozoa. J. Exp. Zool. 286, 513–522. () [DOI] [PubMed] [Google Scholar]
- 142.Robitaille P-ML, Mumford KG, Brown GG. 1987. 31P nuclear magnetic resonance study of trout spermatozoa at rest, after motility, and during short-term storage. Biochem. Cell Biol. 65, 474–485. ( 10.1139/o87-061) [DOI] [Google Scholar]
- 143.Smith MB, Babcock DF, Lardy HA. 1985. A 31P NMR study of the epididymis and epididymal sperm of the bull and hamster. Biol. Reprod. 33, 1029–1040. ( 10.1095/biolreprod33.5.1029) [DOI] [PubMed] [Google Scholar]
- 144.Lynch MJ, Masters J, Pryor JP, Lindon JC, Spraul M, Foxall PJD, Nicholson JK. 1994. Ultra high field NMR spectroscopic studies on human seminal fluid, seminal vesicle and prostatic secretions. J. Pharm. Biomed. Anal. 12, 5–19. ( 10.1016/0731-7085(94)80004-9) [DOI] [PubMed] [Google Scholar]
- 145.Hamamah S, Seguin F, Bujan L, Barthelemy C, Mieusset R, Lansac J. 1998. Quantification by magnetic resonance spectroscopy of metabolites in seminal plasma able to differentiate different forms of azoospermia. Hum. Reprod. 13, 132–135. ( 10.1093/humrep/13.1.132) [DOI] [PubMed] [Google Scholar]
- 146.Tomlins AM, Foxall PJ d, Lynch MJ, Parkinson J, Everett JR, Nicholson JK. 1998. High resolution 1H NMR spectroscopic studies on dynamic biochemical processes in incubated human seminal fluid samples. Biochim. Biophys. Acta—Gen. Subj. 1379, 367–380. ( 10.1016/S0304-4165(97)00116-5) [DOI] [PubMed] [Google Scholar]
- 147.Sharma U, Chaudhury K, Jagannathan NR, Guha SK. 2001. A proton NMR study of the effect of a new intravasal injectable male contraceptive RISUG on seminal plasma metabolites. Reproduction 122, 431–436. ( 10.1530/rep.0.1220431) [DOI] [PubMed] [Google Scholar]
- 148.Gupta A, Mahdi AA, Ahmad MK, Shukla KK, Bansal N, Jaiswer SP, Shankhwar SN. 2011. A proton NMR study of the effect of Mucuna pruriens on seminal plasma metabolites of infertile males. J. Pharm. Biomed. Anal. 55, 1060–1066. ( 10.1016/j.jpba.2011.03.010) [DOI] [PubMed] [Google Scholar]
- 149.Gupta A, Mahdi AA, Ahmad MK, Shukla KK, Jaiswer SP, Shankhwar SN. 2011. 1H NMR spectroscopic studies on human seminal plasma: a probative discriminant function analysis classification model. J. Pharm. Biomed. Anal. 54, 106–113. ( 10.1016/j.jpba.2010.07.021) [DOI] [PubMed] [Google Scholar]
- 150.Gupta A, Mahdi AA, Shukla KK, Ahmad MK, Bansal N, Sankhwar P, Sankhwar SN. 2013. Efficacy of Withania somnifera on seminal plasma metabolites of infertile males: a proton NMR study at 800 MHz. J. Ethnopharmacol. 149, 208–214. ( 10.1016/j.jep.2013.06.024) [DOI] [PubMed] [Google Scholar]
- 151.Bonechi C, Collodel G, Donati A, Martini S, Moretti E, Rossi C. 2015. Discrimination of human semen specimens by NMR data, sperm parameters, and statistical analysis. Syst. Biol. Reprod. Med. 61, 353–359. ( 10.3109/19396368.2015.1054003) [DOI] [PubMed] [Google Scholar]
- 152.Zhang X, Diao R, Zhu X, Li Z, Cai Z. 2015. Metabolic characterization of asthenozoospermia using nontargeted seminal plasma metabolomics. Clin. Chim. Acta 450, 254–261. ( 10.1016/j.cca.2015.09.001) [DOI] [PubMed] [Google Scholar]
- 153.Darbandi M, et al. 2019. Reactive oxygen species-induced alterations in H19-Igf2 methylation patterns, seminal plasma metabolites, and semen quality. J. Assist. Reprod. Genet. 36, 241–253. ( 10.1007/s10815-018-1350-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mumcu A, Karaer A, Dogan B, Tuncay G. 2019. Metabolomics analysis of seminal plasma in patients with idiopathic Oligoasthenoteratozoospermia using high-resolution NMR spectroscopy. Andrology 8, 450–456. ( 10.1111/andr.12707) [DOI] [PubMed] [Google Scholar]
- 155.Mehrparvar B, Chashmniam S, Nobakht F, Amini M, Javidi A, Minai-Tehrani A, Arjmand B, Gilany K. 2020. Metabolic profiling of seminal plasma from teratozoospermia patients. J. Pharm. Biomed. Anal. 178, 112903 ( 10.1016/j.jpba.2019.112903) [DOI] [PubMed] [Google Scholar]
- 156.Ebrahimi F, Ibrahim B, Teh CH, Murugaiyah V, Chan KL. 2017. NMR-based plasma metabolomic discrimination for male fertility assessment of rats treated with Eurycoma longifolia extracts. Syst. Biol. Reprod. Med. 63, 179–191. ( 10.1080/19396368.2017.1295332) [DOI] [PubMed] [Google Scholar]
- 157.Barberet J, et al. 2019. Can novel early non-invasive biomarkers of embryo quality be identified with time-lapse imaging to predict live birth? Hum. Reprod. 34, 1439–1449. ( 10.1093/humrep/dez085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mishkovsky M, Frydman L. 2008. Progress in hyperpolarized ultrafast 2D NMR spectroscopy. ChemPhysChem 9, 2340–2348. ( 10.1002/cphc.200800461) [DOI] [PubMed] [Google Scholar]
- 159.Adams RW.2014. Pure shift nmr spectroscopy. In eMagRes, pp. 295–310. Chichester, UK: John Wiley & Sons.
- 160.Zangger K. 2015. Pure shift NMR. Prog. Nucl. Magn. Reson. Spectrosc. 86–87, 1–20. ( 10.1016/j.pnmrs.2015.02.002) [DOI] [PubMed] [Google Scholar]
- 161.Corcoran O, Spraul M, Corcoran O. 2003. LC–NMR– MS in drug discovery. Drug Discov. Today 8, 624–631. [DOI] [PubMed] [Google Scholar]
- 162.Walker GS, Connell TNO. 2008. Comparison of LC-NMR and conventional NMR for structure elucidation in drug metabolism studies. Exp. Opin. Drug Metab. Toxicol. 4, 1295–1305. ( 10.1517/17425255.4.10.1295) [DOI] [PubMed] [Google Scholar]
- 163.Garrod S, Humpfer E, Spraul M, Connor SC, Polley S, Connelly J, Lindon JC, Nicholson JK, Holmes E. 1999. High-resolution magic angle spinning 1H NMR spectroscopic studies on intact rat renal cortex and medulla. Magn. Reson. Med. 41, 1108–1118. () [DOI] [PubMed] [Google Scholar]
- 164.Morvan D, Demidem A, Papon J, De Latour M, Claude Madelmont J.. 2002. Melanoma tumors acquire a new phospholipid metabolism phenotype under cystemustine as revealed by high-resolution magic angle spinning proton nuclear magnetic resonance spectroscopy of intact tumor samples. Cancer Res. 62, 1890–1897. [PubMed] [Google Scholar]
- 165.Hurd RE, Yen Y-F, Chen A.. 2010. Hyperpolarized nuclear magnetic resonance spectroscopy: a new method for metabolomic research. In Methodologies for metabolomics (eds Lutz N, Sweedler J, Wevers R), pp. 446–471. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 166.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K.. 2003. Increase in signal-to-noise ratio of >10 000 times in liquid-state NMR. Proc. Natl Acad. Sci. USA 100, 10 158–10 163. ( 10.1073/pnas.1733835100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lerche MH, Karlsson M, Ardenkjær-Larsen JH, Jensen PR.. 2019. Targeted metabolomics with quantitative dissolution dynamic nuclear polarization. In NMR-based metabolomics: methods and protocols (eds Gowda G, Raftery D), pp. 385–393. Cham, Switzerland: Springer. [DOI] [PubMed] [Google Scholar]
- 168.Christensen CE, Karlsson M, Winther JR, Jensen PR, Lerche MH. 2014. Non-invasive in-cell determination of free cytosolic [NAD+]/[NADH] ratios using hyperpolarized glucose show large variations in metabolic phenotypes. J. Biol. Chem. 289, 2344–2352. ( 10.1074/jbc.M113.498626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lin E. 2012. Novel drug therapies and diagnostics for personalized medicine and nanomedicine in genome science, nanoscience, and molecular engineering. Pharm. Regul. Aff. Open Access 1, 1000e116–1000e117. ( 10.4172/2167-7689.1000e116) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.