Abstract
N6-methyladenosine (m6A) is the most abundant inner RNA modification in eukaryotes. Due to the development of RNA sequencing technology, the distribution pattern of m6A in the transcriptome has been uncovered. Dynamically, the reversible N6-methylation is mediated by two types of proteins, which are classified as “writers” and “erasers”. Under the association of specific co-factors, writers show spatiotemporal N6-methyltransferase activity. Mechanically, m6A can be recognized by “reader” proteins or can directly modify RNA conformation, and it widely affects gene expression by mediating RNA stability, translation, splicing and export. m6A is involved in a series of physiology processes. Dysregulation of m6A is gradually defined as the pathogenesis of some diseases, e.g., cancer and cardiovascular disease. Therefore, a good understanding of m6A is essential for molecular biology and pathology research. In this article we systemically present an overview of the functions and mechanisms of identified m6A regulators. The discovered biological and pathological processes affected by m6A are also summarized. We hope that readers with related research interests benefit from our review.
Keywords: Diseases, Epigenetics, Gene expression, m6A, m6A regulator, RNA
Introduction
Strict regulation of gene expression is indispensable for development and physiological processes. Numerous protein and nucleic acid factors have been identified to affect gene expression at distinct stages of RNA processing, such as transcription, splicing and maturation, exportation and protein translation. In recent years, it has been realized that the most abundant inner chemical RNA modification, N6-methyladenosine (m6A), has novel regulatory roles in gene expression. m6A was first characterized in the 1970s in eukaryotes ranging from yeast to mammals.1, 2, 3, 4 In the following 40 years, although several components of m6A methyltransferase or demethylase have been identified, such as methyltransferase-like protein 3/14 (METTL3-14), WTAP (Wilms tumour 1-associating protein) and fat mass and obesity-associated protein (FTO), the detailed regulatory mechanisms of these proteins have not been described.5, 6, 7, 8, 9 In 2012, two independent groups assayed transcriptomic m6A modification sites by means of an m6A immunoprecipitation with next-generation sequencing (MeRIP-Seq).10,11 According to their data, more than 12,000 N6-methylation peaks exist in over 7000 mRNAs and long non-coding RNAs (lncRNA) of human and mouse. These m6A sites share a consensus motif of “RRACH” (R = A or G; H = A, U, or C) and are enriched in the regions of the 3′ UTR, long internal exons or near stop codons. Moreover, the existence of intronic m6A sites suggests that the m6A modification might happen before pre-mRNA splicing. Most of the m6A sites are conserved between human and mouse.10 Intriguingly, the level of m6A modification of partial m6A sites is different in normal tissues and tumours.11 Moreover, silencing the methyltransferase METTL3 not only causes a significant decrease of the total m6A level but also affects gene expression and induces apoptosis of HepG2 cells.10 These results infer a potential association among m6A, RNA processing and cell fate. Based on the foundational impact provided by the m6A mapping approach, the upstream regulatory and downstream functional mechanisms have been gradually uncovered in the past several years: m6A is reversibly managed and maintained in dynamic homeostasis (Fig. 1). RNAs are N6-methylated by the methyltransferase complex, and the central components of the complex are called “writers”, while the major m6A demethylases are called “erasers”. N6-methylated RNAs will be recognized and will interact with a series of “reader” proteins, which are involved in RNA processing, leading to alteration of gene expression. In addition to polyadenylate-tailed mRNA and lncRNA, m6A has also been detected in microRNAs (miRNAs), circular RNAs (circRNAs) and ribosomal RNAs (rRNA) to carry out novel roles (described below). It has also been found that m6A mediates gene expression, widely affecting physiological and pathological processes. Here, we summarize the latest progress from studies of the m6A regulatory system. The significant physiological and pathological mechanisms in which m6A is involved are also presented.
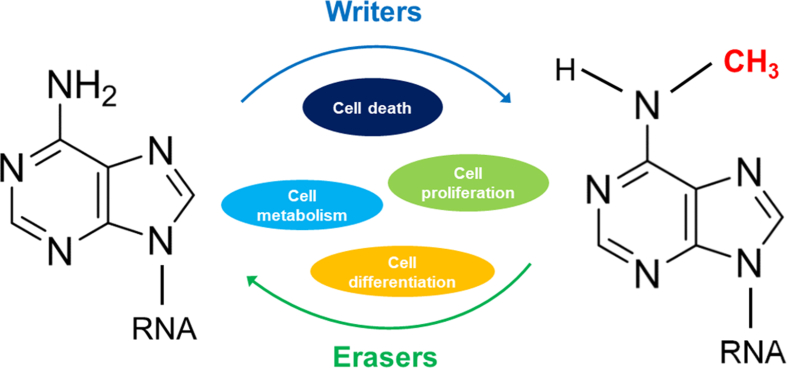
Figure 1.
N6-methylation is reversible. The dynamic homeostasis of m6A that regulated by writers and erasers involves in cellular physiological processes including cell proliferation, differentiation, metabolism and death.
RNAs are reversibly m6A-modified through a series of enzymatic systems
m6A is one of the few reversible inner RNA modifications. Methylation and demethylation of m6A are enzymatically regulated. N6-methyltransferation involves two methyltransferase complexes. The approximately 200-kDa complex, which consists of METTL3 and METTL14, is defined as methyltransferase A (MT-A).5, 6, 7 MT-A performs the majority of the catalytic activity. The other complex, which is approximately 800 kDa in size, is defined as methyltransferase B (MT-B) and might be the accessorial fraction required for cellular localization, RNA targeting and recruitment.5 N6-demethylation is mainly controlled by FTO and ALKBH5. Although the exact components and structures of the methyltransferase complexes are not fully characterized, the central “writers” or “erasers” and their roles have been identified.
METTL3: the catalytic core
It was not known how RNAs were N6-methylated until MT-A was isolated from the nucleus of HeLa cells in 1997.6 METTL3 was the only characterized subunit of MT-A at that time. METTL3 belongs to the S-adenosyl-l-methionine (SAM)-dependent methyltransferase (SAM MTase) superfamily, which contains a SAM-binding domain and transfers the methyl group of SAM to the adenosine of the target molecule. METTL3 is highly conserved among eukaryotes from yeast to mammals.7 Among all the identified subunits of the m6A writer complex, METTL3 performs fundamental N6-methylase catalytic activity. Genomic deletion of METTL3 results in the complete loss of m6A in the transcriptome.12 Generally, METTL3 is concentrated in the nucleus speckle and catalyses N6-methylation of RNAs, while cytoplasmic METTL3 has also been detected in several cancer cell lines.6,13,14 Cytoplasmic METTL3 functions in a methylation-independent manner: it can interact with eukaryotic translation initiation factor 3 subunit H (EIF3H) and the 3′ UTR of target mRNAs, forming a METTL3-EIF3H-mRNA loop to facilitate translation.13,14
METTL14: the N6-methyltransferase-inactive collaborator of METTL3
METTL14, a homologue of METTL3, is another component of the MT-A complex.7 These two SAM MTases form a heterodimer and co-localize in the nucleus speckle.15 METTL14 was first characterized as a catalytic subunit of the methyltransferase complex that methylates distinct m6A sites from METTL3. Crudely purified METTL14 showed N6-methylase activity in vitro.15 However, sequence analysis suggested that METTL14 only contains a disrupted SAM-binding domain and has no catalytic activity.16 The later structural analysis not only revealed the interaction of METTL3 and METTL14 but also revealed the fact that METTL14 lacks a SAM-binding domain.17 Indeed, silencing of METTL14 largely decreases the level of m6A.15 In addition, compared with the METTL3-METTL14 heterodimer, the catalytic activity of monomeric METTL3 was rather weaker in vitro.15,18 This finding indicates that METTL14 may act as a catalytic enhancer of METTL3. At present, METTL14 is proposed to be the stabilizer, activator and RNA binding scaffold of METTL3.17,18
WTAP: the crucial partner for localization of the catalytic core
WTAP was initially identified as a protein associated with Wilms' tumour suppressor WT1 by a yeast two-hybrid assay.9 Because of its ubiquitous expression and nuclear localization, WTAP was also previously considered to be a house-keeping or RNA-processing related protein. Following studies demonstrated that WTAP regulates the alternative splicing of pre-mRNA and the stability of mRNA.19,20 WTAP also suppresses the expression of WT1-target genes by interacting with WT1.21 In 2014, WTAP was found to be involved in m6A regulation. Pull-down assays showed that WTAP binds both METTL3 and METTL14.22 Knockdown of WTAP causes a significant decrease in the level of m6A. As WTAP lacks N6-methyltransferase activity and shares consensus target m6A sites with METTL3, it is suggested that WTAP helps METTL3-METTL14 localize to nucleus speckles and recruit other N6-methyltransferase components.15 Silencing of WTAP disrupts the nucleus speckle-localization of METTL3-METTL14.
Other writers with accessory functions
In recent years, other associated writers have gradually been discovered: In 2013, Horiuchi et al identified a series of WTAP-interacting proteins. In addition to METTL3 (described as MTA-70), protein virilizer homologue (VIRMA, also known as KIAA1429), KIAA0853 (also known as Zinc finger CCCH domain-containing protein 13, ZC3H13), RNA-binding protein 15 (RBM15), E3 ubiquitin-protein ligase HAKAI (also known as Casitas B-lineage lymphoma-transforming sequence-like protein 1, Cbll1) and two arginine/serine-rich domain-containing proteins, BCLAF1 and THRAP3 are included in the WTAP-binding protein complex. These WTAP-binding proteins are related with the alternative splicing of pre-mRNA.23 This study preliminarily screened out possible candidates associated with the N6-methyltransferase complex. Further research indicated that VIRMA, ZC3H13, RBM15 and HAKAI are involved in N6-methylation: VIRMA may be involved in the selective N6-methylation of sites in 3′ UTRs and located near stop codons in mRNAs.24 ZC3H13 acts as a connector between WTAP and RBM15.25 ZC3H13 is also essential for the nuclear localization of other writers. Silencing of ZC3H13 leads to cytoplasmic localization of VIRMA and HAKAI. RBM15 interacts with WTAP in a ZC3H13 dependent manner.26 RBM15 functions as a connecter between catalytic writers and readers. As an RNA binding protein (RBP), RBM15 recruits specific target RNAs to catalytic centres and guides the reader to selectively recognize m6A sites.27 HAKAI ordinarily functions as an E3 ubiquitin ligase.28 The detailed roles of HAKAI in N6-methylation are still unclear.
Catalytic N6-methylases with novel targets and organization
With the exception of METTL3, there are also N6-methylases unrelated to MT complexes for specific types or structures of RNAs. METTL16, a homologue of METTL3, can N6-methylate a conserved hairpin of MAT2A mRNA and U6 snRNA with stimulation of SAM. Further exploration revealed that the methylation sites of METTL16 are enriched in introns and are not conserved, which is distinct from the METTL3-modulated pattern.29 The detailed mechanisms of METTL16 still need to be explored. Zinc finger CCHC domain-containing protein 4 (ZCCHC4) has been reported to be an N6-methylase for 28S rRNA. Loss of ZCCHC4 globally inhibits protein translation.30 The proteomic association and roles involved in N6-methylation of ZCCHC4 are unclear.
Mammalian m6A erasers
To date, FTO and ALKBH5 are the only two characterized N6-demethylases. Both of these N6-demethylases are members of the alpha-ketoglutarate-dependent dioxygenase AlkB-like superfamily. These AlkB-like dioxygenases have a catalytic domain that is homologous to that in Fe-2OG dioxygenases and catalyses the hydroxylation of the target methyl by using 2-oxoglutarate and oxygen molecular as substrates. The hydroxymethyl is unstable when it is connected to a nitrogen atom, resulting in demethylation.31,32
It has been demonstrated that FTO plays multiple catalytic roles and has multiple substrates. FTO was initially discovered to catalyse the demethylation of 3-methylthymidine (3mT) in single-stranded DNA and 3-methyluracil (3mU) in single-stranded RNA.8,33 In 2011, the N6-demethylase activity of FTO was characterized in vitro.34 FTO was the first identifiedm 6A eraser. A recent study discovered that FTO can demethylase the N6,2-O-dimethyladenosine (m6Am) in the 5′ cap of mRNAs.35 The crosstalk among these types of methylation deserves to be explored. Immunostaining indicated that FTO is concentrated in nucleus speckles, suggesting that FTO might demethylate nuclear m6A-modified RNAs.34 Cytoplasmic FTO and its high N6-demethylase activity were also observed in certain types of cells, such as leukaemia cells.36 An in vivo study showed that FTO might not mediate N6-demethylation in the early embryonic stage. The m6A level in FTO-knockout mouse embryos was not significantly affected.35 These data suggest that FTO may spatiotemporally catalyse N6-demethylation.
The N6-demethylase activity of ALKBH5 was discovered immediately after FTO in 2013. Modifying the expression of ALKBH5 affects the level of m6A in certain types of cells.37 In addition, ALKBH5 deficiency in mice is not lethal and only increases the apoptotic rate of testicular cells and leads to defects in spermatogenesis.37 These data suggest that ALKBH5 selectively demethylates m6A, similar to FTO. The demethylation process mediated by ALKBH5 may be necessary for RNA processing, which regulates testis development and male fertility. On the other hand, unlike FTO, ALKBH5 has specific nuclear localization and N6-demethylase activity.35,37 Loss of function analysis showed that ALKBH5 mediates RNA export in HeLa cells and RNA splicing in sperm cells in an m6A-demethylase activity-dependent manner, perhaps due to the interaction between ALKBH5 and splicing factors.37
Selective N6-methylaiton in the transcriptome
Although the “RRACH” motif appears frequently in transcriptomes, the distribution of m6A sites is enriched in specific regions, including long internal exons, the 3′-UTR and regions near stop codons of mRNAs. Intronic N6-methylation also exists in pre-mRNA.10,11 Moreover, quite a large portion of m6A sites exhibit spatiotemporally specific patterns. Since the catalytic core, METTL3/14, is ubiquitously expressed and has not been demonstrated to independently recognize specific regions, selective N6-methylation is believed to be carried out by some associated components. In this section we would like to discuss the demonstrated and potential mechanisms involved in selective N6-methylation.
VIRMA or RNA polymerase II regulates N6-methylation in the 3′-UTR and near stop codons
Evidence has suggested that some accessary writers function as guides to recruit the N6-methyltransfase complex to regions in the 3′ UTR and near stop codons. VIRMA is the largest characterized component of the N6-methyltransferase complex and is indicated to be the scaffold that bridges the N6-methyltransferase complex and target RNAs. MeRIP-Seq data in HeLa cells indicate that knockdown or depletion of VIRMA significantly disrupts the distribution of m6A sites in the 3′-UTR and near stop codons without altering the total m6A level, revealing that VIRMA plays critical roles in 3′-UTR- and stop codon-specific targeting.24 However, the detailed mechanism of the selective targeting mediated by VIMRA is unclear. VIRMA-associated cleavage and polyadenylation specificity factor subunit 5 and 6 (CPSF5 and CPSF6), which are the components of the CFIm complex, are able to bind to “UGUA” elements in the 3′-UTR of pre-mRNAs. It has been observed that VIRMA and CPSF5 share a substantial overlap of targets, which implies that the CFIm complex might be involved in selective N6-methylation. Nevertheless, silencing of CPSF5 and CPSF6 only increase the density of N6-methylation in the 3′ UTR. Perhaps other subunits of the CFIm complex mediate this process.
Another possible mechanism is that selective N6-methylation in the 3′ UTR and near stop codons might be co-transcriptionally regulated. RNA polymerase II (Pol II) has been reported to interact with the N6-methyltransfase complex. A Pol II retardation-induced transcription pause increases the recruitment of m6A writers and the level of m6A.38 It could be suggested that the localization of transcription termination signals near the corresponding region of the 3′ UTR and stop codons might result in the accumulation of m6A sites.
Potential transcriptional and post-transcriptional mechanisms of N6-methylation in long internal exons
Exonic N6-methylation might be correlated with some exon-associated factors. Generally, the corresponding positions in the chromatin of long exons show higher histone methylation levels than other regions of mRNA.38 It is intriguing that histone methylation of H3K36me3 was reported to be related with N6-methylation. METTL14 can interact with H3k36me3 in chromatin. Knockdown of histone-lysine N-methyltransferase SETD2 decreases the m6A level in mRNA by approximately 40%. Therefore, it was proposed that H3k36me3 guides N6-methylation in the corresponding positions of nascent RNAs, which leads to a high N6-methylation density in long exons.39 However, the correlation between H3K36me3 sites and N6-methylaiton sites has not been confirmed.
It is known that various RBPs bind specific motifs in pre-mRNAs, mediating RNA processing post-transcriptionally. Long internal exons have a high density of the RBPs binding motif. Before discussing the association between RBPs and long internal exon-specific N6-methylation, we must make clear whether N6-methylation also occurs in the post-transcriptional period. Notably, a study of m6A in circRNAs performed by Zhou et al may provide some clues. They observed that among a total of 1191 detected circRNAs in human embryonic stem cells, 304 circRNAs were generated from genes that encoded a mRNA without the m6A signal. The rest of the 887 circRNAs were generated from genes that encoded N6-methylated mRNAs, and the m6A sites of 392 circRNAs were distributed in different exons from the exons in the cognate N6-methylated mRNAs, displaying somewhat distinct distribution patterns.40 circRNAs are generated from pre-mRNA via back-splicing post-transcriptionally, which results in the circularization of exons and pairing of flanking introns, forming a lariat structure.41, 42 The distinct m6A distribution pattern in mRNA and circRNAs suggests that RNAs are also N6-methylated post-transcriptionally. Therefore, the N6-methyltranferase-associated RBPs were possibly able to guide the selective N6-methylation. For instance, RBM15, an accessary m6A writer mentioned above, is also an RBP. RBM15 has been demonstrated to be involved in N6-methylation of lncRNA XIST. It could be observed from the CLIP sequencing data that an abundance of RBM15 binding sites were adjacent to N6-methylation sites in exons. Knockdown of RBM15 reduced the level of m6A in mRNA,27 suggesting that RBM15 may recruit the N6-methyltransfase complex to its targets.
Conformational regulation of RNA G-quadruplexes in intronic N6-methylation
G-quadruplexes (G4s) are a type of non-canonical secondary structure in DNA or RNA that form by guanine-rich sequences. These sequences are defined as potential G-quadruplex-forming sequences (PQSs) and are characterized by 4 guanine clusters separated by loops.43 It has been reported that G4s can modulate DNA methylation.44 Recently, the potential correlation between m6A and G4s also emerged from the epitranscriptomic analysis performed by Burrow's group. Initially, several instances of colocalization of m6A and PQSs in virus were explored.45 At the time of the writing of this review, they further discovered that m6A in intron splice sites of human pre-mRNAs were frequently localized in PQS regions.46 These observations suggest that selective N6-methylation might partially result from G4s. Since G4s sequences have been demonstrated to restrict DNA methylation by binding the methyltransferase DNMT1,44 it is likely that intronic RNA G4s are able to modulate local m6A by recruiting m6A writers in a similar way. Nevertheless, it was observed that non-intronic N6-methylation sites did not show a strong correlation with G4s.46 Intronic m6A prefers to be added at SAG consensus motifs,47 which may be indicate that these are the limiting elements involved in the m6A modification. It is possible that some G4s interact with intermediates that can recognize the SAG motifs and act as guides for the m6A writers. However, this hypothesis should be further confirmed by mutational analysis. Additionally, the colocalization of m6A and G4s need to be widely verified in other species.
Transcription factors and histone methylation are associated with transcript-specific N6-methylation
It has been observed that the m6A peaks are always concentrated in partial transcripts from accumulated MeRIP-Seq data, which implies that the N6-methyltransfases are recruited to specific transcripts during transcription or post-transcription periods. Histone methylations and transcription factors are two types of essential transcriptional regulators. In addition to histone methylation, as discussed above, transcription factors are also indicated to be involved in selective N6-methylation and to guide N6-methylation co-transcriptionally. For instance, the SMAD family member 2/3 bound METTL3/14 and co-transcriptionally recruits METTL3/14 to its target transcripts.48 METTL3 interacts with selective gene promotors by associating with the CCAAT enhancer binding protein zeta.49 At the post-transcriptional level, RBP is one of the potential regulators of transcript-specific N6-methylation. Other related factors remain to be detected.
Chemical modification affects the activity of N6-methyltransfase
Most m6A sites exhibit spatiotemporally specific patterns, and the level of m6A also dynamically fluctuates during different developmental or biological stages. As m6A is enzymatically regulated, the activity of the N6-methyltransfase is closely related to the diversity of the level of m6A. The catalytic activity of METTL3/14 has been demonstrated to be regulated by a variety of chemical modifications. For instance, small ubiquitin-related modifier 1 (SUMO1) inhibits the N6-methyltransferase activity of METTL3 by SUMOylating the lysine residues of METTL3 without affecting its protein level or its association with other N6-methyltransferase components. This SUMOylation can be decreased by sentrin-specific protease 1.50 The phosphorylation of residue Ser-399 of METTL14 is crucial for its interaction with METTL3. Mechanically, the phosphorylated Ser-399 in METTL14 forms a salt bridge with ‘Arg-471’ of METTL3. Dephosphorylation of Ser-399 disrupts the interaction of METTL3 and METTL14, resulting in the inactivation of N6-methyltransfase.17 Proteomic analysis also revealed phosphorylation sites in WTAP. However, it is unclear whether the phosphorylation in WTAP is related to localization and interaction with the N6-methyltransfase complex.51, 52, 53 The mechanisms affecting the level of m6A writers have not been clearly reported. Although the expression of writers is dysregulated in some diseases, the details of this dysregulation are unknown.
The cooperation of a series of factors dynamically modulate N6-methylation and lead to distinct N6-methylation patterns in various types of cells and stages. More underlying factors and mechanisms remain to be explored and verified. In addition, it is unclear whether an m6A eraser and N6-demethylation are involved in the formation of the m6A distribution pattern. Analysis of the associations of erasers might be able to provide answers.
Recognition of m6A and its effects on RNA processing and gene expression
m6A controls the fate of RNAs. Aberrant expression levels of writers or erasers result in the dysregulation of RNA degradation, exportation, translation and more. In most cases, N6-methylation does not affect RNA features directly (Fig. 2). Readers are able to recognize m6A and regulate the stability, exportation and translation of methylated RNAs. The YTH (YT521-B homology) domain containing protein family (YTHDF) is a group of evolutionarily conserved m6A readers. The YTH domain has been suggested to bind RNA in an m6A-dependent manner.54, 55, 56 The family members YTHDF1-3 contain long, low-complexity regions enriched with Pro-Xn-Gly motifs.57 The association of N6-methylated RNAs promotes the phase separation of YTHDFs. Conversely, YTHDFs lead N6-methylated RNAs to diverse compartments in the cytosol, which then undergo specific processes.58 YTHDF1 is transported to stress granules by the guidance of the bound N6-methylated mRNAs. In stress granules, YTHDF1 recruits and interacts with EIF3 to form a loop structure composed of mRNAs and translational factors. This association further enhances ribosome loading and translation initiation of target mRNAs.59 Distinct from YTHDF1, YTHDF2 affects the stability of target RNAs. The interaction of N6-methylated RNAs with YTHDF2 promotes the YTHDF2-RNA complex to localize in processing bodies. The CCR4-NOT deadenylation complex and decapping complex are recruited by YTHDF2 and degenerate N6-methylated RNAs.60,61 The third member of the YTHDF family, YTHDF3, has been reported to serve as the functional partner of the other YTHDFs in HeLa cells. It was revealed that YTHDFs share a considerable amount of target transcripts and interact with each other, while YTHDF3 was the earliest member to recognize and bind N6-methylated RNAs. YTHDF3 associates with the translation initiation complex, facilitates the activity of YTHDF1 and subsequently increases the efficiency of YTHDF2, indicating that YTHDF3 seems to be the deliverer of N6-methylated RNAs. Insufficient YTHDF3 decreases the m6A affinity of YTHDF1 and YTHDF2.62,63 In addition, Yang et al also reported the novel finding that YTHDF3 can interact with eukaryotic translation initiation factor 4 gamma 2 (EIF4G2) and recruit translation initiation cooperators to the target m6A sites in circular RNAs. This process drives the 5′ cap independent translation of circular RNAs.64 These functional studies of YTHDF3 not only revealed the translation regulation manner of YTHDF3 but also indicated that the members of the YTHDF family are interrelated counterparts of the m6A metabolic machinery. However, structural analysis might still be the most efficient means to detect the underlying domains in YTHDFs that are crucial for the spatiotemporal distinction of phase separation and target specify.
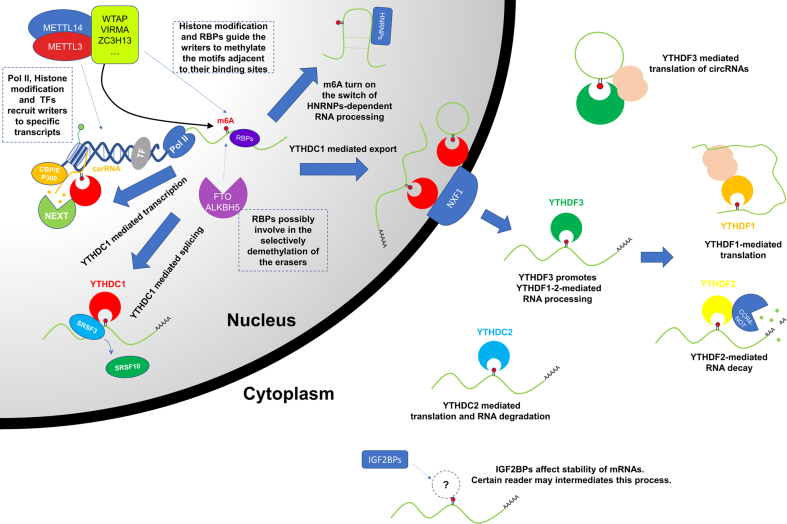
Figure 2.
Schematic of N6-methylation/demethylation and m6A-regulated RNA processing. RNAs are N6-methylated by the writers and reversely N6-demethlyated by the erasers. The processes of methylation/demethylation may be guided by specific transcriptional factors, RBPs and histone modifications. N6-methylated RNAs are further recognized and processed by a serious of readers and associated factors.
YTH domain-containing protein 1 (YTHDC1) is another cluster of readers that are localized in the nucleus.65 YTHDC1 is functionally related to RNA maturation. When pre-mRNA is transcribed, YTHDC1 promotes exon-inclusion by recruiting serine/arginine-rich splicing factor 3 (SRSF3) to the target pre-mRNA and displacing the interacted SRSF10.66 Furthermore, the interaction of YTHDC1, SRSF3 and nuclear RNA export factor 1 (NXF1) promotes nuclear export of N6-methylated mRNAs.67 Chen et al demonstrated that YTHDC1 facilitates the export of the N6-methylated circular RNA circNSUN2.68 This research direction was significant for completing our understanding of the splicing and export mechanisms of circular RNA. Further explorations of the associated splicing and export factors were expected. YTHDC1 was also found to interact with the N6-methylated nuclear lncRNA XIST and to enhance XIST-mediated X chromosome silencing. In addition to XIST, other N6-methylated nuclear lncRNAs, such as NEAT1 and MALAT1, bind to YTHDC1.27 However, the effect of this binding is unknown. Recently, YTHDC1 was detected to be involved in m6A-mediated transcription regulation. Nuclear chromosome-associated regulatory RNAs (carRNAs), including promoter-associated RNA (paRNA), enhancer RNA (eRNA) and RNA transcribed from transposable elements (repeats RNA), can recruit CBP/EP300 and YY1 to promote chromatin opening and transcription. YTHDC1 interacts with N6-methylated carRNAs and degrades them via association with nuclear exosome targeting complex (NEXT), resulting in selective inhibition of transcription.69
YTHDC2 is the largest YTH protein, and it contains multiple helicase domains and two ankyrin repeats. Correspondingly, YTHDC2 plays a translation regulatory role, similar to 3′–5′ RNA helicases.70 YTHDC2 has also been indicated to affect mRNA stability by associating with XRN1 exoribonuclease.71 However, the effect of YTHDC2 is relatively weak because of its limitations in m6A binding. YTHDC2 has been shown to have a distinct m6A target pattern and lower affinity for N6-methylated RNAs than other YTH readers.56,57,71 Moreover, YTHDC2-deficiency only causes a spermatogenesis defect.71,72 A CLIP study suggested that YTHDC2 does not interact with N6-methylated RNAs in HEK293 cells.27,57 It is possible that YTHDC2 mediates m6A in a novel and tissue-specific way.
m6A are believed to be related to the maturation of miRNAs. m6A in pri-miRNAs improves DGCR8 binding and the level of mature miRNAs. The promotion induced by m6A occurs in a global and non-cell-type specific manner.73 Since DGCR8 cannot specifically recognize m6A, ubiquitously expressed nuclear m6A readers may act as cooperators for the recruitment of N6-methylated pri-miRNA, such as YTHDC1.
Therefore, we can summarize that m6A functions by methylation, recognition and processing of RNA in general. Following writer-regulated methylation, readers further specifically deal with N6-methylated RNAs. The distinct structures of readers determine their localization, target points and associated cofactors. In addition to RNA processing, the associated cofactors might also reversibly affect readers by selectively binding m6A. As a result, the fate of RNAs is organically regulated.
In a few cases, an RNA structure modified by m6A would function directly by altering the interactions of RNAs and proteins. Several members of the heterogeneous nuclear ribonucleoprotein (HNRNP) family, including HNRNPC, HNRNPA2B1 and HNRNPG, which are involved in RNA processing, are affected by m6A in this way.74, 75, 76 For example, HNRNPC binds the single-stranded poly-U motif in pre-mRNA and regulates pre-mRNA processing. Liu et al reported that m6A destabilized the U-A pairing that could expose the single-stranded U-enriched region in pre-mRNA from UA duplexes and facilitate the binding of HNRNPC.75
Insulin-like growth factor 2 mRNA-binding proteins 1-3 (IGF2BP 1-3) have also been reported to bind and stabilize N6-methylated mRNAs.77 However, it seems that IGF2BPs cannot specifically recognize m6A. It is possible that IGF2BPs indirectly interact with N6-methylated mRNAs depending on other cofactors.
Biological functions and dysregulation of m6A in diseases
Abundant m6A controls gene expression throughout the lifespan of an organism. It has been well proven that the highly regulated stages of m6A modification, recognition and processing are indispensable for diverse developmental and physiological activities. Conversely, once writers, erasers or readers become disorganized, the induced unbalanced gene expression causes disease or other biological defects (Table 1). In this part, we summarize representative studies in biology and pathology.
Table 1.
m6A-involved biological and pathological processes.
| Physiological and pathological processes | Critical m6A regulators | Representative target genes/transcripts | Pathogeny | References |
|---|---|---|---|---|
| Embryonic development | METTL3, YTHDF2, YTHDC1 | carRNAs | – | 12,18,69,79 |
| Haematopoietic stem transition | METTL3, YTHDF2 | notch1a | – | 80 |
| Neural stem cell maintenance | METTL3/14 | pax6, neurogenin-2, cbp, p300 | – | 81,82 |
| Myelination of axons | METTL14 | neurofascin 155 | – | 83 |
| Male fertility | ALKBH5, YTHDC2 | Transcriptomic research | – | 37,72,85 |
| Female fertility | YTHDF2, YTHDC2 | smc3, cep76 | – | 71,72,86 |
| Dopaminergic midbrain circuitry | FTO | drd3, kcnj6, grin1 | – | 89 |
| Learning and memory | YTHDF1 | Transcriptomic research | – | 90 |
| T cell homeostasis | METTL3 | socs1, socs3, cish | – | 91 |
| Anti-virus | ALKBH5 | OGDH | – | 92 |
| Hepatocellular carcinoma | METTL3, YTHDF2 | SOC2 | Upregulation of METTL3 | 93 |
| Endometrial cancer | METTL3/14, YTHDF1/2 | PHLPP2, PRR5, PRR5L, mTOR | METTL3 deficiency, METTL14 mutation | 94 |
| Glioblastoma | ALKBH5 | FOXM1 | Upregulation of ALKBH5 | 95 |
| Acute myeloid leukaemia | METTL14, FTO | MYB/MYC, ABS2, RARA | Upregulation of METTL14 and FTO | 36,96 |
| Ischaemic cardiac injury | METTL3 | TFEB | Upregulation of METTL3 | 97 |
| Cardiac hypertrophy | METTL3 | Transcriptomic research | Upregulation of METTL3 | 98 |
| Heart failure | ALKBH5 | serca2a | Downregulation of ALKBH5 | 99 |
m6A is essential for selective gene expression in development and cell differentiation
Cell differentiation is a milestone of development. m6A was initially considered to be a significant epigenetic regulator of embryonic stem cell (ESC) differentiation. By means of MeRIP-Seq, high levels of m6A were detected in the transcriptomes of mammalian ESCs that were especially enriched in the transcripts of genes critical for pluripotency. Decreasing the level of m6A via knockout of METTL3 led to the abolished differentiation of ESCs and embryonic lethality in mice. Pluripotency genes such as NANOG and oct4 showed excessive levels in the status of METTL3 deficiency.12,18,78 Later, m6A was indicated to be the signal for YTHDF2 to degrade specific mRNAs in zebrafish embryo development.79 This result corresponds with the previously observed abnormally sustained mRNAs induced by METTL3-KO. In addition to its YTHDF2-dependent “turn-off” role of gene expression, m6A fulfilled YTHDC1-dependent transcriptional inhibition in the earlier stage of embryonic development (see the details of the mechanism in the previous section).69 These data demonstrate that m6A controls selective gene expression in embryonic development at both the transcriptional and post-transcriptional levels.
In addition, m6A is involved in secondary cell differentiation. For instance, m6A mediates the transition from haemogenic endothelial cells to haematopoietic stem and progenitor cells in zebrafish. During haematopoietic development, notch1a mRNAs are N6-methylated by METTL3 and then decayed by YTHDF2. The inhibition of notch signalling facilitates the generation of haematopoietic stem and progenitor cells.80 m6A also maintains the self-renewal and prevented premature differentiation of neural stem cells. During neurogenesis, the mRNAs of genes related to neuronal differentiation and development, such as pax6 and neurogenin-2 and histone H3K27 acetyltransferases, including cbp and p300, are N6-methylated. The expression of differentiation-related genes is inhibited by m6A-directed mRNA decay and a decrease of H3K27-ac.81,82 At the subsequent stage of the myelination of axons, m6A mediates the splicing of global RNAs in the oligodendrocyte-lineage, affecting the expression of genes that are essential for myelination, such as neurofascin 155. Knock-out of METTL14 retards the specialization of oligodendrocyte precursor cells to mature oligodendrocytes, indicating that m6A acts as a “switch” for gene expression during development and cell differentiation.83
m6A regulates germ cell maturity and fertility
In 2013, Zheng et al discovered that ALKBH5 modulates spermatogenesis. Among eight different organs, including the heart, brain, gonadal fat pads, liver, kidney, spleen, lung and testis, the testis showed the highest expression level of ALKBH5 mRNAs. ALKBH5-KO mice were able to survive to adulthood. However, male mice were infertile and had a significantly undersized testis, malformed spermatozoa, maturation arrest and apoptotic spermatocytes. Although differential expression of genes related to spermatogenesis and apoptosis was detected in ALKBH5-deficency testis, the detailed mechanisms of ALKBH5 in spermatogenesis were unclear at that time.37 In regards to the transcriptome, mRNA shortening at the period of spermiogenesis is necessary for mRNA metabolism efficiency.84 Tang et al put forward the hypothesis that ALKBH5 is involved in this process. They observed that m6A was enriched in the long 3′UTR of mRNAs during spermiogenesis. Via demethylation of ALKBH5, these mRNAs with a long 3′UTR avoid decay and are spliced to short forms.85
Although m6A mediates female fertility, the regulatory mechanism is distinct from that found in spermatogenesis. During the period of MII oocyte production, YTHDF2 plays the critical role of eliminating N6-methylated maternal RNAs, which is an important process for oocyte maturation and early zygotic development. Loss of YTHDF2 would result in female-specific infertility.86
The m6A reader YTHDC2 is a special mediator of spermatogenesis and oogenesis.71,72 YTHDC2 mRNAs are highly expressed in the testis and increased in sperm cell meiosis. Knockout of YTHDC2 in mice impairs testis and ovary development. Mechanically, YTHDC2 improves the translation of its target mRNAs, including spermatogenesis-related smc3 and centriole reduplication-related Cep76, during meiosis.71,72
m6A affects neural activities
The mammalian nervous system is an accurately organized control system that regulates behaviour, metabolism, and other functions. Various neural actions are further regulated by complex molecular mechanisms that are still under exploration. As the most abundant RNA modification, m6A also exists in the transcriptome of neural cells. The detailed roles of m6A in neural activity are being gradually revealed. Defects of the m6A eraser FTO was observed to be associated with abnormal phenotypes, such as attention deficit disorder, growth retardation, aberrant energy expenditure and locomotor activity.87,88 Hess et al revealed that FTO mediates the conduction of dopaminergic signalling, which controls a serious of neural activities. Demethylase-inactivation of FTO impairs D2-like receptor-regulated neural and behavioural reactions under the stimulation of cocaine. MeRIP-Seq indicated that knockout of FTO increases m6A levels in mRNAs in the mouse midbrain and striatum. The mRNA levels of genes related to dopaminergic signalling, including drd3, kcnj6 and grin1, were upregulated; but conversely their protein levels were decreased.89 Shi et al showed that m6A affects learning and memory in a YTHDF1-dependent manner. By neuronal stimulation, YTHDF1 facilitates protein translation of target m6A mRNAs to promote synaptic transmission in the mouse hippocampus. Deficiency of YTHDF1 impairs learning and memory.90
m6A involves in immunity
m6A has been indicated to modulate gene expression during the maturation of T cells. JAK1/3-STAT5 signalling is critical for T cell differentiation and proliferation, which is inhibited by SOCS family proteins. Through stimulation by IL-7, the mRNAs of SOCS family genes, including socs1, socs3 and cish, were N6-methylated and degraded. JAK1/3-STAT5 signalling was disinhibited and activated. METTL3-deficiency disrupts native T cell homeostasis.91 An m6A-dependent antiviral mechanism was also revealed. The ALKBH5 protein is inactivated by altered methylation in mouse primary peritoneal macrophages when suffering from infection of vesicular stomatitis virus. Inhibition of ALKBH5 increases the N6-methylation level of α-ketoglutarate dehydrogenase (OGDH) mRNAs. OGDH catalyses the synthesis of itaconate, which is required for viral replication. m6A destabilizes OGDH mRNAs and decreases the level of the OGDH protein. Viral replication is, hence, retarded by itaconate deficiency.92
Aberrant m6A is an oncogenic event
The molecular essence of cancer is characterized by the induction of oncogenes and inhibition of tumour suppressor genes. Accumulated studies have demonstrated that aberrant m6A regulation is one of the pathogeneses of cancers. In some situations, abnormal N6-methylase or N6-demethylase activities dysregulate cancer-related gene expression. For instance, METTL3 is significantly increased in human hepatocellular carcinoma. Increased METTL3 results in high N6-methylation and YTHDF2-controlled degradation of tumour suppressor SOCS2 mRNA. Downregulation of SOCS2 promotes the proliferation and invasion of tumour cells.93 Mutation of METTL14 and METTL3-insufficiency have been detected in endometrial cancer. The decrease of N6-methylation activates tumorigenic AKT signalling through two ways: First, dephosphorylation of the S473 residue of AKT(S473) by phosphatase PHLPP2 is decreased. Low levels of m6A on PHLPP2 mRNA inhibits the promotion of translation managed by YTHDF1. On the contrary, the phosphorylation of AKT by serine/threonine-protein kinase complex mTORC2 is increased. YTHDF2 targets the N6-methylated mRNAs of mTORC2 components, including PRR5, PRR5L and mTOR. The decrease of N6-methylase activity inhibits their degradation. Therefore, phosphorylation of AKT is facilitated and tumorigenic signalling is activated.94 In glioblastoma stem-like cells, ALKBH5 is highly expressed. The transcription factor FOXM1 is beneficial for the proliferation of glioblastoma stem-like cell and tumorigenicity. FOXM1 mRNA is targeted and demethylated by ALKBH5. The interaction of ALKBH5 and FOXM1 mRNA relies on lncRNA FOXM1-AS. N6-demethylated FOXM1 mRNA recruits HuR to increase its stability and protein expression.95 Intriguingly, upregulation of METTL14 and FTO have both been detected in acute myeloid leukaemia (AML). Although the global m6A level is increased in the transcriptome of AML cells, METTL14 and FTO have both been shown to be oncogenic factors.36,96 Weng et al revealed that METTL14 increases the N6-methylation, stability and translation efficiency of MYB and MYC mRNAs. MYB/MYC signalling inhibits myeloid differentiation and promotes the proliferation of AML cells.96 Li et al demonstrated that FTO demethylates the mRNAs of ASB2 and RARA, which regulate myeloid differentiation. This process causes transcripts to be unstable and inhibits protein translation of ASB2 and RARA. Thus, FTO aggravates AML progress.36 Both METTL14 and FTO are negatively regulated by myeloid differentiation agents. SPI1 has been found to be a transcription repressor of METTL14.96 The mRNA level of FTO is decreased by All-trans retinoic acid.36 However, their cooperating m6A readers have not been identified. Data indicate that YTHDFs are not responsible for the mRNA stability or the translation of the targets of FTO and METTL14 in AML.
Excessive m6A is correlated with heart disease
Heart disease is the most lethal disease worldwide. The pathogenesis of heart disease still has not been fully understood. From the 2010s, studies of the post-translational regulation mechanisms of heart disease began to emerge. Certain cardiac non-coding RNAs have been identified and considered to be therapeutic targets. In 2018, the post-transcriptional roles of m6A in a serious cardiac pathology were revealed.97, 98, 99 It was discovered that elevated N6-methylase activity aggravates ischaemic cardiac injury. When cardiomyocytes or the myocardium was challenged with oxidative stress, such as hypoxia/reoxygenation (H/R) and ischaemia/reperfusion (I/R), the mRNA and protein levels of METTL3 were significantly induced. The increased METTL3 targeted pre-mRNAs of the transcription factor EB (TFEB), which is critical for the transcription of lysosomal genes and the regulation of autophagy. Then, the N6-methylated pre-mRNAs of TFEB were bound and degraded by HNRNPD, resulting in the inhibition of cardiomyocyte autophagy and increasing apoptosis. This m6A-related degradation of TFEB pre-mRNA was negatively regulated by ALKBH5. Reversely, TFEB could promote the transcription of ALKBH and suppress METTL3 at the post-transcriptional level, forming a feedback loop.97 Mature mammalian cardiomyocytes are terminally differentiated cells with negligible proliferation. Cardiomyocytes that die from I/R are replaced by fibroblasts, which lack the capacity to contract. Meanwhile cardiac hypertrophy is triggered to adapt the haemodynamic change. Pressure overload and energy deficiency of cardiomyocytes eventually lead to cell death and heart failure. During myocardial remodelling, m6A upregulation has been indicated to play important roles. Dorn et al observed that m6A levels on polyA+ RNAs were elevated in FBS-induced hypertrophic neonatal rat cardiomyocytes. Overexpression of METTL3 in neonatal cardiomyocytes increased the cell surface area. On the contrary, silencing of METTL3 resulted in a smaller cell size. In vivo experiments showed that both a cardiac-specific transgene (cTG) and knockout (cKO) of METTL3 did not significantly affect heart development or cardiac function in juvenile mice. However, 8-month-old METTL3 cTG mice exhibited cardiac hypertrophy, while 8-month-old METTL3 cKO mice suffered heart failure. These results suggest that METTL3 is involved in the stress adaption of heart. However, the details of its mechanisms are unclear. In addition, m6A mapping data and expression profiles of m6A regulators during the stages of mouse embryonic heart development were not shown in this research. These are crucial for rigorously revealing the roles of m6A in heart development.98 Mathiyalagan et al found that the transcriptomic m6A level was remarkably elevated in ischemia-induced mammalian heart failure. Decreased FTO mRNA and protein levels were detected throughout the process of heart failure. MeRIP-Seq indicated that the mRNAs of some Ca2+ homeostasis and contraction-regulation genes, such as serca2a, were hypermethylated, leading to degradation of mRNA and decrease in translation. Decreased expression of serca2a resulted in dysregulation of myocardial remodelling. Replenishment of FTO could demethylate serca2a mRNA and rescue the failing heart. In vitro experiments showed that hypoxia could inhibit the expression of FTO mRNA and protein. In addition, METTL14 has been observed to have elevated protein levels without an mRNA increase in heart failure, whether METTL14 involved in the process is unknown.99
Perspectives
Since m6A was globally characterized in transcriptomic by RIP-Seq in 2012,10,11 numerous studies on m6A have been performed. It is exciting that the mechanisms of methylation/demethylation, recognition and RNA processing have basically been revealed. Increasing pathological and physiological events have been demonstrated to be regulated by m6A. These research achievements have enriched medical theory and deepened our understanding of disease progression.
According to present studies, we can conclude that N6-methylation and demethylation act in a tissue- and developmental stage-specific pattern. For instance, N6-methylation is generally predominant in embryonic development,12,18,69,78 and deficiency of m6A erasers does not lead significant embryonic defects.35,37 However, in some tissues or types of cells, such as the brain, testis, ovary and heart, N6-demethylation is physiologically indispensable.37,71,72,85,97,99 To spatiotemporally understand N6-methylation and demethylation, several uncertain points about m6A still remain. First, the details of the associated factors of m6A writers/erasers are needed. Although the writers and erasers possess N6-methylation/demethylation activities, they have no potency to distinguish specific targeting motifs among thousands of transcripts. The association of certain transcription factors, histone modifications and RBPs have been detected, which suggests these proteins might be involved in target selection.23,24,27,39,48,49 The details of the proteomic interactions and analysis of the target sites of the writers/erasers in different types of cells and development stages are necessary. Second, information regarding the expression regulation and modification of the writers/erasers is needed. Dynamic equilibrium between N6-methylation and demethylation is critical, as aberrant levels of writers or erasers would cause various defects or disease. It was discovered that writers and erasers are regulated at the mRNA and protein levels.93, 94, 95, 96, 97,99 However, most of the upstream modulators remain to be explored. In addition, distinct subcellular localization of writers and erasers exist in certain types of cells.13,14,36 Cytoplasmic localization might result in some novel functions. Perhaps protein modifications affect the subcellular localization of writers and erasers. Next, the mechanisms of differential recognition of N6-methylated transcripts need to be ascertained. Commonly, readers determine the fate of N6-methylated transcripts. Structure distinctions, cellular localization and external stress may affect the selective targeting of readers.54,56, 57, 58
Aberrant m6A levels in some types of cancers might be applied to diagnosis. Combining immunohistochemistry with histological analysis of the level of m6A would facilitate more accurate diagnoses. In addition, the activity of erasers has been found to be regulated by various reagents. For example, the activity of FTO is activated by ascorbate and inhibited by N-oxalylglycine, fumarate, succinate, fluorescein derivatives, meclofenamic acid and R-2-hydroxyglutarate.8,100, 101, 102 Both FTO and ALKBH5 need Fe2+ to act as cofactor.103,104 R-2-hydroxyglutarate has been especially indicated to inhibit leukaemia by affecting FTO/m6A/MYC/CEBPA signalling.102 The therapeutic potency of the other activators and inhibitors remains to be verified.
In conclusion, developing technology and deepening research have led us to realize the significance of m6A. Dynamic and integral mapping of m6A sites and the proteomic interaction involving each m6A regulator is expected to uncover the panorama of m6A.
Conflict of interest
The authors have declared that no competing interest exists.
Acknowledgements
This work was supported by Taishan Scholar Program of Shandong Province (tsqn201812044), Natural Science Foundation of Shandong Province (JQ201815) and National Natural Science Foundation of China (81900259).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Wei C.M., Gershowitz A., Moss B. Methylated nucleotides block 5ʹ terminus of HeLa-cell messenger-RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 2.Rottman F.M., Desrosiers R.C., Friderici K. Nucleotide methylation patterns in eukaryotic mRNA. Prog Nucleic Acid Res Mol Biol. 1976;19:21–38. doi: 10.1016/s0079-6603(08)60906-x. [DOI] [PubMed] [Google Scholar]
- 3.Wei C.M., Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 4.Schibler U., Kelley D.E., Perry R.P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- 5.Narayan P., Rottman F.M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- 6.Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M. Purification and Cdna cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 7.Bujnicki J.M., Feder M., Radlinska M., Blumenthal R.M. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m6A methyltransferase. J Mol Evol. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- 8.Gerken T., Girard C.A., Tung Y.C. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Little N.A., Hastie N.D., Davies R.C. Identification of WTAP, a novel Wilms' tumour 1-associating protein. Hum Mol Genet. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- 10.Dominissini D., Moshitch-Moshkovitz S., Schwartz S. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 11.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3ʹ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geula S., Moshitch-Moshkovitz S., Dominissini D. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 13.Choe J., Lin S., Zhang W. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Yue Y., Han D. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer L.M., Zhang D., Aravind L. Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. BioEssays. 2016;38:27–40. doi: 10.1002/bies.201500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Feng J., Xue Y. Structural basis of N(6)-adenosine methylation by the METTL3–METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortega A., Niksic M., Bachi A. Biochemical function of female-lethal (2)D/Wilms' tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J Biol Chem. 2003;278(5):3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- 20.Horiuchi K., Umetani M., Minami T. Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci U S A. 2006;103(46):17278–17283. doi: 10.1073/pnas.0608357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Small T.W., Bolender Z., Bueno C. Wilms' tumor 1-associating protein regulates the proliferation of vascular smooth muscle cells. Circ Res. 2006;99(12):1338–1346. doi: 10.1161/01.RES.0000252289.79841.d3. [DOI] [PubMed] [Google Scholar]
- 22.Ping X.L., Sun B.F., Wang L. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horiuchi K., Kawamura T., Iwanari H. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288(46):33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue Y., Liu J., Cui X. VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knuckles P., Lence T., Haussmann I.U. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen J., Lv R., Ma H. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69:1028–1038. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil D.P., Chen C.K., Pickering B.F. m6A RNA methylation promotes XIST- mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita Y., Krause G., Scheffner M. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E- cadherin complex. Nat Cell Biol. 2002;4:222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- 29.Pendleton K.E., Chen B., Liu K. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma H., Wang X., Cai J. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15(1):88–94. doi: 10.1038/s41589-018-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clifton I.J., McDonough M.A., Ehrismann D., Kershaw N.J., Granatino N., Schofield C.J. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem. 2006;100(4):644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 32.Falnes P.O., Johansen R.F., Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 33.Jia G., Yang C.G., Yang S. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia G., Fu Y., Zhao X. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauer J., Luo X., Blanjoie A. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541(7637):371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z., Weng H., Su R. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng G., Dahl J.A., Niu Y. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slobodin B., Han R., Calderone V. Transcription impacts the efficiency of mRNA translation via Co-transcriptional N6-adenosine methylation. Cell. 2017;169(2):326–337. doi: 10.1016/j.cell.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang H., Weng H., Zhou K. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature. 2019;567:414–419. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou C., Molinie B., Daneshvar K. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20(9):2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeck W.R., Sorrentino J.A., Wang K. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Cammas A., Millevoi S. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017;45(4):1584–1595. doi: 10.1093/nar/gkw1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao S.Q., Ghanbarian A.T., Spiegel J. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol. 2018;25(10):951–957. doi: 10.1038/s41594-018-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleming A.M., Nguyen N.L.B., Burrows C.J. Colocalization of m6A and G-quadruplex-forming sequences in viral RNA (HIV, zika, hepatitis B, and SV40) suggests topological control of adenosine N6-methylation. ACS Cent Sci. 2019;5(2):218–228. doi: 10.1021/acscentsci.8b00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jara-Espejo M., Fleming A.M., Burrows C.J. Potential G-quadruplex forming sequences and N6-methyladenosine colocalize at human pre-mRNA intron splice sites. ACS Chem Biol. 2020;15(6):1292–1300. doi: 10.1021/acschembio.0c00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Louloupi A., Ntini E., Conrad T., Ørom U.A.V. Transient N-6-methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 2018;23(12):3429–3437. doi: 10.1016/j.celrep.2018.05.077. [DOI] [PubMed] [Google Scholar]
- 48.Bertero A., Brown S., Madrigal P. The SMAD2/3 interactome reveals that TGFb controls m6A mRNA methylation in pluripotency. Nature. 2018;555:256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbieri I., Tzelepis K., Pandolfini L. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du Y., Hou G., Zhang H. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018;46:5195–5208. doi: 10.1093/nar/gky156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H., Di Palma S., Preisinger C. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12(1):260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 52.Olsen J.V., Vermeulen M., Santamaria A. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104) doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 53.Huttlin E.L., Jedrychowski M.P., Elias J.E. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theler D., Dominguez C., Blatter M., Boudet J., Allain F.H. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res. 2014;42:13911–13919. doi: 10.1093/nar/gku1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo S., Tong L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc Natl Acad Sci U S A. 2014;111:13834–13839. doi: 10.1073/pnas.1412742111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem. 2015;290:24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patil D.P., Pickering B.F., Jaffrey S.R. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 2017;28:113–117. doi: 10.1016/j.tcb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ries R.J., Zaccara S., Klein P. m6A enhances the phase separation potential of mRNA. Nature. 2019;571:424–428. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X., Zhao B.S., Roundtree I.A. N(6)-Methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X., Lu Z., Gomez A. N6-Methyladenosine- dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du H., Zhao Y., He J. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi H., Wang X., Lu Z. YTHDF3 facilitates translation and decay of N6-methyladenosine- modified RNA YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li A., Chen Y.S., Ping X.L. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Y., Fan X., Mao M. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z., Theler D., Kaminska K.H. The YTH domain is a novel RNA binding domain. J Biol Chem. 2010;285(19):14701–14710. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao W., Adhikari S., Dahal U. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61(4):507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Roundtree I.A., Luo G.Z., Zhang Z. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6 doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen R.X., Chen X., Xia L.P. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu J., Dou X., Chen C. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367(6477):580–586. doi: 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wojtas M.N., Pandey R.R., Mendel M., Homolka D., Sachidanandam R., Pillai R.S. Regulation of m6A transcripts by the 3′→5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell. 2017;68(2):374–387. doi: 10.1016/j.molcel.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 71.Kretschmer J., Rao H., Hackert P., Sloan K.E., Höbartner C., Bohnsack M.T. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′-3′ exoribonuclease XRN1. RNA. 2018;24:1339–1350. doi: 10.1261/rna.064238.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsu P.J., Zhu Y., Ma H. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6-Methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu N., Zhou K.I., Parisien M., Dai Q., Diatchenko L., Pan T. N6-Methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N(6)-Methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu B., Su S., Patil D.P. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9:420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang H., Weng H., Sun W. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances Mrna stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Batista P.J., Molinie B., Wang J. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao B.S., Wang X., Beadell A.V. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542(7642):475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C., Chen Y., Sun B. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549(7671):273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y., Li Y., Yue M. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21(2):195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoon K.J., Ringeling F.R., Vissers C. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171(4):877–889. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu H., Dzhashiashvili Y., Shah A. m6A mRNA methylation is essential for oligodendrocyte maturation and CNS myelination. Neuron. 2020;105(2):293–309. doi: 10.1016/j.neuron.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bao J., Vitting-Seerup K., Waage J. UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3′UTR transcripts. PLoS Genet. 2016;12:e1005863. doi: 10.1371/journal.pgen.1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang C., Klukovich R., Peng H. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2018;115(2):E325–E333. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ivanova I., Much C., Di Giacomo M. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67(6):1059–1067. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choudhry Z., Sengupta S.M., Grizenko N. Association between obesity-related gene FTO and ADHD. Obesity. 2013;21(12):E738–E744. doi: 10.1002/oby.20444. [DOI] [PubMed] [Google Scholar]
- 88.Fischer J., Koch L., Emmerling C. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 89.Hess M.E., Hess S., Meyer K.D. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16(8):1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 90.Shi H., Zhang X., Weng Y.L. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563(7730):249–253. doi: 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li H.B., Tong J., Zhu S. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y., You Y., Lu Z. N6-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science. 2019;365(6458):1171–1176. doi: 10.1126/science.aax4468. [DOI] [PubMed] [Google Scholar]
- 93.Chen M., Wei L., Law C.T. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 94.Liu J., Eckert M.A., Harada B.T. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20(9):1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang S., Zhao B.S., Zhou A. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation Program. Cancer Cell. 2017;31(4):591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weng H., Huang H., Wu H. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22(2):191–205. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Song H., Feng X., Zhang H. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15(8):1419–1437. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dorn L.E., Lasman L., Chen J. The N6-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. 2019;139(4):533–545. doi: 10.1161/CIRCULATIONAHA.118.036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mathiyalagan P., Adamiak M., Mayourian J. FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2018;139(4):518–532. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang T., Hong T., Huang Y. Fluorescein derivatives as bifunctional molecules for the simultaneous inhibiting and labeling of FTO protein. J Am Chem Soc. 2015;137(43):13736–13739. doi: 10.1021/jacs.5b06690. [DOI] [PubMed] [Google Scholar]
- 101.Huang Y., Yan J., Li Q. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43(1):373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Su R., Dong L., Li C. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172(1–2):90–105. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Han Z., Niu T., Chang J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464(7292):1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 104.Aik W., Scotti J.S., Choi H. Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42(7):4741–4754. doi: 10.1093/nar/gku085. [DOI] [PMC free article] [PubMed] [Google Scholar]