Abstract
Background
Tumor-associated neutrophils (TANs) have been a research hotspot in recent years. However, the role and relevant mechanisms of TANs in the tumor microenvironment (TME) have not yet been elucidated.
Method
The ribonucleic acid (RNA) expression levels of fucosyltransferase 4 (FUT4) and elastase, neutrophil expressed (ELANE) in samples from The Cancer Genome Atlas (TCGA) (n=4,538) were analyzed. Receiver operating characteristic (ROC) curves were used to calculate the critical cutoff values, and different data were defined as high and low expression. The tumor microenvironment immune type (TMIT) was defined according to the activation state of TAN, and the samples were classified into three TMITs based on their cut-off values. Mutational datasets and overall survival were compared according to the TMITs.
Results
The prognostic significance of FUT4, ELANE, and myeloperoxidase (MPO) was different among the 15 cancers, and the prognostic significance of different TMITs varied across the different tumors. Compared with the other groups, TMIT 3 had a favorable prognostic effect, which was most prominent in lung adenocarcinoma (LUAD) [hazard ratio (HR) =0.292, 95% confidence interval (CI): 0.185–0.459, P<0.001].
Conclusions
Our study demonstrated that highly-activated TANs predicted a favorable prognosis in humans using genomic analyses for the first time. This provides a realistic basis for further exploring the role of TANs in the immune microenvironment and provides real world data for tumor immunotherapy.
Keywords: Tumor microenvironment immune type (TMIT), neutrophils, mRNA sequencing, mutation burden
Introduction
Neutrophils participate in defense mechanisms that protect the host against injury and infection. The recent discovery of novel tumor-infiltrating myeloid cells in the tumor microenvironment (TME) has garnered widespread attention. In the TME, neutrophils in the blood enter the tumor tissue through the vascular wall via upregulation of chemotactic substances that are produced by tumor tissues. These infiltrating neutrophils are called tumor-associated neutrophils (TANs) (1). Numerous murine studies have shown that neutrophils in the TME have two biological functions (2). Considering the different effects of TANs on tumor progression, studies have proposed both anti-tumor and tumor-promoting properties, known as N1 and N2 types, respectively (3).
In human tumors, TANs account for a large part of the immune infiltration of a variety of cancers including lung cancer, stomach cancer, and kidney cancer (4). TANs were found to be associated with prognosis in some of these studies. Immunohistochemistry has confirmed that TAN infiltration in various tumors indicates a poor prognosis, suggesting a tumor-promoting phenotype, although the mechanisms remain largely unclear. Tazzyman showed that neutrophils increase the proliferation of lung carcinoma cells in vitro (5). An early study nicely demonstrated that depletion of neutrophils can have an inhibitory effect on tumor growth, indicating that the tumor requires TAN for rapid growth (6). Some scholars believe that the neutrophil phenotype is related to its degree of activation; that is, when the degree of activation is high, the N1 phenotype is expressed, and conversely when the activation is low, the N2 phenotype is expressed (7). Recent evidences over the functional plasticity of cancer-related neutrophils have alleviated the conflicting reports regarding neutrophil functions in cancer. However, this conclusion lacks a real-world evidential basis.
Cluster of differentiation 15 (CD15), a common marker synthesized by fucosyltransferase 4 (FUT4) and used to identify human neutrophils, works as an effective molecule for exploring the immunological roles of neutrophils in the TME (8). FUT4 is a key gene for synthesizing tumor-associated carbohydrate antigen including Lewis A, B, X and Y (9), which catalyzes the transfer of l -fucose from GDP-fucose to the substrates. FUT4 is highly expressed in several types of cancers, including leukemia, gastric and breast cancers, and is positively correlated with tumor progression (10). Few evidence of the association between FUT4 and cancer prognosis has been reported. Previous research has confirmed that CD15 + TAN can be used as an independent adverse prognostic indicator for cancer (11). Neutrophil elastase was first described as a serine protease stored in azurophilic granules of neutrophils. While the enzyme is abundantly present within mature neutrophils, mRNA transcripts encoded by the ELANE gene (12). TAN relies on various enzymes; predominantly neutrophil elastase (NE) and myeloperoxidase (MPO). When neutrophils are activated, NE is released, causing decomposition of surrounding proteins and tissue destruction. MPO is another functional and activation marker of neutrophils, and its level and activity change represent the function and activity state of neutrophils. The role of ELANE and MPO are not completely understood, and research on the TAN functional phenotype at the genetic level is still lacking.
In human tumors, TANs account for a large part of the immune infiltration of many cancers. An increasing number of studies have recently examined the prognostic value of TANs infiltration in various types of cancer, but the utility of this value as a prognostic tool is controversial. Considering the clinical relevance of the multifaceted TANs-associated immune response, a comprehensive immunogenomic analysis of TANs is crucial to deepen our understanding of the prognostic value of TANs. In the present study, we analyzed real-world tumor patients from The Cancer Genome Atlas (TCGA) database to explore the prognostic value of TAN activation status. Our study aims to assess the following: (I) the prognostic value of each gene (FUT4, ELANE, and MPO); (II) the prognostic value of different immunophenotypes by combining gene definition based on immunological typing of TAN activation status; and (III) the best indicator for evaluating immune typing. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6629).
Methods
Processing of genomic data from TCGA project
In this study, we used publicly available data sets from TCGA data portal (https://portal.gdc.cancer.gov). The following solid tumor types were selected: bladder urothelial carcinoma (BLCA, n=412), breast cancer (BRCA, n=1,067), cervical cancer (CESC, n=307), glioblastoma multiforme (GBM, n=159), kidney renal papillary cell carcinoma (KIRP, n=291), lung adenocarcinoma (LUAD, n=501), esophageal carcinoma (ESCA, n=185), head and neck squamous cell carcinoma (HNSC, n=528), brain lower grade glioma (LGG, n=513), colorectal and rectal cancer (COADREAD, n=615), skin cutaneous melanoma (SKCM, n=469), adrenocortical carcinoma (ACC, n=92), uterine corpus endometrial carcinoma (UCEC, n=176), pancreatic adenocarcinoma (PAAD, n=185), and prostate adenocarcinoma (PRAD, n=500).
Samples of the 15 aforementioned cancer types (N=6,000) were included in the analysis. The data sets comprised messenger ribonucleic acid sequencing (mRNA-seq) data from TCGA tumor samples (13,14). All samples were assayed by RNA-seq, as described by the TCGA Research Network (15). Gene expression values were represented as RNA-Seq by Expectation Maximization (RSEM) data normalized within each sample to the upper quartile of total reads. The total number of somatic mutations was adopted to assess the mutational burden, as it is significantly correlated with the number of non-synonymous mutations. Multiple somatic mutations including non-synonymous mutations, insertion-deletion mutations, and silent mutations were each counted and summated. Germline mutations without somatic mutations were excluded (16).
Clinical and mutational data were also obtained from TCGA. The expression levels of elastase, neutrophil expressed (ELANE), MPO, and FUT4 were measured using mRNA expression (RNA Seq V2 RSEM). We classified TCGA samples of each cancer type into three tumor microenvironment immune types (TMITs) by merging the mRNA expression levels of FUT4 and ELANE, or FUT4 and MPO as follows: TMIT 1, low activation TAN phenotype, high FUT4 expression, and low ELANE/MPO expression; TMIT 2, moderately activated TAN phenotype, low FUT4 expression and low ELANE/MPO expression or high FUT4 expression and high ELANE/MPO expression; TMIT 3, highly activated TAN phenotype, low FUT4 expression and high ELANE/MPO expression. Receiver operating characteristic (ROC) curves were constructed using Statistical Package for the Social Sciences (SPSS) software to determine the optimal cutoff values of high and low expression of FUT4/ELANE/MPO. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analyses of genomic data
We divided the 6,000 samples from the 15 cancer types into three groups. The prognostic significance of the three TMITs was estimated using Kaplan-Meier plots (log-rank test) and Cox proportional hazards regression analysis. Multivariate Cox regression analyses were performed to determine whether there was a significant association between clinicopathological characteristics and the ability to predict specific TMITs. We performed a nonparametric test to confirm whether the number of mutations varied between different immune groups. Statistical significance was set as P<0.05. All statistical analyses and data presentations were performed using SPSS, version 21.0 (IBM, Armonk, NY, USA). The R Project for Statistical Computing was used for TCGA data download.
Results
Distribution of FUT4/ELANE/MPO expression and mutational burden across TCGA cancer types
We analyzed 6,000 tumor samples from 15 cancer types included in the TCGA dataset. Patient characteristics from TCGA are summarized in Table S1. The cutoff values of FUT4, MPO, and ELANE expression for each cancer type were calculated using ROC curves, and were 0.581, 190.416, and 6.5643, respectively. These cutoffs varied according to tumor type (Table 1).
Table 1. The cutoff values of FUT4, ELANE, and MPO for all solid tumors.
| Cancer type | Cutoff value (using ROC curve) | |||||||
|---|---|---|---|---|---|---|---|---|
| FUT4 | MPO | ELANE | ||||||
| Value | Percent of samples (from top), % | Value | Percent of samples (from top), % | Value | Percent of samples (from top), % | |||
| GBM | 136.59 | 52.83 | 0.52 | 76.10 | 1.49 | 56.60 | ||
| ESCA | 2706.58 | 30.81 | 12.02 | 25.95 | 1.75 | 54.05 | ||
| CESC | 864.70 | 16.94 | 3.40 | 54.72 | 2.03 | 27.36 | ||
| BRCA | 412.29 | 41.14 | 10.19 | 44.61 | 5.75 | 29.71 | ||
| LUAD | 535.49 | 37.33 | 15.41 | 49.50 | 8.90 | 34.73 | ||
| HNSC | 133.64 | 60.98 | 1.18 | 78.03 | 0.52 | 33.52 | ||
| LGG | 141.30 | 33.14 | 0.35 | 95.32 | 0.35 | 76.41 | ||
| SKCM | 142.50 | 63.11 | 6.60 | 25.59 | 0.34 | 35.39 | ||
| ACC | 152.96 | 14.13 | 0.45 | 43.48 | 4.14 | 39.13 | ||
| UCEC | 219.01 | 27.27 | 8.88 | 30.11 | 0.70 | 21.59 | ||
| BLCA | 315.76 | 44.90 | 1.98 | 76.21 | 0.13 | 71.36 | ||
| PAAD | 845.99 | 28.65 | 6.10 | 50.81 | 1.94 | 47.57 | ||
| PRAD | 191.23 | 19.80 | 2.12 | 52.60 | 1.16 | 66.80 | ||
| KIRP | 289.23 | 47.42 | 1.11 | 82.13 | 0.33 | 43.64 | ||
| COAD-READ | 2094.32 | 50.89 | 27.08 | 7.48 | 0.95 | 57.24 | ||
FUT4, fucosyltransferase 4; ELANE, elastase, neutrophil expressed; MPO, myeloperoxidase; BLCA, bladder urothelial carcinoma; BRCA, breast cancer; CESC, cervical cancer; GBM, glioblastoma multiforme; KIRP, kidney renal papillary cell carcinoma; LUAD, lung adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LGG, brain lower grade glioma; COADREAD, colorectal and rectal cancer; SKCM: skin cutaneous melanoma; ACC, adrenocortical carcinoma; UCEC, uterine corpus endometrial carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma.
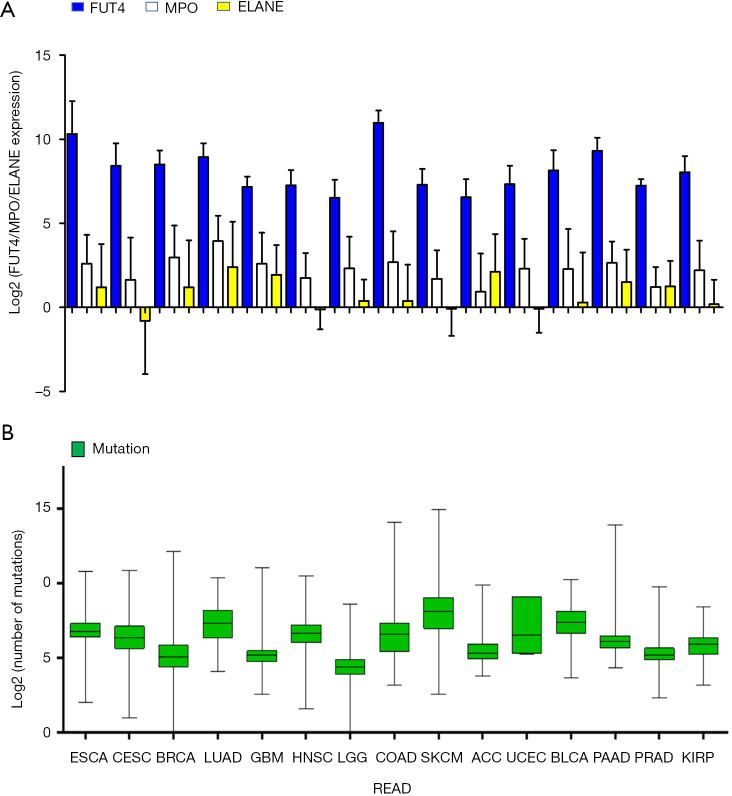
We performed a log2 transformation on the gene expression of FUT4, MPO, and ELANE, and based on this, we analyzed the correlation of the expression levels of the three genes. The expression levels of FUT4 and ELANE (P=0.001, R2 =0.132) or FUT4 and MPO (P<0.001, R2 =0.228) were obviously irrelevant, despite a considerable proportion of tumor samples being classified as TMIT 2. Figure 1A presents FUT4/MPO/ELANE expression of the 15 TCGA cancer types. Figure 1B shows the log2-transformed values of the number of mutations of the cancer types.
Figure 1.
Distribution of FUT4/MPO/ELANE expression and mutation burden and across TCGA cancer types. (A) FUT4/MPO/ELANE expression according to 15 TCGA cancer types; (B) the log2-transformed values of the number of mutations according to cancer type. FUT4, fucosyltransferase 4; MPO, myeloperoxidase; ELANE, elastase, neutrophil expressed; TCGA, The Cancer Genome Atlas.
Distribution and clinical implication of TMITs across different cancer types
We then divided the expression of FUT4/MPO/ELANE into high and low subgroups using the ROC cutoff values for each tumor type. The survival analysis of the three genes and immunophenotyping in all samples according to the different subgroups are shown in Figure S1. We found that both high and low expression of FUT4/MPO/ELANE exhibited different effects on overall survival.
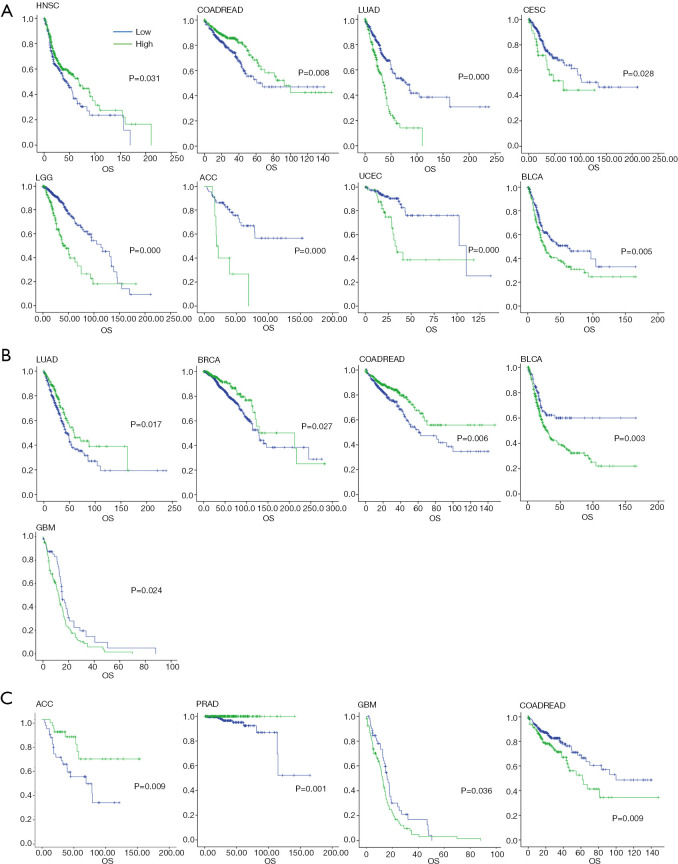
High expression of FUT4 suggested good prognosis in HNSC (HR =0.745, 95% CI: 0.57–0.974, P=0.031) and COADREAD (HR =0.626, 95% CI: 0.441–0.890, P=0.008). But prognosis was bad in LUAD (HR =2.380, 95% CI: 1.768–3.204, P<0.001), CESC (HR =1.808, 95% CI: 1.057–3.093, P=0.028), LGG (HR =2.755, 95% CI: 1.932–3.927, P<0.001), ACC (HR =4.435, 95% CI: 1.934–10.250, P<0.001), BLCA (HR =1.513, 95% CI: 1.128–2.029, P=0.005), and UCEC (HR =3.235, 95% CI: 1.611–6.499, P<0.001) (Figure 2A).
Figure 2.
Overall survival analysis across TCGA cancer types. (A) Kaplan-Meier analysis of FUT4 in HNSC, COADREAD, LUAD, CESC, LGG, ACC, BLCA, and UCEC; (B) Kaplan-Meier analysis of ELANE in LUAD, BRCA, and COADREAD. Low expression of ELANE exhibited a good prognosis in GBM and BLCA; (C) Kaplan-Meier analysis of MPO subgroups. High expression of MPO suggested a good prognosis in ACC and PRAD, while low expression of MPO showed better prognosis in GBM and COADREAD. HNSC, head and neck squamous cell carcinoma; COADREAD, colorectal and rectal cancer; LUAD, lung adenocarcinoma; CESC, cervical cancer; LGG, brain lower grade glioma; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; UCEC, uterine corpus endometrial carcinoma; ELANE, elastase, neutrophil expressed; BRCA, breast cancer; GBM, glioblastoma multiforme; FUT4, fucosyltransferase 4; ELANE, elastase, neutrophil expressed; MPO, myeloperoxidase.
For ELANE subgroups, high expression had a good prognosis in LUAD (HR =0.68, 95% CI: 0.493–0.937, P=0.017), BRCA (HR =0.646, 95% CI: 0.437–0.955, P=0.027), and COADREAD (HR =0.614, 95% CI: 0.433– 0.871, P=0.006). Furthermore, low expression of ELANE also exhibited a good prognosis in GBM (HR =1.540, 95% CI: 1.056–2.248, P=0.024) and BLCA (HR =1.738, 95% CI: 1.201–2.514, P=0.003) (Figure 2B). Moreover, high expression of ELANE also had a good prognosis tendency in CESC, ESCA, HNSC, LGG, ACC, PRAD, PAAD, UCEC, and SKCM (P>0.05; Figure S2).
In MPO subgroups, high expression suggested good prognosis in ACC (HR =1.671, 95% CI: 1.129–2.473, P=0.009) and PRAD (HR =0.014, 95% CI: 0.000–2.387, P=0.001). Also, low expression of MPO exhibited better prognosis in GBM (HR =1.499, 95% CI: 1.023–2.196, P=0.036) and COADREAD (HR =3.235, 95% CI: 1.611– 6.499, P=0.009) (Figure 2C).
Similar to previous classifications, we divided all tumor samples into three groups of TMIT, which represent different activation statuses. TMIT 1 is characterized by the recruitment of a large number of neutrophils, however, there are few neutrophils with high activity in this group. Among all the evaluated samples, in the FUT4/ELANE classification, 1,651 (19.89%) were classified as TMIT 1, which indicated a low proportion of high-activity TANs. The proportions of TMIT 2 were 61.39%, and the proportions of TMIT 3 were 18.71%.
Immunophenotyping of FUT4 combined with MPO was used to classify the TME. In all of the tumor samples, the proportions of TMIT 1/2/3 were 29.02%, 63.06%, and 7.91%, respectively. The distributions of TMIT groups for the 15 cancer types are shown in Figure S3. In most tumor types, TMIT 2 containing two cases had the highest proportion. The TMIT 3 group exhibited a similar proportion to the TMIT 1 in two grouping methods. However, the proportion of TMIT 3 in LGG, BLCA, and PRAD exceeded TMIT 2 in the FUT4/ELANE groups.
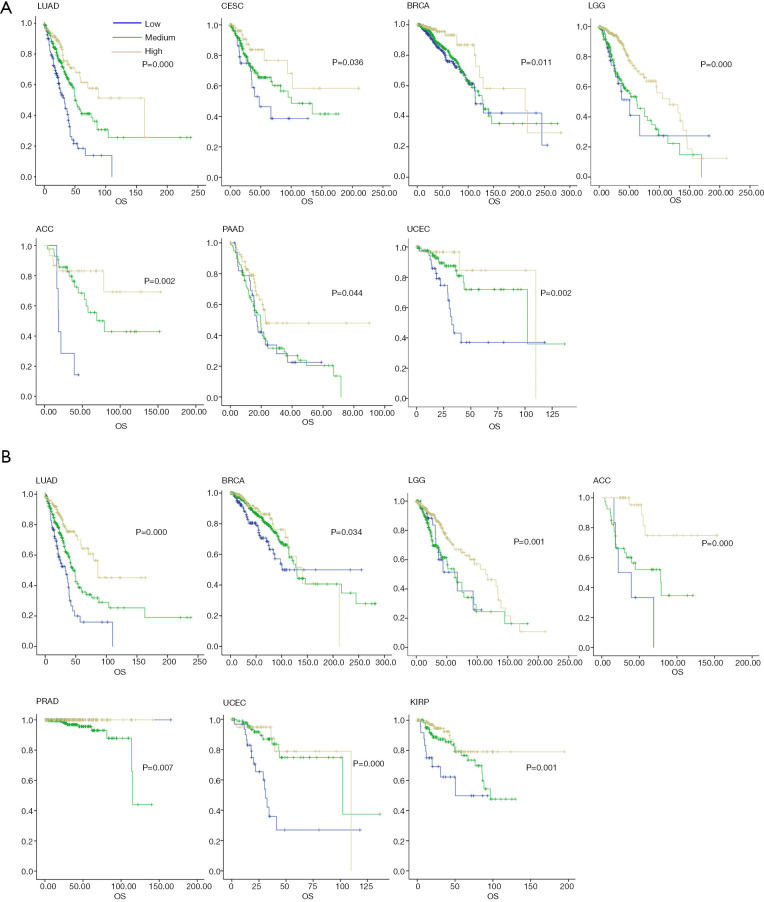
An overall survival analysis across these TCGA cancer types according to the TMITs is shown in Figure 3. The prognostic significance of different TMITs varied in different tumors. TMIT 3 had the most favorable prognostic effect compared with the other groups. TMIT 3 of the FUT4 and ELANE groups had a good prognosis in LUAD (HR =0.292, 95% CI: 0.185–0.459, P<0.001), CESC (HR =0.416, 95% CI: 0.231–0.750, P=0.036), BRCA (HR =0.333, 95% CI: 0.142–0.783, P=0.011), LGG (HR =0.360, 95% CI: 0.205–0.632, P<0.001), ACC (HR =0.133, 95% CI: 0.04–0.423, P=0.002), PAAD (HR =0.511, 95%CI: 0.284–0.92, P=0.044), and UCEC (HR =0.204, 95% CI: 0.059–0.707, P=0.002) (Figure 3A), and had a good prognosis tendency in ESCA, BLCA, PRAD, COADREAD (P>0.05, Figure S4A). TMIT3 of FUT4 and MPO groups had a good prognosis in LUAD, BRCA, LGG, ACC, PRAD, UCEC, and KIRP (P<0.05, Figure 3B), and had a good prognosis tendency in ESCA, BLCA, PAAD, and SKCM (P>0.05; Figure S4B).
Figure 3.
Overall survival analysis according to tumor microenvironment immune types (TMITs) across TCGA cancer types. (A) TMIT 3 of FUT4 and ELANE groups had a good prognosis in LUAD, CESC, BRAD, LGG, ACC, PAAD, and UCEC. TMIT 3 of FUT4 and MPO groups showed a good prognosis in LUAD, BRCA, LGG, ACC, PRAD, UCEC, and KIRP; (B) LUAD, lung adenocarcinoma; CESC, cervical cancer; BRCA, breast cancer; LGG, brain lower grade glioma; ACC, adrenocortical carcinoma; PAAD, pancreatic adenocarcinoma; UCEC, uterine corpus endometrial carcinoma; KIRP, kidney renal papillary cell carcinoma; TCGA, The Cancer Genome Atlas; FUT4, fucosyltransferase 4; ELANE, elastase, neutrophil expressed.
Univariable Cox regression analysis of immunophenotyping across these TCGA cancer types is shown in Table 2. Patients in the TMIT 3 group had a favorable trend compared to those in TMITs 1 and 2. In LGG (FUT4/ELANE: HR =0.360, P<0.001; FUT4/MPO: HR =0.496, P=0.025) and ACC (FUT4/ELANE: HR =0.185, P=0.007; FUT4/MPO: HR =1.000, P=0.032), we achieved good analysis results. Multivariate analysis confirmed that the prognostic significance of different TMITs varied in different tumors. The risk of LGG (HR =0.353, P<0.001) and SKCM (HR =1.073, P=0.775) decreased in the FUT4/ELANE group. There was no significant difference in LUAD and PAAD. By analyzing the number of mutations in the grouping of the two immune microenvironments, we found that among the 15 tumors types, there were statistically significant correlations between the mutation data and the immunophenotyping of five tumor types in each of the two immunological types. The results of the other tumors are counted in Table 3.
Table 2. Univariate and multivariate Cox regression analysis of immunophenotyping in 15 tumors.
| Cancer type | Groups of FUT4/ELANE | Groups of FUT4/MPO | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | ||||||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95%C I) | P | ||||
| HNSC | |||||||||||
| TMIT1 | 1.000 | 0.423 | * | 1.000 | 0.010 | 1.000 | 0.007 | ||||
| TMIT2 | 1.189 (0.895–1.581) | 0.233 | 2.190 (1.115–4.302) | 0.023 | 2.556 (1.124–5.814) | 0.025 | |||||
| TMIT3 | 1.244 (0.789–1.962) | 0.348 | 2.794 (1.394–5.600) | 0.004 | 3.166 (1.368–7.326) | 0.007 | |||||
| COADREAD | |||||||||||
| TMIT1 | 1.000 | 0.604 | * | 1.000 | 0.004 | 1.000 | 0.013 | ||||
| TMIT2 | 0.826 (0.541–1.261) | 0.377 | 3.708 (1.158–11.874) | 0.027 | 3.381 (1.047–10.919) | 0.042 | |||||
| TMIT3 | 0.976 (0.613–1.554) | 0.918 | 5.723 (1.772–18.483) | 0.004 | 4.384 (1.326–14.494) | 0.015 | |||||
| LGG | |||||||||||
| TMIT1 | 1.000 | <0.001 | 1.000 | <0.001 | 1.000 | 0.001 | 1.000 | <0.001 | |||
| TMIT2 | 0.914 (0.534–1.565) | 0.743 | 0.847 (0.494–1.453) | 0.547 | 1.003 (0.549–1.836) | 0.991 | 1.124 (0.612–2.065) | 0.705 | |||
| TMIT3 | 0.360 (0.205–0.632) | <0.001 | 0.353 (0.200–0.622) | <0.001 | 0.500 (0.272–0.922) | 0.026 | 0.496 (0.268–0.918) | 0.025 | |||
| ACC | |||||||||||
| TMIT1 | 1.000 | 0.002 | 1.000 | <0.001 | 1.000 | 0.002 | 1.000 | 0.032 | |||
| TMIT2 | 0.240 (0.090–0.641) | 0.004 | 0.580 (0.190–1.770) | 0.339 | 0.528 (0.195–1.429) | 0.208 | 0.322 (0.105–0.990) | 0.048 | |||
| TMIT3 | 0.130 (0.040–0.423) | 0.001 | 0.071 (0.016–0.325) | 0.001 | 0.100 (0.027–0.376) | 0.001 | 0.185 (0.055–0.630) | 0.007 | |||
| KIRP | |||||||||||
| TMIT1 | 1.000 | 0.907 | * | 1.000 | 0.003 | 1.000 | 0.001 | ||||
| TMIT2 | 1.176 (0.569–2.428) | 0.662 | 0.411 (0.190–0.887) | 0.023 | 0.392 (0.173–0.888) | 0.025 | |||||
| TMIT3 | 1.141 (0.482–2.700) | 0.764 | 0.213 (0.088–0.516) | 0.001 | 0.166 (0.066–0.416) | <0.001 | |||||
| SKCM | |||||||||||
| TMIT1 | 1.000 | 0.066 | 1.000 | 0.023 | 1.000 | 0.408 | * | ||||
| TMIT2 | 0.748 (0.566–0.989) | 0.041 | 0.680 (0.503–0.919) | 0.012 | 0.938 (0.716–1.288) | 0.640 | |||||
| TMIT3 | 1.122 (0.705–1.785) | 0.627 | 1.073 (0.662–1.739) | 0.775 | 0.641 (0.297–1.268) | 0.187 | |||||
| ESCA | |||||||||||
| TMIT1 | 1.000 | 0.379 | * | 1.000 | 0.138 | 1.000 | 0.051 | ||||
| TMIT2 | 0.685 (0.348–1.350) | 0.275 | 0.595 (0.349–1.015) | 0.057 | 0.479 (0.258–0.889) | 0.020 | |||||
| TMIT3 | 0.585 (0.272–1.256) | 0.169 | 0.598 (0.267–1.341) | 0.212 | 0.450 (0.186–1.088) | 0.076 | |||||
| LUAD | |||||||||||
| TMIT1 | 1.000 | <0.001 | 1.000 | <0.001 | 1.000 | <0.001 | 1.000 | <0.001 | |||
| TMIT2 | 0.489 (0.354–0.675) | <0.001 | 0.494 (0.355–0.687) | <0.001 | 0.564 (0.398–0.800) | 0.001 | 0.598 (0.419–0.854) | 0.005 | |||
| TMIT3 | 0.292 (0.185–0.459) | <0.001 | 0.296 (0.187–0.468) | <0.001 | 0.284 (0.180–0.447) | <0.001 | 0.285 (0.179–0.452) | <0.001 | |||
| GBM | |||||||||||
| TMIT1 | 1.000 | 0.473 | * | 1.000 | 0.749 | * | |||||
| TMIT2 | 0.919 (0.576–1.465) | 0.723 | 1.189 (0.734–1.926) | 0.482 | |||||||
| TMIT3 | 1.177 (0.712–1.946) | 0.524 | 1.206 (0.710–2.048) | 0.488 | |||||||
| CESC | |||||||||||
| TMIT1 | 1.000 | 0.042 | 1.000 | 0.034 | 1.000 | 0.176 | 1.000 | 0.034 | |||
| TMIT2 | 0.618 (0.335–1.140) | 0.123 | 0.440 (0.230–0.844) | 0.013 | 0.505 (0.239–1.067) | 0.073 | 0.376 (0.171–0.825) | 0.015 | |||
| TMIT3 | 0.333 (0.142–0.783) | 0.012 | 0.335 (0.139–0.805) | 0.014 | 0.702 (0.349–1.413) | 0.321 | 0.626 (0.302–1.298) | 0.208 | |||
| UCEC | |||||||||||
| TMIT1 | 1.000 | 0.004 | * | 1.000 | <0.001 | * | |||||
| TMIT2 | 0.349 (0.168–0.725) | 0.005 | 0.236 (0.109–0.509) | <0.001 | |||||||
| TMIT3 | 0.204 (0.059–0.707) | 0.012 | 0.206 (0.074–0.568) | 0.002 | |||||||
| BRCA | |||||||||||
| TMIT1 | 1.000 | 0.014 | 1.000 | 0.006 | 1.000 | 0.036 | 1.000 | 0.041 | |||
| TMIT2 | 0.867 (0.614–1.222) | 0.414 | 0.855 (0.605–1.206) | 0.371 | 0.636 (0.429–0.942) | 0.024 | 0.629 (0.424–0.932) | 0.021 | |||
| TMIT3 | 0.416 (0.231–0.750) | 0.004 | 0.418 (0.232–0.753) | 0.004 | 0.538 (0.314–0.920) | 0.024 | 0.533 (0.311–0.912) | 0.022 | |||
| PAAD | |||||||||||
| TMIT1 | 1.000 | 0.048 | 1.000 | 0.020 | 1.000 | 0.155 | * | ||||
| TMIT2 | 0.990 (0.602–1.628) | 0.969 | 0.948 (0.576–1.560) | 0.832 | 0.713 (0.431–1.180) | 0.189 | |||||
| TMIT3 | 0.551 (0.306–0.991) | 0.047 | 0.511 (0.284–0.920) | 0.025 | 0.589 (0.344–1.010) | 0.054 | |||||
| BLCA | |||||||||||
| TMIT1 | 1.000 | 0.894 | * | 1.000 | 0.488 | * | |||||
| TMIT2 | 1.112 (0.656–1.885) | 0.694 | 0.858 (0.461–1.596) | 0.628 | |||||||
| TMIT3 | 1.052 (0.607–1.821) | 0.857 | 0.733 (0.386–1.392) | 0.342 | |||||||
| PRAD | |||||||||||
| TMIT1 | 1.000 | 0.553 | * | 1.000 | 0.275 | 1.000 | 0.273 | ||||
| TMIT2 | 13,085.559 (0.000–2.337) | 0.952 | 64.632 (0.000–29,479,564.780) | 0.531 | 65.241 (0.000–28,718,151.430) | 0.529 | |||||
| TMIT3 | 6,472.443 (0.000–1.157) | 0.956 | 0.991 (0.000–1,350,060.570) | 0.999 | 0.989 (0.000–1,305,099.117) | 0.999 | |||||
*, not in the final step of multivariate analysis. FUT4, fucosyltransferase 4; ELANE, elastase, neutrophil expressed; MPO, myeloperoxidase; BLCA, bladder urothelial carcinoma; BRCA, breast cancer; CESC, cervical cancer; GBM, glioblastoma multiforme; KIRP, kidney renal papillary cell carcinoma; LUAD, lung adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LGG, brain lower grade glioma; COADREAD, colorectal and rectal cancer; SKCM, skin cutaneous melanoma; ACC, adrenocortical carcinoma; UCEC, uterine corpus endometrial carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; CI, confidence interval; HR, hazard ratio; TMIT, tumor microenvironment immune type.
Table 3. Analysis of the relationship between tumor mutation data and immune typing.
| Number of somatic mutations | FUT4/ELANE groups | FUT4/MPO groups | |||
|---|---|---|---|---|---|
| r | P | r | P | ||
| ACC | −0.194 | 0.088 | −0.328 | 0.003 | |
| HNSC | 0.097 | 0.106 | 0.072 | 0.228 | |
| COADREAD | −0.092 | 0.118 | −0.063 | 0.363 | |
| LGG | −0.104 | 0.080 | −0.071 | 0.229 | |
| PRAD | −0.082 | 0.186 | −0.178 | 0.004 | |
| KIRP | 0.089 | 0.251 | 0.090 | 0.245 | |
| SKCM | −0.063 | 0.232 | −0.123 | 0.020 | |
| ESCA | −0.255 | 0.001 | −0.243 | 0.002 | |
| LUAD | −0.051 | 0.455 | 0.104 | 0.122 | |
| GBM | 0.046 | 0.593 | −0.129 | 0.133 | |
| CESC | 0.043 | 0.555 | 0.129 | 0.075 | |
| UCEC | −0.632 | 0.252 | 0.105 | 0.866 | |
| BRCA | −0.186 | <0.001 | −0.095 | 0.003 | |
| PAAD | −0.243 | 0.003 | −0.135 | 0.105 | |
| BLCA | −0.039 | 0.664 | 0.078 | 0.379 | |
FUT4, fucosyltransferase 4; ELANE, elastase, neutrophil expressed; MPO, myeloperoxidase; BLCA, bladder urothelial carcinoma; BRCA, breast cancer; CESC, cervical cancer; GBM, glioblastoma multiforme; KIRP, kidney renal papillary cell carcinoma; LUAD, lung adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LGG, brain lower grade glioma; COADREAD, colorectal and rectal cancer; SKCM, skin cutaneous melanoma; ACC, adrenocortical carcinoma; UCEC, uterine corpus endometrial carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma.
Enrichment analysis
We also performed enrichment analysis to determine the biological correlation of the gene expressions. Metascape was used for enrichment analysis. In tumors with high expression of the ELANE gene predicting good prognosis, these analyses revealed enriched terms that were related to complement activation of the classical pathway, protein digestion and absorption, immunoglobulin production, etc. (Figure S5). Under the same immune microenvironment group, the common elements for highly expressed genes of the five aforementioned types of tumors are shown in Figure S6. We observed 68 common elements in “COADREAD” and “LUAD”, 29 common elements in “COADREAD” and “ESCA”, 31 common elements in “COADREAD” and “BRCA”, and 24 common elements in “LUAD” and “BRCA”.
Discussion
The present study investigated the prognostic role of TAN-related genes in various tumors at the genomic level for the first time. We confirmed that the activation status of neutrophils is related to the functional phenotype. We also compared the prognostic value of different enzyme combinations. By using a large number of solid tumor samples from TCGA, we presented several key aspects for classifying the TME based on FUT4 and ELANE/MPO mRNA expression. We determined the optimal cutoff values for FUT4/ELANE/MPO expression using SPSS ROC curves. Our results suggest that the prognostic value of neutrophils should be comprehensively interpreted along with other TME factors across different cancer types.
TANs play a key role in the TME, and their synthetic secreted protease is closely related to tumor proliferation and metastasis. The number of infiltrating neutrophils in tumor tissues is positively correlated with tumor lymph node metastasis and tumor stage. Shen et al. examined the prognostic value of neutrophils in patients with cancer by meta-analysis, and reported that the increase in the number of infiltrating neutrophils in tumors was significantly associated with poor prognosis in cancer patients. However, TAN enlargement in bowel cancer suggested a good prognosis (4). For lung cancer surgery patients, TANs stimulate T cell proliferation and interferon-γ (IFN-γ) release. The interaction between neutrophils and activated T cells leads to a significant upregulation of costimulatory molecules on the surface of TANs, and this positive feedback loop enhances T cell proliferation, thereby inhibiting the survival of tumor cells (17). Using the Lox-Stop-Lox-Kirsten rat sarcoma viral oncogene homolog (LSL-Kras) model of lung adenocarcinoma, Alyssa D. Gregory has recently shown that NE promotes tumor growth. However, the result of in vitro studies revealed that NE exerts differential effects on tumor cells in a concentration-dependent fashion. These studies reported that treatment with modest levels of NE led to proliferation of A549 lung epithelial tumor cells, while higher doses led to cell death. MPO protein levels are also positively associated with canine mammary tumor (CMT) development (18). Droeser suggested that the infiltration MPO-positive neutrophils was specifically associated with favorable prognosis in colorectal cancer patients (19). In human lung cancer specifically, research found a striking heterogeneity in the presence of MPO-positive TAN. Most previous studies have suggested that TAN infiltration into tumor tissues often indicates a poor prognosis.
Our study found that the prognosis of FUT4, which predicts neutrophil infiltration, varies by human tumor type. We also observed that high expression of ELANE was indicative of a good prognosis in 12 types of solid tumor (from TCGA dataset). Only GBM and BLCA had low expression and a good prognosis (P<0.05). Furthermore, high or low expression of MPO in these 15 tumor types exhibited the same prognostic value. We observed that ELANE and MPO gene levels differ in the prognosis of human tumors, which is consistent with previous animal experiments at the protein level.
Our results are generally consistent with those observed in previous studies evaluating the dual role of neutrophils. Although our genetic analysis suggested that the prognostic value of each gene varied greatly, we used immunophenotyping to show consistent prognostic value in multiple tumors It has been demonstrated in a variety of tumors that highly-activated TANs exhibit an anti-tumor phenotype, which is inconsistent with the current immunohistochemical TAN-promoting tumor phenotype. From a genomics perspective, the highly activated TMIT 3 is a small proportion in the TME; most TANs are partially or lowly activated. Since the existing research does not further classify TANs according to the activation state at both the protein and gene levels, most studies report that TANs are predominantly tumor-promoting, which does not conflict with our results, and also reinforces the significance of our research.
Although we have discovered the anti-tumor properties of neutrophils, we are still unclear about the mechanism of action of N1 TANs. Sun et al. found that the cytotoxicity of neutrophils and the ability to activate natural killer (NK) cells were significantly enhanced after TNF-α and IFN-γ treatment, thereby inhibiting tumor growth. Shrestha et al. recently reported that angiotensin-converting enzyme inhibitors and angiotensin II receptor antagonists can induce neutrophils to exhibit anti-tumor phenotypes and functions, thereby inhibiting tumor growth (20). In addition, lipid A can induce neutrophils to exert anti-tumor effects, and can produce better anti-tumor effects when combined with the chemotherapy drug, oxaliplatin (21). Using a mouse model of lung cancer to block transforming growth factor beta (TGF-β) signaling with TGF-β receptor inhibitors, Fridlender et al. found that the number of infiltrating TANs in mouse tumors increased significantly, tumor growth slowed, and anti-tumor effects of CD8+ T cells increased compared with mice that did not use TGF-β receptor inhibitors (3). TANs acquire an activated phenotype with high phagocytic activity, these were shown to secrete a large panel of cytokines and chemokines, with a preponderance of proinflammatory cytokines, whereas the expression of anti-inflammatory cytokines was found low. However, in contact with activated T cells, TANs isolated from lung cancer upregulated many costimulatory molecules. These could explain the ability of TANs to stimulate both CD4+ and CD8+ T-cell proliferation (22). In our study, the RNA-seq data was obtained from the various cancer cells and surrounding tissues, which was advantageous in assessing the infiltration of neutrophils. The above research is consistent with our assumptions suggesting that TMIT may be useful in predicting response in different cancer types.
The accumulating data showing in animal models that TANs can be manipulated to acquire more cytotoxic phenotypes has raised the questions whether TANs in cancer patients may be a new possible target for cancer immunotherapy. We speculate that TMIT 3 tumor immune microenvironments are most likely to benefit from attracting T cell infiltration into the TME. TMIT 1 and 2 lack high-activity neutrophils or only exhibit a low percentage of highly-active neutrophils, and thus, it is unlikely that stimulating T cell proliferation will lead to a T cell-mediated cancer response. Tumor and microenvironment-derived actors can induce local aggregation of neutrophils to tumors and promote the malignant progression of tumors, and specific antibodies can antagonize neutrophil chemotaxis and exert an anti-tumor effect. The important role of neutrophils in tumors suggests that targeted neutrophils may potentially offer anti-tumor value.
We propose that classification of the TME into three TMITs may be a novel and useful model for better efficacy. Although genetic profiling provides predictive information through changes in mRNA levels, there is a need for a quantitative assessment in biopsies to analyze the changes of corresponding functional enzymes during neoplastic development. By analyzing the changes in enzyme expression and the mechanisms of action, we can further analyze the value of its markers for cancer detection and monitoring.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This project was funded by the National Natural Science Foundation of China (grant no. 81672974, and no. 81602719) and the Science and Technology Development Plan of Shandong Province (No. 2017GSF18111).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6629
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6629). ZJ serves as an unpaid editorial board member (or Section Editor) of Annals of Translational Medicine from September/2020 to August/2021. The authors have no other conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Cortez-Retamozo V, Etzrodt M, Newton A, et al. Origins of tumor-associated macrophages and neutrophils. Proc Natl Acad Sci U S A 2012;109:2491-6. 10.1073/pnas.1113744109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandau S. The dichotomy of neutrophil granulocytes in cancer. Semin Cancer Biol 2013;23:139-40. 10.1016/j.semcancer.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 3.Fridlender ZG, Sun J, Kim S, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell 2009;16:183-94. 10.1016/j.ccr.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen M, Hu P, Donskov F, et al. Tumor-associated neutrophils as a new prognostic factor in cancer: a systematic review and meta-analysis. PLoS One 2014;9:e98259. 10.1371/journal.pone.0098259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tazzyman S, Barry ST, Ashton S, et al. Inhibition of neutrophil infiltration into A549 lung tumors in vitro and in vivo using a CXCR2-specific antagonist is associated with reduced tumor growth. Int J Cancer 2011;129:847-58. 10.1002/ijc.25987 [DOI] [PubMed] [Google Scholar]
- 6.Pekarek LA, Starr BA, Toledano AY, et al. Inhibition of tumor growth by elimination of granulocytes. J Exp Med 1995;181:435-40. 10.1084/jem.181.1.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Carlo E, Forni G, Lollini P, et al. The intriguing role of polymorphonuclear neutrophils in antitumor reactions. Blood 2001;97:339-45. 10.1182/blood.V97.2.339 [DOI] [PubMed] [Google Scholar]
- 8.Hiramatsu S, Tanaka H, Nishimura J, et al. Neutrophils in primary gastric tumors are correlated with neutrophil infiltration in tumor-draining lymph nodes and the systemic inflammatory response. BMC Immunol 2018;19:13. 10.1186/s12865-018-0251-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escrevente C, Machado E, Brito C, et al. Different expression levels of alpha3/4 fucosyltransferases and Lewis determinants in ovarian carcinoma tissues and cell lines. Int J Oncol 2006; 29:557-66. [PubMed] [Google Scholar]
- 10.Aziz F, Gao W, Yan Q, et al. Fucosyltransferase-4 and Oligosaccharide Lewis Y Antigen as potentially Correlative Biomarkers of Helicobacter pylori CagA Associated Gastric Cancer. Pathol Oncol Res 2017; 23:173-9. 10.1007/s12253-016-0122-1 [DOI] [PubMed] [Google Scholar]
- 11.Jensen HK, Donskov F, Marcussen N, et al. Presence of intratumoral neutrophils is an independent prognostic factor in localized renal cell carcinoma. J Clin Oncol 2009;27:4709-17. 10.1200/JCO.2008.18.9498 [DOI] [PubMed] [Google Scholar]
- 12.Sinha S, Watorek W, Karr S, et al. Primary structure of human neutrophil elastase. Proc Natl Acad Sci U S A 1987;84:2228-32. 10.1073/pnas.84.8.2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer C, Diehn M, Alizadeh AA, et al. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics 2006;7:115. 10.1186/1471-2164-7-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ock CY, Keam B, Kim S, et al. Pan-Cancer Immunogenomic Perspective on the Tumor Microenvironment Based on PD-L1 and CD8 T-Cell Infiltration. Clin Cancer Res 2016;22:2261-70. 10.1158/1078-0432.CCR-15-2834 [DOI] [PubMed] [Google Scholar]
- 17.Governa V, Trella E, Mele V, et al. The Interplay Between Neutrophils and CD8(+) T Cells Improves Survival in Human Colorectal Cancer. Clin Cancer Res 2017;23:3847-58. 10.1158/1078-0432.CCR-16-2047 [DOI] [PubMed] [Google Scholar]
- 18.Wu CC, Chang SC, Zeng GY, et al. Proteome Analyses Reveal Positive Association of COL2A1, MPO, TYMS, and IGFBP5 with Canine Mammary Gland Malignancy. Proteomics Clin Appl 2019;13:e1800151. 10.1002/prca.201800151 [DOI] [PubMed] [Google Scholar]
- 19.Droeser RA, Hirt C, Eppenberger-Castori S, et al. High myeloperoxidase positive cell infiltration in colorectal cancer is an independent favorable prognostic factor. PLoS One 2013;8:e64814. 10.1371/journal.pone.0064814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrestha S, Noh JM, Kim SY, et al. Angiotensin converting enzyme inhibitors and angiotensin II receptor antagonist attenuate tumor growth via polarization of neutrophils toward an antitumor phenotype. Oncoimmunology 2016;5:e1067744. 10.1080/2162402X.2015.1067744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seignez C, Martin A, Rollet CE, et al. Senescence of tumor cells induced by oxaliplatin increases the efficiency of a lipid A immunotherapy via the recruitment of neutrophils. Oncotarget 2014;5:11442-51. 10.18632/oncotarget.2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J Clin Invest 2014;124:5466-80. 10.1172/JCI77053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as