Abstract
Background
Hepatocellular carcinoma (HCC) is understood to be an immunogenic tumor caused by chronic liver disease. Emerging research has indicated close interaction between various immune cells and tumor cells. Immunophenotyping, which has shown potential predictive value for the prognosis of various human malignancies, might allow responsive and non-responsive patients to be identified based on the extent and distribution of immune cell infiltration. Several novel immunotherapeutic approaches have been trialed and have shown promising efficacy. However, the efficacy of immunotherapies in HCC is limited by several factors. This study aimed to investigate tumor-infiltrating immune cells in HCC.
Methods
Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) allows immune cell profiling analysis by deconvolution of gene expression microarray data. In this study, we analyzed the proportions of immune cells in 14 paired samples of HCC tissues obtained from GSE84402 in Gene Expression Omnibus (GEO) database.
Results
In the 14 paired samples, HCC tissues showed significant infiltration by regulatory T cells (Tregs), activated natural killer (NK) cells, and M0 macrophages (P<0.001, P=0.007 and P=0.001, respectively), which were validated in CIBERSORT with the P value set at ≤0.05. In four paired samples identified from those selected by CIBERSORT, HCC tissues were found to have significant Treg and activated NK cell infiltration compared to non-tumor tissues (P=0.007 and P=0.015, respectively). Additionally, Pearson correlation analysis revealed Tregs to be positively correlated with activated NK cells (Correlation coefficient =0.41).
Conclusions
HCC tumor tissues were markedly infiltrated by Tregs and activated NK cells, which should be considered as candidate therapeutic targets in HCC multidisciplinary treatments.
Keywords: Hepatocellular carcinoma (HCC), regulatory T cells (Treg), natural killer cells (NK cells), tumor-infiltrating immune cells
Introduction
Hepatocellular carcinoma (HCC) is understood to be an immunogenic tumor caused by chronic liver disease. HCC develops when the liver becomes chronically inflamed and the intra-hepatic immunosuppressive microenvironment occurs (1-3). Emerging research has indicated close interaction between various immune cells and tumor cells. Many molecular mechanisms involved in the biological properties of tumor cells during hepatocarcinogenesis also have considerable implications on the immune system (4).
Although a variety of tumor-associated antigens are expressed in HCC, no immune therapies have proved beneficial in the treatment of HCC (5). Several novel immunotherapeutic approaches have been trialed and have shown promising efficacy (6,7). However, the efficacy of immunotherapies in HCC is limited by several factors. More focused studies into how tumor response can be accurately assessed and predicted, as well as how the immunosuppressive effects of the tumor microenvironment can be overcome, are urgently needed (8,9). Immunophenotyping, which has shown potential predictive value for the prognosis of various human malignancies (10,11), might allow responsive and non-responsive patients to be identified based on the extent and distribution of immune cell infiltration (10,12,13). Bioinformatic analysis of tumor-infiltrating immune cells in HCC tissues have been addressed in previous studies (14,15). In a study reported by Rohr-Udilova et al., total B cells, memory B cells, T follicular helper cells and M1 macrophages were strongly infiltrated into HCC (14). Since the efficacy of immunotherapy for HCC is not satisfactory (16), the supplementary assessment of tumor-infiltrating immune cells should be addressed further.
Recent reports have demonstrated that deconvolution of gene expression data by Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) can provide crucial information regarding immune cell composition in HCC (14,17). In this analysis, we selected microarray dataset GSE84402 with 14 paired tumor and the corresponding non-cancerous tissues and aimed to investigate the relative proportions and distribution of tumor-infiltrating immune cells in HCC patients using CIBERSORT. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5830).
Methods
Data source
The GSE84402 series comprising 14 pairs of HCC tissues and the corresponding non-cancerous tissues was obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Double-stranded complementary DNA (cDNA) was synthesized using 5 µg of total RNA from each sample, before amplification, labeling with biotin, and hybridization to GeneChip Human Genome U133 Plus 2.0 Array (18). As declared by Wang et al., ethical approval was obtained from the Zhongshan Hospital Research Ethics Committee, and written informed consent was obtained from each patient (18).
The GSE84402 series matrix file and platform documents (GPL570) were downloaded. The Perl programming language was used to translate gene probe IDs to gene symbol ID. The limma package (19,20) in R program was used to normalized the gene expression levels of the tumor and non-tumor tissue samples from HCC patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
CIBERSORT analysis
Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT), created by Newman et al., is an analytical tool that allows the abundance of member cell types in a mixed cell population to be estimated using gene expression data (17). CIBERSORT gene signature matrix, termed LM22, contains 547 genes and distinguishes 22 human hematopoietic cell phenotypes, including 7 T cell types, naïve and memory B cells, plasma cells, NK cells, and myeloid subsets. CIBERSORT was used to obtain the immune cell profile of each sample from GSE84402, with the number of permutations set at 100 (17). Table 1 shows the complete samples from GSE84402. For each sample, 22 types of immune cell, along with CIBERSORT metrics including the Pearson correlation coefficient, CIBERSORT P value, and root mean squared error (RMSE), were quantified. The CIBERSORT P value represents the statistical significance of the deconvolution results across all cell subsets and can be used to filter out deconvolution with less significant fitting accuracy. From the samples analyzed, 8 non-tumor samples and 7 tumor samples were identified when a CIBERSORT P value ≤0.05 was required. Table 2 lists the samples selected. SPSS 23.0 software (SPSS Inc., Chicago, USA) was employed to compare CIBERSORT immune cell fractions between tumor and non-tumor tissues from HCC patients.
Table 1. Comparison of CIBERSORT immune cell fractions between tumor and non-tumor tissues from HCC patients.
| Immune cell type | CIBERSORT fraction in % of all infiltrating immune cells (mean ± SD) | ||
|---|---|---|---|
| Tumors (n=14) | Non-tumors (n=14) | P value | |
| T cells CD8 | 0.140±0.071 | 0.192±0.073 | 0.075 |
| T cells CD4 naive | 0.0±0.0 | 0.0±0.0 | 1.0 |
| T cells CD4 memory resting | 0.071±0.082 | 0.088±0.066 | 0.553 |
| T cells CD4 memory activated | 0.002±0.008 | 0.0±0.001 | 0.408 |
| T cells follicular helper | 0.074±0.030 | 0.066±0.030 | 0.545 |
| T cells regulatory (Tregs) | 0.066±0.040 | 0.011±0.017 | <0.001 |
| T cells gamma delta | 0.004±0.009 | 0.012±0.017 | 0.109 |
| B cells naive | 0.013±0.017 | 0.020±0.027 | 0.454 |
| B cells memory | 0.022±0.026 | 0.008±0.016 | 0.133 |
| Plasma cells | 0.065±0.025 | 0.061±0.022 | 0.658 |
| NK cells resting | 0.008±0.016 | 0.006±0.009 | 0.709 |
| NK cells activated | 0.040±0.021 | 0.017±0.019 | 0.007 |
| Macrophages M0 | 0.043±0.041 | 0.0±0.003 | 0.001 |
| Macrophages M1 | 0.090±0.030 | 0.090±0.033 | 0.965 |
| Macrophages M2 | 0.143±0.081 | 0.169±0.054 | 0.351 |
| Monocytes | 0.033±0.024 | 0.056±0.034 | 0.056 |
| Dendritic cells resting | 0.074±0.059 | 0.043±0.024 | 0.085 |
| Dendritic cells activated | 0.003±0.007 | 0.0±0.0 | 0.160 |
| Mast cells resting | 0.003±0.011 | 0.001±0.003 | 0.505 |
| Mast cells activated | 0.081±0.055 | 0.125±0.144 | 0.318 |
| Eosinophils | 0.0±0.0 | 0.0±0.0 | 1.0 |
| Neutrophils | 0.026±0.020 | 0.033±0.015 | 0.313 |
Italic P values indicate significance between individual groups. HCC, hepatocellular carcinoma.
Table 2. Comparison of CIBERSORT-selected immune cell fractions between tumor and non-tumor tissues from HCC patients.
| Immune cell type | CIBERSORT fraction in % of all infiltrating immune cells (mean ± SD) | ||
|---|---|---|---|
| Tumors (n=8) | Non-tumors (n=7) | P value | |
| T cells CD8 | 0.164±0.066 | 0.198±0.080 | 0.422 |
| T cells CD4 naive | 0.0±0.0 | 0.0±0.0 | 1.0 |
| T cells CD4 memory resting | 0.043±0.065 | 0.058±0.047 | 0.64 |
| T cells CD4 memory activated | 0.005±0.011 | 0.001±0.002 | 0.372 |
| T cells follicular helper | 0.069±0.033 | 0.077±0.031 | 0.675 |
| T cells regulatory (Tregs) | 0.052±0.035 | 0.008±0.009 | 0.007 |
| T cells gamma delta | 0.007±0.011 | 0.020±0.019 | 0.152 |
| B cells naive | 0.018±0.018 | 0.023±0.030 | 0.479 |
| B cells memory | 0.017±0.028 | 0.0±0.001 | 0.132 |
| Plasma cells | 0.069±0.031 | 0.064±0.028 | 0.759 |
| NK cells resting | 0.005±0.012 | 0.002±0.004 | 0.578 |
| NK cells activated | 0.042±0.027 | 0.015±0.019 | 0.05 |
| Macrophages M0 | 0.047±0.049 | 0.0±0.0 | 0.025 |
| Macrophages M1 | 0.104±0.029 | 0.094±0.039 | 0.605 |
| Macrophages M2 | 0.126±0.053 | 0.155±0.050 | 0.326 |
| Monocytes | 0.031±0.028 | 0.055±0.033 | 0.184 |
| Dendritic cells resting | 0.082±0.065 | 0.035±0.019 | 0.09 |
| Dendritic cells activated | 0.003±0.006 | 0.0±0.001 | 0.31 |
| Mast cells resting | 0.0±0.0 | 0.002±0.004 | 0.369 |
| Mast cells activated | 0.093±0.059 | 0.154±0.183 | 0.444 |
| Eosinophils | 0.0±0.0 | 0.0±0.0 | 1.0 |
| Neutrophils | 0.021±0.010 | 0.032±0.012 | 0.097 |
Italic P values indicate significance between individual groups. HCC, hepatocellular carcinoma.
The pheatmap package in R program, (https://cran.r-project.org/web/packages/pheatmap/) was used to perform heatmap analysis of immune cells in tumor and non-tumor tissues. Correlation analysis of immune cells was conducted using the corrplot package (https://cran.r-project.org/web/packages/corrplot/index.html). Four paired samples from the samples selected by CIBERSORT were used to calculate the difference between candidate infiltrating immune cells.
Results
Comparison of CIBERSORT immune cell fractions
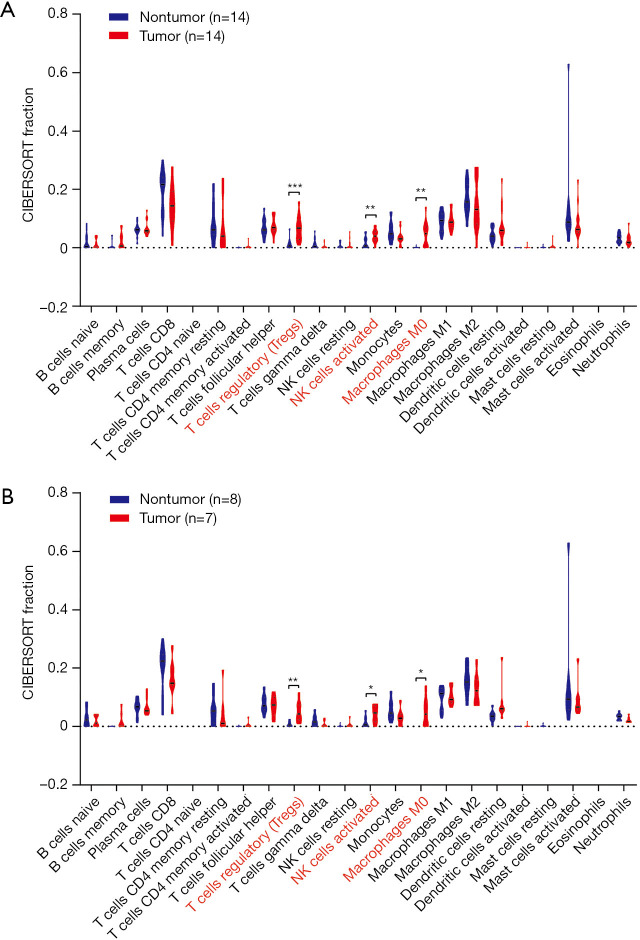
The CIBERSORT immune cell fractions of the tumor and non-tumor tissues from HCC patients from GSE84402 were compared (Table 1). The tumor tissues showed significant infiltration by regulatory T cells (Tregs), activated natural killer (NK) cells, and M0 macrophages compared to non-tumor tissues (P<0.001, P=0.007, and P=0.001, respectively, Table 1 and Figure 1A).
Figure 1.
CIBERSORT immune cell fractions of tumor and non-tumor tissues from all samples (A) and selected samples (B) in hepatocellular carcinoma patients in the GSE84402 series. *, P<0.05; **, P<0.01; ***, P<0.001.
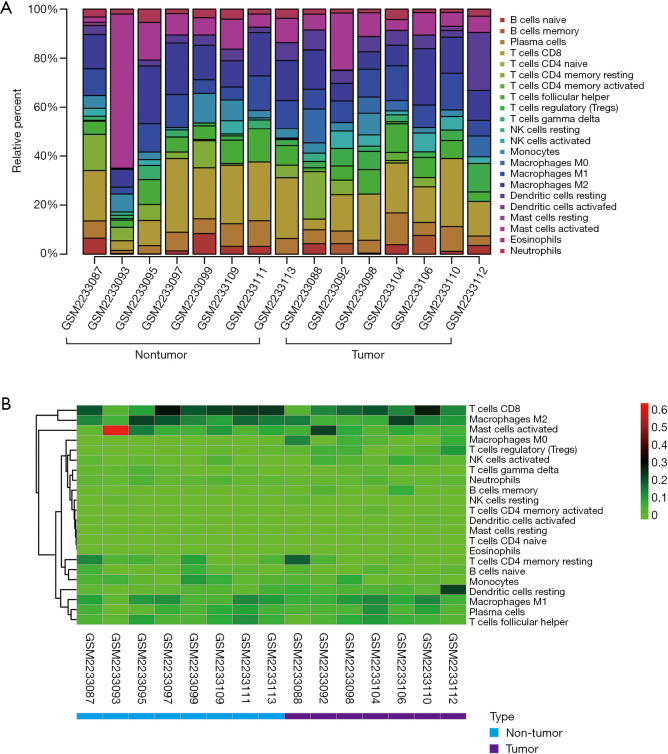
From the samples analyzed, 8 non-tumor and 7 tumor samples were selected when the CIBERSORT P value was set at ≤0.05. Table 2 compares the CIBERSORT immune cell fractions of the tumor and non-tumor tissues from HCC patients. Consistent with above, there was significant infiltration by regulatory T cells, activated NK cells, and M0 macrophages in HCC tumor tissues (P=0.007, P=0.05, and P=0.025, respectively; Table 2 and Figure 1B). The landscape and heatmap of immune cells in the tumor and non-tumor tissue samples are presented in Figure 2.
Figure 2.
Landscape (A) and heatmap (B) of immune cells between tumor and non-tumor samples in samples selected by CIBERSORT.
Comparison of CIBERSORT immune cell fractions in paired samples
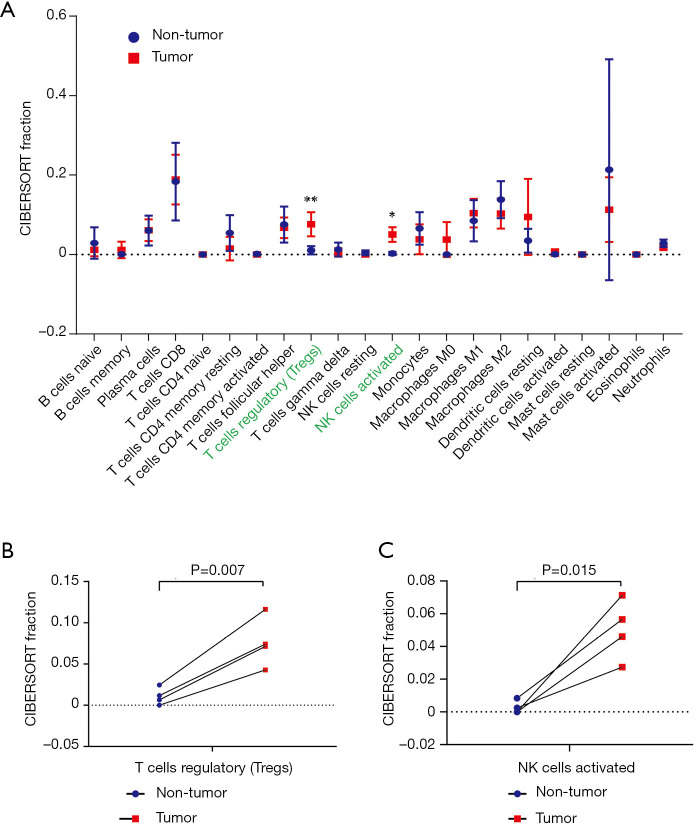
Four paired tumor and non-tumor samples from HCC patients (GSM2233092/GSM2233093, GSM2233098/GSM2233099, GSM2233110/GSM2233111, and GSM2233112/GSM2233113) were identified from the samples selected by CIBERSORT. As shown in Figure 3, infiltration by regulatory T cells and activated NK cells was more significant in tumor tissues than in non-tumor tissues (P<0.01 and P<0.05, respectively; Figure 3A). Moreover, the levels of regulatory T cells and activated NK cells were significantly increased in each tumor tissue sample compared with those in the corresponding non-tumor tissue sample (P=0.007 and P=0.015, respectively; Figure 3B,C).
Figure 3.
Validation for CIBERSORT immune cell fractions in four pairs of tumor and non-tumor tissues. *, P<0.05; **, P<0.01.
Correlation of immune cells
Figure 4 shows a correlation map displaying the Pearson correlation values for each comparison between the immune cells in the CIBERSORT-selected samples. As shown in Figure 4, regulatory T cells had a positive Pearson correlation value of 0.77 with resting dendritic cells and 0.46 with macrophages M0 cells. Furthermore, activated NK cells had positive Pearson correlation values of 0.67 and 0.41 with memory B cells and regulatory T cells, respectively.
Figure 4.
Correlation of immune cells in the samples selected by CIBERSORT.
Discussion
Several computational tools, including microarray microdissection with analysis of differences (MMAD) (1,21), linear least-square regression (LLSR) (22) and digital sorting algorithm (DSA) (23), have been used to deconvolute complex gene expression profiles mixtures to infer cellular composition. However, their sensitivities to experimental noise and cell types, and high unknown mixture content limited the utility for tumor infiltrating immune cells assessment. CIBERSORT is a widely useful approach for high throughput characterization of tumor infiltrating immune cells assessment from complex tissues (24).
Tumor-infiltrating immune cells have been associated with the aggressiveness of many cancers (25,26), and their prognostic correlation with HCC has been extensively investigated (14,27,28). In HCC patients, abundant T lymphocytes (29,30), B cells (31), NK cells (32), and dendritic cells (33,34) have been found to contribute to favorable prognosis. In contrast, high levels of regulatory T cells (35), neutrophils (36,37), monocytes (38), and CXCR3+ subtype B cells (39) have been associated with poor outcomes in HCC patients. Moreover, a link has been reported between the proportions of different types of macrophages and the prognosis of HCC (40,41). Therefore, uncovering the diverse composition of tumor-infiltrating immune cells in tumor immunology might be helpful in overcoming the limitations and barriers faced by immunotherapies in HCC (42).
In this analysis, we found significant infiltration by Tregs and activated NK cells in HCC tumor tissues. In the GSE84402 series, 13 out of 14 cases were HBsAg positive. Consistent with our findings, a previous study revealed that Tregs in tumor tissues displayed higher frequencies and more suppressive phenotypic functions than those in peritumoral and peripheral tissues (43). The higher Treg levels in patients with chronic hepatitis B (CHB) exerted a suppressive effect on the specific immune responses induced by HBV antigens, as well as by HCC tumor antigens. Furthermore, Tregs could inhibit tumor immuno-surveillance against HCC, which potentially plays a role in the immunopathogenesis from CHB to HCC (44). Tregs have crucial involvement in forming and maintaining the hepatic inhibitory microenvironment, as well as in the development of cirrhosis, the transformation of cirrhosis to HCC, and HCC progression and metastasis (45). Moreover, various studies have demonstrated that high quantities of tumor-infiltrating Tregs are considered to be an unfavorable prognostic indicator for advanced tumor biological behaviors and poor survival in HCC patients (46-49).
NK cell regulation of antigen-specific T lymphocytes occurs during viral infection. When NK cells become depleted, the number of antigen-specific T lymphocytes increases; therefore, these NK cells may reduce T cell levels, leading to a weaker antitumor immune response (50). NK cell activator has been shown to induce liver inflammation by significantly increasing the levels of infiltrating lymphocytes, along with concurrently increasing apoptosis and proliferation of hepatocytes; consequently, epithelial-to-mesenchymal transition of hepatocytes is accelerated (51). Decreased NK cell counts have also been reported in HCC patients (52). NK cell deficiency is believed to be a key mechanism underlying tumor cell evasion of the host’s immune system. In HCC, NK cells appear to have reduced function. A growing bank of evidence suggests that impaired NK cell function leads to the body’s failure to eliminate tumor cells, which indicates that tumor cells could be killed more effectively through enhancing the activity of dysfunctional NK cells (32,53). Our results also showed that Tregs were positively correlated with activated NK cells. Current evidence showed that Tregs are closely linked to the homeostasis of NK cells and attenuate the sensitivity of target cells through minimizing interleukin (IL)-2 to NK cells (50). In contrast, in chronic myeloid leukemia, the degree of NK cell differentiation was closely and inversely correlated with the proportion of Treg cells (54). Considered these conflicting results regarding the correlation of tumor-infiltrating NK cells with Tregs, more mechanism investigations on tumor-infiltrating NK cells in HCC development should be carried out in future.
Our study has some limitations. The primary is that this study is based on relatively small samples, the external validation of our results should be considered. Secondly, the associations between Tregs and NK cells and prognosis in HCC patients were not addressed in this study. Thirdly, this study was a preliminary bioinformatic analysis, no experimental data was available. Even though, the CIBERSORT approach to analyzing HCC samples from the GSE84402 series revealed that tumor tissues were markedly infiltrated by Tregs and activated NK cells. Therefore, these cells should be considered as candidate therapeutic targets in multidisciplinary treatment for HCC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (81803901).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5830
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5830). The authors have no conflicts of interest to declare.
(English Language Editor: J. Reynolds)
References
- 1.Mahipal A, Tella SH, Kommalapati A, et al. Immunotherapy in hepatocellular carcinoma: is there a light at the end of the tunnel? Cancers (Basel) 2019;11:1078. 10.3390/cancers11081078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buonaguro L, Mauriello A, Cavalluzzo B, et al. Immunotherapy in hepatocellular carcinoma. Ann Hepatol 2019;18:291-7. 10.1016/j.aohep.2019.04.003 [DOI] [PubMed] [Google Scholar]
- 3.Tan KW, Chacko AM, Chew V. PD-1 expression and its significance in tumour microenvironment of hepatocellular carcinoma. Transl Gastroenterol Hepatol 2019;4:51. 10.21037/tgh.2019.06.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang X, Shu Z, Zhang W, et al. Clinical significance of the immune cell landscape in hepatocellular carcinoma patients with different degrees of fibrosis. Ann Transl Med 2019;7:528. 10.21037/atm.2019.09.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prieto J, Melero I, Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2015;12:681-700. 10.1038/nrgastro.2015.173 [DOI] [PubMed] [Google Scholar]
- 6.Okusaka T, Ikeda M. Immunotherapy for hepatocellular carcinoma: current status and future perspectives. ESMO Open 2018;3:e000455. 10.1136/esmoopen-2018-000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siu EH, Chan AW, Chong CC, et al. Treatment of advanced hepatocellular carcinoma: immunotherapy from checkpoint blockade to potential of cellular treatment. Transl Gastroenterol Hepatol 2018;3:89. 10.21037/tgh.2018.10.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu W, Liu K, Chen M, et al. Immunotherapy for hepatocellular carcinoma: recent advances and future perspectives. Ther Adv Med Oncol 2019;11:1758835919862692. 10.1177/1758835919862692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian M, Shi Y, Liu W, et al. Immunotherapy of hepatocellular carcinoma: strategies for combinatorial intervention. Sci China Life Sci 2019;62:1138-43. 10.1007/s11427-018-9446-2 [DOI] [PubMed] [Google Scholar]
- 10.Gnjatic S, Bronte V, Brunet LR, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer 2017;5:44. 10.1186/s40425-017-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mlecnik B, Bindea G, Angell HK, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016;44:698-711. 10.1016/j.immuni.2016.02.025 [DOI] [PubMed] [Google Scholar]
- 12.Vilain RE, Menzies AM, Wilmott JS, et al. Dynamic Changes in PD-L1 Expression and Immune Infiltrates Early During Treatment Predict Response to PD-1 Blockade in Melanoma. Clin Cancer Res 2017;23:5024-33. 10.1158/1078-0432.CCR-16-0698 [DOI] [PubMed] [Google Scholar]
- 13.Ribas A, Shin DS, Zaretsky J, et al. PD-1 Blockade Expands Intratumoral Memory T Cells. Cancer Immunol Res 2016;4:194-203. 10.1158/2326-6066.CIR-15-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohr-Udilova N, Klinglmuller F, Schulte-Hermann R, et al. Deviations of the immune cell landscape between healthy liver and hepatocellular carcinoma. Sci Rep 2018;8:6220. 10.1038/s41598-018-24437-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X, Shu Z, Zhang W, et al. Clinical significance of the immune cell landscape in hepatocellular carcinoma patients with different degrees of fibrosis. Ann Transl Med 2019;7:528. 10.21037/atm.2019.09.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizukoshi E, Kaneko S. Immune cell therapy for hepatocellular carcinoma. J Hematol Oncol 2019;12:52. 10.1186/s13045-019-0742-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer 2017;16:136. 10.1186/s12943-017-0680-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diboun I, Wernisch L, Orengo CA, et al. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 2006;7:252. 10.1186/1471-2164-7-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liebner DA, Huang K, Parvin JD. MMAD: microarray microdissection with analysis of differences is a computational tool for deconvoluting cell type-specific contributions from tissue samples. Bioinformatics 2014;30:682-9. 10.1093/bioinformatics/btt566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbas AR, Wolslegel K, Seshasayee D, et al. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One 2009;4:e6098. 10.1371/journal.pone.0006098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong Y, Wan YW, Pang K, et al. Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinformatics 2013;14:89. 10.1186/1471-2105-14-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. 10.1007/978-1-4939-7493-1_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960-4. 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 26.Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 2015;26:259-71. 10.1093/annonc/mdu450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foerster F, Hess M, Gerhold-Ay A, et al. The immune contexture of hepatocellular carcinoma predicts clinical outcome. Sci Rep 2018;8:5351. 10.1038/s41598-018-21937-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiao YW, Chiu LT, Chen CH, et al. Tumor-Infiltrating Leukocyte Composition and Prognostic Power in Hepatitis B- and Hepatitis C-Related Hepatocellular Carcinomas. Genes (Basel) 2019;10:630. 10.3390/genes10080630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson ED, Enriquez HL, Fu YX, et al. Tumor masses support naive T cell infiltration, activation, and differentiation into effectors. J Exp Med 2010;207:1791-804. 10.1084/jem.20092454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garnelo M, Tan A, Her Z, et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut 2017;66:342-51. 10.1136/gutjnl-2015-310814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi JY, Gao Q, Wang ZC, et al. Margin-infiltrating CD20(+) B cells display an atypical memory phenotype and correlate with favorable prognosis in hepatocellular carcinoma. Clin Cancer Res 2013;19:5994-6005. 10.1158/1078-0432.CCR-12-3497 [DOI] [PubMed] [Google Scholar]
- 32.Sung PS, Jang JW. Natural killer cell dysfunction in hepatocellular carcinoma: pathogenesis and clinical implications. Int J Mol Sci 2018;19:3648. 10.3390/ijms19113648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai XY, Gao Q, Qiu SJ, et al. Dendritic cell infiltration and prognosis of human hepatocellular carcinoma. J Cancer Res Clin Oncol 2006;132:293-301. 10.1007/s00432-006-0075-y [DOI] [PubMed] [Google Scholar]
- 34.Ninomiya T, Akbar SM, Masumoto T, et al. Dendritic cells with immature phenotype and defective function in the peripheral blood from patients with hepatocellular carcinoma. J Hepatol 1999;31:323-31. 10.1016/S0168-8278(99)80231-1 [DOI] [PubMed] [Google Scholar]
- 35.Fu J, Xu D, Liu Z, et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 2007;132:2328-39. 10.1053/j.gastro.2007.03.102 [DOI] [PubMed] [Google Scholar]
- 36.Kuang DM, Zhao Q, Wu Y, et al. Peritumoral neutrophils link inflammatory response to disease progression by fostering angiogenesis in hepatocellular carcinoma. J Hepatol 2011;54:948-55. 10.1016/j.jhep.2010.08.041 [DOI] [PubMed] [Google Scholar]
- 37.Li YW, Qiu SJ, Fan J, et al. Intratumoral neutrophils: a poor prognostic factor for hepatocellular carcinoma following resection. J Hepatol 2011;54:497-505. 10.1016/j.jhep.2010.07.044 [DOI] [PubMed] [Google Scholar]
- 38.Ji J, Eggert T, Budhu A, . Hepatic stellate cell and monocyte interaction contributes to poor prognosis in hepatocellular carcinoma. Hepatology 2015;62:481-95. 10.1002/hep.27822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu RX, Wei Y, Zeng QH, et al. Chemokine (C-X-C motif) receptor 3-positive B cells link interleukin-17 inflammation to protumorigenic macrophage polarization in human hepatocellular carcinoma. Hepatology 2015;62:1779-90. 10.1002/hep.28020 [DOI] [PubMed] [Google Scholar]
- 40.Li JQ, Yu XJ, Wang YC, et al. Distinct patterns and prognostic values of tumor-infiltrating macrophages in hepatocellular carcinoma and gastric cancer. J Transl Med 2017;15:37. 10.1186/s12967-017-1139-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu XD, Zhang JB, Zhuang PY, et al. High expression of macrophage colony-stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. J Clin Oncol 2008;26:2707-16. 10.1200/JCO.2007.15.6521 [DOI] [PubMed] [Google Scholar]
- 42.Mossanen JC, Tacke F. Role of lymphocytes in liver cancer. Oncoimmunology 2013;2:e26468. 10.4161/onci.26468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Chen P, Liao R, et al. Intratumoral regulatory T cells with higher prevalence and more suppressive activity in hepatocellular carcinoma patients. J Gastroenterol Hepatol 2013;28:1555-64. 10.1111/jgh.12202 [DOI] [PubMed] [Google Scholar]
- 44.Zhang HH, Mei MH, Fei R, et al. Regulatory T cells in chronic hepatitis B patients affect the immunopathogenesis of hepatocellular carcinoma by suppressing the anti-tumour immune responses. J Viral Hepat 2010;17 Suppl 1:34-43. 10.1111/j.1365-2893.2010.01269.x [DOI] [PubMed] [Google Scholar]
- 45.Li W, Han J, Wu H. Regulatory T-cells promote hepatitis B virus infection and hepatocellular carcinoma progression. Chronic Dis Transl Med 2016;2:67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu JF, Ding YH, Ying XH, et al. Regulatory T cells, especially ICOS(+) FOXP3(+) regulatory T cells, are increased in the hepatocellular carcinoma microenvironment and predict reduced survival. Sci Rep 2016;6:35056. 10.1038/srep35056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun L, Xu G, Liao W, et al. Clinicopathologic and prognostic significance of regulatory T cells in patients with hepatocellular carcinoma: a meta-analysis. Oncotarget 2017;8:39658-72. 10.18632/oncotarget.17340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Liu T, Tang W, et al. Hepatocellular Carcinoma Cells Induce Regulatory T Cells and Lead to Poor Prognosis via Production of Transforming Growth Factor-beta1. Cell Physiol Biochem 2016;38:306-18. 10.1159/000438631 [DOI] [PubMed] [Google Scholar]
- 49.Shi C, Chen Y, Chen Y, et al. CD4(+) CD25(+) regulatory T cells promote hepatocellular carcinoma invasion via TGF-beta1-induced epithelial-mesenchymal transition. Onco Targets Ther 2018;12:279-89. 10.2147/OTT.S172417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Juengpanich S, Shi L, Iranmanesh Y, et al. The role of natural killer cells in hepatocellular carcinoma development and treatment: A narrative review. Transl Oncol 2019;12:1092-107. 10.1016/j.tranon.2019.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Hao X, Sun R, et al. Natural Killer Cell-Derived Interferon-Gamma Promotes Hepatocellular Carcinoma Through the Epithelial Cell Adhesion Molecule-Epithelial-to-Mesenchymal Transition Axis in Hepatitis B Virus Transgenic Mice. Hepatology 2019;69:1735-50. 10.1002/hep.30317 [DOI] [PubMed] [Google Scholar]
- 52.Zahran AM, Abdel-Meguid MM, Ashmawy AM, et al. Frequency and Implications of Natural Killer and Natural Killer T Cells in Hepatocellular Carcinoma. Egypt J Immunol 2018;25:45-52. [PubMed] [Google Scholar]
- 53.Liu P, Chen L, Zhang H. Natural Killer Cells in Liver Disease and Hepatocellular Carcinoma and the NK Cell-Based Immunotherapy. J Immunol Res 2018;2018:1206737. 10.1155/2018/1206737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Najima Y, Yoshida C, Iriyama N, et al. Regulatory T cell inhibition by dasatinib is associated with natural killer cell differentiation and a favorable molecular response-The final results of the D-first study. Leuk Res 2018;66:66-72. 10.1016/j.leukres.2018.01.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as