Abstract
Background
Intervertebral disc degeneration (IVDD) is regarded as the leading cause of low back pain, resulting in disability and a heavy burden on public health. Several studies have unveiled that long noncoding RNAs (lncRNAs) play a key role in the pathogenesis and progression of IVDD. In this study, we aimed to investigate the biological function and latent molecular mechanism of the lncRNA FAM83H antisense RNA 1 (FAM83H-AS1) in IVDD development.
Methods
Firstly, we established an IVDD model in rats using advanced glycation end products (AGEs) intradiscal injection. Subsequently, gain-of-function assays were conducted to investigate the role of FAM83H-AS1 in the progression of IVDD. Bioinformatics analysis, RNA pull down assay and rescue experiments were employed to shed light on the molecular mechanism underlying FAM83H-AS1 involving in IVDD.
Results
Our findings verified that AGEs treatment aggravated IVDD damage, and FAM83H-AS1 was downregulated in the IVDD group. Additionally, overexpression of FAM83H-AS1 contributed to the growth of nucleus pulposus (NP) cells and ameliorated IVDD injury. It was revealed that FAM83H-AS1 possessed the speculated binding sites of miR-22-3p. More importantly, we confirmed that FAM83H-AS1 functioned as a sponge of miR-22-3p in IVDD. Lastly, we demonstrated that miR-22-3p mediated the impact of FAM83H-AS1 on cell proliferation, ECM degradation, and inflammation.
Conclusions
Our study indicated that FAM83H-AS1 relieved IVDD deterioration through sponging miR-22-3p, and provides novel insights into the mechanisms underlying FAM83H-AS1 in IVDD progression.
Keywords: Intervertebral disc degeneration (IVDD), long noncoding RNA (lncRNA), FAM83H-AS1, miR-22-3p
Introduction
Low back pain is a leading cause of disability, with approximately 70–85% of people suffering from low back pain during their lifetime (1,2). Intervertebral disc degeneration (IVDD) is widely acknowledged as the leading contributor to low back pain, which not only has deleterious effects on quality of life, but also leads to a heavy financial burden (3,4). Although it is widely accepted that IVDD is a biological process of disc aging, many IVDD patients exhibit accelerated disc degeneration attributable to various risk factors, such as genetics, obesity, rapid increase in weight, smoking, and lack of physical activity (5-8). According to the severity of the IVDD, invasive or non-invasive treatment strategies can be applied. Non-invasive treatments such as pharmacological treatment or acupuncture have a limit and do not help to resolving severe conditions. Advanced invasive and regenerative strategies hold great promise, such as total disc replacement, cartilaginous endplates decalcification, tissue engineering, biomaterials and cell therapy (9). In addition, combined strategies using cell-seeded scaffolds for fully regeneration of nucleus pulposus or annulus fibrosus cells have been extensive studied in recent years (10). To help developing new treatment strategies, it is crucial to elucidate the specific pathogenesis of IVDD.
The main characteristics of IVDD include reduction of IVD height, loss of water and extracellular matrix (ECM) content, structural defects, chronic inflammation, and metabolic changes (11-13). Advanced glycation end products (AGEs) are generated by non-enzymatic reactions between free amino groups of macromolecules and reducing sugars, also known as the Maillard reactions (14). It is reported that the accumulation of AGEs commonly occurs in aging and degenerative tissues, and AGEs are strongly associated with metabolic dysfunction, inflammation, and endoplasmic reticulum (ER) stress, particularly in collagen-rich tissues (15,16). Emerging evidence demonstrates that AGE deposition gives rise to structural disruption and the progression of IVDD (17). A myriad of literature demonstrates that AGEs function as a vital mediator in the development of IVDD through regulating a wide range of processes, such as ER stress, inflammation, and oxidative stress.
An increasing number of investigations have shown that long noncoding RNAs (lncRNAs) play a crucial role in the pathogenesis and progression of multiple diseases, including IVDD (18,19). For example, knockdown of the lncRNA ANRIL alleviated myocardial cell apoptosis in acute myocardial infarction through regulation of IL-33/ST2 (20). Furthermore, the lncRNA HCG18 promoted the progression of IVDD via sponging miR-146a-5p and modulating TRAF6 expression (21). Silencing lncRNA LOC101928963 suppressed cell proliferation and induced apoptosis in spinal cord glioma through binding to PMAIP1 (22). Given that abnormally expressed lncRNAs act as pivotal modulators in IVDD, understanding the function and underlying mechanisms of lncRNAs in the development of IVDD may be beneficial to the improvement of IVDD treatments.
Family with sequence similarity 83 member H (FAM83H) is initially identified as a protein essential for dental enamel formation, and mutations in FAM83H is associated with developing nonsyndromic amelogenesis imperfect (23). Moreover, FAM83H is found to stabilize β-catenin, maintain the formation of desmosomes, and involve in the tumorigenesis of a variety of cancers such as osteosarcomas, hepatocellular carcinoma and gastric cancer (24-26). FAM83H antisense RNA 1 (FAM83H-AS1), also termed onco-lncRNA-3, is located in the key region of FAM83H antisense region 8q24 and is 12,198 nucleotides in length (27). Dysregulated FAM83H-AS1 has been reported to exert oncogenic activities in a wide range of malignant tumors (28-30). More importantly, a recent study revealed that FAM83H-AS1 expression was positively associated with the grade of disc degeneration, and FAM83H-AS1 facilitated the growth of nucleus pulposus (NP) cells and ECM deposition (31). Hence, in this study, we aimed to further explore the unknown molecular mechanisms governing FAM83H-AS1 in IVDD.
In the present study, we established an IVDD model via AGE treatment, then probed the biological role of lncRNA FAM83H-AS1 in IVDD. Our findings revealed that upregulation of FAM83H-AS1 relieved IVDD injury. Mechanistically, FAM83H-AS1 attenuated ECM degradation and inflammation-related pain in IVDD via sponging miR-22-3p. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7056).
Methods
Animal models
Forty Sprague-Dawley (SD) rats (three-month-old and 250 g weight each) were used in this study. All procedures and experiments were performed under a project license SYXK (Jing) 2016-0043 in accordance with the ethical standards of the Ethics Committee of Chinese-Japanese Friendship Hospital. The surgical procedures were carried out as described previously (32). Rats were anesthetized by 2% (w/v) pentobarbital (40 mg/kg). Following confirmation of the IVDs (Co7/8, Co8/9 and Co9/10), rats were randomly divided into control group, IVDD-2 µg AGEs group, IVDD-10 µg AGEs group and IVDD-50 µg AGEs group (n=5 for each group). In the control group were treated with PBS via intradiscal injection, and IVDD model mice were injected with 2, 10 or 50 µg/mL AGEs. Each segment of the IVDs was administered with a total injection volume of 4 µL of PBS or AGEs solution. The injections were administered once a week for 1 month. To evaluate the effect of FAM83H-AS1 overexpression in vivo, rats were divided Sham + OV-NC, IVDD + OV-NC and IVDD + OV-FAM83H-AS1 group (n= 5 for each group). Sham + OV-NC group was normal rats received surgery and injection with empty lentivirus control, IVDD + OV-NC and IVDD + OV-FAM83H-AS1 group were IVDD rats injected with empty lentivirus control or FAM83H-AS1 expression lentivirus. The disc samples were analyzed by H&E staining 4 weeks post-surgery.
Pain behavior measurement
In view of the decline of paw withdrawal threshold that manifests with heightened pain sensitivity, the hindpaw mechanical hyperalgesia test was utilized to estimate the severity of pain which was related to IVDD in rats. The assay was conducted in wire mesh-floored cages and rats were acclimatized to cages 20 minutes before measurement. Calibrated von Frey filaments (Stoelting, USA) ranging from 0.4 to 15.0 g were then applied to the plantar hindpaw with enough pressure to bend the filament. A 50% force withdrawal threshold was dependent on the mean of 3 trials for each side of hindpaws.
Isolation and culture of NP cells
Lumbar discs were separated after rats were euthanized, and NP tissues were isolated from lumbar discs under a dissecting microscope and maintained in 0.1% collagenase and 2 U/mL hyaluronidase at room temperature for 4 hours. Following digestion, tissues were placed in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, then maintained at 37 °C in a humid incubator with 5% CO2. NP cells were collected from the explants after 1 week. Harvested NP cells were then counted and seeded into 10 cm culture plates at the appropriate density. The morphology of cells from the second passage had no obvious changes compared to the primary cells (passage 0). Thus, second passage cells were adopted for all subsequent experiments. NP cells were maintained in complete medium under 5% CO2 at 37 °C and fresh medium was replaced every other day.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA extraction from NP tissues and cells was performed using Trizol reagent (Invitrogen, USA) based on the manufacturer’s instructions. After RNA concentration was determined, total RNA was reverse transcribed to cDNA using the Reverse Transcription Kit (Invitrogen). SYBR Green PCR Kit (Applied Biosystems) was then used to conduct PCR on the 7000 Sequence Detection System (Applied Biosystems). U6 and GAPDH served as reference standards for normalization, and the 2−ΔΔCt method was utilized to calculate the relative gene expression. The primers for qRT-PCR were listed in Table 1.
Table 1. Primers of indicated genes used in qRT-PCR (quantitative real-time PCR).
| Gene name | Sense primer | Anti-sense primer |
|---|---|---|
| FAM83H-AS1 | 5'-TAGGAAACGAGCGAGCCC-3' | 5'-GCTTTGGGTCTCCCCTTCTT-3' |
| miR-22-3p | 5'-GGGAAGCTGCCAGTTGAAG-3' | 5'-GTGCGTGTCGTGGAGTCG-3' |
| U6 | 5'-TAGGAAACGAGCGAGCCC-3' | 5'-GCTTTGGGTCTCCCCTTCTT-3' |
| GAPDH | 5'-TGCCCAGAACATCATCCCT-3' | 5'-GGTCCTCAGTGTAGCCCAAG-3' |
Cell proliferation assays
Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was used to detect the viability of NP cells, based on the manufacturer’s instructions. Following treatment with PBS or AGEs, transfected NP cells were seeded into 96-well plates and cultured at 37 °C. At the indicated time points, each well was supplemented with 10 µL of CCK-8 reagent. After an additional 4 hours incubation at 37 °C, absorbance at 450 nm was measured with a spectrophotometer (BioTek, USA).
For 5-ethynyl-2’-deoxyuridine (EdU) incorporation assay, Click iT™ EdU cell proliferation assay kit (Invitrogen) was used in accordance with the manufacturer’s instructions. NP cells were inoculated into 96-well plates, treated with 50 µM EdU for 2 hours, immobilized in 4% polyoxymethylene, followed by permeabilization with 0.5% Triton X-100 and counterstaining with DAPI. The images were captured with a florescence microscope (Olympus, Japan).
Flow cytometry analysis
For quantification of F4/80+CD11C+ macrophages in NP tissues, FACSCalibur flow cytometry (Beckman Coulter, USA) was performed. The macrophages were incubated with fluorescent antibodies for CD11C and F4/80 (Cell Signaling Technology, USA), then analyzed by flow cytometry.
Immunofluorescence staining
After washing 3 times with PBS, macrophages were fixed in 4% formaldehyde, permeabilized with 0.5% Triton X-100, and blocked in 5% BSA. Subsequently, cells were incubated with a primary antibody against CD68 (Cell Signaling Technology) at 4 °C overnight, then probed with a fluorochrome-labeled secondary antibody at 37 °C for 2 hours. Macrophages were visualized with a fluorescence microscope.
Western blot
Total protein was isolated from tissues using lysis buffer (Beyotime, China). The equivalent protein samples were separated on 10% SDS-PAGE gels, then transferred to PVDF membranes (Bio-Rad, USA). Following blocking in 5% skim milk, membranes were incubated overnight with primary antibodies at 4 °C, appropriate secondary antibodies for 1 hour at 37 °C, then treated with Amersham ECL Western Blotting System (Amersham Pharmacia, USA). The following primary antibodies were used: ADAMTS-4 (Abcam# ab219548), MMP-3 (Cell signaling#14351), collagen II (Thermal Fisher #MA5-13026), aggrecan (Thermal Fisher #MA5-16888), IL-23 (Thermal Fisher # 16-7123-81), IL-10 (Thermal Fisher # MA5-23796), and GAPDH (Cell signaling #5174). GAPDH was used as an internal control.
RNA pull down assay
Biotinylated wild type and mutant FAM83H-AS1 were synthesized with T7 RNA polymerase (Roche, USA) and the Biotin RNA Labeling Mix (Roche) according to the manufacturer’s instructions. Cell extracts were incubated with biotin-labeled RNAs and streptavidin agarose beads (Invitrogen) for 1 hour at 37 °C. RNAs were eluted from beads, purified, then detected by qRT-PCR.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (IBM, Chicago, IL, USA), and experimental data were expressed as mean ± SD from at least 3 independent assays. Comparisons between two groups were analyzed using Student’s t-test, and comparisons between multiple groups were analyzed by one-way analysis of variance (ANOVA). P<0.05 was regarded as statistically significant.
Results
AGEs treatment exacerbated IVDD in a rat model in vivo
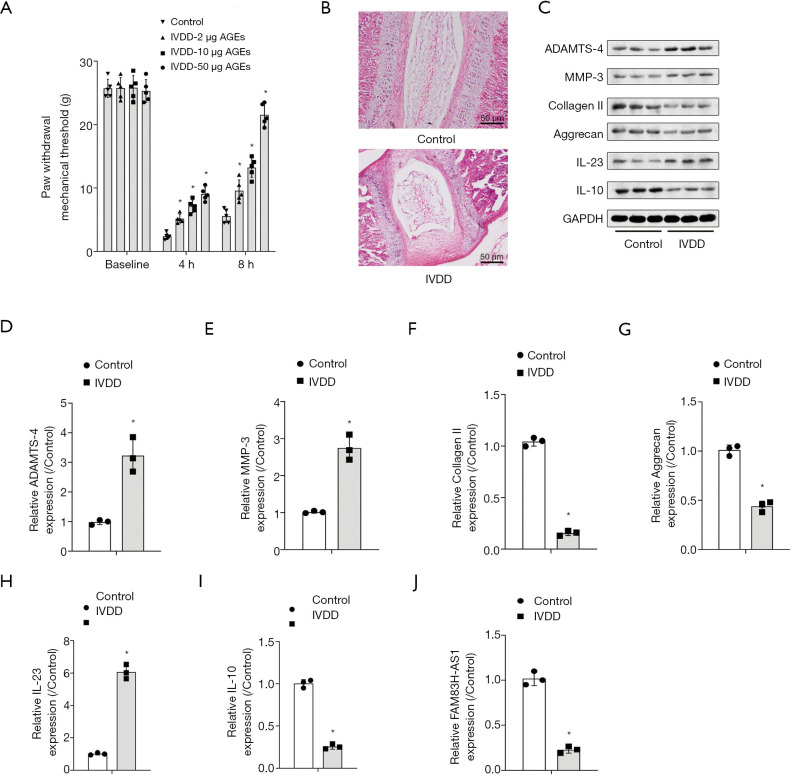
To establish the IVDD model, rats were injected with AGEs. We observed that intradiscal injection of AGEs resulted in a decrease in mechanical withdrawal thresholds and mechanical hyperalgesia, which appeared to be both dose-dependent and time-dependent (Figure 1A). Rats from the IVDD-50 µg AGEs group were used for further study. Results of HE staining demonstrated that AGEs treatment led to IVDD injury (Figure 1B). Western blot demonstrated that administration of AGEs significantly augmented the expression of ADAMTS-4 and MMP-3, and diminished collagen II and aggrecan levels, suggesting that AGEs treatment aggravated the degradation of the ECM (Figure 1C,D,E,F,G). Additionally, the increase in IL-23 expression and the reduction in anti-inflammatory cytokine IL-10 levels were ascribed to AGEs intradiscal injection (Figure 1H,I). qRT-PCR analysis unveiled that FAM83H-AS1 expression decreased significantly in the IVDD group compared with the control group (Figure 1J). Overall, AGEs injection triggered IVDD damage in a rat model.
Figure 1.
AGEs treatment exacerbated IVDD in a rat model in vivo. Rats were divided into two groups: control group and IVDD model group. Rats in the control group were treated with PBS, and IVDD model rats were inoculated with AGEs. (A) Results of pain behavior measurement between groups; (B) HE staining images of IVDs in control and IVDD-50 µg AGEs rats; (C) Western blot was performed to compare ECM-related protein expression levels and inflammation between control and IVDD-50 µg AGEs group rats; (D,E,F,G,H,I) the quantitative analysis of corresponding protein expression levels; (J) FAM83H-AS1 expression was examined by qRT-PCR in control and IVDD-50 µg AGEs group mice. All experimental data are shown as mean ± SD from at least 3 independent assays. *, P<0.05 vs. control group. AGEs, advanced glycation end products; IVDD, intervertebral disc degeneration.
Overexpression of FAM83H-AS1 promoted NP cell proliferation, alleviated ECM degradation and prevented IVDD progression in vivo
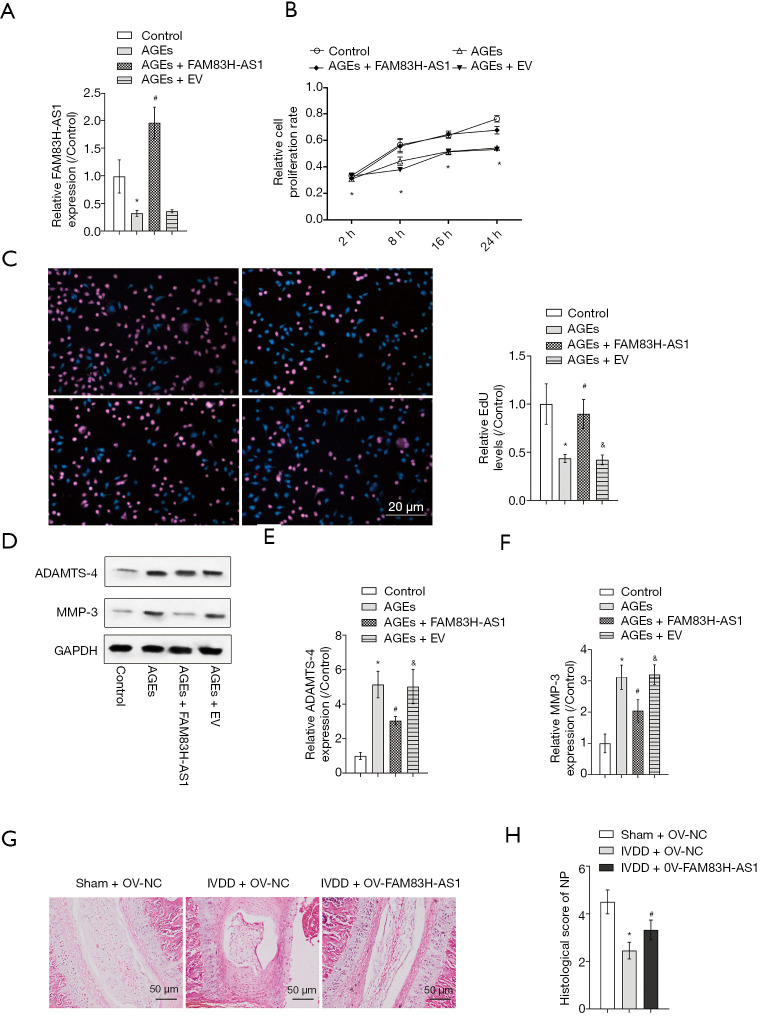
In order to investigate the role of FAM83H-AS1 in IVDD, we overexpressed FAM83H-AS1 in AGEs-induced NP cells, and transfection efficacy was verified by qRT-PCR (Figure 2A). The CCK-8 assay and EdU assay unveiled that ectopic expression of FAM83H-AS1 resulted in the recovery of inhibited cell proliferation caused by AGEs treatment (Figure 2B,C). In agreement with the above findings, western blot analysis demonstrated that overexpression of FAM83H-AS1 reduced the protein expression levels of ADAMTS-4 and MMP-3 in the IVDD group (Figure 2D,E,F). To investigate the function of FAM83H-AS1 in vivo, we performed a disc puncture procedure to model IVDD in rats and overexpressed FAM83H-AS1 by lentivirus. As shown in Figure 2G, the IVDD + OV-NC group showed obviously more degeneration than the IVDD + OV-FAM83H-AS1 group, indicating that FAM83H-AS1 overexpression prevented IVDD progression. In addition, there was a significantly difference of histological score between the IVDD + OV-NC group and IVDD + OV-FAM83H-AS1 group (Figure 2H). Based on the aforementioned findings, we concluded that overexpression of FAM83H-AS1 ameliorated IVDD injury through regulating NP cell growth and ECM degradation.
Figure 2.
Overexpression of FAM83H-AS1 promoted NP cell proliferation, alleviated ECM degradation and prevent IVDD progression in vivo. NP cells isolated from IVD tissues were treated with 50 µg/mL AGEs, followed by transfection with FAM83H-AS1 plasmid or empty vector. NP cells treated with PBS served as the control. (A) The efficiency of FAM83H-AS1 overexpression was verified by qRT-PCR analysis; (B) CCK-8 assay was performed to assess the viability of NP cells; (C) cell proliferation capacity was also determined using the EdU incorporation assay; (D,E,F) the protein expression of ADAMTS-4 and MMP-3 was measured by western blot. (G) Representative H&E staining of disc samples from different experimental groups at 4 weeks post-surgery. (H) Histological scores at 4 weeks post-surgery in three groups. All experimental data are shown as mean ± SD from at least 3 independent assays. *, P<0.05 vs. control group; #, P<0.05 vs. AGEs group; &, P<0.05 vs. IVDD + FAM83H-AS1 group. ECM, extracellular matrix; NP, nucleus pulposus; AGEs, advanced glycation end products; qRT-PCR, quantitative real-time PCR.
FAM83H-AS1 functioned as a sponge of miR-22-3p
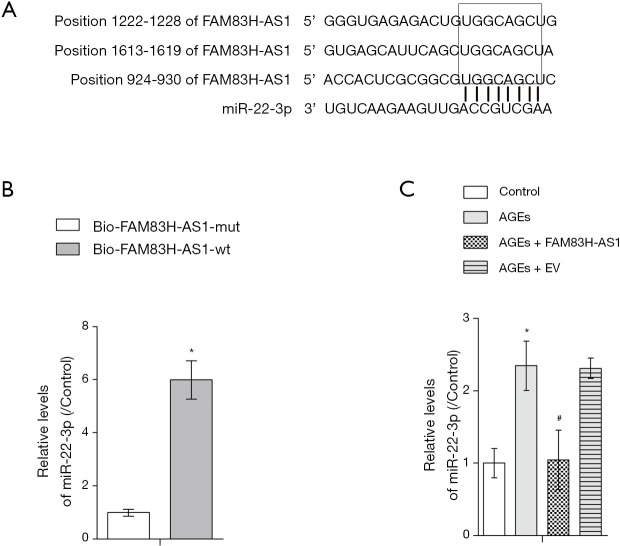
With the aid of bioinformatics analysis, we found that FAM83H-AS1 harbored the potential binding sites for miR-22-3p (Figure 3A). RNA pull down assay demonstrated that miR-22-3p was only abundantly enriched in complexes pulled down by wild type FAM83H-AS1 in contrast with mutant FAM83H-AS1 precipitates, validating that miR-22-3p directly bound to FAM83H-AS1 (Figure 3B). Results of qRT-PCR assay showed that the expression of miR-22-3p was enhanced by AGEs treatment, and that the restoration of miR-22-3p levels occurred due to overexpression of FAM83H-AS1 (Figure 3C). Overall, FAM83H-AS1 negatively regulated miR-22-3p expression via directly binding with it.
Figure 3.
FAM83H-AS1 functioned as a sponge of miR-22-3p. (A) The putative binding sites between FAM83H-AS1 and miR-22-3p revealed through bioinformatics analysis; (B) RNA pull down assay validated the binding specificity of miR-22-3p to FAM83H-AS1; (C) qRT-PCR detection of miR-22-3p expression after different treatments. All experimental data are shown as mean ± SD from at least 3 independent assays. *, P<0.05 vs. control group; #, P<0.05 vs. AGEs group. qRT-PCR, quantitative real-time PCR.
FAM83H-AS1 regulated IVDD through targeting miR-22-3p
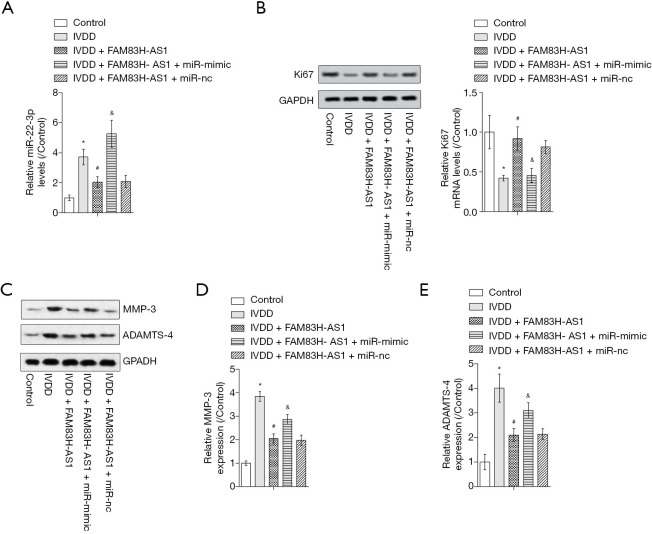
To confirm whether the function of FAM83H-AS1 was mediated by miR-22-3p, we carried out rescue experiments. As shown in Figure 4A, the increased miR-22-3p expression in the IVDD model group was recovered by overexpression of FAM83H-AS1, then elevated by upregulation of miR-22-3p. Western blot revealed that the decline in Ki67 levels caused by IVDD was reversed by ectopic expression of FAM83H-AS1, and overexpression of miR-22-3p contributed to the restoration of Ki67 expression (Figure 4B). Moreover, elevated ADAMTS-4 and MMP-3 expression resulting from IVDD was reduced by FAM83H-AS1 upregulation, and subsequent overexpression of miR-22-3p partially counteracted the effects of FAM83H-AS1 on the protein levels of ADAMTS-4 and MMP-3 (Figure 4C,D,E). Taken together, these findings indicated that FAM83H-AS1 functioned as a regulator of IVDD via modulation of miR-22-3p.
Figure 4.
FAM83H-AS1 regulated IVDD through targeting miR-22-3p. (A) The level of miR-22-3p in different treatment groups was measured by qRT-PCR assay; (B) the protein expression of Ki67 as determined by western blot; (C,D,E) Western blot was carried out for evaluation of ADAMTS-4 and MMP-3 protein levels. All experimental data are shown as mean ± SD from at least 3 independent assays. *, P<0.05 vs. control group; #, P<0.05 vs. IVDD group; &, P<0.05 vs. IVDD + FAM83H-AS1 group. IVDD, intervertebral disc degeneration; qRT-PCR, quantitative real-time PCR.
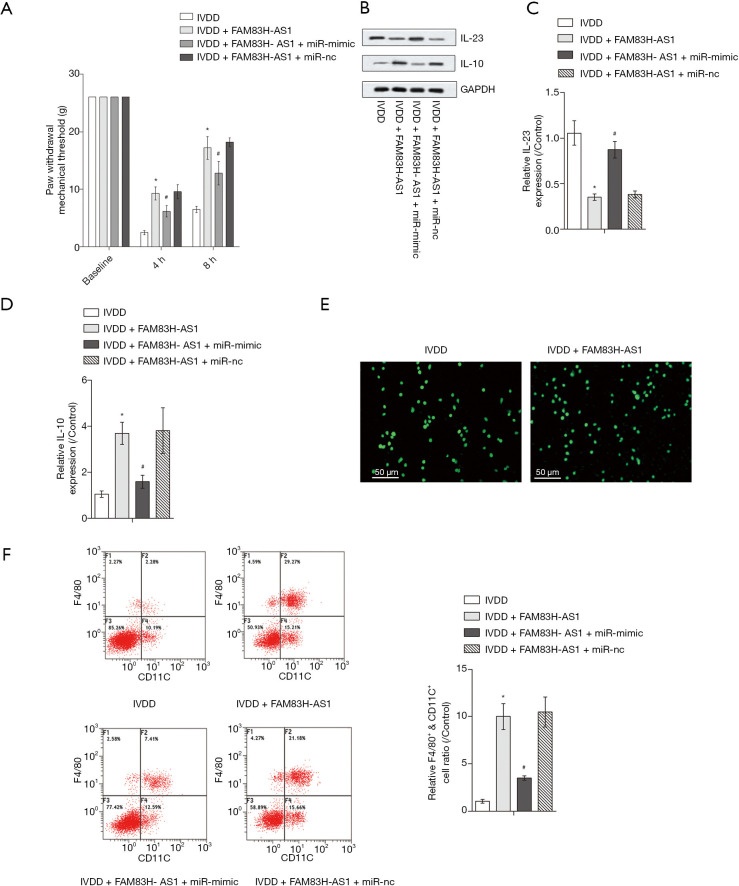
Upregulation of miR-22-3p partially abolished the impact of FAM83H-AS1 on inflammatory pain
Next, we explored the role of FAM83H-AS1/miR-22-3p in inflammatory pain. The IVDD-induced mechanical hyperalgesia was lowered due to highly expressed FAM83H-AS1 and overexpression of miR-22-3p, which led to the recovery of the mechanical withdrawal threshold (Figure 5A). Likewise, IL-23 levels were diminished and the expression of IL-10 was increased in IVDD model mice when FAM83H-AS1 was overexpressed. Subsequently, upregulation of miR-22-3p abrogated the function of FAM83H-AS1 in the expression of IL-23 and IL-10 (Figure 5B,C,D). Cells extracted from IVD tissues were morphologically consistent with macrophages, and the significant expression of CD68 further validated that macrophages were successfully isolated (Figure 5E). Flow cytometry analysis demonstrated that the proportion of macrophages positive for F4/80+CD11C+ in the IVDD group was elevated by overexpression of FAM83H-AS1, and miR-22-3p upregulation recovered the ratio of positive F4/80+CD11C+ macrophages (Figure 5F). Overall, FAM83H-AS1 ameliorated inflammatory responses in IVDD through sponging miR-22-3p.
Figure 5.
Upregulation of miR-22-3p partially abolished the impact of FAM83H-AS1 on inflammatory pain. (A) The paw withdrawal thresholds of rats in different treatment groups; (B,C,D) Western blot was performed to measure the protein expression of IL-23 and IL-10 after different treatments; (E) the immunofluorescence assay was conducted to detect CD68 expression in macrophages; (F) the positive F4/80+CD11C+ macrophages were analyzed by flow cytometry. All experimental data are shown as mean ± SD from at least 3 independent assays. *, P<0.05 vs. IVDD group; #, P<0.05 vs. IVDD + FAM83H-AS1 group. qRT-PCR, quantitative real-time PCR; IVDD, intervertebral disc degeneration.
Discussion
Accumulating evidence suggests that lncRNAs play a critical role in IVDD progression through modulating various cellular processes, such as autophagy, senescence, proliferation, and ECM synthesis (33,34). A previous study demonstrated that lncRNA FAM83H-AS1 contributes to NP cell growth through the regulation of the Notch signaling pathway (31). Furthermore, FAM83H-AS1 functions as an oncogene in a diverse range of cancers through promoting cell proliferation, metastasis, and radioresistance, while suppressing cell apoptosis (35-37). In this study, we applied AGEs intervertebral injection to establish the IVDD rat model. Our findings confirmed that AGEs treatment resulted in IVDD damage. Additionally, FAM83H-AS1 was significantly downregulated in the IVDD group, indicating its potential role in IVDD progress. NP cells that generate collagen and proteoglycan aggrecan to construct the complex ECM are of great importance in maintaining healthy discs (13). We found that overexpression of FAM83H-AS1 increased NP cell growth and attenuated ECM degradation, suggesting that FAM83H-AS1 exerted a suppressive role in the development of IVDD.
Recently, the role of miR-22-3p has been explored in several diseases. It has been reported that miR-22-3p functions as a tumor suppressor gene in human cancers and inhibits cell proliferation and enhanced chemosensitivity in malignancies (38,39). Moreover, numerous studies reveal the potential of miR-22-3p in various orthopedic disorders. For example, Perez-Sanchez et al. demonstrated that high expression of miR-22-3p is correlated with disease activity and structural damage in patients with ankylosing spondylitis (40). Furthermore, miR-22-3p was found to be significantly downregulated in osteoporosis, resulting from impaired WNT signaling through analysis of miRNA profile (41). Consistently, Wu et al. further indicated that miR-22-3p slows the progression of osteogenesis via controlling cell growth (42). More importantly, it has been shown that miR-22 participates in the regulation of H2O2-induced cell senescence, growth, and ECM synthesis (43). Until now, the association between miR-22-3p and FAM83H-AS1 in IVDD pathogenesis had not yet been elucidated. In our study, we found that FAM83H-AS1 acted as a molecular sponge for miR-22-3p. We then investigated the participation of FAM83H-AS1/miR-22-3p in the progression of IVDD. Our results suggested that FAM83H-AS1 executed its regulatory effects on ECM degradation and NP cell growth in IVDD development through the suppression of miR-22-3p.
Several studies have revealed that macrophages exhibit overt heterogeneity in phenotype and function, and local environmental factors determine the function and activation states of macrophages (44,45). According to their activation state, macrophages are segmented into a classically activated (M1) phenotype and alternatively activated (M2) phenotype. M1 macrophages secrete high levels of pro-inflammatory cytokines including IL-6, IL-12, and TNF-α, while M2 macrophages predominantly secrete anti-inflammatory factors, such as IL-10 and CCL18 (46). In this study, we found that IL-10 was decreased while IL-23 were increased in IVDD group, however this was reversed by FAM83H-AS1 overexpression. In addition, restore miR-22-3p expression abolished the effects on IL-10 and IL-23 caused by FAM83H-AS1, indicating that miR-22-3p mediated the regulatory role of FAM83H-AS1 in IL-23 and IL-10 expression and macrophage phenotypes in the IVDD model. These results indicated that FAM83H-AS1 promoted macrophage polarization to inhibit inflammation in IVDD through targeting miR-22-3p.
Conclusions
In summary, our study unraveled the biological function and novel molecular mechanism of FAM83H-AS1 in IVDD. We demonstrated that FAM83H-AS1 relieved IVDD damage through sponging miR-22-3p. To the best of our knowledge, this study is the first to shed light on the association between FAM83H-AS1 and miR-22-3p. Our findings may provide a promising therapeutic strategy for IVDD.
Supplementary
The article’s supplementary files as
Acknowledgments
We deeply appreciate all participants.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures and experiments were performed under a project license SYXK (Jing) 2016-0043 in accordance with the ethical standards of the Ethics Committee of Chinese-Japanese Friendship Hospital.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7056
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7056
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7056). The authors have no conflicts of interest to declare.
(English Language Editor: C. Betlazar-Maseh)
References
- 1.GBD 2016 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017;390:1211-59. 10.1016/S0140-6736(17)32154-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argyriou AA, Anastopoulou GG, Bruna J. Inconclusive evidence to support the use of minimally-invasive radiofrequency denervation against chronic low back pain. Ann Transl Med 2018;6:127. 10.21037/atm.2018.01.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung KM, Karppinen J, Chan D, et al. Prevalence and pattern of lumbar magnetic resonance imaging changes in a population study of one thousand forty-three individuals. Spine (Phila Pa 1976) 2009;34:934-40. 10.1097/BRS.0b013e3181a01b3f [DOI] [PubMed] [Google Scholar]
- 4.Gore M, Sadosky A, Stacey BR, et al. The burden of chronic low back pain: clinical comorbidities, treatment patterns, and health care costs in usual care settings. Spine (Phila Pa 1976) 2012;37:E668-77. 10.1097/BRS.0b013e318241e5de [DOI] [PubMed] [Google Scholar]
- 5.Battié MC, Videman T, Kaprio J, et al. The Twin Spine Study: contributions to a changing view of disc degeneration. Spine J 2009;9:47-59. 10.1016/j.spinee.2008.11.011 [DOI] [PubMed] [Google Scholar]
- 6.Samartzis D, Karppinen J, Mok F, et al. A population-based study of juvenile disc degeneration and its association with overweight and obesity, low back pain, and diminished functional status. J Bone Joint Surg Am 2011;93:662-70. 10.2106/JBJS.I.01568 [DOI] [PubMed] [Google Scholar]
- 7.Williams FM, Popham M, Sambrook PN, et al. Progression of lumbar disc degeneration over a decade: a heritability study. Ann Rheum Dis 2011;70:1203-7. 10.1136/ard.2010.146001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battié MC, Videman T, Levalahti E, et al. Genetic and environmental effects on disc degeneration by phenotype and spinal level: a multivariate twin study. Spine (Phila Pa 1976) 2008;33:2801-8. 10.1097/BRS.0b013e31818043b7 [DOI] [PubMed] [Google Scholar]
- 9.van Uden S, Silva-Correia J, Oliveira JM, et al. Current strategies for treatment of intervertebral disc degeneration: substitution and regeneration possibilities. Biomater Res 2017;21:22. 10.1186/s40824-017-0106-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowles RD, Setton LA. Biomaterials for intervertebral disc regeneration and repair. Biomaterials 2017;129:54-67. 10.1016/j.biomaterials.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams MA, Roughley PJ. What is intervertebral disc degeneration, and what causes it? Spine (Phila Pa 1976) 2006;31:2151-61. 10.1097/01.brs.0000231761.73859.2c [DOI] [PubMed] [Google Scholar]
- 12.Le Maitre CL, Hoyland JA, Freemont AJ. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis Res Ther 2007;9:R77. 10.1186/ar2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts S, Evans H, Trivedi J, et al. Histology and pathology of the human intervertebral disc. J Bone Joint Surg Am 2006;88 Suppl 2:10-4. [DOI] [PubMed] [Google Scholar]
- 14.Chan CM, Huang DY, Huang YP, et al. Methylglyoxal induces cell death through endoplasmic reticulum stress-associated ROS production and mitochondrial dysfunction. J Cell Mol Med 2016;20:1749-60. 10.1111/jcmm.12893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y, Wang Y, Zhang Y, et al. Advanced glycation end products regulate anabolic and catabolic activities via NLRP3-inflammasome activation in human nucleus pulposus cells. J Cell Mol Med 2017;21:1373-87. 10.1111/jcmm.13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang CK, Wang CC, Lu TF, et al. Involvement of Endoplasmic Reticulum Stress, Autophagy, and Apoptosis in Advanced Glycation End Products-Induced Glomerular Mesangial Cell Injury. Sci Rep 2016;6:34167. 10.1038/srep34167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Illien-Jünger S, Lu Y, Qureshi SA, et al. Chronic ingestion of advanced glycation end products induces degenerative spinal changes and hypertrophy in aging pre-diabetic mice. PLoS One 2015;10:e0116625. 10.1371/journal.pone.0116625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Sun X, Wang H, et al. Long non-coding RNAs in non-small cell lung cancer: functions and distinctions from other malignancies. Transl Cancer Res 2019;8:2636-53. 10.21037/tcr.2019.10.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Huang X, Hu F, et al. LncRNA ANRIL knockdown relieves myocardial cell apoptosis in acute myocardial infarction by regulating IL-33/ST2. Cell Cycle 2019;18:3393-403. 10.1080/15384101.2019.1678965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xi Y, Jiang T, Wang W, et al. Long non-coding HCG18 promotes intervertebral disc degeneration by sponging miR-146a-5p and regulating TRAF6 expression. Sci Rep 2017;7:13234. 10.1038/s41598-017-13364-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng YJ, Liang TS, Wang J, et al. Silencing lncRNA LOC101928963 Inhibits Proliferation and Promotes Apoptosis in Spinal Cord Glioma Cells by Binding to PMAIP1. Mol Ther Nucleic Acids 2019;18:485-95. 10.1016/j.omtn.2019.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W, Yang M, Wang C, et al. Evolutionary analysis of FAM83H in vertebrates. PLoS One 2017;12:e0180360. 10.1371/journal.pone.0180360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussein UK, Ha SH, Ahmed AG, et al. FAM83H and SCRIB stabilize beta-catenin and stimulate progression of gastric carcinoma. Aging (Albany NY) 2020;12: 11812-34. 10.18632/aging.103351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim KM, Hussein UK, Park SH, et al. FAM83H is involved in stabilization of beta-catenin and progression of osteosarcomas. J Exp Clin Cancer Res 2019;38:267. 10.1186/s13046-019-1274-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KM, Park SH, Bae JS, et al. FAM83H is involved in the progression of hepatocellular carcinoma and is regulated by MYC. Sci Rep 2017;7:3274. 10.1038/s41598-017-03639-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baratieh Z, Khalaj Z, Honardoost MA, et al. Aberrant expression of PlncRNA-1 and TUG1: potential biomarkers for gastric cancer diagnosis and clinically monitoring cancer progression. Biomark Med 2017;11:1077-90. 10.2217/bmm-2017-0090 [DOI] [PubMed] [Google Scholar]
- 28.Ma YK, Shen TH, Yang XY. Upregulation of LncRNA FAM83H-AS1 in hepatocellular carcinoma promotes cell proliferation, migration and invasion by Wnt/beta-catenin pathway. Eur Rev Med Pharmacol Sci 2019;23:7855-62. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Cui J, Wang Y, et al. FAM83H-AS1 is upregulated and predicts poor prognosis in colon cancer. Biomed Pharmacother 2019;118:109342. 10.1016/j.biopha.2019.109342 [DOI] [PubMed] [Google Scholar]
- 30.Da J, Liu P, Wang R, et al. Upregulation of the long non-coding RNA FAM83H-AS1 in gastric cancer and its clinical significance. Pathol Res Pract 2019;215:152616. 10.1016/j.prp.2019.152616 [DOI] [PubMed] [Google Scholar]
- 31.Wei R, Chen Y, Zhao Z, et al. LncRNA FAM83H-AS1 induces nucleus pulposus cell growth via targeting the Notch signaling pathway. J Cell Physiol 2019;234:22163-71. 10.1002/jcp.28780 [DOI] [PubMed] [Google Scholar]
- 32.Chen D, Xia D, Pan Z, et al. Metformin protects against apoptosis and senescence in nucleus pulposus cells and ameliorates disc degeneration in vivo. Cell Death Dis 2016;7:e2441. 10.1038/cddis.2016.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Lv G, Li J, et al. LncRNA-RP11-296A18.3/miR-138/HIF1A Pathway Regulates the Proliferation ECM Synthesis of Human Nucleus Pulposus Cells (HNPCs). J Cell Biochem 2017;118:4862-71. 10.1002/jcb.26166 [DOI] [PubMed] [Google Scholar]
- 34.Zhan S, Wang K, Xiang Q, et al. lncRNA HOTAIR upregulates autophagy to promote apoptosis and senescence of nucleus pulposus cells. J Cell Physiol 2020;235:2195-208. 10.1002/jcp.29129 [DOI] [PubMed] [Google Scholar]
- 35.Bi Y.Y, Shen G, Quan Y, et al. Long noncoding RNA FAM83H-AS1 exerts an oncogenic role in glioma through epigenetically silencing CDKN1A (p21). J Cell Physiol 2018;233:8896-907. 10.1002/jcp.26813 [DOI] [PubMed] [Google Scholar]
- 36.Dou Q, Xu Y, Zhu Y, et al. LncRNA FAM83H-AS1 contributes to the radioresistance, proliferation, and metastasis in ovarian cancer through stabilizing HuR protein. Eur J Pharmacol 2019;852:134-41. 10.1016/j.ejphar.2019.03.002 [DOI] [PubMed] [Google Scholar]
- 37.Shan H, Yang Y, Zhu X, et al. FAM83H-AS1 is associated with clinical progression and modulates cell proliferation, migration, and invasion in bladder cancer. J Cell Biochem 2019;120:4687-93. 10.1002/jcb.27758 [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Cheng M, Mi L, et al. Mir-22-3p Enhances the Chemosensitivity of Gastrointestinal Stromal Tumor Cell Lines to Cisplatin through PTEN/PI3K/Akt Pathway. Iran J Allergy Asthma Immunol 2018;17:318-25. 10.18502/ijaai.v17i4.91 [DOI] [PubMed] [Google Scholar]
- 39.Jiang W, Han X, Wang J, et al. miR-22 enhances the radiosensitivity of small-cell lung cancer by targeting the WRNIP1. J Cell Biochem 2019;120:17650-61. 10.1002/jcb.29032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Sanchez C, Font-Ugalde P, Ruiz-Limon P, et al. Circulating microRNAs as potential biomarkers of disease activity and structural damage in ankylosing spondylitis patients. Human Molecular Genetics 2018;27:875-90. 10.1093/hmg/ddy008 [DOI] [PubMed] [Google Scholar]
- 41.Mäkitie RE, Hackl M, Niinimaki R, et al. Altered MicroRNA Profile in Osteoporosis Caused by Impaired WNT Signaling. J Clin Endocrinol Metab 2018;103:1985-96. 10.1210/jc.2017-02585 [DOI] [PubMed] [Google Scholar]
- 42.Wu C, Zheng Z, Ren W, et al. Mm9_circ_009056 enhances osteogenesis by targeting BMP7 via CGRP-mediated miR-22-3p. Biochem Biophys Res Commun 2018;501:199-205. 10.1016/j.bbrc.2018.04.215 [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Zou M, Li J, et al. LncRNA H19 targets miR-22 to modulate H2O2-induced deregulation in nucleus pulposus cell senescence, proliferation, and ECM synthesis through Wnt signaling. J Cell Biochem 2018;119:4990-5002. 10.1002/jcb.26738 [DOI] [PubMed] [Google Scholar]
- 44.Gordon S, Taylor P.R. Monocyte and macrophage heterogeneity. Nat Rev Immunol 2005;5:953-64. 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- 45.Mantovani A, Sica A, Sozzani S, et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004;25:677-86. 10.1016/j.it.2004.09.015 [DOI] [PubMed] [Google Scholar]
- 46.Liu G, Yang H. Modulation of macrophage activation and programming in immunity. J Cell Physiol 2013;228:502-12. 10.1002/jcp.24157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as