Abstract
Background
Oral squamous cell carcinoma (OSCC) is a highly heterogeneous neoplasm where the identification of heterogeneity is a critical clinical need to improve treatment planning and prognosis prediction. Utilizing the Hyperion imaging system to carry out high-dimensional proteomics analysis on the heterogeneity of tumor samples, this study aims to detect and analyze the heterogeneity of OSCC without lymph node metastasis and explore potential contributing factors for poor prognosis of early-stage OSCC.
Methods
We collected tumor tissue samples from four OSCC patients at the T1N0M0 stage, who presented with similar clinical manifestations. Patient formalin-fixed, paraffin-embedded (FFPE) tissue sections were prepared and stained using a panel of 26 immune or tumor-related antibodies. Different metal tags were assigned to each antibody. The stained cells were then detected and analyzed by the Hyperion imaging system.
Results
Tumor samples of four OSCC patients presenting with similar clinical characteristics at the T1N0M0 stage had different cell subtypes, including CD4+ T cells, CD8+ T cells, CD19+ B cells, CD11c+ dendritic cells, CD56+ natural killer cells, granulocytes, etc. More immunosuppressive cells were found in the sample of patient 1. We propose that differences in the tumor microenvironment of samples may contribute to different patients’ prognosis in the future.
Conclusions
High-dimensional proteomics analyses using the Hyperion imaging system help identify and analyze the tumor microenvironment heterogeneity of OSCC. Our study now presents this valuable resource and explains the potential reasons behind early OSCC patients’ poor prognosis.
Keywords: Oral squamous cell carcinoma (OSCC), hyperion imaging system, heterogeneity, tumor microenvironment, precision medicine
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common cancers of the head and neck (1). Its high rate of mortality and disfigurement leads to heavy global public health concern (2). Despite the rapid developments in surgical-radiological treatments and reconstruction techniques, therapies for OSCC have encountered a bottleneck period while the 5-year survival rate has not significantly improved in the past 20 years (3). One potential reason may be that although the breakthroughs in surgical techniques of OSCC can sufficiently remove the tumor and bring out the improvement of short-term prognosis, the existing tumor microenvironment still contributes to tumor recurrence and metastasis.
The tumor microenvironment (TME) is the environment in which tumor cells develop. It comprises the surrounding immune cells, fibroblasts, blood vessels, signaling molecules, and the extracellular matrix (4). Tumor cells and their surrounding microenvironment interact constantly, and many studies have demonstrated that the components of the tumor microenvironment, including CD8+ T cells (5,6), dendritic cells (7,8), NK cells (9), and macrophages (9), have a close correlation with tumor prognosis.
Currently, many clinical questions involving the prediction of OSCC patients’ prognosis based on the TNM staging system remain unclear. A common phenomenon is that many early-stage OSCC patients (T1N0M0 stage) have poor prognoses despite most having better ones. This indicates an inaccuracy of prognosis prediction for OSCC patients solely depending on TNM classification. As the TNM staging system is based on clinical and pathological characteristics, one possible explanation may be that the tumor microenvironment’s heterogeneity which has profound implications both for tumor development and therapeutic outcomes has been overlooked. Due to limitations in traditional research methodologies, it is relatively hard to analyze multiple tumor microenvironment components and evaluate heterogeneity. However, with the Hyperion imaging system, the world’s first commercially available metal-tagged antibody-directed imaging mass cytometry platform including a Hyperion tissue imager, a Hellos system, and software for data acquisition and viewing, we can now evaluate the tumor microenvironment (10).
This study investigated the heterogeneous tumor microenvironment from samples of four OSCC patients at the T1N0M0 stage who presented with similar clinical manifestations. We also proposed potential factors contributing to different prognosis for OSCC patients despite the same TNM stage. This study is the first of its kind to provide valuable information on the heterogeneity of OSCC tumors, suggesting a new area of focus for the investigation of conflicting prognoses for OSCC patients with the same TNM stage and similar clinical manifestations.
We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7194).
Methods
Sample selecting and clinical features
OSCC patients admitted between Nov 2018 and May 2019 were selected from the Department of Oral and Maxillofacial Surgery, Peking University Hospital of Stomatology. To decrease clinical heterogeneity of patients, we designed the following entry criteria: patients’ age (30–50 years old), gender (male), ethnicity (Asian), tumor location (tongue), tumor size (<2 cm), pathological diagnosis (SCC), tumor differentiation (well-differentiated) and without lymph node metastasis. None of the patients had undergone chemotherapy or radiotherapy before the operation. Formalin-Fixed, Paraffin-Embedded (FFPE) sections were obtained from the Department of Pathology, and the slice thickness was 5 µm. The current study’s procedures were approved by the Institutional Ethics Committee of Clinical Research (Peking University Hospital of Stomatology). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Because of the retrospective nature of the research, the requirement for informed consent was waived.
Panel and antibody conjugation
An antibody panel was designed to target epitopes for OSCC and markers for tumor immune cells and tumor-related antigens and distinguish between tumor cells, intercellular substances, and immune cell types (Table 1). Clone information is also available in Table 1. As outlined in Table 1, CD antibodies were used to visualize immune cells, structural proteins were used to visualize histological landmarks, and the Ki-67 and CK proteins were used to identify tumor cells. All antibodies were conjugated to metal tags according to the manufacturer’s instruction on Hyperion tissue imager.
Table 1. Antibodies targeting the tumor environment of oral squamous cell carcinoma (OSCC).
| Antibody | Label | Clone | Vendor | Catalog |
|---|---|---|---|---|
| CD45 | Er166 | 2D1 | Biolegend | 368502 |
| CD3 | Er170 | Polyclonal, C-Termina | Fluidigm | 3170019D |
| CD4 | Gd156 | EPR6855 | Fluidigm | 3156033D |
| Foxp3 | Gd155 | 206D | Biolegend | 320102 |
| CD8a | Dy162 | D8A8Y | Fluidigm | 3162035D |
| CD11b/Mac-1 | Sm149 | EPR1344 | Fluidigm | 3149028D |
| CD11c | Sm154 | Polyclonal | Fluidigm | 3154025D |
| CD14 | Nd144 | EPR3653 | Fluidigm | 3144025D |
| CD15 | Eu153 | HI98 | Biolegend | 301902 |
| CD16 | Nd146 | EPR16784 | Fluidigm | 3146020D |
| CD19 | Nd142 | 6OMP31 | Fluidigm | 3142014D |
| CD33 | Nd145 | Polyclonal | Fluidigm | 3145017D |
| CD56 | Sm152 | RNL-1 | Abcam | 9018 |
| CD68 | Tb159 | KP1 | Biolegend | 916104 |
| IFN-γ | Sm147 | Polyclonal | Abcam | 9657 |
| HLA-DR | Yb174 | YE2/36 HLK | Fluidigm | 3174023D |
| CD274/PD-L1 | Nd150 | E1L3N | Fluidigm | 3150031D |
| CD273/PD-L2 | Yb172 | 176611 | Fluidigm | 3172028D |
| CD279/PD-1 | Ho165 | EPR4877(2) | Fluidigm | 3165039D |
| CK AE1/AE3 | Dy164 | AE-1/AE-3 | Biolegend | 914204 |
| α-SMA | Pr141 | 1A4 | ebioscience | 14-9760-82 |
| collagen I | Tm169 | 3G3 | Abcam | 88147 |
| CD324/E-Cadherin | Gd158 | 4A2 | Abcam | 231303 |
| Vimentin | Nd148 | RV202 | Abcam | 8978 |
| Ki-67 | Er168 | Ki-67 | Biolegend | 350502 |
| b-catenin | Lu175 | 12F7 | Biolegend | 844603 |
Hyperion tissue imager scan
Images were acquired using a Hyperion imaging system (Fluidigm) (11,12). The largest square area was laser-ablated in a raster pattern at 200 Hz, and the preprocessing of the raw data was completed using a commercial acquisition software (MCD Viewer software, Fluidigm). The MCD Viewer was then used to confirm experimental success and produce high-quality images with selected combinations of markers. All export files were processed for subsequent further analysis.
Data processing and analysis
The original MCD file was converted into a TIFF format by imctools (https://github.com/BodenmillerGroup/imctools). Data were segmented into single cells using Cellprofiler (version 3.1.8) (13). In brief, Fluidigm DNA markers were designed to identify nuclei, while CD3, CD8, CD19, CD56, pan-CK, α-SMA, and other markers could be used to identify cell membranes. The data were then combined and analyzed by Cellprofier to identify single-cell object masks. The masks (TIFF file) containing cell location and boundary were obtained. The single-cell data were censored at the 95th percentile to remove outliers, and z-scored cluster means were visualized as heat maps. For t-SNE and PhenoGraph, the data was normalized by Harmony (14). Clustering Single cells of the large cohort were clustered into groups by their phenotypical similarity using PhenoGraph; unsupervised clustering and aggregation of these clusters were organized into larger groups based on their markers (15). Barnes-Hut t-SNE For visualization, high-dimensional single-cell data were reduced to two dimensions using the nonlinear dimensionality reduction algorithm t-SNE (16,17). We applied the Barnes-Hut implementation of t-SNE to Harmony-normalized data with default parameters (perplexity =42).
Results
Preprocessing of OSCC samples and data acquisition
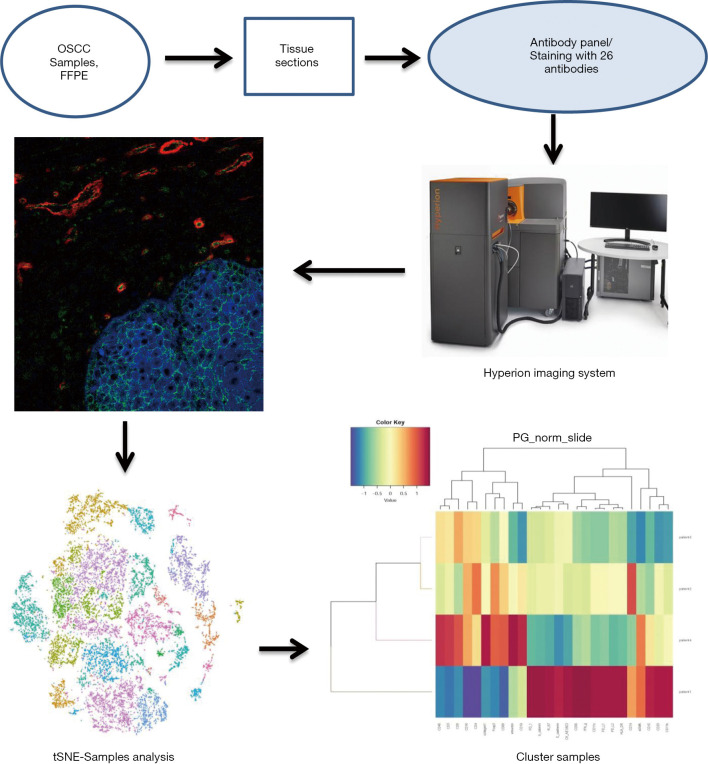
All OSCC patients at the Department of Oral and Maxillofacial Surgery, Peking University Hospital of Stomatology were screened for the study. Six patients met our clinical inclusion criteria; however, one patient was excluded due to unsatisfied tissue sections and another due to being lost in the follow-up time. As a result, four patients with OSCC were included in the study, and their clinical features are shown in Table 2. All patients were young Asian males diagnosed with SCC of the tongue at the T1N0M0 stage, who presented with similar clinical characteristics. A standard procedure was used to obtain FFPE slices from the four included patients. All samples were labeled with a panel of 26 antibodies, and the labeling antibodies were identified and analyzed by the Hyperion imaging system. Twenty-six protein epitope dimensions were used to detect the tumor microenvironment. The workflow is shown in Figure 1.
Table 2. Clinicopathological characteristics of the 4 patients included.
| Study No. | Demographic information | Tumor site | Tumor size | TNM stage | Pathological features | Follow-up (Months) |
|---|---|---|---|---|---|---|
| Patient 1 | Male, 35 years, Asian | tongue | Ulceration, a diameter of 1.0 cm | T1N0M0 | Well-differentiated SCC, no lymph node metastasis, 0.5 cm depth of invasion | No recurrence, 16 months |
| Patient 2 | Male, 37 years, Asian | tongue | Ulceration, a diameter of 1.8 cm | T1N0M0 | Well-moderately differentiated SCC, no lymph node metastasis, 0.5 cm depth of invasion | No recurrence, 19 months |
| Patient 3 | Male, 47 years, Asian | tongue | Ulceration, a diameter of 1.3 cm | T1N0M0 | Well-moderately differentiated SCC, no lymph node metastasis, 0.6 cm depth of invasion | No recurrence, 14 months |
| Patient 4 | Male, 34 years, Asian | tongue | Ulceration, a diameter of 1.8 cm | T1N0M0 | Well-moderately invasion depth SCC, no lymph node metastasis, 0.5 cm depth of invasion | No recurrence, 15 months |
SCC, squamous cell carcinoma.
Figure 1.
Flow chart of sample processing and analytical methods for Hyperion imaging system.
Analysis of immune microenvironment of OSCC by hyperion imaging system
In a tumor microenvironment, tumor cells and immune cells are seen to have mutual interactions. The tumor-infiltrating immune cells directly influence treatments and prognoses of OSCC. Therefore, understanding the difference between immune cell subsets among OSCC patients would provide critical information for scientific progress in immunotherapy and prognosis.
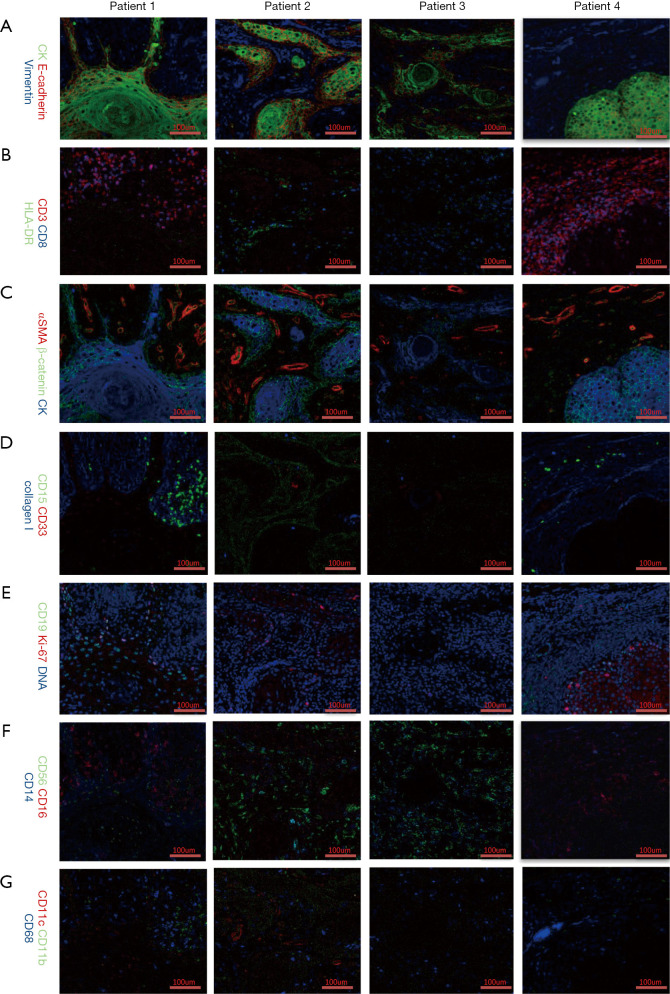
Different markers were observed with the Hyperion imaging system (Figure 2). From the panorama, we found that the composition of the tumor microenvironment and the distribution and expression of different markers were different among patients with the same TNM stage (T1N0M0) and similar clinical features intrinsic heterogeneity of the tumor microenvironment. Further analysis of tumor-related antigens’ markers showed that CK, which was observed in samples of all patients, developed pseudopodia in that of patient one, suggesting movement and escape of tumor cells (Figure 2A,C).
Figure 2.
Hyperion images of immune and tumor-related markers identified on 1 mm × 1 mm formalin-fixed, paraffin-embedded oral squamous cell carcinoma (OSCC) tissue slides.
Some tumor immune cell markers also had a significant numerical advantage in patients. In Figure 2B, CD3 and CD8 populations in patient four were found to outnumber those in other patients, implying that patient four had a better tumor immune microenvironment. Besides, HLA-DR and CD11c were mainly observed in the sample of patient two, while being close to completely absent in those of other patients (Figure 2B,G). Interestingly, CD15 and CD19 were found predominantly in patient one, but absent in other patients (Figure 2D,E), suggesting the existence of more immunosuppressive cells in patient one, including CD19+ B cells, CD15+ granulocytes and neutrophils cells. Notably, the fact that CD56 cells were highly expressed in patient two and patient three hinted a numerical advantage of CD56+ NK cells in patient two and patient three (Figure 2F). Altogether, this data reveals significant heterogeneity, especially with regards to the subsets of CD8+ T cells, CD56+ NK cells, CD19+ B cells, CD15+ granulocytes, and CD11c+ dendritic cells, in the immune microenvironment in different OSCC patients. A thorough grouping and classification of OSCC patients based on such information might help predict the prognosis of OSCC and implement precisely-targeted therapies.
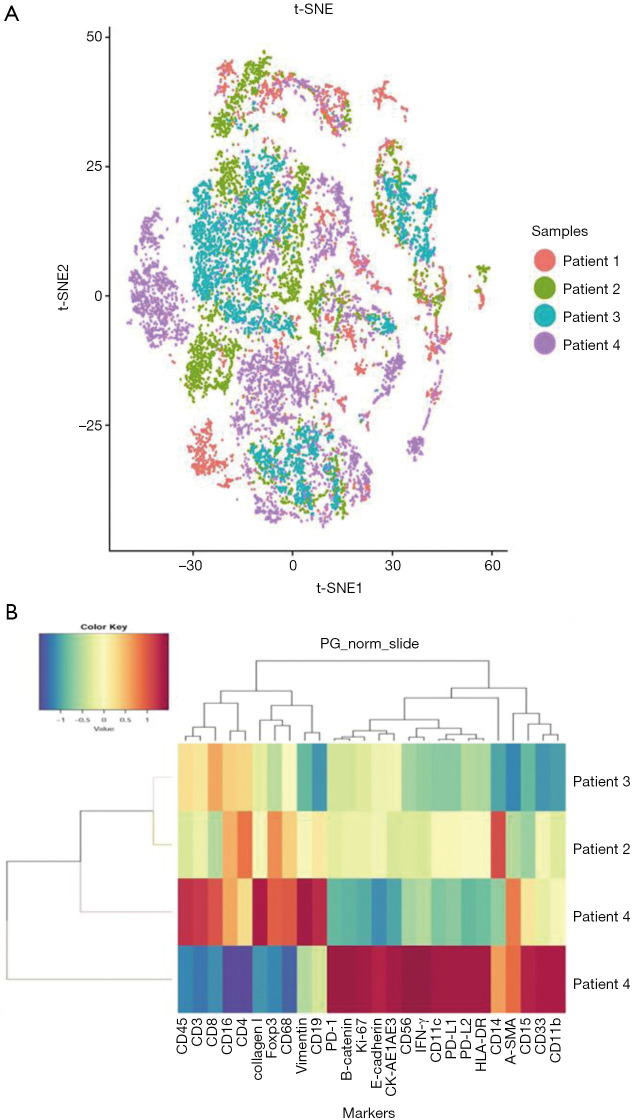
Immune cell subsets and tumor-related antigens in different patients were identified and characterized by tSNE analysis to visualize phenotypic diversity of microenvironment in patients with OSCC. As both overlapping and independently expressed cell subsets were found in samples of the four patients, there was an indication of heterogeneity in their tumor microenvironments (Figure 3A). Based on such differences in microenvironments, cluster analyses of samples were carried out. Patients two, three, and four were seen to have similar tumor microenvironments, but patient one showed significant variations (Figure 3B). As seen in Figure 3B, molecular markers, including CD15, CD33, CD11b, CD14, PD-1, PD-L1, PD-L2, Ki-67, and β-catenin, were more strongly expressed in patient one comparing to other patients. This data indicated a distinctive tumor microenvironment for patient one, which was seen to be more immunosuppressive.
Figure 3.
Cluster analysis of oral squamous cell carcinoma (OSCC) samples. (A) t-SNE descending dimension map of the 4 samples of OSCC. The colors represent different samples; (B) differential expression of tumor microenvironment markers and cluster analysis of the 4 samples.
Discussion
One of the most pivotal reasons for unsuccessful OSCC treatment is a standardized treatment strategy that does not consider the heterogeneous nature of the tumor microenvironment. As a result, some OSCC patients tend to have a less ideal response to uniform treatments and hence worse prognoses than others. While previous literature have demonstrated substantial influences of tumor microenvironment on the prognosis of cancer (18,19), further studies on the heterogeneous features will help improve the understanding and accuracy of prognosis determination for OSCC patients.
Hyperion imaging system is an ideal analytical tool for multi-parametric detection and analyses of heterogeneity in the tumor microenvironment as it allows simultaneous detection of dozens of markers in the same tissue section. It has been successfully applied in numerous areas, showing the composition of many tumor microenvironments (12,20). However, to date, there was no literature on using the Hyperion imaging system to explore the role of intratumoral heterogeneity in OSCC. Therefore, our study aimed to fill in the gap by detecting and analyzing heterogeneity of the tumor microenvironment of OSCC, thereby improving the precision of tumor prognosis using the system.
Our study presented an atlas of immune microenvironment based on a panel of 26 antibodies, as we also identified several major immune cell phenotypes, including CD8+ T cells, CD56+ NK cells, CD11c+ dendritic cells, CD15+ granulocytes, CD19+ B cells, and CK+ tumor cells subsets. Previous studies have demonstrated that cytotoxic CD8+ T cells inhibited tumor growth by targeting antigenic tumor cells (6). Our study found a high expression of CD8+T cells in patient four, suggesting a potentially better prognosis for the patient. Natural killer cells (NK cells) are critical immune cells in the body, yet in published studies, conclusions about their role in oral squamous cell carcinoma had not reached a consensus. Indeed, most authors thought that NK cells’ presence was closely related to the excellent prognosis of OSCC. For example, Agarwal reported that NK cells were significant markers for the good prognosis of OSCC (21). Ikeda further suggested that even a small number of NK cells were still conducive to determining advanced OSCC (22). According to Dovak’s research, radiotherapy could cause a continuous decrease of T cells and B cells in OSCC patients, but the quick recovery of NK cells played a crucial role in the immune maintenance of OSCC patients (23). In our study, the high expression of CD56 in patient two and patient three suggested better prognoses for both patients.
Dendritic cells (DC) were discovered by Canadian scientist Ralph m. Steinman, in 1973. It is the most professional antigen-presenting cells (APC) with the strongest function. Previous studies have reported that high expression of dendritic cells is an important marker for a good prognosis of OSCC (24, 25), while cost suggested that the large population of dendritic cells in the tissue of lower lip might contribute to an excellent prognosis for labial cancer (26). As we found a high expression of dendritic cells in patient two, it would not be surprising that he may have a good prognosis.
Alternatively, previous studies have reported that B cells, granulocytes and neutrophils were related to poor prognosis (27-29). As their immunosuppressive cells were more highly expressed in patient one, this patient might have a worse prognosis than other patients.
Based on its data, we could speculate that although all patients were presenting with similar characteristics at the same stage of T1N0M0, patient one might have a different prognosis due to more immunosuppressive cells in his tumor microenvironment.
In this study, we also performed bioinformatics analysis to predict the prognosis of OSCC, which might provide useful information for developing individualized immunotherapy. The results suggested variable expression of immune cell populations might explain differences in prognoses for OSCC patients despite their same TNM stage and similar clinical features. Furthermore, basic information for screening drugs for targeted therapy was provided, thus greatly improving OSCC treatments’ individualization.
As this is the first study to analyze the heterogeneity of the microenvironment of OSCC with the use of the Hyperion imaging system, more investigations are required to address the current limitations. While most of the current data were descriptive in small sample sizes, some markers or subsets’ expression tendency would require further validation. Secondly, the presented immune cell phenotypes were based on computational bioinformatics analysis and have not been verified by basic experiments. The cost to use the Hyperion imaging system was also relatively expensive for practical use for individual patients. For these reasons, future studies would be required to identify these phenotypes’ roles and increase the sample size with a low-cost approach.
Conclusions
A high-dimensional Hyperion imaging system contributes to the analysis of OSCC heterogeneity. Additionally, it can be used to help predict the prognosis of OSCC. Our study provides a valuable method for analyzing tumor heterogeneity and explains potential factors leading to different prognoses for OSCC patients despite the same TNM stage and similar clinical characteristics.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This research was funded by the Youth Program of National Natural Science Foundation of China (Grant No. 82002878) and Peking University Hospital of Stomatology Young Foundation (Grant No. YS020219).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The current study’s procedures were approved by the Institutional Ethics Committee of Clinical Research (Peking University Hospital of Stomatology). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Because of the retrospective nature of the research, the requirement for informed consent was waived.
Reporting Checklist: The authors have completed the Materials Design Analysis Reporting (MDAR) reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7194
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7194
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7194). The authors have no conflicts of interest to declare.
(English Language Editor: C. Betlazar-Maseh)
References
- 1.Marur S, Forastiere AA. Head and Neck Squamous Cell Carcinoma: Update on Epidemiology, Diagnosis, and Treatment. Mayo Clin Proc 2016;91:386-96. 10.1016/j.mayocp.2015.12.017 [DOI] [PubMed] [Google Scholar]
- 2.Raman P, Gayathri PS. Dentist's Delay or Dexterity to Diagnose the Deadly: A Clinico-radiological Series of Oral Malignancies Exhibiting Varied Presentations in the Tamil Nadu Rural Belt. Cureus 2019;11:e4051. 10.7759/cureus.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Dijk BA, Brands MT, Geurts SM, et al. Trends in oral cavity cancer incidence, mortality, survival and treatment in the Netherlands. Int J Cancer 2016;139:574-83. 10.1002/ijc.30107 [DOI] [PubMed] [Google Scholar]
- 4.Li H, Wang Y, Zhang M, et al. The high expression of TNF-α and NF-κB in tumor microenvironment predicts good prognosis of patients with BCLC-0-B hepatocellular carcinoma. Transl Cancer Res 2019;8:532-41. 10.21037/tcr.2019.03.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostroumov D, Fekete-Drimusz N, Saborowski M, et al. CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell Mol Life Sci 2018;75:689-713. 10.1007/s00018-017-2686-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amicone L, Marchetti A. Microenvironment and tumor cells: two targets for new molecular therapies of hepatocellular carcinoma. Transl Gastroenterol Hepatol 2018;3:24. 10.21037/tgh.2018.04.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung NC, Lee JH, Chung KH, et al. Dendritic Cell-Based Immunotherapy for Solid Tumors. Transl Oncol 2018;11:686-90. 10.1016/j.tranon.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabado RL, Balan S, Bhardwaj N. Dendritic cell-based immunotherapy. Cell Res 2017;27:74-95. 10.1038/cr.2016.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadler-Olsen E, Wirsing AM. Tissue-infiltrating immune cells as prognostic markers in oral squamous cell carcinoma: a systematic review and meta-analysis. Br J Cancer 2019;120:714-27. 10.1038/s41416-019-0409-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson HW, Fischer JR, Zanotelli VRT, et al. The single-cell pathology landscape of breast cancer. Nature 2020;578:615-20. 10.1038/s41586-019-1876-x [DOI] [PubMed] [Google Scholar]
- 11.Chang Q, Ornatsky OI, Siddiqui I, et al. Imaging Mass Cytometry. Cytometry A 2017;91:160-9. 10.1002/cyto.a.23053 [DOI] [PubMed] [Google Scholar]
- 12.Allo B, Lou X, Bouzekri A, et al. Clickable and High-Sensitivity Metal-Containing Tags for Mass Cytometry. Bioconjug Chem 2018;29:2028-38. 10.1021/acs.bioconjchem.8b00239 [DOI] [PubMed] [Google Scholar]
- 13.Carpenter AE, Jones TR, Lamprecht MR, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006;7:R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289-96. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine JH, Simonds EF, Bendall SC, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 2015;162:184-97. 10.1016/j.cell.2015.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Maaten L, Hinton G. Visualizing Data using t-SNE. Journal of Machine Learning Research 2008;9:2579-605. [Google Scholar]
- 17.van der Maaten L. Barnes-Hut-SNE. Available online: https://www.researchgate.net/publication/234119714_Barnes-hut-sne
- 18.Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-830.e14. 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson-Tessi M, Gillies RJ, Gatenby RA, et al. Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res 2015;75:1567-79. 10.1158/0008-5472.CAN-14-1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerdtsson E, Pore M, Thiele JA, et al. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg Sci Phys Oncol 2018;4:015002. 10.1088/2057-1739/aaa013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal R, Chaudhary M, Bohra S, et al. Evaluation of natural killer cell (CD57) as a prognostic marker in oral squamous cell carcinoma: An immunohistochemistry study. J Oral Maxillofac Pathol 2016;20:173-7. 10.4103/0973-029X.185933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda T, Seki S, Fujiwara M, et al. Low-risk population among patients with tumor-node-metastasis stage III/IV oral squamous cell carcinoma. Oncol Lett 2017;14:3711-6. 10.3892/ol.2017.6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dovšak T, Ihan A, Didanovič V, et al. Effect of surgery and radiotherapy on complete blood count, lymphocyte subsets and inflammatory response in patients with advanced oral cancer. BMC Cancer 2018;18:235. 10.1186/s12885-018-4136-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jardim JF, Gondak R, Galvis MM, et al. A decreased peritumoral CD1a+ cell number predicts a worse prognosis in oral squamous cell carcinoma. Histopathology 2018;72:905-13. 10.1111/his.13415 [DOI] [PubMed] [Google Scholar]
- 25.Kikuchi K, Kusama K, Taguchi K, et al. Dendritic cells in human squamous cell carcinoma of the oral cavity. Anticancer Res 2002;22:545-57. [PubMed] [Google Scholar]
- 26.Costa NL, Gonçalves AS, Martins AF, et al. Characterization of dendritic cells in lip and oral cavity squamous cell carcinoma. J Oral Pathol Med 2016;45:418-24. 10.1111/jop.12380 [DOI] [PubMed] [Google Scholar]
- 27.Najmeh S, Cools-Lartigue J, Rayes RF, et al. Neutrophil extracellular traps sequester circulating tumor cells via β1-integrin mediated interactions. Int J Cancer 2017;140:2321-30. 10.1002/ijc.30635 [DOI] [PubMed] [Google Scholar]
- 28.Peltanova B, Raudenska M, Masarik M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: a systematic review. Mol Cancer 2019;18:63. 10.1186/s12943-019-0983-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cools-Lartigue J, Spicer J, McDonald B, et al. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J Clin Invest 2013;123:3446-58. 10.1172/JCI67484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as