Abstract
Attachment of microorganisms to natural or artificial surfaces and the development of biofilms are complex processes which can be influenced by several factors. Nevertheless, our knowledge on biofilm formation in karstic environment is quite incomplete. The present study aimed to examine biofilm development for a year under controlled conditions in quasi-stagnant water of a hydrothermal spring cave located in the Buda Thermal Karst System (Hungary). Using a model system, we investigated how the structure of the biofilm is formed from the water and also how the growth rate of biofilm development takes place in this environment. Besides scanning electron microscopy, next-generation DNA sequencing was used to reveal the characteristic taxa and major shifts in the composition of the bacterial communities. Dynamic temporal changes were observed in the structure of bacterial communities. Bacterial richness and diversity increased during the biofilm formation, and 9–12 weeks were needed for the maturation. Increasing EPS production was also observed from the 9–12 weeks. The biofilm was different from the water that filled the cave pool, in terms of the taxonomic composition and metabolic potential of microorganisms. In these karstic environments, the formation of mature biofilm appears to take place relatively quickly, in a few months.
Subject terms: Biofilms, Microbial communities, Microbial ecology, Ecological modelling
Introduction
Thermal karst environments provide diverse habitats for microorganisms having planktonic or biofilm forming lifestyles. Attachments of microorganisms to natural or artificial surfaces (e.g., rocks, glass slides) and the development of biofilms are rather complex processes which can be influenced by several factors1. Nevertheless, our understanding of these systems is incomplete to date, only a few studies have focused on the bacterial metagenome of cave speleothems2–4. Nitrogen-based primary production strategy was detected based on the metabolic profiling of the speleothems of the Weebubbie Cave (Australia) and Kartchner Caverns (Arizona, USA)2,4. However, metagenomic study of highly acidic (pH 0–1) snottites showed that Acidithiobacillus species, which made up more than 70% of the community, were involved in the primary production in the Frasassi cave (Italy). It has also been confirmed that these organisms use the Sox system for the oxidation of reduced sulfur compounds4.
In the last decade, several microbiological studies have been conducted in the Buda Thermal Karst System (BTKS) which is one of the few hydrogeological systems in the world where springs, caves and the effects of hydrothermal fluids on carbonate rocks can be recently examined. Studying the prokaryotes inhabiting the BTKS have been brought new insight into the microbially influenced hypogenic speleogenesis5–8. All these studies were based on samples from spring caves. The presence of complex meso- and thermophilic biofilm microbial communities based on the activity of a special and interconnected autotrophic sulfur and nitrogen prokaryotic metabolic network can be assumed in the studied systems5–8. However, the phases of biofilm formation, the potential influencing factors and the duration of biofilm maturation is not yet clarified.
Therefore, the aim of this study was to monitor the biofilm development in the quasi-stagnant thermal water of Rudas-Török (RT) spring cave (a part of the BTKS) using an in situ experimental model system. High-throughput next-generation DNA sequencing was used to reveal the characteristic taxa and major shifts in the composition of the bacterial communities, and electron microscopy was applied to observe the morphological changes in the structure of biofilms. As a result, the succession of biofilm formation was revealed under controlled conditions. Site-based long-term monitoring of the cave biofilms, the planktonic bacterial community and the physical and chemical parameters of the pool water provide new information on biofilm formation in thermal karst systems.
Materials and methods
Description of the sampling sites
The RT spring cave is located in the southern part of the BTKS close to the Danube (Fig. 1A) and offers an ideal opportunity to conduct an in situ model experiment. Here, the discharged thermal fluids mainly consist of upwelling thermal waters mainly meteoric origin karst water with less, additional saline basinal fluids9. The chemical parameters of the thermal spring are stable and influenced only by the water level of the Danube10. The water level in the spring cave is controlled also by an overflowing system and quasi-stagnant.
Figure 1.
Geographic location of the sampling site (A). The arrangement of the in situ experimental model system in the RT spring cave (B,C).
In order to explore the formation of microbial biofilm under natural conditions, an in situ model system was set up in the RT spring cave (Fig. 1B). The most appropriate place for our experiment was the cave pool (size: 3 m long × 2 m wide). It is an easy-to-access, undisturbed environment and closed from the public visitors. A wooden slat was placed over the approximately 0.5 m deep pool about 30 cm and surface disinfected polypropylene test tube racks containing sterile glass slides were fixed on it using fishing line (Fig. 1C). For the biofilm formation, glass slides were immersed into the water-filled cave pool 10 cm below the water surface. Glass slide samples were taken aseptically every three-weeks during the 30 weeks of sampling period, in three replicates. A pool water sample from the Rudas–Török spring cave (RTW) was also taken at the beginning of the experiment. The RTW sample was collected 10 cm below the water surface from the immediate vicinity of the glass slides without agitation the pool water before sampling. An additional biofilm sample was taken one year after the start of the experiment with three replicates The glass slides collected during the sampling were separately immersed into bottles containing sterile water and transported to the laboratory under refrigerated conditions. Samples were stored at 6–8 °C until being processed in the laboratory within 4 h.
Determination of physical and chemical parameters of the water
During the in situ experiment, the pH, temperature, specific electrical conductivity and dissolved oxygen concentration of the pool water were measured using a WTW MultiLine P 8211 multi-parameter portable meter next to the test tube racks containing the glass slides. In addition, specific electrical conductivity, temperature and water level was measured continuously by a DATAQUA datalogger and was compared with the actual level of the Danube river. All other parameters were measured in the laboratory as previously described6,7.
Scanning electron microscopy
To study the biofilms developed on the glass slides, approx. 1 × 1 cm2 quadrants were cut for scanning electron microscopy (SEM) with a glass cutter and placed in glutaraldehyde (5% in 0.1 M phosphate buffer) for 3–4 h at room temperature. All subsequent steps were the same as previously described in Anda et al.7. The samples were examined using a Zeiss EVO MA 10 scanning electron microscope at an accelerating voltage of 10 kV.
Next-generation amplicon sequencing
Approximately 1000 ml of the water sample was filtered through a 0.22 μm pore diameter polycarbonate filter (Millipore). The biofilm samples were removed from the glass slides with sterile razor blades into sterile water and compacted by centrifugation (15,000×g for 2 min, centrifuge: Z33MK-2 (Hermle, Germany)). Total genomic DNA was extracted from the samples using Ultra Clean Soil DNA Isolation Kit (MO BIO Laboratories Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions with the exception that cell disruption step was carried out by shaking at 30 Hz for 2 min in a Mixer Mill MM301 (Retsch, Haan, Germany).
The PCR amplification and sequencing of the 16S rRNA gene, the subsequent bioinformatic analysis and taxonomic assignments were performed as described in detail by Szabó et al.11, with the exception of taxonomic assignment which was made using the ARB-SILVA SSU 138 database as reference12. Analysis scripts are also available as Supplementary Material of the aforementioned paper. To obtain the same number of sequences per sample for beta diversity evaluations and to compare alpha diversity, subsampling sample reads to the smallest data set (n = 2954), calculation of richness (sobs—number of OTUs at a 97% similarity threshold, Chao1—Chao 1 index and ACE—Abundance-based Coverage Estimator) and diversity estimators (Simpson’s inverse and Shannon indices) using 1000 iterations were executed using the mothur software13.Non-metric multidimensional scaling (NMDS) ordination was carried out in R14 using the ‘vegan’ package15, SIMPER test was performed using the Past3 program16. Raw sequence data can be accessed under the NCBI BioProject ID PRJNA483930.
Results
Physical–chemical parameters of the pool water in the spring cave
During the one year monitoring period, the temperature of the water in the Rudas-Török spring cave ranged from 34.3 to 38.2 °C (Suppl. Fig. S1). Continuous decline in temperature was observed from late September 2015 to late January and an increasing trend was detected from the end of January until September 2016.
The specific electrical conductivity was 1715.1 ± 7.2 µS cm−1. The water level in the spring cave was an average 0.5 ± 0.01 m, while the water level of River Danube was 231.6 ± 1.08 m a.s.l. (Suppl. Fig. S1).
The accuracy for the water level is ± 0.1%, for the temperature is 0.1 °C and for the specific electrical conductivity is 1%.
The pH and dissolved oxygen content of the cave water is shown in Suppl. Fig. S2. The pH values were between 6.0 and 6.6 during the measurement period, while the dissolved oxygen content ranged between 0.2 and 1.2 mg L−1.
Relatively high sulfate (309 mg L−1) concentration was measured in the RT spring cave’s water taken at the beginning of the experiment. The total organic carbon content and the total nitrogen concentration of the thermal water was 6.2 mg L−1 and 0.9 mg L−1. Concentrations of NH4+-N, NO3–N, and NO2–N were 0.2 mg L−1, < 1 mg L−1, and 0.2 mg L−1, respectively. These results were also related to the values previously reported by Borsodi et al.6 and Enyedi et al.17.
Scanning electron microscopic observations of biofilm structure
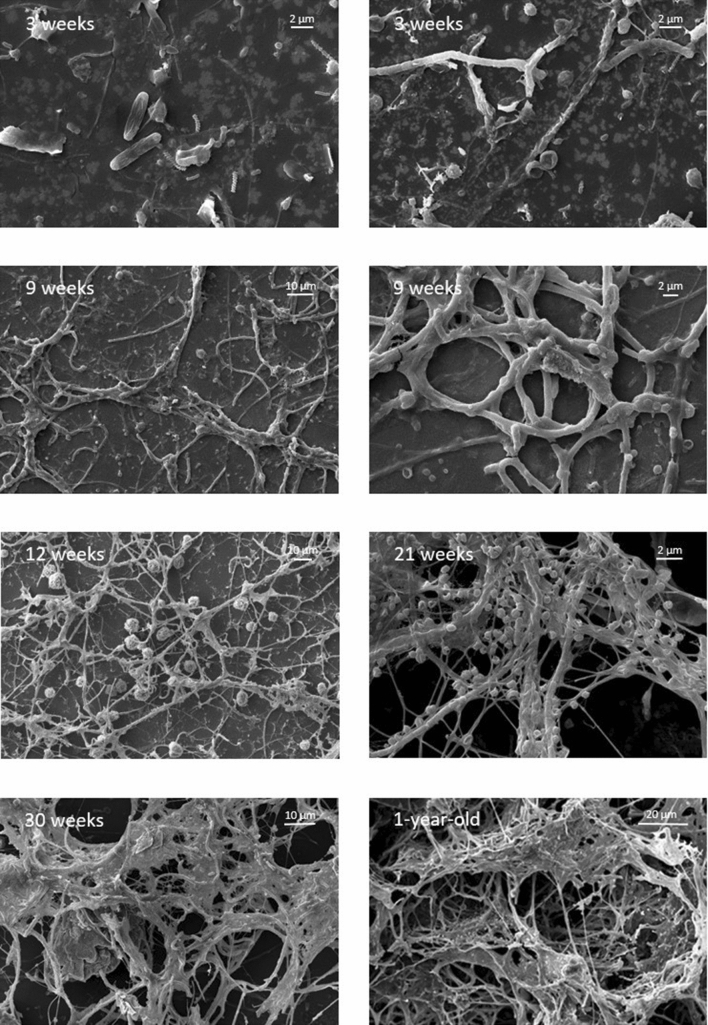
Only few, filamentous bacteria with different lengths, cocci, rod- and spiral-shaped bacteria, typical of Nitrospira were observed on the high-resolution SEM images of the three-week biofilm sample (Fig. 2). An increasing number of cells with predominantly filamentous morphology were detected in the case of the six and nine-week biofilm samples (Fig. 2). In addition to filamentous bacteria of various thickness, cell aggregate-forming rods and cocci of different size embedded in (most probably) calcium carbonate minerals accumulating exopolysaccharides were also observed. These/such minerals were also well visible on the SEM image of twelve-week biofilm samples (Fig. 2). The cells also occurred in embedded mucous material. This mucus, which appeared on the six-week biofilm sample for the first time, was the extracellular polymer matrix (EPS) secreted by the biofilm forming bacteria. The multilayer network architecture of microbial biofilm formed mainly by long, filamentous bacteria can be seen on the twenty-one and twenty-four-week biofilm images (Fig. 2).
Figure 2.
SEM micrographs of biofilm samples developing on the glass slides during a one year period (3 weeks (A,B), 9 weeks (C,D), 12 weeks (E), 21 weeks (F), 30 weeks (G), 1-year-old biofilm (H)) in the RT thermal spring. Scale bar (A,B,D,F) 2 µm, (C,E,G) 10 µm, (H) 20 µm.
Detected cells exhibited predominantly filamentous morphology with different thickness and appearance and minerals (e.g., rhomboidal calcite) were embedded in large amounts of mucous material on the SEM images of twenty-seven, thirty-week and one-year biofilm samples (Fig. 2).
Bacterial taxonomic composition during biofilm maturation
Pyrosequencing approach based on the variability of the V3-V4 region of the 16S rRNA gene was used to reveal the characteristic taxa and major shifts in the composition of the developing biofilm bacterial communities. A total of 88,000 high-quality bacterial 16S rRNA gene sequences were obtained from the 12 samples (Table 1).
Table 1.
Bacterial species richness (ACE and Chao1) and diversity indices (inverse Simpsons and Shannon–Wiener) calculated from NGS data.
| Sample ID | No. of sequencesa | Sobs | Chao1 | ACE | Shannon | Inverse Simpson |
|---|---|---|---|---|---|---|
| RTW | 2954 (6536) | 46 ± 2.5 | 59 ± 8.2 | 65 ± 10.7 | 0.4 ± 0.02 | 1.1 ± 0.01 |
| 3 weeks | 2954 (2954) | 143 | 162 | 173 | 3.5 | 17.1 |
| 6 weeks | 2954 (9010) | 176 ± 5.3 | 240 ± 21.8 | 266 ± 37.9 | 3.3 ± 0.03 | 10.5 ± 0.3 |
| 9 weeks | 2954 (5986) | 198 ± 4.9 | 260 ± 20.7 | 273 ± 27.9 | 3.8 ± 0.02 | 17.4 ± 0.5 |
| 12 weeks | 2954 (7291) | 172 ± 5.6 | 243 ± 24.4 | 265 ± 39.9 | 3.6 ± 0.02 | 17.4 ± 0.4 |
| 15 weeks | 2954 (13,152) | 190 ± 6.2 | 269 ± 24.9 | 287 ± 40.2 | 3.6 ± 0.03 | 14.1 ± 0.5 |
| 18 weeks | 2954 (8225) | 193 ± 5.7 | 273 ± 24.9 | 320 ± 49.7 | 3.7 ± 0.02 | 17.8 ± 0.5 |
| 21 weeks | 2954 (8555) | 199 ± 6.3 | 288 ± 28.4 | 360 ± 52.5 | 3.7 ± 0.03 | 14.6 ± 0.5 |
| 24 weeks | 2954 (5102) | 204 ± 4.6 | 263 ± 18.4 | 302 ± 37.8 | 3.8 ± 0.02 | 18.3 ± 0.5 |
| 27 weeks | 2954 (4800) | 202 ± 4.5 | 253 ± 15.7 | 266 ± 14.6 | 3.8 ± 0.02 | 18.9 ± 0.4 |
| 30 weeks | 2954 (8352) | 216 ± 6.5 | 311 ± 28.3 | 351 ± 51.2 | 4.0 ± 0.02 | 25.6 ± 0.7 |
| 1 year | 2954 (3431) | 187 ± 2.2 | 213 ± 6.9 | 222 ± 6.1 | 3.9 ± 0.01 | 23.7 ± 0.3 |
aNumbers in parentheses stand for the total number of sequences obtained with NGS; for calculating richness estimators and diversity indices, read numbers were subsampled to the read number of the sample having the lowest sequence count.
The values of the species estimation indices such as the number of OTUs, Chao1 and ACE (frequency-based coverage estimation index) were the lowest in the water sample, and in the third-, sixth- and twelfth-week biofilm samples (Table 1). The highest number of OTUs was observed in the 30th week biofilm sample. Both Shannon and inverse Simpson diversity indices were the lowest in the water sample, while their values showed more or less increasing tendency during the maturation of the biofilm. The three-week biofilm was made by 143 OTUs, the 30-week biofilm by 214, and the one-year biofilm by 187 OTUs. In contrast, 47 OTUs were found in the water sample. Only 8 OTUs were shared between the 3 and 30 week, one-year-old biofilm and thermal water samples, and an additional 20 OTUs were shared among the biofilm samples (Suppl Fig. S3). Many OTUs were specific to only one sample, only to the developing or only to the mature biofilm.
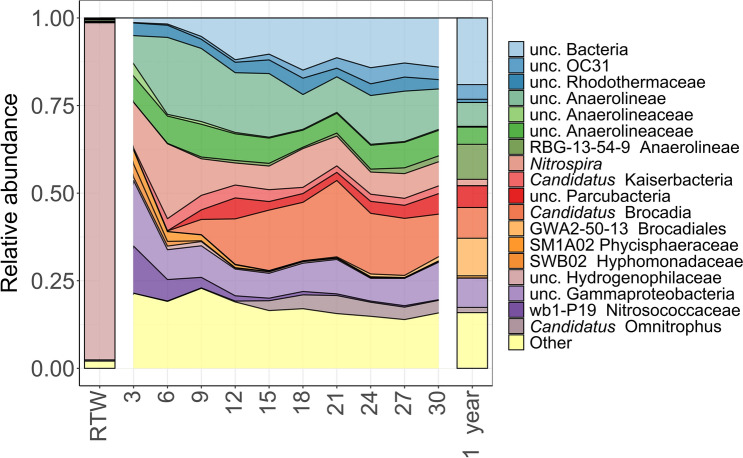
The reads were classified into altogether 40 different bacterial phyla in the water and biofilm samples. The taxonomic diversity at phylum level reached the maximum in the third week of the biofilm formation. Members of the phyla Chloroflexi, Patescibacteria, Planctomycetota, Proteobacteria and Nitrospirota, Bacteroidota and an unclassified group of Bacteria proved to be most abundant in the biofilm samples, but their relative abundance changed at different rates during the one year period. Some genus level bacterial taxa (e.g., unclassified Anaerolineaceae) were abundant throughout the whole studied period, while the abundance of others (e.g., unclassified Parcubacteria, Candidatus Brocadia, unclassified Bacteria) fluctuated (Fig. 3).
Figure 3.
Percentile distribution of amplicon sequences on genus level revealed from the RTW sample and changes in the bacterial composition of biofilm communities in the one year period based on the 16S rRNA gene amplicon sequence data. Genera having < 2% relative abundance are combined in the “Other” category.
The phylum Chloroflexi dominated the biofilms during the whole studied period with an average of 23.9% relative abundance. The relative abundance of an unclassified Anaerolineaceae (Chloroflexi) increased from 8.1 to 21.9%. Nitrospira (Nitrospirota) proved to be the most abundant genus (21.4%) in the sixth week, while its relative abundance was only 1.8% in the one-year biofilm sample. During the studied period, the abundance of the phylum Patescibacteria showed a slight increase, although the highest relative abundance of the phylum (14.3%) was detected in the twelfth week. The relative abundance of this phylum Planctomycetota showed a significant increase over one year, while the members of the phylum were almost completely absent for the first six weeks. Candidatus Brocadia (Planctomycetota) was the most dominant taxon (21.7%) at the genus level in the twenty-first week. Several phylotypes of the phylum Proteobacteria were found in the biofilm samples, however, the relative abundance has significantly changed at the class level in the examined period. The most noticeable change was detected in the case of the Gammaproteobacteria. The representatives of the class were dominant in the initial period and their proportion decreased significantly during the year. In the biofilm samples, their relative abundance of reads representing unclassified Bacteria ranged between 1.3% and 19%, and their proportion increased in parallel with the maturation of the biofilm.
The water sample of the spring cave was dominated by an unclassified Hydrogenophilaceae (96.1% relative abundance). OTUs assigned to Hydrogenophilaceae were also detected in some of the biofilm samples but only with low relative abundance (0.1–1.2%). It was even more interesting that only one OTU (OTU3—unclassifiedHydrogenophilaceae) was present almost exclusively in the water sample. This OTU showed the highest sequence similarity with bacterial species characterized by sulfur oxidation metabolism (93.4% sequence similarity with Sulfuritortus calidifontis). The partial 16S rRNA gene sequence was compared with EzBioCloud database entries18.
OTU91 (unclassified Anaerolineaceae) contributed the most to the sample type separation of the RTB sample (biofilm developed for years on the rock of the RT spring cave) based on SIMPER analysis, while the most abundant unclassified Hydrogenophilaceae (OTU3) in the pool water (Fig. 4). In the examined one-year period, the most dynamic changes were observed at OTU level. Certain OTUs were present only at the beginning of the studied period (OTU62—Candidatus Kaiserbacteria) or were only dominant in the first weeks, (OTU11—wb1-P19 (Nitrosococcaceae), OTU18—unclassified Gammaproteobacteria), other OTUs appeared only later in the biofilm (e.g. OTU2—Candidatus Brocadia, OTU7—unclassified Bacteria, OTU10—unclassified Bacteria).
Figure 4.
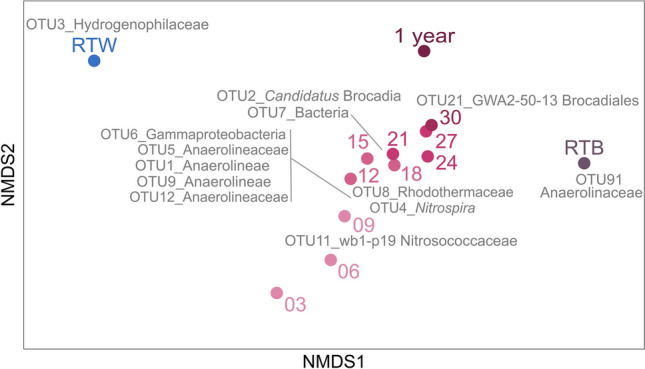
NMDS ordination based on Bray–Curtis distance of the bacterial OTUs from biofilm developed for years on the rock (RTB), glass slides (3–30 week and 1 year) and water samples (RTW) of the RT spring (stress: 0.03). Based on SIMPER analysis, OTUs responsible for 60% dissimilarity among samples are shown in gray. Figure is based on the combination the results of this study and Enyedi et al. (2019).
Discussion
Microorganisms inhabiting biofilms form structurally and functionally well-organized communities where the interactions among the participants can be extremely complex1. The in situ model systems arranged in controlled, natural and quasi-stagnant physical–chemical conditions are important to understand the changes that take place in the development of biofilms. Therefore, the present study performed in the RT spring cave highlighted some interesting phenomena on biofilm formation in thermal karst systems.
In the initial stage of biofilm formation, primary colonizing microorganisms generally bind through specific and non-specific interactions to a conditioning film adhering to solid surfaces, consisting of various organic materials19. It was also clearly visible on the SEM images (Fig. 2) made at the beginning of the model experiment. In the next stage of biofilm formation, microbial cells embed into an EPS. The increasing EPS production was observed from the 9–12 weeks during the experiment of biofilm development in the RT spring cave. The EPS consists of polysaccharides, glycoproteins, exoenzymes and nucleic acids that attach directly to the surface, also known as a substrate20,21.
The microscopically observed biofilm maturation correlated well with species richness estimators and diversity indices. The number of OTUs greatly increased during the first nine weeks when the morphological diversity of the biofilm also became more and more complex. Most of the dynamic changes observed at the OTU level happened until the twelfth week, so we could state that nine to twelve weeks were needed for the maturation of the biofilm. The greatest difference between the species richness estimators was among the water and biofilm samples.
In an earlier study, a natural biofilm sample (RTB) developed for years on the rock of the RT spring cave17 was analyzed. The taxonomic composition of bacteria inhabiting the natural and experimental biofilms was similar. The most abundant community members of the RTB as well as the 3–30 weeks and one-year in situ model biofilm samples were also the same, showing no difference or selection between the glass slide and the carbonate rock as a surface substrate. Based on our previous results17, unforeseen taxonomic bacterial diversity was obtained from these highly radioactive environments (600 ± 21 Bq/L radon concentration in the case of Rudas-Török spring cave) based on next-generation sequencing data, containing mainly unclassified bacteria affiliated with low level similarity to cultured bacterial taxa. The surprisingly high diversity suggests that the microorganisms living here are well adapted to this extreme environment.
As regards the taxonomic diversity, Proteobacteria was detected with relative high abundance both in the water and biofilm samples (Fig. 3). It could be presumed based on the previous studies, because members of this phylum were frequently detected as constituents of bacterial communities in different cave samples6,22–26. The water sample of the RT spring cave was dominated almost exclusively by an unclassified Hydrogenophilaceae (OTU3) that showed the highest sequence similarity with chemo-lithotrophic sulfur-oxidizing bacteria (Fig. 3). OTU3 (with its 96.1% relative abundance) could be considered as the potential ‘core OTU' of the thermal water. To our knowledge, this is the first report about such extraordinary high relative abundance of an OTU in the thermal waters of karst caves. Deja-Sikora et al.27 reported a similar phenomenon, the dominance of one unclassified Betaproteobacteria OTU affiliated with Comamonadaceae (abundance ranging from 1.7 to 57.8%) in sulfide-rich waters of the Carpathian Foredeep. The authors assumed that members of Comamonadaceae were likely to represent archetypal microbial species in those waters. The dominant read of OTU3 originated from the discharging deep thermal waters, however, was almost completely absent from the biofilm samples of the spring cave. This finding was surprising because the role of bacteria arriving with the discharging water sample was hypothesized in the formation of the biofilm. Among Proteobacteria, an unclassified Gammaproteobacteria was the most abundant in the biofilm samples (Fig. 3), nevertheless, no more information is known about these unclassified phylotypes.
The phylum Chloroflexi was dominant in the biofilms throughout the studied period with the members of unclassified Anaerolineaceae (Fig. 3). These Gram-negative, filamentous, thermophilic and strictly anaerobic, chemo-organotrophic organisms28 can serve as the basis of the biofilm formation in the RT spring cave. The non-cultivated members of the class Dehalococcoidi have fermentative metabolism and can use N-acetylglucosamine under anoxic conditions (using nitrate as electron acceptor). N-acetylglucosamine, which forms the backbone of the murein of most bacterial cell walls, is released continuously when cells are destroyed29. Presumably, the representatives of these filamentous Chloroflexi can be the first adherent organisms according to the SEM images (Fig. 2), and the low oxygen level in the spring cave may have favored their reproduction. The OTUs assigned to the phylum Chloroflexi were also frequent not only in the biofilm formed on the glass slides but in the biofilm developed on the rock surface of the RT spring cave as well17.
Representatives of the genus Nitrospira (Nitrospirota) were also present throughout the experiment. Their highest proportion were observed in the sixth week of the biofilm formation (Fig. 3). The characteristic cell shape typical for the genus Nitrospira has been observed in the three-week biofilm sample on the SEM images, as well (Fig. 2). Nitrogen is frequently a limited nutrient source in caves; therefore, the importance of the nitrogen cycle has been emphasized in other studies25,30. Chemolithotrophic autotrophic prokaryotes, including nitrifiers, play a key role in the primary production of cave environments31. The presence of ammonia-oxidizing Nitrosospira and nitrite-oxidizing Nitrospira and Nitrobacter were revealed previously from the deposits of the cave wall of the western Loess Plateau of China32 and the presence of these organisms were detected in the caves and spring caves of the BTKS5–8, as well. Through their activity, nitrite-oxidizing aerobic chemolithotrophic bacteria may contribute to the low nitrite concentration values, which were also measured in the cave waters of the BTKS.
A possible reason for the low ammonia content in the BTKS is the oxidation of ammonia, in which members of both the Archaea and Bacteria may be involved. The ammonia-oxidizing archaea (AOA) organisms belonging to the phylum Thaumarchaeota appeared in high proportion in the archaeal clone libraries created from biofilms originated from the caves and spring caves of the BTKS6,7. For the members of the Archaea, an increasing temporal trend was observed in the biofilms from the fifteenth weeks, although the primer-pair which was used for amplification is rather Bacteria-specific33. The diversity and importance of Archaea in karst cave environments, in contrast to the Bacteria, is largely unexplored2,3,34. In the study of the speleothems of the Weebubbie Cave (Nullarbor karst, Australia) and Kartchner Caverns (Arizona, USA), the authors also demonstrated the importance of members of Archaea, especially the ammonia-oxidizing Thaumarchaeota2,3,34. Our findings may confirm the hypothesis that AOA organisms could have an important role in the nitrification process in the RT spring cave as well.
The members of the phylum Planctomycetota (Candidatus Brocadia) proved to be dominant in the biofilm samples (Fig. 3). Representatives of the ‘Candidatus Brocadia’ may participate also in the local nitrogen cycle by the anaerobic oxidation of ammonia (anammox) combined with nitrite reduction that results in the formation of elemental nitrogen35. The anaerobic ammonia-oxidizing bacteria grow very slowly, the fastest growing species also have a 10-day generation time36, which may be associated with the fact that the relative abundance of the phylum showed a significant increase only from the sixth week of biofilm formation.
Representatives of the phylum Patescibacteria (unclassified Parcubacteria) were found in high proportions in the biofilm samples (Fig. 3). These organisms were mostly observed in anoxic environments37, their presence can be associated with the low dissolved oxygen values in the RT spring cave. The members of the Parcubacteria have small genome size (< 1.1 Mbp) and based on this, it can be assumed that they form symbiotic relationships in the biofilm38.
Unclassified bacterial reads were also occurred in a high proportion in the biofilm samples and their relative abundance showed a significant increase during a year. In a metagenomics study of carbonate caves in the Kartchner Caverns, pyrosequencing resulted nearly 400,000 partial 16S rRNA sequence data in the case of the 10 examined samples. Unfortunately, most of the taxa obtained from the cave could not be identified, whereas the vast majority of prokaryotic taxa in the absence of a cultured representative is still unknown to science2.
In conclusion, the in situ experiment performed in this study allowed us to examine the development of biofilm over a year under controlled but natural and near constant physical–chemical conditions. From the adhesion of the first microbial cells, biofilm differed fundamentally from the pool water of the spring cave. At least nine weeks were needed for the development of a mature biofilm regarding the morphological complexity and taxonomic diversity. The prokaryotes involved in the aerobic and anaerobic nitrification processes were characteristic in the biofilm samples, in addition to the anaerobic fermentative and filamentous Chloroflexi, in contrast to the water sample where the dominance of an uncultivated member of the family Hydrogenophilaceae was observed.
Supplementary Information
Acknowledgements
The authors are thankful to Károly Márialigeti for his valuable help in the establishment of the model system, to Viktória Burkus MSc student for field work and Mihály Óvári for the additional help with the physical and chemical parameters.
Author contributions
D.A. wrote the manuscript and A.K.B. coordinated the study. D.A. and P.K.B. carried out the sampling and determined physicochemical parameters. A.Sz. and D.A. performed DNA isolation, amplification, sequencing, bioinformatic analyses and the statistical analyses. J.M. and É.Á. performed the SEM analysis. A.K.B., T.F., J.M. and J.M.Sz. gave advice on the interpretation of the results; provided critical feedback and helped to shape the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the Hungarian Scientific Research Fund (NKFI) Grant NK101356. The work of D. A. was supported by the ÚNKP-18-3-III-ELTE-597 New Excellence Program of the Ministry of Human Capacities. T. F. was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences (Grant No. BO/00837/20/8).
Data availability
Raw sequence data were submitted to NCBI under BioProject ID PRJNA483930.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dóra Anda, Email: andadora@caesar.elte.hu.
Andrea K. Borsodi, Email: borsodi.andrea@ttk.elte.hu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-78759-4.
References
- 1.Simões M, Simões LC, Vieira MJ. A review of current and emergent biofilm control strategies. LWT-Food Sci. Technol. 2010;43:573–583. doi: 10.1016/j.lwt.2009.12.008. [DOI] [Google Scholar]
- 2.Ortiz M, et al. Making a living while starving in the dark: metagenomic insights into the energy dynamics of a carbonate cave. ISME J. 2014;8:478–491. doi: 10.1038/ismej.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tetu SG, et al. Life in the dark: metagenomic evidence that a microbial slime community is driven by inorganic nitrogen metabolism. ISME J. 2013;7:1227–1236. doi: 10.1038/ismej.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones DS, et al. Community genomic analysis of an extremely acidophilic sulfur-oxidizing biofilm. ISME J. 2012;6:158–170. doi: 10.1038/ismej.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borsodi AK, et al. Biofilm bacterial communities inhabiting the cave walls of the Buda Thermal Karst System, Hungary. Geomicrobiol. J. 2012;29:611–627. doi: 10.1080/01490451.2011.602801. [DOI] [Google Scholar]
- 6.Borsodi AK, et al. Biofilm forming bacteria and archaea in thermal karst springs of Gellért Hill discharge area (Hungary) J. Basic Microb. 2018;58:928–937. doi: 10.1002/jobm.201800138. [DOI] [PubMed] [Google Scholar]
- 7.Anda D, et al. Thermophilic prokaryotic communities inhabiting the biofilm and well water of a thermal karst system located in Budapest (Hungary) Extremophiles. 2015;19:787–797. doi: 10.1007/s00792-015-0754-1. [DOI] [PubMed] [Google Scholar]
- 8.Anda D, et al. Comparison of bacterial and archaeal communities from different habitats of the hypogenic Molnár János Cave of the Buda Thermal Karst System (Hungary) J. Cave Karst Stud. 2017;79:113–121. doi: 10.4311/2015MB0134. [DOI] [Google Scholar]
- 9.Mádl-Szőnyi J, Erőss A, Tóth Á. Fluid flow systems and hypogene karst of the Transdanubian Range, Hungary—with special emphasis on Buda Thermal Karst. In: Klimchouk A, Palmer AN, De Waele J, Auler AS, Audra P, editors. Hypogene Karst Regions and Caves of the World. Cham: Springer; 2017. pp. 267–278. [Google Scholar]
- 10.Erőss A, et al. Radionuclides as natural tracers for the characterization of fluids in regional discharge areas, Buda Thermal Karst, Hungary. J. Hydrol. 2012;426:124–137. doi: 10.1016/j.jhydrol.2012.01.031. [DOI] [Google Scholar]
- 11.Szabó A, et al. Soda pans of the Pannonian steppe harbor unique bacterial communities adapted to multiple extreme conditions. Extremophiles. 2017;21:639–649. doi: 10.1007/s00792-017-0932-4. [DOI] [PubMed] [Google Scholar]
- 12.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Team, R. C . R: A Language and Environment for Statistical Computing, Version 3.0.2 (Computer Software) Vienna: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 15.Oksanen J, et al. The vegan package. Community Ecol. Package. 2007;10:631–637. [Google Scholar]
- 16.Hammer Ø, Harper DA, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;41:9. [Google Scholar]
- 17.Enyedi NT, et al. Radioactive environment adapted bacterial communities constituting the biofilms of hydrothermal spring caves (Budapest, Hungary) J. Environ. Radioact. 2019;203:8–17. doi: 10.1016/j.jenvrad.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Yoon SH, et al. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/S0966-842X(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 20.Decho AW. Microbial exopolymer secretions in ocean environments: their role(s) in food webs and marine processes. Oceanogr. Mar. Biol. 1990;28:73–153. [Google Scholar]
- 21.Wingender J, Neu TR, Flemming HC. What are bacterial extracellular polymeric substances? In: Wingender J, Neu TR, Flemming HC, editors. Microbial Extracellular Polymeric Substances Characterization, Structure and Function. Berlin: Springer; 1999. pp. 1–19. [Google Scholar]
- 22.Porter ML, Engel AS, Kane TC, Kinkle BK. Productivity-diversity relationships from chemolithoautotrophically based sulfidic karst systems. Int. J. Speleol. 2009;38:27–40. doi: 10.5038/1827-806X.38.1.4. [DOI] [Google Scholar]
- 23.Macalady JL, et al. Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl. Environ. Microb. 2006;72:5596–5609. doi: 10.1128/AEM.00715-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Northup DE, et al. Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. Environ. Microbiol. 2003;5:1071–1086. doi: 10.1046/j.1462-2920.2003.00500.x. [DOI] [PubMed] [Google Scholar]
- 25.Holmes AJ, et al. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 2001;3:256–264. doi: 10.1046/j.1462-2920.2001.00187.x. [DOI] [PubMed] [Google Scholar]
- 26.Barton HA. Starving artists: bacterial oligotrophic heterotrophy in caves. In: Engel AS, editor. Microbial Life of Cave Systems. Berlin/Boston: Walter de Gruyter GmbH; 2015. pp. 79–104. [Google Scholar]
- 27.Deja-Sikora E. Comamonadaceae OTU as a remnant of an ancient microbial community in sulfidic waters. Microb. Ecol. 2019;78:85–101. doi: 10.1007/s00248-018-1270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada T, et al. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int. J. Syst. Evol. Microb. 2006;56:1331–1340. doi: 10.1099/ijs.0.64169-0. [DOI] [PubMed] [Google Scholar]
- 29.Kindaichi T, Yuri S, Ozaki N, Ohashi A. Ecophysiological role and function of uncultured Chloroflexi in an anammox reactor. Water Sci. Technol. 2012;66:2556–2561. doi: 10.2166/wst.2012.479. [DOI] [PubMed] [Google Scholar]
- 30.Marks CR, Stevenson BS, Rudd S, Lawson PA. Nitrospira dominated biofilm within a thermal artesian spring: a case for nitrification-driven primary production in a geothermal setting. Geobiology. 2012;10:457–466. doi: 10.1111/j.1472-4669.2012.00335.x. [DOI] [PubMed] [Google Scholar]
- 31.Ortiz M, Neilson JW, Legatzki A, Maier RM. Bacterial and archaeal diversity on cave speleothem and rock surfaces: a carbonate cave case study from Kartchner Caverns. In: Engel AS, editor. Microbial Life of Cave Systems. Berlin/Boston: Walter de Gruyter GmbH; 2015. pp. 105–123. [Google Scholar]
- 32.Wu Y, et al. Profiling bacterial diversity in a limestone cave of the western Loess Plateau of China. Front. Microbiol. 2015;6:244. doi: 10.3389/fmicb.2015.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legatzki A, et al. Bacterial and archaeal community structure of two adjacent calcite speleothems in Kartchner Caverns, Arizona, USA. Geomicrobiol. J. 2011;28:99–117. doi: 10.1080/01490451003738465. [DOI] [Google Scholar]
- 35.Kuypers MM, et al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature. 2003;422:608–611. doi: 10.1038/nature01472. [DOI] [PubMed] [Google Scholar]
- 36.Fuerst JA, Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 2011;9:403–413. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- 37.Nelson WC, Stegen JC. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 2015;6:713. doi: 10.3389/fmicb.2015.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hedlund BP, Dodsworth JA, Murugapiran SK, Rinke C, Woyke T. Impact of single-cell genomics and metagenomics on the emerging view of extremophile “microbial dark matter”. Extremophiles. 2014;18:865–875. doi: 10.1007/s00792-014-0664-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data were submitted to NCBI under BioProject ID PRJNA483930.