SUMMARY
Motile cilia are cellular beating machines that play a critical role in mucociliary clearance, cerebrospinal fluid movement and fertility. In the airways, the hundreds of motile cilia present on the surface of a multiciliated epithelia cell beat coordinately to protect the epithelium from bacteria, viruses and harmful particulates. During multiciliated cell differentiation, motile cilia are templated from basal bodies each extending a basal foot, an appendage linking motile cilia together to ensure coordinated beating. Here, we demonstrate that among the many motile cilia of a multiciliated cell, a hybrid cilium with structural features of both primary and motile cilia is harboured. The hybrid cilium is conserved in mammalian multiciliated cells, originates from parental centrioles and its cellular position is biased and dependent on ciliary beating. We further show that the hybrid cilium emerges independently of other motile cilia, and functions in regulating basal body alignment.
Graphical Abstract
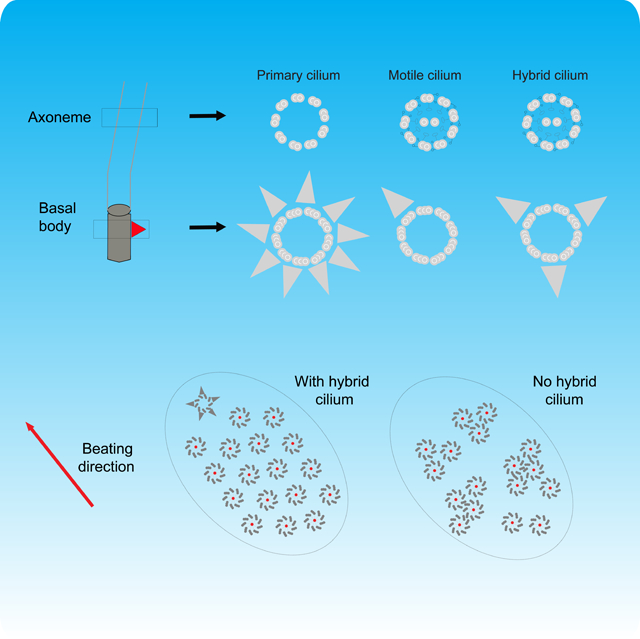
eTOC:
Liu, Nguyen et al. show that among hundreds of cilia of a multiciliated cell, there is one that differs from the others in structure, origin and cellular position, which participates in basal body alignment.
INTRODUCTION
Motile cilia are beating organelles found on the surface of specialized epithelial cells that generate the propulsive force required for critical physiological activities such as mucociliary clearance, ependymal cerebrospinal fluid circulation and ovum directional movement in the fallopian tubes (Reiter and Leroux, 2017; Spassky and Meunier, 2017). In the airways, the hundreds of motile cilia within one multiciliated cell beat cooperatively to remove bacteria, viral particles and environmental pollutants, thereby helping the immune system clearing infections and ensuring lung health (Lucas et al., 2020; Reiter and Leroux, 2017).
To beat in coordination, motile cilia rely on the basal foot, a conical structure attached to the basal body on one end, and to the microtubule cytoskeleton on the other, thereby linking motile cilia together in a network (Clare et al., 2014; Kunimoto et al., 2012; Vladar et al., 2012). The hundreds of motile cilia on the surface of a multiciliated cell are thought to be identical to one another (Jain et al., 2010; Spassky and Meunier, 2017), templated from basal bodies each with one basal foot. When motile cilia beat coordinatively, basal bodies are aligned in rows with their basal feet pointing toward the direction of ciliary beating (Boisvieux-Ulrich et al., 1985; Frisch and Farbman, 1968; Gibbons, 1961), a phenomenon termed rotational polarity (Marshall and Kintner, 2008; Mitchell et al., 2007).
In airway cells, loss of basal feet results in disorganization of the microtubule apical network, irreversible disorientation of basal bodies and loss of motile cilia coordination (Herawati et al., 2016). In mice (Kunimoto et al., 2012), this leads to respiratory manifestations consistent with primary ciliary dyskinesia (PCD), a human rare disease characterized by chronic airway infections, bronchiectasis and frequently associated with conductive hearing loss, male infertility, heterotaxy and cardiac malformations (Kunimoto et al., 2012; Lucas et al., 2020).
In addition to multiciliated cells, the basal foot is also present in cells protruding a primary cilium. Its suggested function is to keep the primary cilium submerged in certain cell types by linking the basal body to Golgi, in concert with the pool of centrosomal proximal end proteins. In turn this controls ectopic Shh-signalling activation (Galati et al., 2016; Mazo et al., 2016). Different from motile cilia, the basal foot of the primary cilium has multiple copies per basal body and it is thought to originate from nine (or less depending on the cell type) subdistal appendages, mother centriole-associated structures contributing to the organization of the interphase microtubule array (Chong et al., 2020; Paintrand et al., 1992; Uzbekov and Alieva, 2018).
We have recently resolved the molecular architecture of the basal foot in situ by super-resolution microscopy in primary and motile cilia (Nguyen et al., this issue). In the process of determining the molecular map of basal foot in motile cilia, we discovered the presence of a “hybrid” cilium in multiciliated cells. This hybrid cilium is characterized by a basal body containing multiple basal feet, resembling a primary cilium, but expressing proteins and with an axonemal ultrastructure typical of motile cilia. Here we characterize hybrid cilium structure, conservation, formation and function in airway multiciliated cells.
RESULTS
Identification of a Hybrid Cilium in Airway Multiciliated Cells
Previous studies have indicated that the basal foot shows both structural and numerical differences in the two ciliary systems: the most obvious one is that primary cilia have multiple basal feet per basal body, while motile cilia have one basal foot per basal body (Boisvieux-Ulrich et al., 1985; Frisch and Farbman, 1968; Gibbons, 1961; Kunimoto et al., 2012; Mitchell et al., 2007). While imaging basal foot proteins in human airway multiciliated cells, it was therefore surprising to observe a ring-like pattern of basal foot proteins in addition to the expected dot pattern for a single basal foot (Figures 1A, 1B and 1C, see boxed regions). This arrangement suggested the presence of a basal body in a multiciliated cell similar to that of a primary cilium (Figure 1B).
Figure 1. Super-resolution reveals a basal body with multiple basal feet in airway multiciliated cells.
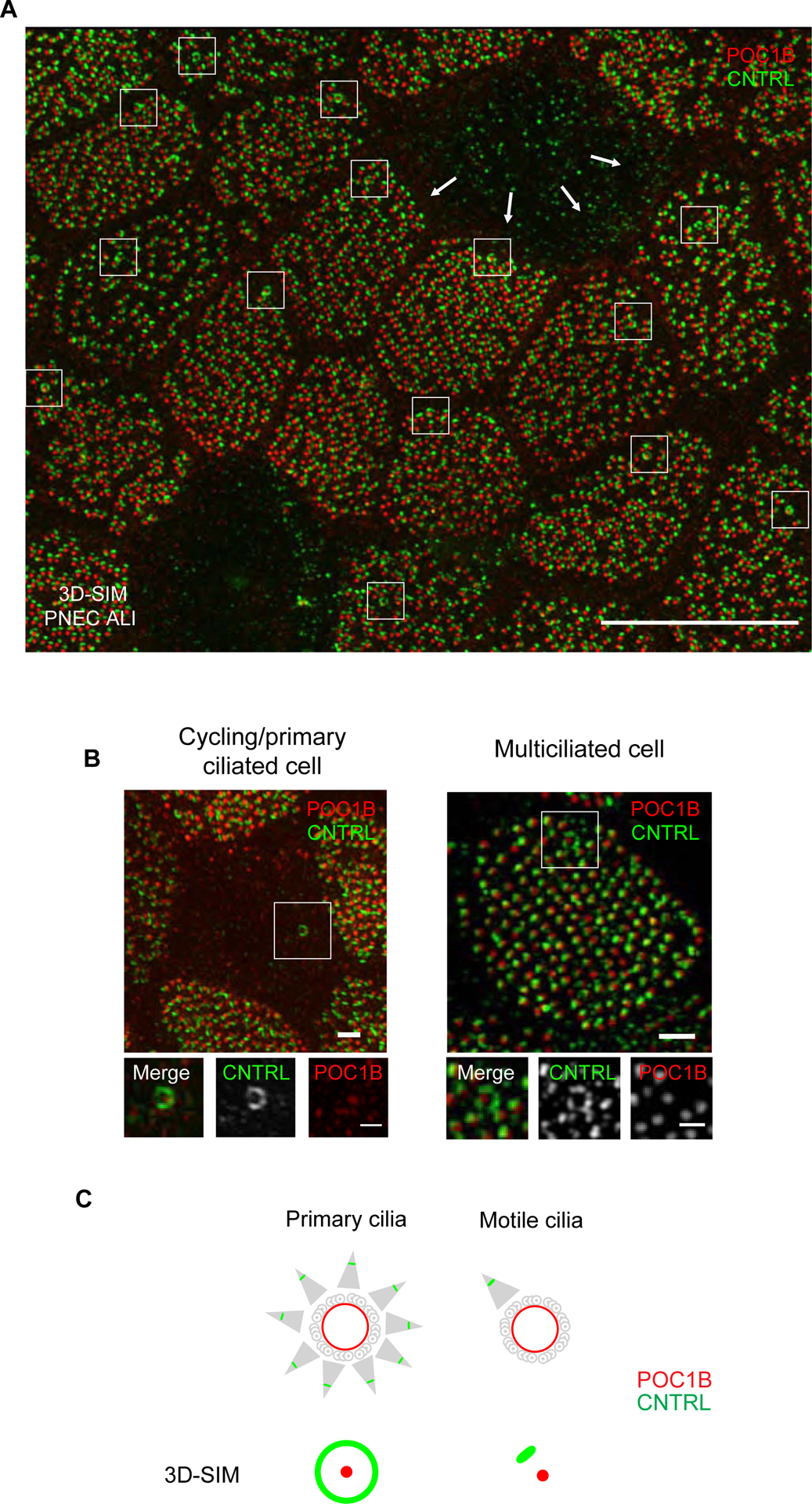
(A) 3D-SIM volume maximum intensity projection of large field of view of nasal primary airway multiciliated cell (PNEC) grown in air liquid interphase labeled with anti-CNTRL (green) and anti-POC1B (red) antibodies. Note the ring-like pattern of CNTRL localization encircling the basal body labeled by POC1B (boxed areas). Scale bar represents 10 μm. ALI, air liquid interface. (B) 3D-SIM volume maximum intensity projection of an airway cycling/primary ciliated cell (left) or nasal airway multiciliated cell (right) grown in ALI labeled with anti-CNTRL (green) and anti-POC1B (red) antibodies. Scale bars represent 1 μm and 500 nm (boxed areas). (C) Cartoon representation of basal body-basal foot structure by TEM in cells with primary or motile cilia (upper panel) and by 2 colour 3D-SIM imaging of basal body and basal foot (lower panel). By 3D-SIM microscopy the basal body protein POC1B appears as a dot while the basal foot protein CNTRL appears as a dot in motile cilia, but as a ring in primary cilia. Red represents localization of POC1B and green represents localization of CNTRL. See also Figure S1.
To validate this initial observation, we collected micrographs of primary nasal human multiciliated cells grown in an air-liquid interface co-labelled with antibodies recognizing different basal foot (CNTRL, CEP128) and basal body (POC1B) proteins by 3D structured illumination microscopy (3D-SIM) (Figures 1B and S1). This analysis demonstrated that the ring pattern resulted from a single basal body with multiple basal feet, and not from multiple basal bodies clustering together as in the compound cilium, a membrane-delimited structure made of multiple motile cilia clustered together, which is frequently found in airway cells after injury (McAuley and Anand, 1998).
Imaging of freshly isolated nasal epithelial human cells established that this basal body is also present in tissue ex vivo and not only in primary cell culture systems (Figure 2A). To confirm its presence by other imaging methods, we used stochastic optical resolution fluorescence microscopy (STORM), a technique reaching routinely ~25 nm in plane resolution, as well as transmission electron microscopy (TEM) to examine sections of airway cells differentiated in vitro. Both imaging methods confirmed the presence of this basal body (Figures 2B and 2C). TEM and 3D-SIM also showed that it templates a cilium (Figures 2D and 2E).
Figure 2. The basal body extrudes a cilium and is conserved in different mammalian primary cells, tissues and species.
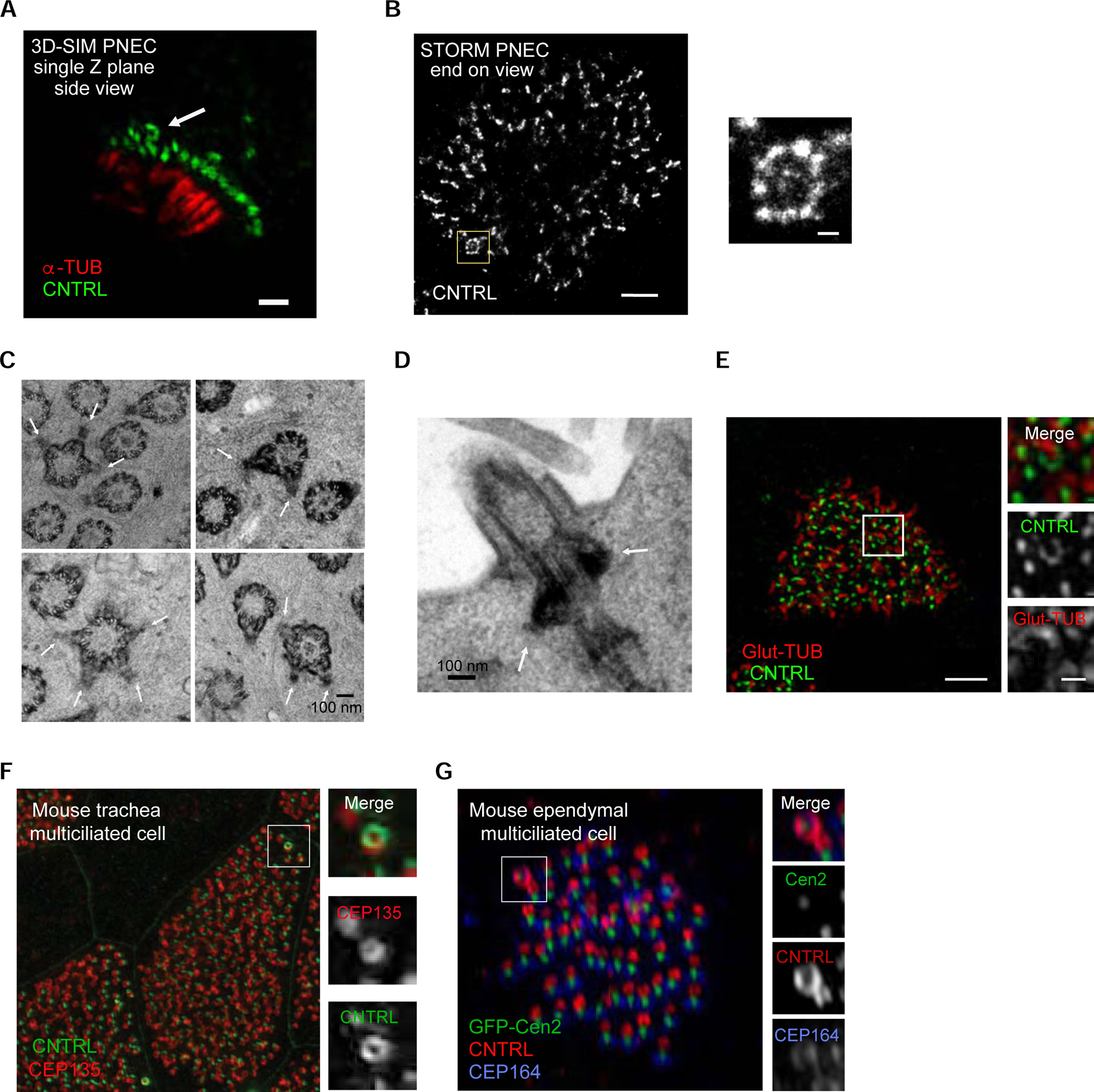
(A) 3D-SIM single-plane image of a human nasal airway multiciliated cell freshly isolated from a healthy individual, labeled with anti-CNTRL (green) and anti-alpha-tubulin (red) antibodies, showing the presence of the basal body with multiple basal feet. Scale bar represents 1 μm. (B) Left: STORM micrograph of airway multiciliated cell labeled with anti-CNTRL antibody, showing a distinct ring-like distribution of CNTRL (boxed area). Right: High-magnification view of boxed area. Scale bars represent 1 μm (left) and 100 nm (right). (C) Collage of representative TEM micrographs showing basal bodies harbouring multiple basal feet in ALI-cultured human airway multiciliated cells (see white arrows). Scale bar represents 100 nm. (D) TEM micrograph showing axoneme emanating from basal bodies harbouring multiple basal feet (in white arrows) in human airway multiciliated cell. Scale bar represents 1 μm. (E) Left: 3DSIM volume maximum intensity projection of an ALI-cultured human airway multiciliated cell labeled with anti-CNTRL (green) and anti-Glut-TUB (red) antibodies. Note the axoneme emanating from ring-like structure labeled with CNTRL (boxed area). Right: High-magnification view of boxed area with individual channels. Scale bars represent 2 μm (left) and 500 nm (right). (F) 3D-SIM volume maximum intensity projection of mouse tracheal multiciliated cell (ALI D20), labeled with anti-CNTRL (green) and anti-CEP135 (red) antibodies. Right: High-magnification view of boxed area with individual channels. Scale bar represents 2 μm. (G) 3D-SIM volume maximum intensity projection of adult mouse ependymal multiciliated cells (P16), labeled with GFP-Centrin2 (basal body), anti-CNTRL (red) and anti-CEP164 (blue, distal appendage protein) antibodies. Right: High-magnification view of boxed area labeled in left. Scale bar represents 1 μm and 200 nm.
To assess the conservation of the hybrid cilium beyond airway epithelium, multiciliated cells differentiated from mouse tracheal and brain ependymal tissue progenitors were examined. Using 3D-SIM, a basal body associated with multiple basal feet was observed in each cell type, showing that the hybrid cilium is an organelle present in multiple tissues that have multiciliated cells, both in mouse and human (Figures 2F and 2G).
The Cilium Discovered has Features of Primary and Motile Cilia
Next, we asked which type of cilium was templated from this basal body with multiple basal feet. To address this question, we examined its ultrastructure by Focused Ion Beam-Scanning Electron Microscopy (FIB-SEM), an EM technique capable of imaging large volumes, several cells wide, with isotropic resolution (Figure 3A) (Kizilyaprak et al., 2019). Analysis of tomographic sections confirmed that this basal body templates a bona fide cilium (consistent with Figures 2D and 2E) and showed that it contains a central pair of microtubules (a structure required for in plane beating) as the surrounding motile cilia in the same multiciliated cell (Figures 3B and 3C and Videos S1 and S2). 3D-SIM micrographs of airway multiciliated cells labeled with anti-radial spoke head (RSPH4A) (Frommer et al., 2015) and nexin-dynein regulatory complex (GAS8) (Olbrich et al., 2015) antibodies further demonstrated that this cilium harbours components of the ciliary beating machinery (Figure 3D).
Figure 3. The cilium has hybrid features between primary and motile cilia.
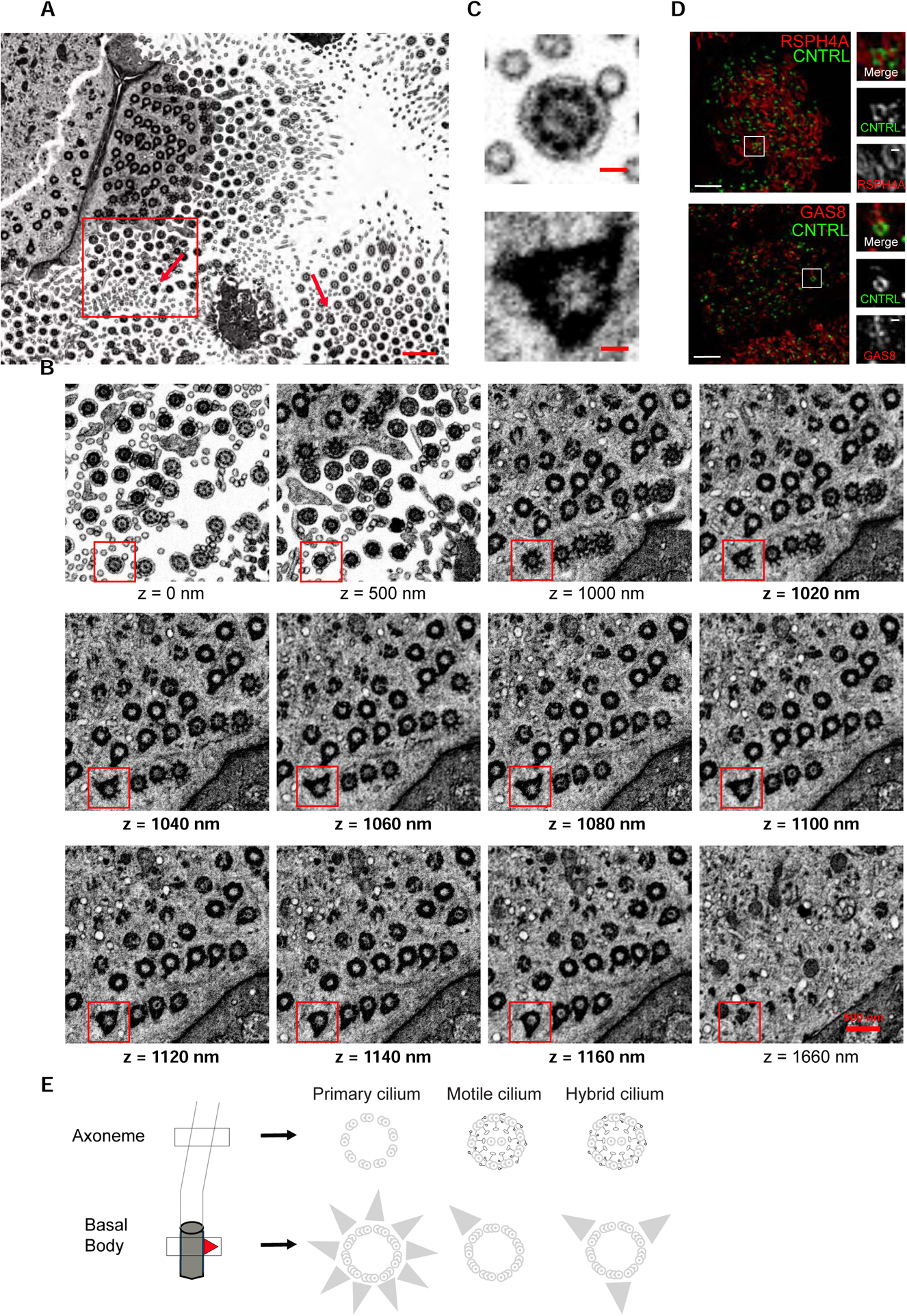
(A) Representative section from FIB-SEM tomogram of human primary nasal multiciliated cells. Arrows indicate basal bodies with multiple basal feet. Scale bar represents 1 μm. (B) High-magnification view of boxed area in (A) at different z positions of the tomogram from z=0 nm to z=1660 nm. Note the hybrid cilium axoneme and central pair (z=0 nm), transition fibers (z=1000–1040 nm), multiple basal feet (z=1160–1160 nm) and the absence of the endocytic pocket (z=1660 nm). Scale bar represents 500 nm. (C) A high-magnification view of boxed area in (B) highlighting the basal body with a central pair and multiple basal feet. Scale bar represents 100 nm. (D) 2D projection micrographs of 3D-SIM volume of human airway multiciliated cells (left), and high-magnification views of boxed areas (right), labeled with anti-CNTRL (green), anti-RSPH4A (red, top) and anti-GAS8 (red, bottom) antibodies. Scale bars represent 2 μm. (E) Cartoon representation of hybrid cilium structure relative to the primary cilium and motile cilium. The axoneme extends from the basal body (grey), each with a basal foot (red triangle). The basal body of the primary cilium presents multiple basal feet, while the one of the motile cilium has one basal foot. In regard to the axoneme, the motile cilium presents outer dynein arms, inner dynein arms, nexin-dynein regulatory complexes, radial spokes and central pair complexes (black) , which are critical for in plane ciliary beating. The hybrid cilium has features of both the primary and motile cilium, namely multiple basal feet and protein complexes critical for ciliary beating.
Altogether, our data show that in human airway multiciliated cells, not all the cilia are identical, and that a single cilium presents hybrid features of both primary and motile cilia, namely multiple basal feet and proteins associated with the ciliary beating machinery (Figure 3E).
The Hybrid Cilium Originates from Parental Centrioles
A long-standing question in the field of multiciliated cell development has been the fate of parental centrioles after multiciliogenesis, which are initially present in precursor basal stem cell as a mother-daughter engaged pair in the centrosome (Jain et al., 2010). The presence of multiple basal feet typical of primary cilia suggested the possibility that the hybrid cilium originates from the mother centriole, which in mouse airway multiciliated cells first templates the primary cilium present during the early stages of differentiation (Jain et al., 2010), then is resorbed before centriole amplification through the canonical and deuterosome pathway (Al Jord et al., 2014; Vladar and Stearns, 2007; Zhao et al., 2013).
To first test whether the mother and daughter centrosomal centrioles are retained in differentiated multiciliated cells, we performed a pulse-chase experiment allowing to label differentially centrosomal centrioles from newly formed basal bodies in mouse cultured ependymal cells (Figure S2). Time-lapse monitoring of RFP-Cen1 centrosomal centrioles in cells from transgenic mice expressing Cen2-EGFP, which labeled newly formed basal bodies (Higginbotham et al., 2004) showed that centrosomal centrioles are retained within the newly formed basal body patch and are capable of growing cilia (Figure S2, Video S3). However, we could not confirm whether the centrosomal centrioles present multiple basal feet by 3D-SIM due to the low number of cells with centrioles expressing both RFP-Cen1 and Cen2-GFP and their oblique orientation at the surface of ependymal cells differently from airway cells.
To further test our hypothesis that the hybrid cilium is derived from the mother centriole, we then used centrinone A, a small molecule drug blocking canonical centriole duplication by inhibiting Plk4, a kinase critical for the early stages of centriole duplication, to deplete cells of parental centrioles (Figure 4A) (Wong et al., 2015). As expected, basal cells isolated from mouse trachea treated with centrinone A showed a reduction in centrioles before airway cell differentiation (Figure S3) (Nanjundappa et al., 2019). When cells were treated with centrinone A either only during basal cell expansion or also throughout differentiation of mouse trachea airway cells (MTEC), drug treatment did not impact multiciliated cell formation (Figure S3), as previously described (Nanjundappa et al., 2019). However, centrinone A treatment caused a reduction in hybrid cilia number in the mature multiciliated cell stage, indicating that the hybrid cilium originates from parental centrioles, most likely the mother that initially harbours subdistal appendages/basal feet (Figures 4B, 4C and 4D).
Figure 4. Hybrid cilium originates from parental centrioles.
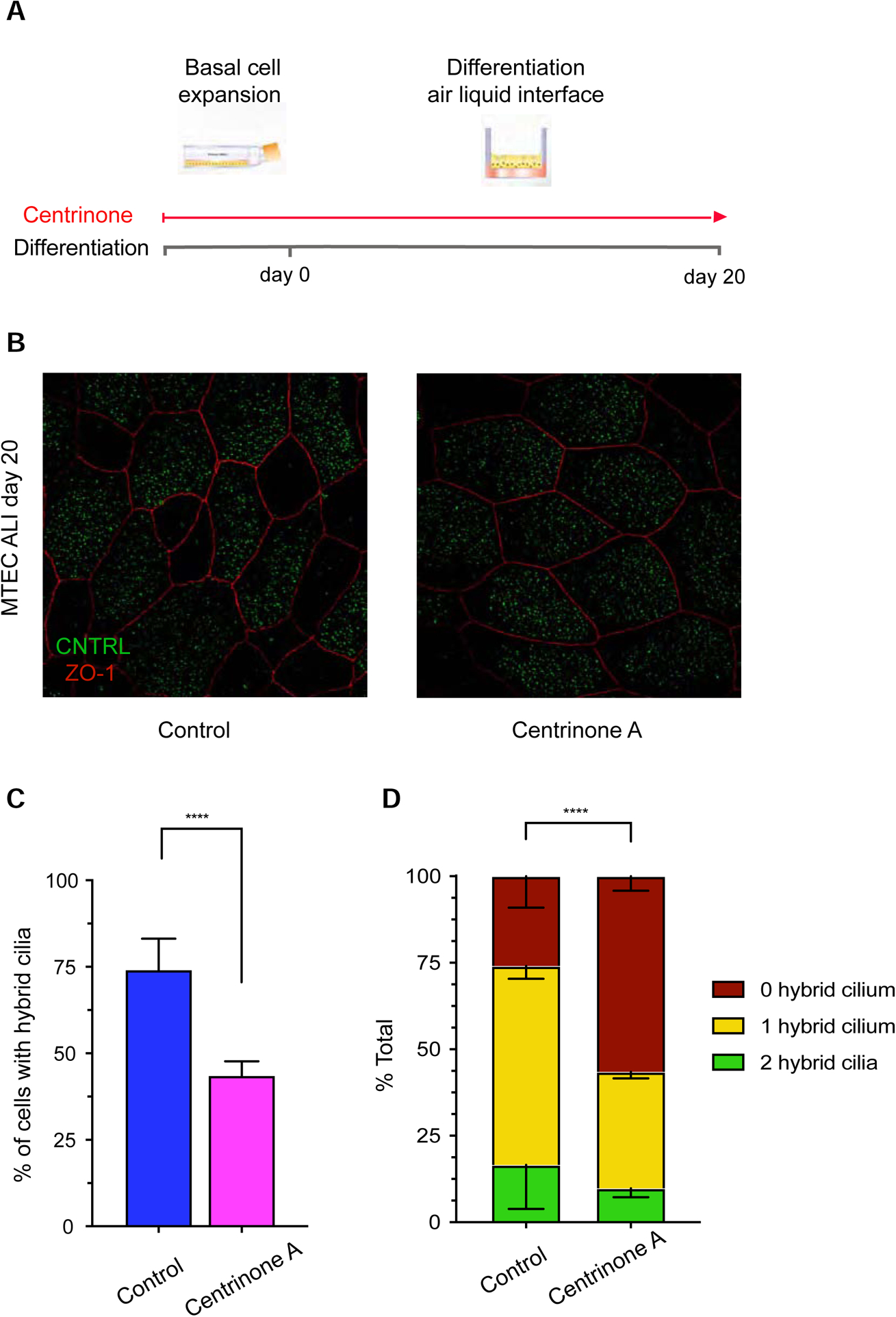
(A) Cartoon representation of centrinone A treatment in mouse tracheal multiciliated cells. (B) 2D projection micrograph of 3D-SIM volume of representative example of mouse tracheal multiciliated cells at ALI D20 treated with DMSO control or centrinone A labeled with anti-CNTRL (green) and anti-ZO-1 (red) antibodies. Arrowheads indicate CNTRL rings. Scale bar represents 5 μm. (C) Bar graphs representing percentage of cells with hybrid cilium in DMSO control (blue) and centrinone A-treated (pink) cells treated during basal cell expansion and throughout differentiation; n>800 over three independent biological replicates. Data are represented as mean ± SD. Statistical analysis was done using Cochran-Mantel-Haenszel test. **** p<0.0001. (D) Bar graph representing percentage of cells with none (red), one (yellow) or two (green) hybrid cilia in ALI D20 mouse tracheal multiciliated cells treated with DMSO control (left) or centrinone A (right); n>800 over three independent biological replicates. Data are represented as mean ± SD. Statistical analysis was done using Chi-square test. See also Figures S2 and S3.
Hybrid Cilium Formation is Independent from Other Motile cilia
Despite sharing the same cellular environment during differentiation, the hybrid cilium origin and structure are different from all other motile cilia. This suggested the possibility that hybrid cilium formation is determined independently of other motile cilia in the same multiciliated cell.
Interestingly, previous studies have established that mutations in certain human disease genes causing PCD are characterized by a lack, or a diminished number, of motile cilia. This phenotype, named oligocilia or reduced generation of multiple cilia (RGMC) is determined by loss-of-function mutations in PCD genes (Lucas et al., 2020) encoding transcription factors and regulators of the multiciliogenesis program such as the nuclear protein MCIDAS (Boon et al., 2014; Stubbs et al., 2012), the transcription factor FOXJ1 (Gomperts et al., 2004; Wallmeier et al., 2019) and the transcriptional regulator CCNO (Funk et al., 2015; Wallmeier et al., 2014) (Figure 5A). In particular, multiciliated cells isolated from patients with mutations in CCNO present on their surface mainly one cilium per cell, occasionally two or sparse cilia ((Wallmeier et al., 2014) and our observations). MCIDAS loss-of-function patient and KO mice cells can also present one basal body/cilium per cell, albeit less frequently than in CCNO mutations (Boon et al., 2014; Lu et al., 2019). In the case of CCNO mutant cells, this solitary cilium beats, indicating that it has acquired motility machinery (Wallmeier et al., 2014).
Figure 5. Hybrid cilium formation is independent from other motile cilia in airway multiciliated cells.
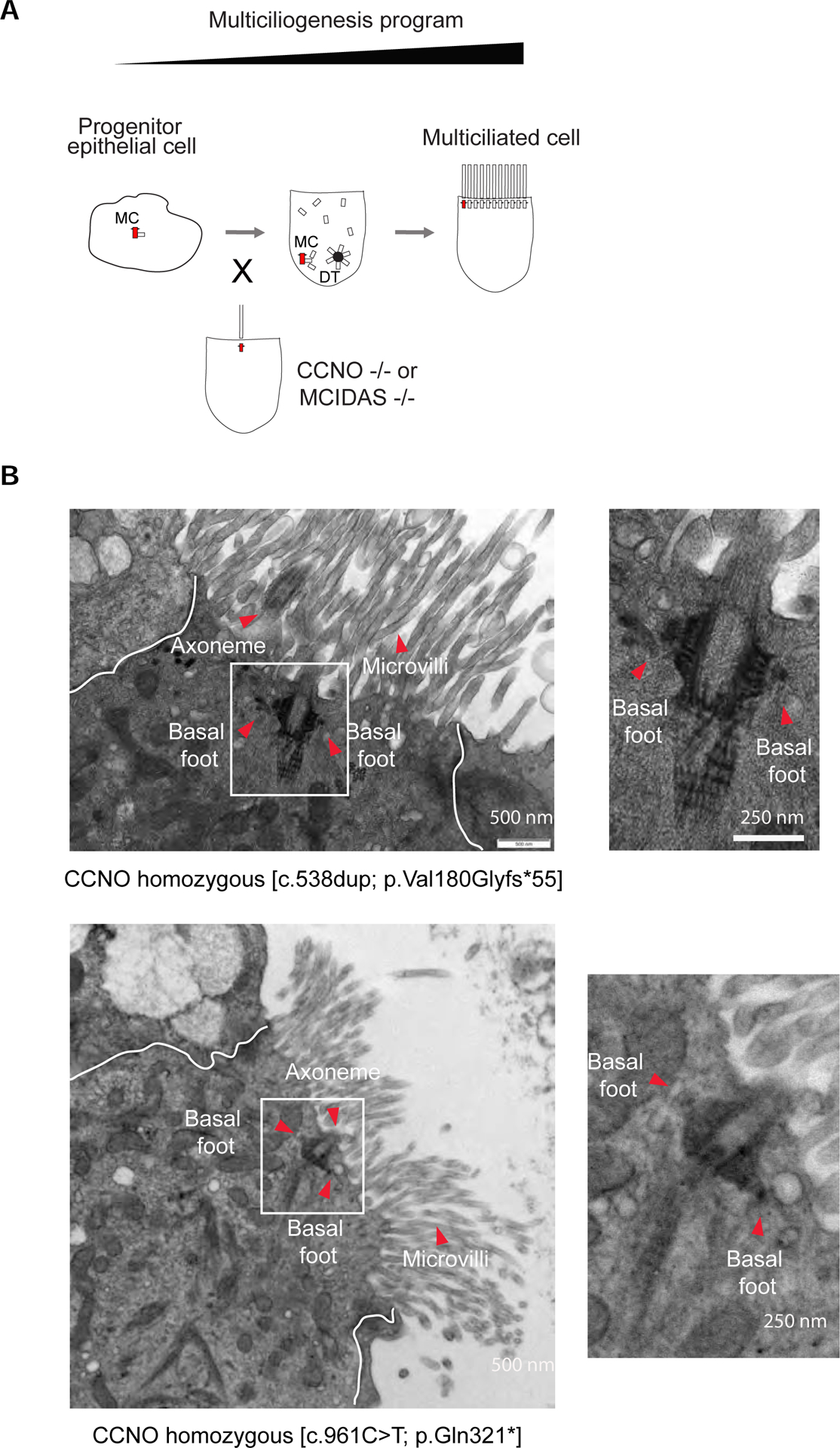
(A) Cartoon depicting a simplified version of the multiciliogenesis cellular program. Note CCNO and MCIDAS loss of function mutations lead to oligocilia phenotype. MC, mother centriole. DT, deuterostome. (B) Left: TEM micrographs of a human airway multiciliated cells isolated from nasal epithelium of PCD patients with CCNO loss of function early termination mutations. Boxed areas represent basal bodies with multiple basal feet. Red arrowheads indicate axoneme, basal feet and microvilli. Right: High-magnification views of boxed area. Scale bars represent 500 nm (left) and 250 nm (right).
To test the hypothesis that the hybrid cilium is in fact the single cilium reaching the surface of multiciliated cells in oligocilia/RGMC patients, we recruited two individuals with different loss-of-function mutations in CCNO and analysed their cells extensively using TEM sections. Our data clearly showed that cells from PCD patients with rare CCNO mutations present a cilium with multiple basal feet per basal body as in the hybrid cilium (Figure 5B).
Altogether, our data show that the hybrid cilium and “CCNO cilium” share the same structure, thereby suggesting that hybrid cilium formation is determined even in the absence of other motile cilia on the surface of airway multiciliated cells.
Hybrid Cilium Position is Biased Toward the Direction of Beating and Functions in Basal Body Alignment
It has previously been shown that during multiciliated cell differentiation, motile cilia first generate fluid flow, then fine-tune their position within a cell to generate a more effective directional flow and mucociliary transport (Guirao et al., 2010; Mitchell et al., 2007). Since an established basal foot function in multiciliated cells is to link basal bodies together through microtubule cables (Kunimoto et al., 2012), we hypothesized that the hybrid cilium could function to align basal bodies in the direction of flow.
To test this hypothesis, we first evaluated its relative position in cells. We developed a MATLAB image analysis script locating the position of the hybrid cilium relative to beating direction, which was measured by basal body-basal foot rotational polarity in multiciliated cells (Figure 6A). This analysis confirmed quantitatively that the hybrid cilium is positioned preferentially in the front end relative to the beating direction as suggested by our 3D-SIM images (Figure 6B, see also Figure 1A). To establish whether this positional bias was dependent on ciliary beating, we analysed cells from three PCD patients with independent loss of function mutations in outer dynein arm proteins (two patients with mutations in DNAH5 (PCD 1 and PCD 3) and one with DNAH11 (PCD 2), (Key Resource Table); both proteins are critical for ciliary motility, but their loss of function does not impact basal body formation (Knowles et al., 2012; Omran et al., 2000)). As expected, PCD patients’ cells exhibited normal ciliation, impaired ciliary beating and reduced rotational polarity (Figure S4) (Knowles et al., 2012; Liu et al., 2020; Omran et al., 2000). Notably, the hybrid cilium from PCD patient cells was found to be more “centred” in the cell (Figures 6B and S4), similar to the position of centrosomes in cycling cells. To further confirm the “biased” location of the hybrid cilium within a multiciliated cell, we assessed its position relative to the cell centroid and membrane, irrespective of motile cilia beating direction. Consistent with our previous analysis, the hybrid cilium in cells from PCD patients was located closer to the cell centre than in healthy controls (Figures 6C and S4).
Figure 6. Hybrid cilium position is biased in the direction of beating and dependent on flow.
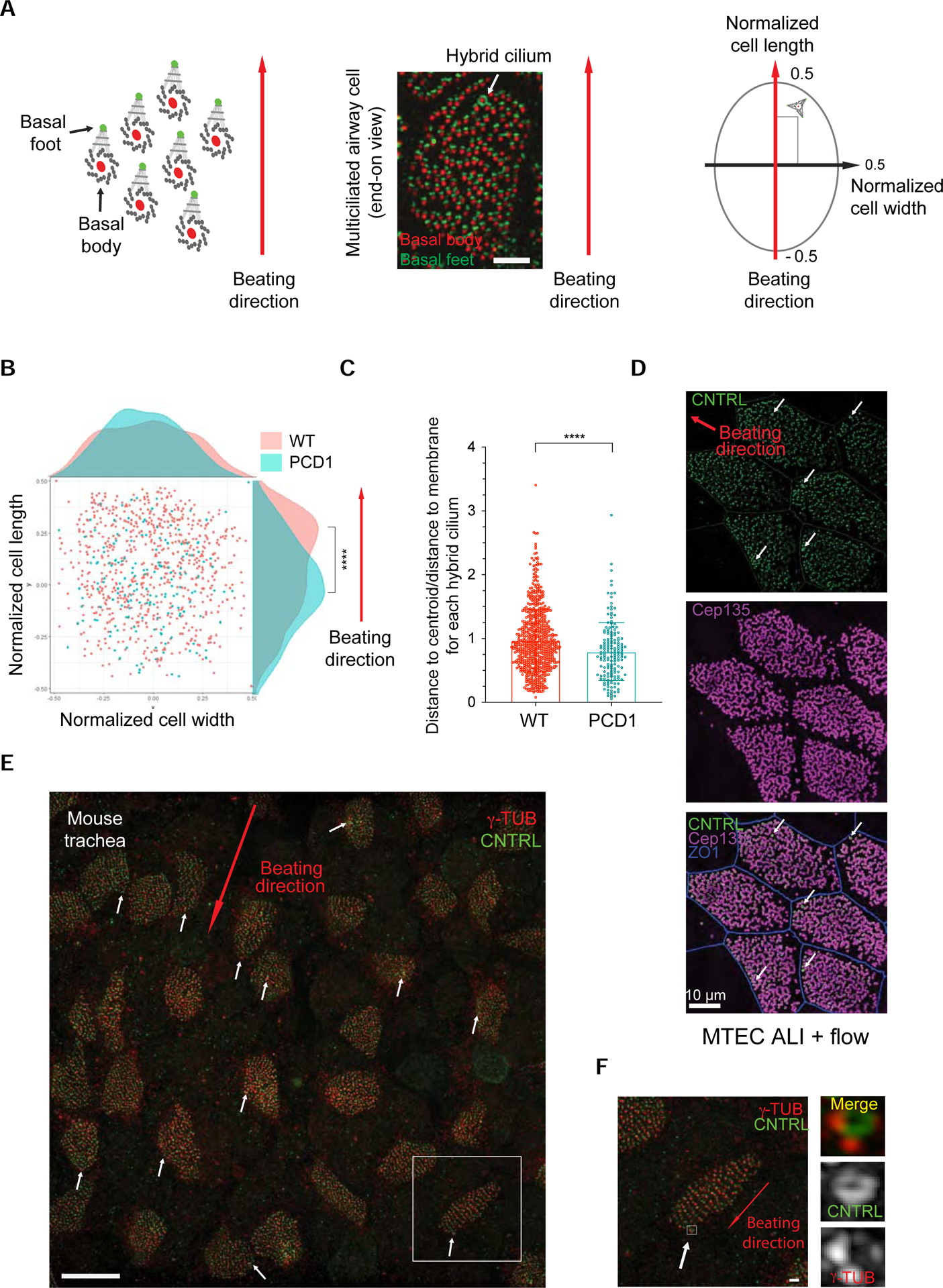
(A) Left: Cartoon depiction of the strategy for analysis of the position of the hybrid cilium relative to cilia beating direction. Middle: representative 3DSIM image used for data analysis. Scale bar represents 5μm. Right: MATLAB-based analysis strategy to assess rotational polarity in multiciliated cells. The position is calculated relative to coordinates (−0.5<x,y<+0.5) obtained by normalizing the cell length and width to 1. Cell length (y-axis) is assigned as parallel to the direction of ciliary beating measured by rotational polarity of basal bodies-basal feet pairs, cell width (x-axis) is assigned as perpendicular to the direction of ciliary beating. (B) Scatterplot showing the cumulative distribution of hybrid cilia along normalized cell width and cell length in human airway multiciliated cells from three healthy individuals (red) or one PCD patient with immotile cilia (green) caused by loss-of-function mutations in DNAH5 (p.[(Arg478*) and intronic mutation abolishing accepted splice-site for exon 68]), indicating a positional bias toward the direction of ciliary beating (See also two other PCD patients in Figure S8). Each dot represents a hybrid cilium in a cell; Healthy n=694, PCD1 =165, **** p<0.0001, student’s t-test. (C) Scatterplot-bar graphs showing distribution of ratio of hybrid cilium-cell centroid distance to hybrid cilium-cell membrane shortest distance of human airway multiciliated cells from healthy individuals or PCD1 patient. Each dot represents a hybrid cilium in a cell; Healthy n=694, PCD n=277, p<0.0001, student’s t-test. **** p<0.0001. Data are represented as mean ± SD. (D) 3D-SIM volume maximum intensity projection of MTEC labeled with anti-CNTRL (green) antibody, anti-CEP135 (magenta) and anti-ZO1 (blue) subjected to rotational movement to facilitate flow generation in transwells. Arrows indicate CNTRL rings. MTEC ALI, mouse trachea epithelia cells growing at air liquid interface. (E) 3D-SIM volume maximum intensity projection of mouse tracheal tissues labelled with anti-CNTRL (green) antibody and anti-γTub (red). Arrow indicates beating direction. Scale bar represents 10 μm. (F) High-magnification view of boxed area in (E). Note hybrid cilium basal body presenting multiple basal feet harbouring γ-Tubulin labeling. Scale bar represents 1 μm. See also Figure S4.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-CEP128 | Abcam | Ab11879 |
| Rabbit anti-ODF2 | Abcam | Ab43840 |
| Rabbit anti-ODF2 | Gift from Kyung Lee lab at NIH | N/A |
| Rabbit anti-CNTRL | Santa Cruz | Sc-135020 |
| Mouse anti-CNTRL | Atlas Antibodies | HPA020468 |
| Rabbit anti-CNTRL | Santa Cruz | Sc-365521 |
| Rabbit anti-CNTRL | Atlas Antibodies | HPA051583 |
| Rabbit anti-POC1B | Invitrogen | PA5–24495 |
| Rat anti-POC1B | Gift from Tomer Avidor-Reiss lab at University of Toledo | N/A |
| Mouse anti-polyglutamylation modification | Adipogen | AG-20B-0020-C1(GT355) |
| Mouse anti-TUBA4A(a-tubulin) | Sigma-Aldrich | T9026(DM1A) |
| Mouse anti-TUBA4A(a-tubulin-FITC conjugated) | Sigma-Aldrich | F2168(DM1A) |
| Mouse anti-g-tubulin | Sigma-Aldrich | T6557 |
| Rabbit anti-RSPH4A | Atlas Antibodies | HPA031196 |
| Rabbit anti-GAS8 | Atlas Antibodies | HPA041311 |
| Mouse anti-acetylated a-tubulin | Sigma-Aldrich | T7451 |
| Rat anti-ZO-1 | Gift from Moe Mahjoub lab at Washington University | N/A |
| Rabbit-Arl13B | Gift from William Trimble Lab at the Hospital for Sick Children | |
| Goat anti rabbit IgG Alex fluor 488 | Thermo Fisher | A11034 |
| Goat anti mouse IgG Alex fluor555 | Thermo Fisher | A21424 |
| Goat anti rabbit IgG Alexa fluor 405 | Thermo Fisher | A-31556 |
| Goat anti rabbit F(ab')2 Alexa fluor 647 | Thermo Fisher | A21246 |
| HRP-conjugated anti-IgG antibodies | Cell Signaling | 7074, 7076 |
| Biological Samples | ||
| Mouse tracheal epithelia cells | This study | N/A |
| Mouse ependymal epithelia cells | This study | N/A |
| Human airway multiciliated cells from patient PCD1(DNAH5: allele1: intronic, abolishes accepted splice site for exon68, allele2:p.Arg478*) | This study | N/A |
| Human airway multiciliated cells from patient PCD2 (DNAH11: allele1: pCys4286*, allele2:pIle4122Ser) | This study | N/A |
| Human airway multiciliate cells from patient PCD3(DNAH5, homogenous, pPhe634fs*2) | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM/F-12 | Thermofisher | 11320033 |
| PneumaCult-Ex medium | Stem Cell Technologies | 5008 |
| PneumaCult-ALI medium | Stem Cell Technologies | 05001 |
| Paraformaldehyde | Electron Microscopy Sciences | 15710 |
| Centrinone A | from Moe Mahjoub lab at Washington University | N/A |
| RIPA lysis buffer | Pierce, Thermo Fisher Scientific | 89900 |
| Critical Commercial Assays | ||
| APEX Antibody labeling kit | Thermo Fisher Scientific | A10470, A10468 |
| Novex ECL Chemiluminescent Substrate Kit | Invitrogen | WP20005 |
| RNeasy Micro Kit | QIAGEN | 7400 |
| SuperScript III First-Strand Synthesis System for RT-PCR | Invitrogen | 18080–051 |
| Gibson Assemby Kit | New England Biolabs | E2611S |
| Experimental Models: Organisms/Strains | ||
| Mouse strain: CEN2-GFP | The Jackson Laboratory; Higginbotham et al., 2004 | CB6-Tg(CAG-EGFP/CETN2)3–4Jgg/J |
| Oligonucleotides | ||
| Primers for EGFP semi quantitiative RT-PCR: FOR: AGAAGAACGGCATCAAGGTG; | This study | N/A |
| Primers for EGFP semi quantitiative RT-PCR: REV: GAACTCCAGCAGGACCATGT’ | This study | N/A |
| Primers for tagRFP semi quantitiative RT-PCR: FOR: AACACCGAGATGCTGTACCC; | This study | N/A |
| Primers for tagRFP semi quantitiative RT-PCR: REV: ACGTAGGTCTCTTTGTCGGC | This study | N/A |
| Primers for Cyclophilin semi quantitiative RT-PCR: FOR: ACCCCACCGTGTTCTTCGAC; | This study | N/A |
| Primers for Cyclophilin semi quantitiative RT-PCR: REV: CATTTGCCATGGACAAGATG | This study | N/A |
| Recombinant DNA | ||
| CMV-TagRFP-Cen1 plasmid | Gift from Xavier Morin lab at CNRS | N/A |
| Software and Algorithms | ||
| FIJI-ImageJ | Schneider et al., 2012 | https://imagej.net/Fiji |
| MATLAB | MathWorks | Version 2019b https://www.mathworks.com/products/get-matlab.html |
| ELYRA PS.1 | Carl Zeiss Microscopy | Version PS.1 |
| ZEN Black | Carl Zeiss Microscopy | Version 8.1 |
| Excel | Microsoft | Version 2013 |
| Prism | GraphPad | Version 8 https://www.graphpad.com/scientific-software/prism/ |
| STORM image drift correction and binning | This study | https://drive.google.com/open?id=11fuWn7kmZ-loCn79CKChJI5FeMme0fDU |
| Auto Slice & View G3 | FEI Company, Hillsboro, OR USA | https://www.fei.com/document/introducing-auto-slice-and-view-G3/#gsc.tab=0 |
| Amira | FEI Company, Hillsboro, OR USA | 6.0.1 |
| Metamorph | Molecular Devices | N/A |
| Hybrid cilia position analysis | This study | https://drive.google.com/open?id=182KAccJf6YC69WbovKgTwtg62Y5DTadA |
| Basal body alignment analysis | This study | https://drive.google.com/open?id=182KAccJf6YC69WbovKgTwtg62Y5DTadA |
Finally, we sought to determine whether directional flow generated during multiciliated cell differentiation helps to position the hybrid cilium. In order to mimic conditions in the airway, mouse tracheal progenitor cells were subjected to mechanical agitation to induce directional surface flow during their differentiation, thereby promoting basal body and cellular alignment (Guirao et al., 2010). Indeed, analyses of mouse tracheal cells confirmed that the hybrid cilium becomes positioned in the front end of the cells toward the direction of flow (Figure 6D), consistent with what was observed in human primary cultures (Figures 1A and 6B) and mouse tracheal tissue explant (Figure 6E). Altogether these data show that the hybrid cilium position is biased towards the direction of beating and that its position is influenced by external flow and ciliary motility.
Interestingly, labelling of mouse trachea with microtubule nucleating and anchoring complex protein γ-Tubulin showed that multiple basal feet of the hybrid cilium are competent for linkage to the microtubules network (Figures 6E and 6F). To better understand the function of the hybrid cilium, mouse tracheal cells were then treated with centrinone A to inhibit hybrid cilia formation, and cells were subjected to directional flow as described above. The chemical approach was the only available method for loss of function studies, since hybrid cilium protein composition is not yet known and therefore no targets for selective genetic removal during ciliogenesis are available. To ensure maximal hybrid cilia reduction, centrinone A was used throughout mouse tracheal airway multiciliated cell differentiation, a treatment that did not impact multiciliated cell formation (Nanjundappa et al., 2019). As previously observed, airway cells in both DMSO and centrinone A-treated conditions showed similar ciliation levels and overall comparable tissue level organization, with some noticeable differences in cellular alignment (Nanjundappa et al., 2019) (Figures 7A–7D). However, while cells treated with vehicle control showed basal bodies locally aligned in rows as expected (Figures 7A and 7B), cells depleted of the hybrid cilium show impaired basal body alignment as evidenced by the staining of basal body component CEP135 (Figures 7C and 7D).
Figure 7. Hybrid cilium is critical for establishing basal body alignment.
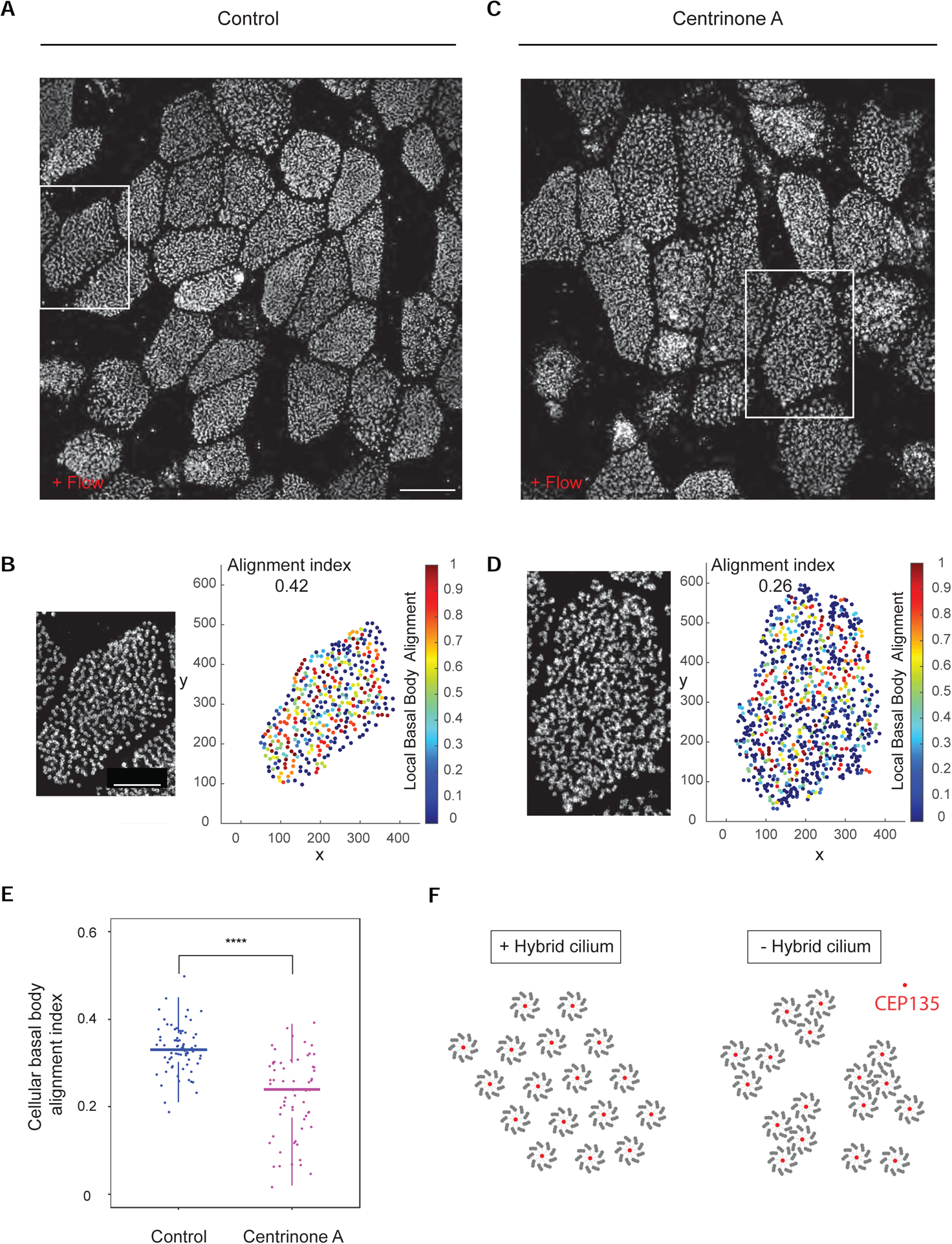
(A) 3D-SIM volume maximum intensity projection of mouse tracheal multiciliated cell (ALI D20), treated with DMSO (control) from basal cells expansion throughout ALI differentiation labeled with anti-CEP135 antibodies. Scale bar represents 10 μm. (B) Left: High-magnification view of boxed area in (A). Right: relative basal body local and global alignment measurements. (C) 3D-SIM volume maximum intensity projection of mouse tracheal multiciliated cell (ALI D20), treated centrinone A (right) from basal cells expansion throughout ALI differentiation labeled with anti-CEP135 antibodies. Scale bar represents 10 μm. (D) Left: High-magnification view of boxed area in (C). Right: relative basal body local and global alignment measurements. (E) Quantification of cellular basal body alignment index in cells treated with DMSO or centrinone A throughout multiciliated cells differentiation (n=70 over 3 independent biological replicates, total 63 cells. DMSO=0.33±0.05; centrinone A=0.23±0.09). Statistical analysis was done using unpaired t-test. Data are represented as mean ± SD. **** p<0.0001. (F) A model showing function of the hybrid cilia. When hybrid cilium is present, basal bodies are well aligned. However, when hybrid cilium is removed by centrinone A, basal body alignment in the cell is disrupted and basal bodies tend to group into clusters. Red present basal body protein CEP135, surrounded by 9 axonemal microtubule triplets.
To quantify the changes in basal body organization, the positions of basal bodies within each cell were analysed for local alignment and then averaged to a global alignment index for each cell with custom MATLAB scripts, similar to previously described methods (Herawati et al., 2016). This quantitative analysis demonstrates that hybrid cilium depletion leads to a significant reduction in alignment index for the multiciliated cells (mean=0.23±0.09 as compared to 0.33±0.05, n=63 (DMSO); n=70 cells (Centrinone A)), suggesting an overall basal body organization disruption at the cellular level (Figures 7B, 7D and 7E). Altogether, our data show that the hybrid cilium regulates basal body alignment in multiciliated cells (Figure 7F).
DISCUSSION
Our study led to the surprising discovery that a different cilium is harboured among the hundreds of motile cilia in a differentiated multiciliated cell. The hybrid cilium has features of motile and primary cilia, including multiple basal feet as found in primary cilia, and a central pair apparatus and proteins required for ciliary beating as in motile cilia. Airway cells lacking hybrid cilium differentiate normally, extrude motile cilia, but lack effective basal body alignment.
Our data reveal the fate of mother centriole in mature multiciliated cells, a major unanswered question in this field. In airway cells, during the early stages of differentiation - before Foxj1 expression - the mother centriole templates a primary cilium that is subsequently resorbed before centriole amplification (Jain et al., 2010). After this step, its role has remained mysterious. Here, we show that one of the parental centrioles, likely the mother, resurfaces to generate the hybrid cilium harboured among motile cilia. In agreement with results in airway cells, pulse-chase experiments in mouse cultured ependymal cells showed that centrosomal centrioles are retained within the newly formed basal body patch and are capable of growing cilia.
In airway cells differentiated in vitro, we observed a subpopulation of cells with two hybrid cilia, but less frequently so in ex vivo trachea and primary nasal airway cells. Our data suggest that mother centriole is the origin of the hybrid cilium, however we cannot rule out that a disengaged daughter centriole might reach the surface of multiciliated cells and express multiple basal feet. An alternative possibility is that basal stem cell duplication driven in vitro during population expansion might lead to an increase in cells with two mother centrioles due to mitotic division defects.
Notably, analysis of PCD patients with CCNO mutations leading to oligocilia demonstrates that hybrid cilium formation is determined independently of other motile cilia in the same multiciliated cell. This suggests two possible scenarios at play during multiciliated cells differentiation: one possibility is that CCNO patients’ cells have lost the capacity to duplicate basal bodies of motile cilia, but are capable of executing the rest of the motile cilia formation program (including transport, docking and extension at the membrane). Supporting this view are previous studies from the Kintner lab in Xenopus that showed that RNA misexpression of FOXJ1 on surface epithelial cells, in a mutant background inhibiting motile cilia formation, leads to the emergence of a single cilium or biciliated cells on the surface of epithelia. These cilia exhibit a 9+2 axonemal microtubule structure, beat both in a rotational and in plane pattern and are of intermediate length between a primary and motile cilia (Stubbs et al., 2008). Interestingly, though, these cilia of intermediate length do not seem to be present in mature human airway multiciliated cells (our observations). Alternatively, it is possible that a specific molecular program exists that is responsible for hybrid cilium formation, which is separate (at least in part), whose drivers and components remain unknown.
Interestingly, PCD patients with oligocilia mutations appear to have worse clinical outcomes than patients with mutations in most of the PCD genes leading to ciliary immotility (Boon et al., 2014; Wallmeier et al., 2014; Wallmeier et al., 2019). This suggests that additional molecular functions, beyond simply ciliary motility, might contribute to the poor clinical outcome.
The hybrid cilium is found to be preferentially positioned towards the direction of ciliary beating. Since effective flow generated by beating motile cilia is required to maintain the position of the hybrid cilium, this suggests that it responds to flow either directly or indirectly. The biased position of the hybrid cilium is reminiscent to that of the primary cilium at the leading edge in the wound region in vascular and bronchial smooth muscle cells, where it is thought to sense extracellular matrix proteins and promote cell migration (Lu et al., 2008; Wu et al., 2009). This biased position also resembles that of primary cilium in radial glia cells, the progenitors of ependymal cells, where it has been shown to regulate translational polarity - a basal body organization specific to ependymal multiciliated cells that is not found in the airway (Mirzadeh et al., 2010).
Our data show that loss of hybrid cilium by centrinone A treatment leads to reduced basal body alignment in airway cells. An interesting mechanistic explanation is that the multiple basal feet of the hybrid cilium function to align the different rows of basal bodies as a rudder of a sailboat; PCP proteins might also be affected by hybrid cilium removal (Mitchell et al., 2009; Vladar et al., 2012).
Notably, basal feet of primary cilia have been previously linked to TGFbeta signalling (Clement et al., 2013; Monnich et al., 2018). It is therefore a possibility that the hybrid cilium might harbour a different complement of signalling molecules compared to motile cilia, which might help explain its specialized function. Our initial efforts to detect primary cilia-specific markers specifically enriched at the hybrid cilium did not produce any valuable insight. Future studies are needed to address whether the hybrid cilium, in addition to being a distinct organelle structurally and functionally, also has specific signalling capabilities.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contract
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contract, Vito Mennella (v.mennella@soton.ac.uk)
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement
Data and Code Availability
The published article includes all datasets generated during this study. Code is available by the Google Drive link specified in method details.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse primary cells
Mouse tracheal epithelia primary cells were acquired from the trachea of C57BL/6 mice at 2–4 months of age, following protocols that are compliant with guidelines of the Institutional Animal Care and Use Committee at Washington University and the National Institutes of Health. For ependymal cell culturing, mouse strain Cen2-GFP (CB6-Tg(CAG-EGFP/CETN2)3–4Jgg/J, The Jackson Laboratory) was used to obtain primary cells, according to the guidelines of the European Community and French Ministry of Agriculture, approved by the Ethic comity Charles Darwin (C2EA-05) and “Direction départementale de la protection des populations de Paris”, (Approval number Ce5/2012/107; APAFiS #9343).
Human primary cells
Healthy volunteers and PCD patients were recruited and airway primary cells were obtained following a protocol approved by Research Ethics Board(REB #1000054690) at the Hospital For Sick Children and adhering to local and national research and ethical approval (Southampton and South West Hampshire Research Ethics 07/Q1702/109). PCD1 is 16 years old female with DNAH5 mutations (c1432C>T&c11571–1G>A). PCD2 is 9 years old female with DNAH11 mutations (c12888T>A&c12365T>G). PCD3 is 12 years old female with DNAH5 mutations (DNAH5 c1901_1902delTC).
METHOD DETAILS
Immortalized and Primary Cell Culture
Human primary nasal airway cells from healthy volunteers and PCD patients were collected with using a cytology brush by a nurse, following a protocol approved by Research Ethics Board(REB #1000054690) at the Hospital For Sick Children and adhering to local and national research and ethical approval (Southampton and South West Hampshire Research Ethics 07/Q1702/109). Airway cells were then expanded in PneumaCult-Ex media, seeded on transwells (Corning HTS Transwell-96 and −24 permeable support; 0.4 μm pore size), and differentiated for at least 21 days following Stem Cell Technologies protocols using PneumaCult-Ex and PneumaCult-ALI media. The media were supplemented with vancomycin, tobramycin, gentamicin and antibiotic-antimycotic antibiotics.
Immunofluorescence and Antibodies
Human nasal and mouse tracheal multiciliated cells from ALI cultures were directly fixed on Transwell filters with either methanol (20 min at −20°C) or 4 % paraformaldehyde (PFA; 10 min at room temperature (RT)). For PFA fixation, cells were subsequently reduced with 0.1% Sodium Borohydride for 7 minutes, then permeabilized with 0.2% Triton X-100 for 25 minutes. Cells were blocked using 5% FBS-containing PBS, incubated with primary antibodies for either 1 hour (RT) or overnight (4°C), and then secondary antibodies conjugated with Alexa FluorAlexafluor −405, −488, −555 orand −647 nm (Thermo Fisher Scientific). When appropriate, cells were stained with directly labeled primary antibodies (prepared using APEX Antibody Labeling Kit, Thermo Fisher Scientific and Mix-n-Stain Antibody Labeling Kit, Sigma-Aldrich). For STORM imaging, Alexa647 conjugated F(ab’)2 fragments were used conjugated. Please refer to key resources table for a list of antibodies used in this study. Trachea labeling was performed essentially as in (Vladar et al., 2015). Briefly, mouse was euthanized by CO2 asphyxiation, douse with 70% ethanol and trachea was isolated. The trachea was placed on ice cold Ham’s F12 +1% pen-strep. The lumen of the trachea was then exposed by cutting it open longitudinally along the dorsal surface using dissecting scissors. The trachea was then fixed by adding −20 °C methanol over the tissue for 10 min, blocked with 10% normal serum in PBST (pH 7.4 +10% Triton X-100), incubated with primary antibodies for 1 hour (RT) and then fluorescent dye conjugated secondary antibodies for 30 minutes (RT). A flat and rectangular area of the trachea was finally cut out and mounted for subsequent imaging.
Super-resolution Imaging
3D-SIM data was collected using ELYRA PS.1 (Carl Zeiss Microscopy) with a Plan-Apochromat 63x or 100x/1.4 Oil immersion objective lens with an additional 1.6x optovar. An Andor iXon 885 EMCCD camera was used to acquire images with 101 nm/pixel z-stack intervals over a 5–10 μm thickness. For each image field, grid excitation patterns were collected for five phases and three rotation angles (−75°; −15°, +45°). The raw data was reconstructed and channel aligned using SIM module of ZEN Black Software (version 8.1). STORM data was collected using PALM mode in ELYRA PS.1 (Carl Zeiss Microscopy) with a Plan-Apochromat 63x or 100x/1.4 Oil immersion objective lens with an additional 1.6x optovar. An Andor iXon 885 EMCCD camera was used to acquire images using TIRF mode. Lasers of wavelength 647 nm and 405 nm were used to activate the fluorophore. Raw data was reconstructed using PALM module of Zen Black Software (version 8.1), with the account for overlapping molecules. Reconstructed data was further processed for drift correction and binning using home-written MATLAB script (can be accessed via the following link: https://drive.google.com/open?id=11fuWn7kmZ-loCn79CKChJI5FeMme0fDU).
Transmission Electron Microscopy (TEM)
ALI filters of fully differentiated human nasal multiciliated cells were fixed in 2% glutaraldehyde in 0.1M sodium cacodylate buffer. Samples were rinsed in 0.1M sodium cacodylate buffer with 0.2M sucrose, post-fixed in 1% OsO4 in 0.1M sodium cacodylate buffer, dehydrated in a graded ethanol series (70%, 90%, 3X 100%), infiltrated with propylene oxide, and embedded in Quetol-Spurr resin. Serial sections (90 nm-thickness) were cut on a Leica Ultracut ultramicrotome, stained with uranyl acetate and lead citrate, and imaged in a FEI Tecnai 20 TEM.
Focused Ion Beam Scanning Electron Microscopy (FIB-SEM)
ALI filters of fully differentiated human nasal multiciliated cells were fixed in 2.5% glutaraldehyde and 0.05% malachite green oxalate in 0.1M sodium cacodylate buffer, rinsed in 0.1M sodium cacodylate buffer, post-fixed in 0.8% potassium ferrocyanide and 1% OsO4 in 0.1M sodium cacodylate buffer. The samples were treated with 1% tannic acid, stained with 0.5% uranyl acetate, followed by dehydration in a graded acetone series (25%, 50%, 75%, 95% and 100%), and embedded in resin. Resin formulation: 18.2% Araldite M (Sigma-Aldrich), 22.7% Epon 812 (Sigma-Aldrich), 54.5% Hardener DDSA (Sigma-Aldrich) and 4.5% DMP-30 (Sigma-Aldrich). FIB-SEM imaging for Sup. Movie 1,2 was performed as described below. Sample blocks for analysis by FIB-SEM were trimmed and mounted on a 45° pre-titled SEM stub and coated with a 4-nm layer of Pt to enhance electrical conductivity. Milling of serial sections and imaging of block face after each Z-slice was carried out with the FEI Helios Nanolab 660 DualBeam using Auto Slice & View G3 ver 1.5.3 software (FEI Company, Hillsboro, OR USA). A block was first imaged to determine the orientation relationship between the block face of ion and electron beams. A protective carbon layer 50 μm long, 8 μm wide and 2 μm thick was deposited on the surface of the region of interest to protect the resin volume from ion beam damage and correct for stage and/or specimen drift, i.e., perpendicular to the image face of the volume to be milled. Trenches on both sides of the region of interest were created to minimize re-deposition during automated milling and imaging. Imaging fiducials were generated for both ion and electron beam imaging and were used to dynamically correct for drift in the x- and y-directions by applying appropriate SEM beam shifts. Ion beam milling was performed at an accelerating voltage 30 kV and beam current of 9.3 nA, stage tilt of 9°, and working distance of 4 mm. With each milling step, 10 nm thickness of the material was removed. Each newly milled block face was imaged with the through-the-lens detector for backscattered electrons (TLD-BSE) at an accelerating voltage of 2 kV, beam current of 0.4 nA, stage tilt of 47°, and working distance of 3 mm. The pixel resolution was 10.3 nm with a dwell time of 30 μs. Pixel dimensions of the recorded image were 1536 × 1024 pixels. Seven hundred and forty-three images were collected and the contrast of the images inversed. Visualization and direct 3-D volume rendering of the acquired dataset was performed with Amira 6.0.1 (FEI Company, Hillsboro, OR USA).
MTEC and ependymal cell experiments
Mouse tracheal epithelia cell (MTEC) cultures were established as previously described (Mahjoub et al., 2010; You et al., 2002). Briefly, C57BL/6 mice were sacrificed at 2–4 months of age, trachea were excised, opened longitudinally to expose the lumen, and placed in 1.5 mg/mL Pronase E in DMEM/F12 medium (Life Technologies) at 4°C overnight. Tracheal epithelial cells were dislodged by gentle agitation and collected in DMEM/F12 with 10% FBS. After centrifugation, cells were treated with 0.5 mg/mL DNase I for 5 min on ice and centrifuged at 4°C for 10 min at 400 g. Cells were resuspended in DMEM/F12 with 10% FBS and plated in a tissue culture dish for 5 h at 37°C with 5% CO2 to adhere contaminating fibroblasts. Non-adhered cells were then collected, concentrated by centrifugation, resuspended in an appropriate volume of mTEC-Plus medium (You et al., 2002), and seeded onto Transwell-Clear permeable filter supports (Corning).
To eliminate parental centrioles, cells were incubated in the presence of 1uM centrinone A (Nanjundappa et al., 2019) for 6 days. Air-liquid interface (ALI) was established 2 days after cells reached confluence by feeding mTEC-Serum-Free medium only in the lower chamber. Cells were cultured at 37°C with 5% CO2, and media replaced every 2 days, and fixed on the indicated days. All chemicals were obtained from Sigma Aldrich unless otherwise indicated. Media were supplemented with 100 U/mL penicillin, 100 mg/mL streptomycin, and 0.25 mg/mL Fungizone (all obtained from Thermo Fisher).
For establishing directional flow in ALI mouse culture, tracheal progenitor cells were isolated and seeded onto 12 mm Transwell-Clear permeable filter supports as described above. To eliminate parental centrioles, cells were incubated in mTEC-Plus medium supplemented with 1uM centrinone A for up to 6–8 days. Air-liquid interface was established two days after cells reached confluence by incubating cells in mTEC-Serum-Free, and centrinone was maintained in the medium throughout the differentiation process. A small volume (100uL) of mTEC-Serum-Free (with or without centrinone) was added to the upper chamber, and culture plates were placed on a nutating shaker (Fisher Scientific). Samples were rotated in a counter-clockwise direction at a speed of one rotation every 3 seconds, yielding an estimated directional flow rate of 12.5mm/sec. Cells were cultured at 37°C with 5% CO2 with media replaced every two days. Cells were fixed and stained for centrioles and basal feet as described.
For ependymal cell culturing, all animal studies were performed in accordance with the guidelines of the European Community and French Ministry of Agriculture and were approved by the Ethic comity Charles Darwin (C2EA-05) and “Direction départementale de la protection des populations de Paris”, (Approval number Ce5/2012/107; APAFiS #9343). The mouse strain, Cen2-GFP (CB6-Tg(CAG-EGFP/CETN2)3–4Jgg/J, The Jackson Laboratory), has already been described by Higginbotham (Higginbotham et al., 2004). For in vivo analysis, animals used were homozygous for the Cen2-GFP. Lateral walls of the lateral brain ventricles were dissected as previously explained (Delgehyr et al., 2015). The tissue was treated with 0.1% triton in BRB80 (80 mM K-Pipes pH6.8; 1 mM MgCl2; 1 mM Na-EGTA) for 1 min prior to fixation and fixed in methanol at −20°C for 10 min. Saturation and antibody incubations were performed in PBS containing 10% FBS and 0.1% triton. Primary antibodies (CNTRL (monoclonal mouse from Santa Cruz) and CEP164) were incubated overnight (4°C). Secondary antibodies conjugated with Alexa Fluor −555 and −647 were incubated for 1h.
For in vitro pulse-chase experiments, cultures were performed as previously described. Transfection of ependymal cell progenitors was performed at 80% of confluency during the proliferation phase with a CMV-TagRFP-Cen1 plasmid (gift from Xavier Morin, ENS, Paris), which codes for human centrin 1 fused to TagRFP under the control of a CMV promoter, using jetPRIME Polyplus kit. Cells (in 25cm3 flask) were transfected with a mix of 0.75μg of DNA, 300μL of jetPRIME Buffer and 1.5μL of jetPRIME transfection reagent in 3 mL of fresh complete medium (DMEM-Glutamax (Thermofisher) containing 10% FBS and 1% Penicillin/Streptomycin). After 4 hours at 37°C in 5% CO2 incubator, the medium was renewed. One day after proliferation, cells were shaken at 250rpm overnight. Cells were plated on coverslips or Labtek chambers slides coated with L-Polylysin (40 μg/ml in pure water) at a density of 0.75 × 104 cells per μl in 20 or 60 μl drops. The medium was then replaced by serum-free DMEM-Glutamax-I 1% P/S, to trigger ependymal differentiation in vitro (DIV0). Cells were either fixed with Paraformaldehyde (4% in PBS) for 10min or used for live imaging between DIV3 and DIV6. Fixed cells were examined with an upright epifluorescence microscope (Zeiss Axio Observer.Z1) equipped with Apochromat X63 (NA 1.4) or X100 (NA 1.4) oil-immersion objectives and a Zeiss Apotome with an H/D grid. Fields of views with cells showing RFP-tagged centrosomal centrioles were acquired using Zen software with 230-nm z-steps and analyzed with image-J.
For live imaging, differentiating ependymal cells with two bright RFP-positive/EGFP positive centrosomal centrioles and RFP-negative/EGFP-positive procentrioles in amplification or growth-phases and filmed using an inverted spinning disk Nikon Ti PFS microscope equipped with an oil-immersion X100 (NA 1.4) objective, an Evolve EMCCD Camera (Photometrics), dpss lasers (491 nm, 561 nm), a motorized scanning deck and an incubation chamber (37°C; 5% CO2; 80% humidity). Laser intensities and image capture times were respectively set to 20%, 50 ms for 488 nm and 25%, 100 ms for 561 nm. Images were acquired with Metamorph software at 60 minutes time interval for 24 hours. Image stacks were recorded with a z-distance of 0.7 mm. Four dimensional (x, y, z, t) time-lapse images were analyzed with Image J.
Semi-quantitative RT-PCR
RNA was purified from 1.5 × 105 cells on coverslips using the RNeasy Micro Kit (QIAGEN, 74000). Reverse transcription was performed using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, 18080–051). PCR was performed on cDNA using the primers 5’-AGAAGAACGGCATCAAGGTG-3’ and 5’-GAACTCCAGCAGGACCATGT-3’ for EGFP, 5’-AACACCGAGATGCTGTACCC-3’ and 5’-ACGTAGGTCTCTTTGTCGGC-3’ for tagRFP and 5’-ACCCCACCGTGTTCTTCGAC-3’ and 5’-CATTTGCCATGGACAAGATG-3’ for cyclophilin. Images of the gels were then analysed on ImageJ. The ratio between EGFP or tagRFP and cyclophilin band intensity were calculated. Quantifications of 3 independent experiments were pooled and plotted.
QUANTIFICATION AND STATISTICAL ANALYSIS
Positional analysis of hybrid cilium and basal body alignment assay
Custom written MATLAB script was used to determine the position of the hybrid cilia in multiciliated cells relative to cilia beating direction (can be accessed via the following link: https://drive.google.com/open?id=182KAccJf6YC69WbovKgTwtg62Y5DTadA). First, intensity thresholds for all channels were chosen for and binary images were generated to identify individual basal body and basal foot objects. Individual cells were outlined via manual cell border drawing. Basal body-basal foot pairs were identified based on the pairwise nearest neighbour search with a distance threshold of ~600 nm. The direction of a single cilium was defined as from the weighted center of the basal body object to that of the paired basal foot. All cilia directions in one cell were determined and the mean direction was regarded as the direction of beating in a cell. The cilia beating angles obtained were transformed into a two-dimensional unit vector: . The resultant vector was the average of all the unit vectors in a cell: . The resultant vector length r was defined as the norm of the resultant vector: r = ∥ṟ∥. The circular standard deviation was defined as: . All the directions in a single cell were also subject to Rayleigh’s test for uniformity distribution. The p-value is calculated as: ; rn = r × n. A p-value < 0.05 indicated that the cilia in the cell are significantly aligned. Aligned vector length was defined to describe the cilia alignment level in a cell with values ranging from 1 to 0, with 1 indicating 100% alignment and 0 indicating no alignment. The mean beating direction of all cilia were defined as the cilia beating direction. The hybrid cilia position relative to the cilia beating direction was measured using the same basal foot and basal body markers in cells whose size is normalized to [−0.5; 0.5] both along the cilia beating direction (regarded as cell length) and the direction perpendicular to it (regarded as cell width).
For analysis of basal body alignment, centers of basal bodies were firstly identified manually. Basal body alignment index for a cell was then calculated using all the basal body coordinates as input by running an algorithm reported previously (Herawati et al., 2016). Briefly, a local basal body alignment index was calculated for each basal body within a cell and the basal body alignment index for a cell is the average of all local values within it. To calculate local basal body alignment index, the neighbouring basal bodies for the basal body were identified firstly by applying a distance threshold (1.3d, d represents the averaged nearest neighbour distance of basal bodies from 3 control images). Next, for ith basal body with n neighbours, local index Ia, i was evaluated using a set of equations shown below. Ia,i value falls between 0 and 1 with 0 as no alignment and 1 as highly aligned.
| equation 1 |
| equation 2 |
| equation 3 |
Basal body alignment level for control and centrinone treatment samples from 3 independent biological replicates was measured. Statistics were performed by student’s t test.
Statistical Analysis
Data was analysed in Microsoft Excel and Prism software. Statistical tests, sample sizes and number of biological or technical replicates were specified in figure legends or method details. Differences were regarded as significant if p < 0.05, unless otherwise stated.
Supplementary Material
Supplemental Video S1. FIB-SEM tomograms of multiple airway multiciliated cells grown in air-liquid interface. 10 nm isotropic resolution. This video is related to Figure 3.
Supplemental Video S2. High magnification of Supplemental Video S1. This video is related to Figure 3.
Supplemental Video S3. Live imaging of TagRFP-Cen1 centrosomal centrioles during centriole amplification in primary cultured ependymal progenitors from Cen2-EFGP mice. This video is related to the Figures 4 and S2, “MTEC and ependymal cell experiments” part of the STAR
Highlights:
Multiciliated cells contain a hybrid cilium with features of primary and motile cilium
The hybrid cilium originates from parental centriole
The hybrid cilium position is biased towards the cilia beating direction
The hybrid cilium functions in basal body alignment
ACKNOWLEDGEMENT
This project is funded by CIHR program grant # 391917 to VM and SD; National Heart, Lung and Blood Institute (R01-HL128370) and the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CDI-CORE-2019-813) to MRM; Z.L. was supported by the SickKids Restracomp Fellowship. The authors acknowledge PCD patients and volunteers for providing nasal cells for this study, Julie Avolio for help with nasal cell scraping. Jia Zhou, Cindy Fang and Jasmine Kang assisted in data analysis. Douglas Holmyard (EM facility, The Hospital for Sick Children) prepared TEM and FIB-SEM samples and helped set up EM imaging. McGill EM facility contributed to FIB-SEM acquisition. We thank Profs Sudipto Roi’s lab, Jeremy Reiter’s lab and Nick Berbari’s lab for experimental help. We thank Profs Bornens, Pelletier, Cheeseman, Kyung Lee, Elsasser, Avidor-Reiss laboratories for sharing antibodies and plasmids.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATIONS OF INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Al Jord A, Lemaitre AI, Delgehyr N, Faucourt M, Spassky N, and Meunier A (2014). Centriole amplification by mother and daughter centrioles differs in multiciliated cells. Nature 516, 104–107. [DOI] [PubMed] [Google Scholar]
- Boisvieux-Ulrich E, Laine MC, and Sandoz D (1985). The orientation of ciliary basal bodies in quail oviduct is related to the ciliary beating cycle commencement. Biol Cell 55, 147–150. [DOI] [PubMed] [Google Scholar]
- Boon M, Wallmeier J, Ma L, Loges NT, Jaspers M, Olbrich H, Dougherty GW, Raidt J, Werner C, Amirav I, et al. (2014). MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nature communications 5, 4418. [DOI] [PubMed] [Google Scholar]
- Chong WM, Wang WJ, Lo CH, Chiu TY, Chang TJ, Liu YP, Tanos B, Mazo G, Tsou MB, Jane WN, et al. (2020). Super-resolution microscopy reveals coupling between mammalian centriole subdistal appendages and distal appendages. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare DK, Magescas J, Piolot T, Dumoux M, Vesque C, Pichard E, Dang T, Duvauchelle B, Poirier F, and Delacour D (2014). Basal foot MTOC organizes pillar MTs required for coordination of beating cilia. Nat Commun 5, 4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement CA, Ajbro KD, Koefoed K, Vestergaard ML, Veland IR, Henriques de Jesus MP, Pedersen LB, Benmerah A, Andersen CY, Larsen LA, et al. (2013). TGF-beta signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep 3, 1806–1814. [DOI] [PubMed] [Google Scholar]
- Delgehyr N, Meunier A, Faucourt M, Bosch Grau M, Strehl L, Janke C, and Spassky N (2015). Ependymal cell differentiation, from monociliated to multiciliated cells. Methods Cell Biol 127, 19–35. [DOI] [PubMed] [Google Scholar]
- Frisch D, and Farbman AI (1968). Development of order during ciliogenesis. Anat Rec 162, 221–232. [DOI] [PubMed] [Google Scholar]
- Frommer A, Hjeij R, Loges NT, Edelbusch C, Jahnke C, Raidt J, Werner C, Wallmeier J, Grosse-Onnebrink J, Olbrich H, et al. (2015). Immunofluorescence Analysis and Diagnosis of Primary Ciliary Dyskinesia with Radial Spoke Defects. Am J Respir Cell Mol Biol 53, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk MC, Bera AN, Menchen T, Kuales G, Thriene K, Lienkamp SS, Dengjel J, Omran H, Frank M, and Arnold SJ (2015). Cyclin O (Ccno) functions during deuterosome-mediated centriole amplification of multiciliated cells. EMBO J 34, 1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati DF, Mitchell BJ, and Pearson CG (2016). Subdistal Appendages Stabilize the Ups and Downs of Ciliary Life. Dev Cell 39, 387–389. [DOI] [PubMed] [Google Scholar]
- Gibbons IR (1961). The relationship between the fine structure and direction of beat in gill cilia of a lamellibranch mollusc. J Biophys Biochem Cytol 11, 179–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts BN, Gong-Cooper X, and Hackett BP (2004). Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. J Cell Sci 117, 1329–1337. [DOI] [PubMed] [Google Scholar]
- Guirao B, Meunier A, Mortaud S, Aguilar A, Corsi JM, Strehl L, Hirota Y, Desoeuvre A, Boutin C, Han YG, et al. (2010). Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat Cell Biol 12, 341–350. [DOI] [PubMed] [Google Scholar]
- Herawati E, Taniguchi D, Kanoh H, Tateishi K, Ishihara S, and Tsukita S (2016). Multiciliated cell basal bodies align in stereotypical patterns coordinated by the apical cytoskeleton. J Cell Biol 214, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Bielas S, Tanaka T, and Gleeson JG (2004). Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res 13, 155–164. [DOI] [PubMed] [Google Scholar]
- Jain R, Pan J, Driscoll JA, Wisner JW, Huang T, Gunsten SP, You Y, and Brody SL (2010). Temporal relationship between primary and motile ciliogenesis in airway epithelial cells. Am J Respir Cell Mol Biol 43, 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilyaprak C, Stierhof YD, and Humbel BM (2019). Volume microscopy in biology: FIB-SEM tomography. Tissue Cell 57, 123–128. [DOI] [PubMed] [Google Scholar]
- Knowles MR, Leigh MW, Carson JL, Davis SD, Dell SD, Ferkol TW, Olivier KN, Sagel SD, Rosenfeld M, Burns KA, et al. (2012). Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure. Thorax 67, 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimoto K, Yamazaki Y, Nishida T, Shinohara K, Ishikawa H, Hasegawa T, Okanoue T, Hamada H, Noda T, Tamura A, et al. (2012). Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell 148, 189–200. [DOI] [PubMed] [Google Scholar]
- Liu Z, Nguyen QPH, Guan Q, Albulescu A, Erdman L, Mahdaviyeh Y, Kang J, Ouyang H, Hegele RG, Moraes T, et al. (2020). A quantitative super-resolution imaging toolbox for diagnosis of motile ciliopathies. Sci Transl Med 12. [DOI] [PubMed] [Google Scholar]
- Lu CJ, Du H, Wu J, Jansen DA, Jordan KL, Xu N, Sieck GC, and Qian Q (2008). Non-random distribution and sensory functions of primary cilia in vascular smooth muscle cells. Kidney Blood Press Res 31, 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Anujan P, Zhou F, Zhang Y, Chong YL, Bingle CD, and Roy S (2019). Mcidas mutant mice reveal a two-step process for the specification and differentiation of multiciliated cells in mammals. Development 146. [DOI] [PubMed] [Google Scholar]
- Lucas JS, Davis SD, Omran H, and Shoemark A (2020). Primary ciliary dyskinesia in the genomics age. Lancet Respir Med 8, 202–216. [DOI] [PubMed] [Google Scholar]
- Mahjoub MR, Xie Z, and Stearns T (2010). Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J Cell Biol 191, 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, and Kintner C (2008). Cilia orientation and the fluid mechanics of development. Curr Opin Cell Biol 20, 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo G, Soplop N, Wang WJ, Uryu K, and Tsou MF (2016). Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia. Dev Cell 39, 424–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JR, and Anand VK (1998). Clinical significance of compound cilia. Otolaryngol Head Neck Surg 118, 685–687. [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z, Han YG, Soriano-Navarro M, Garcia-Verdugo JM, and Alvarez-Buylla A (2010). Cilia organize ependymal planar polarity. J Neurosci 30, 2600–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B, Jacobs R, Li J, Chien S, and Kintner C (2007). A positive feedback mechanism governs the polarity and motion of motile cilia. Nature 447, 97–101. [DOI] [PubMed] [Google Scholar]
- Mitchell B, Stubbs JL, Huisman F, Taborek P, Yu C, and Kintner C (2009). The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr Biol 19, 924–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnich M, Borgeskov L, Breslin L, Jakobsen L, Rogowski M, Doganli C, Schroder JM, Mogensen JB, Blinkenkjaer L, Harder LM, et al. (2018). CEP128 Localizes to the Subdistal Appendages of the Mother Centriole and Regulates TGF-beta/BMP Signaling at the Primary Cilium. Cell Rep 22, 2584–2592. [DOI] [PubMed] [Google Scholar]
- Nanjundappa R, Kong D, Shim K, Stearns T, Brody SL, Loncarek J, and Mahjoub MR (2019). Regulation of cilia abundance in multiciliated cells. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich H, Cremers C, Loges NT, Werner C, Nielsen KG, Marthin JK, Philipsen M, Wallmeier J, Pennekamp P, Menchen T, et al. (2015). Loss-of-Function GAS8 Mutations Cause Primary Ciliary Dyskinesia and Disrupt the Nexin-Dynein Regulatory Complex. American journal of human genetics 97, 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran H, Haffner K, Volkel A, Kuehr J, Ketelsen UP, Ross UH, Konietzko N, Wienker T, Brandis M, and Hildebrandt F (2000). Homozygosity mapping of a gene locus for primary ciliary dyskinesia on chromosome 5p and identification of the heavy dynein chain DNAH5 as a candidate gene. Am J Respir Cell Mol Biol 23, 696–702. [DOI] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, and Bornens M (1992). Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol 108, 107–128. [DOI] [PubMed] [Google Scholar]
- Reiter JF, and Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, and Meunier A (2017). The development and functions of multiciliated epithelia. Nat Rev Mol Cell Biol 18, 423–436. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, and Kintner C (2008). The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet 40, 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, and Kintner C (2012). Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 14, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov R, and Alieva I (2018). Who are you, subdistal appendages of centriole? Open Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Bayly RD, Sangoram AM, Scott MP, and Axelrod JD (2012). Microtubules enable the planar cell polarity of airway cilia. Curr Biol 22, 2203–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, and Stearns T (2007). Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol 178, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallmeier J, Al-Mutairi DA, Chen CT, Loges NT, Pennekamp P, Menchen T, Ma L, Shamseldin HE, Olbrich H, Dougherty GW, et al. (2014). Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation of multiple motile cilia. Nature genetics 46, 646–651. [DOI] [PubMed] [Google Scholar]
- Wallmeier J, Frank D, Shoemark A, Nothe-Menchen T, Cindric S, Olbrich H, Loges NT, Aprea I, Dougherty GW, Pennekamp P, et al. (2019). De Novo Mutations in FOXJ1 Result in a Motile Ciliopathy with Hydrocephalus and Randomization of Left/Right Body Asymmetry. American journal of human genetics 105, 1030–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, et al. (2015). Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Du H, Wang X, Mei C, Sieck GC, and Qian Q (2009). Characterization of primary cilia in human airway smooth muscle cells. Chest 136, 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, and Brody SL (2002). Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol 283, L1315–1321. [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, Xu T, Huang X, Yan X, and Zhu X (2013). The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol 15, 1434–1444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video S1. FIB-SEM tomograms of multiple airway multiciliated cells grown in air-liquid interface. 10 nm isotropic resolution. This video is related to Figure 3.
Supplemental Video S2. High magnification of Supplemental Video S1. This video is related to Figure 3.
Supplemental Video S3. Live imaging of TagRFP-Cen1 centrosomal centrioles during centriole amplification in primary cultured ependymal progenitors from Cen2-EFGP mice. This video is related to the Figures 4 and S2, “MTEC and ependymal cell experiments” part of the STAR
Data Availability Statement
The published article includes all datasets generated during this study. Code is available by the Google Drive link specified in method details.


