Abstract
Frailty is a complex late life phenotype characterized by cumulative declines in multiple physiological systems that increases the risk for disability and mortality. The biological changes associated with aging are risk factors for frailty as well as for complex diseases; whereas longevity is assumed to be an outcome of protective biological mechanisms. Understanding the interplay between biological alterations associated with aging and protective mechanisms associated with longevity in the context of frailty may help guide development of interventions to increase healthspan and promote successful aging. The complexity of these phenotypes and relatively low heritability in studies are the main roadblocks in deciphering genetic mechanisms of these age associated conditions. We review genetic research related to frailty, and discuss the possible intertwined biology of frailty and longevity.
INTRODUCTION
Human life expectancy has been on a phenomenal rise, and globally, the average life expectancy increased by 5.5 years between start of this century and 2016. In 2016, the life expectancy at birth globally was reported to 72 years with a higher life expectancy in women (74.2 years) compared to men (69.8 years) (http://apps.who.int/gho/data/view.main.SDG2016LEXREGv?lang=en). The resultant rise in the aging population across the world has been accompanied by an increase in age associated vulnerabilities to disease. An important focus of current aging research is to develop preventive measures to increase the length of time that a person is healthy rather than just being alive (healthspan).
Aging is a complex multifactorial phenotype,1 with significant heterogeneity in aging process between individuals as well as at the tissue level within an individual.2 The biological hallmarks of aging include genomic instability, loss of proteostasis, telomere attrition, epigenetic alterations, deregulated nutrient-sensing, mitochondrial dysfunction, cellular senescence, altered intercellular communication, and stem cell exhaustion.3 The loss of homeostasis and compromised stress response with aging makes older individuals more susceptible to various disabilities and diseases. There is an increase in the incidence of cardiovascular diseases, dementia, stroke, diabetes, and cancer with advancing age. The evolving field of ‘geroscience’ is based on the concept that aging is the prime risk factor associated with most of these complex diseases, and targeting aging itself is critical to keeping these age-related diseases at bay.4
CONCEPT OF LONGEVITY AND FRAILTY
Longevity is a widely used term for long and extreme lifespan. There is no single universally accepted age cutoff that is used to define longevity. Researchers have used various approaches for identifying individuals to study longevity including centenarians (100 years old), supercentenarians (110 years and older), using predetermined age cutoff (85 or 95 years in various studies) or individuals belonging to the top 10th percentile of survival in population samples.5,6 The biology of longevity is complex with genetic, epigenetic, and environmental factors influencing the attainment of longer lifespan. Family clustering of longevity has been reported;7 the heritability of the longevity trait was 26% for males and 23% in females in a twin study.8 Higher heritability has been reported with increasing parental age and extreme longevity while minimal effect was reported if parents died before the age of 60.9 Heritability estimates in family of centenarians was up to 48% in men and 33% in women.6,10 Yet, more recent studies have contradicted these earlier reports, and point towards assortative mating (individuals with similar phenotypes mate with one another) as a major cause for the inflated heritability estimates of longevity.11 These studies downgrade the heritability estimate to a more modest 10%.11,12 Heritability of aging is modest when compared to those of complex diseases like type 2 diabetes and Alzheimer’s disease.6,13 Genetic studies including Genome Wide Association Studies (GWAS) and candidate gene studies points towards longevity as a polygenic trait influenced by multiple genes.14
Healthspan is defined as years of life free from disabilities and diseases. The concept of healthspan and lifespan are not completely overlapping. An increase in lifespan need not always be accompanied by an increase in healthspan; older individuals may survive in poor health and limited function for many years due to better access to healthcare and improved supportive care. Researchers have emphasized the need to decrease the gap between healthspan and lifespan; adding life to years and not years to life. This window between lifespan and healthspan is characterized by the accumulation of deficits and disabilities, which is clinically identified by the concept of frailty. Frailty is a late life phenotype, associated with low physiologic reserves and increased vulnerability to adverse outcomes such as disability, hospitalization and death.15,16 Frailty is a multifactorial construct and involves physical, cognitive, psychological and social domains.17–19
There is a plethora of frailty measurements, and a single standard frailty scale is yet to be recognized globally.20 Basic criteria to be fulfilled for frailty constructs include accuracy in capturing clinical aspects of frailty as well as reliably predicting frailty related clinical outcomes ranging from falls, mortality to patient response to therapies.21 Biological mechanisms supporting the measure is an important additional quality for frailty construct.20,21 The 2 common approaches used in research studies define frailty either as a clinical syndrome17 or as a cumulative deficit index.22–24 Most large scale genetic as well as expression studies were carried out using one of these 2 frailty definitions. The Cardiovascular Health Study criteria proposed by Fried and colleagues is widely used to operationalize the definition of frailty as a clinical syndrome with attributes selected from existing CHS outcome measures.17 Frailty is operationally defined by the Cardiovascular Health Study criteria based on the following 5 attributes: slow gait speed, weak grip strength, low physical activity, exhaustion and unintentional weight loss. The presence of 3 or more attributes is grouped as frail while the presence of only 1–2 and 0 attributes are grouped as pre-frail and non-frail states, respectively.17 This syndromic definition of frailty has successfully predicted incident falls, hospitalization, disabilities, worsening mobility, surgical outcomes as well as mortality in older adults.17,25,26
Rockwood and colleagues proposed an alternative approach to define frailty as a nonspecific multifactorial state better characterized by the quantity rather than quality of health disorders (called deficits) accumulated by individuals during their life course (e.g., signs, disabilities, symptoms, diseases, laboratory tests, radiographic or electrocardiographic abnormalities).24,27–29 The cumulative frailty index is a continuous measure calculated by dividing the sum of deficits in an individual by the total number of deficits assessed; resulting in values ranging from 0 to 1. Recommended criteria for selecting variables for constructing the cumulative deficit frailty index include that the selected variables should accumulate with age, represent health status, and must represent multiple organ systems.29 Exclusion of variables that saturate early with age such as presbyopia, which is quite common by age 55 years, is recommended. A minimum of 30 variables is recommended for developing the frailty index,24 and it has been shown to predict deteriorating health status, institutionalization, and death in aging populations.24
Twins studies point towards a possible role of genetic components in increasing risk for frailty.30–32 In a study of 3623 individuals from the TwinsUK Adult Twin Registry showed heritability estimate of 25% and 30% respectively for frailty defined using syndromic and cumulative deficit frailty index.32 This study also showed significant genetic (0.57) and environmental (0.44) correlation between syndromic and cumulative FI. Higher intraclass correlation of both frailty measures in monozygotic twins compared to dizygotic twins pointed towards underlying genetics of frailty.32 A study of twins in the United Kingdom showed that 45% of the inter-individual variation in frailty index was heritable while 52% was contributed by the environment.30 In support, a study in a Danish population, the genetic contribution of frailty was estimated to be 43% for an overall Robustness Index Ratio (RIR) variability.31 The RIR construct was created in this study using Hand Grip strength (HG), Mini Mental State Examination (MMSE), Activity of Daily Living, and Self-Reported Health Status. As higher values indicate lower frailty status in this scale, it was termed as RIR. In the Framingham heart study, comparing frailty/prefrailty status vs no frailty in participants aged 60 and older, heritability of the combined trait of prefrailty and frailty was estimated to be 19%.33
In this review we provide an overview of genetic studies of frailty as well as stress the importance of understanding the genetic determinants of longevity, which might play an important role in the pathogenesis of frailty. It is worthwhile mentioning that large scale genetic studies in frailty are very few compared to the massive advances in understanding the genetics of aging and longevity in recent times.
GENETICS OF LONGEVITY
Longevity is a complex phenotype influenced by multiple risk factors and events happening from birth onwards including lifestyle and environmental factors. Though several genetic studies using different strategies have been carried out, the genetics of longevity is still poorly understood due to the extraordinary complexity of the phenotype.34 Studies have shown clustering of long-lived individuals in geographical regions (such as Okinawa in Japan and Sardinia in Italy) pointing towards a role for lifestyle and environment in promoting longevity.35 Family clustering of longevity is observed with 27% of variation in lifespan explained by genetics.36 Most of the genetic studies in this area have focused on individuals with extreme longevity phenotypes like centenarians, supercentenarians or the top 1%–10% longest lived individuals in a given population. Other studies have used various arbitrary cutoff ages to define longevity. A uniform age cut-off is impractical due to the variation in average lifespan of people in different parts of the world. Moreover, secular trends in lifespan are noted with an increase in average lifespan seen over time resulting from improved health care as well as other social factors across the world.37 Another problem faced by genetic research in aging is the variable age cutoff used for control groups. In response, recent studies in large cohorts like UK biobank and LifeGen have used parental longevity as a proxy for the aging phenotype, and have successfully discovered a number of novel loci associated with parental longevity.5,38 This approach is based on the assumption that the apparent health and lifespan advantages in individuals with longer parental longevity may be mediated by inherited genetic factors from their parents.38–40
While multiple studies have been done of different candidate genes and pathways with aging, only 2 genes have shown consistent association with the longevity phenotype; APOE gene that is also implicated in Alzheimer’s disease and cardiovascular diseases and FOXO3 gene belonging to the Forkhead box transcription factor (FOXO) family, which have role of sensors in the insulin\IGF1 signaling.41,42 APOE ε4 allele is associated with increased risk for Alzheimer’s disease and early mortality while ε2 allele is associated with healthy aging and longevity.43,44 Schachter et al. using a candidate gene approach showed that the APOE ε4 allele was less prevalent and ε2 allele frequency was higher in centenarians in France.45 Various candidate gene studies since then have shown the ε4 allele to be associated with decreased odds for longevity.43,44,46 Association of APOE with longevity was also replicated globally across diverse ethnicities in candidate gene studies.43,46,47 FOXO3, belonging to Forkhead Box (FOX) family of transcription factors, plays a role in regulation of autophagy and cell cycle, and animal studies have linked it to longevity.41 FOXOs and its homologues have shown to influence longevity across diverse range of species by its influence in Insulin/IGF1 signaling pathway, which is assumed to be an evolutionary conserved biological pathway affecting longevity.48 The FOXOs gene became a target in human candidate gene association studies based on observations in animal model studies where FOXO homolog DAF-16 in C. elegans and dFOXO in Drosophila have shown to be associated with increased longevity.49 In humans, FOXO3 (alias FOXO3A) encoding the transcription factor forkhead box O-3 (FoxO3) is the most successful candidate gene to be associated with longevity.41,42 These 2 candidate genes were also successfully associated with longevity in a genome wide association study (GWAS) analysis.50 Other genes with inconsistent association with longevity include ACE (angiotensin converting enzyme), APOC1 and APOC3 (Apolipoprotein C), FOXO1, IL6, IL10, KL (Klotho), and CETP (Cholesteryl ester transfer protein).51 Most of these genes have not been replicated across studies, and were not significant in GWAS of longevity phenotypes. As many of these candidate gene studies were done in small samples and fewer Single Nucleotide Polymorphisms (SNPs) genotyped, association by chance cannot be completely ruled out.52
The last decade has seen a surge of GWAS as well as in depth sequencing strategies being adopted in aging research.5,6,53–55 There are several critical reviews that discuss the GWASs of longevity phenotype,6,9,56 and we provide a brief summary (Fig 1). Most genetic variants associated with longevity phenotypes in GWAS have been disease associated variants. These genetic associations point towards causal variants for mortality rather than those that uniquely improve survival and healthy aging. An interesting genetic association with longevity in this regard is the 9p21–23 region, a GWAS hotspot for complex diseases that is also associated with longevity as well as parental longevity.5,53,57 A study looking at the association of genetic variants with living to and beyond the oldest one percentile of survival showed associations with the synonymous variant rs3764814 in USP42 (Ubiquitin carboxyl-terminal hydrolase 42) and the intronic variant rs7976168 in TMTC2 (Transmembrane and tetratricopeptide repeat containing protein2).58 USP42 is involved in transcription regulation targeting p53 by stabilization of p53 at time of stress.58,59 Interleukin-6, a cytokine that is widely associated with most age associated complex diseases, was associated in only one GWAS with longevity; but this association was not replicated in other studies. An IL6 gene polymorphism (rs2069837) was associated with longevity in a Chinese population sample consisting of centenarians and middle age controls.60
Fig 1.
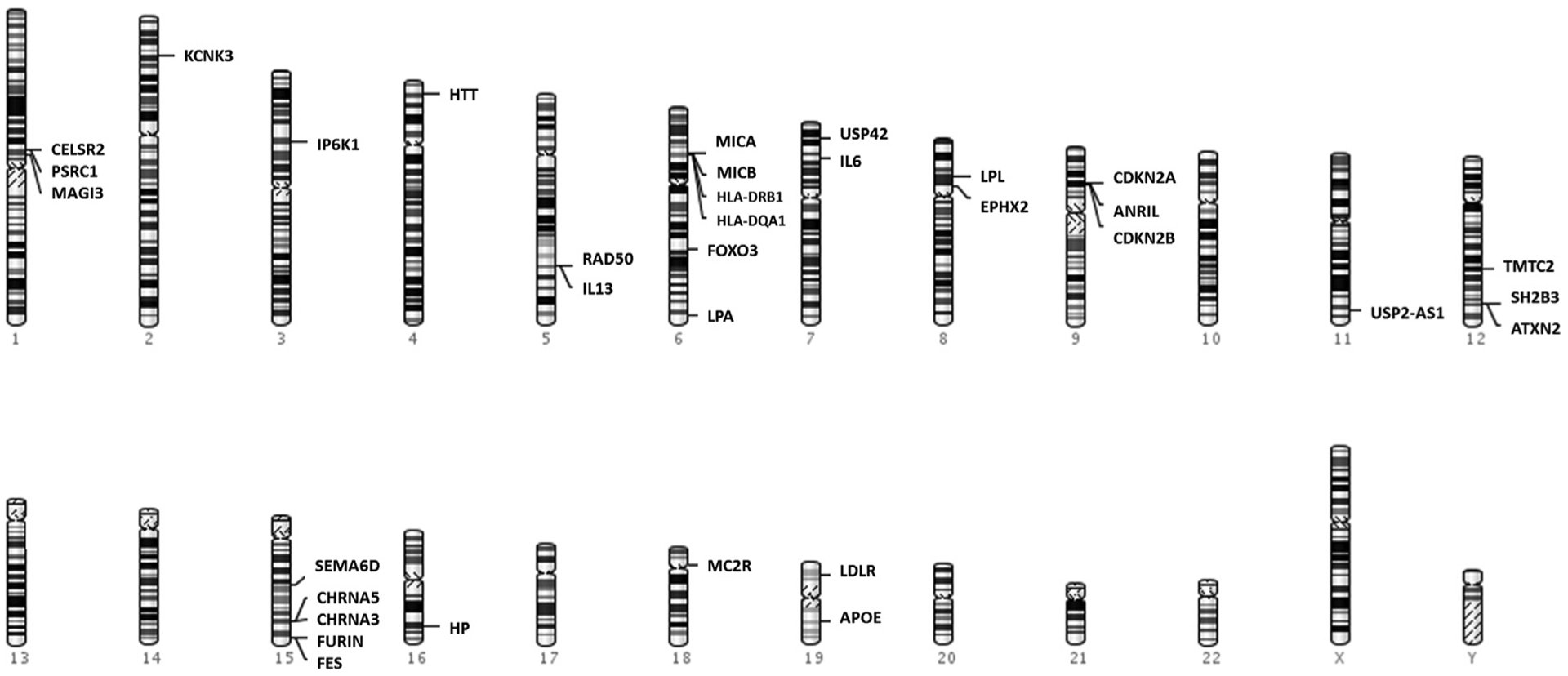
Genes associated with longevity phenotypes in largescale genome wide association studies.
Genome wide association studies of parental longevity in participants in the UK biobank have shown associations with multiple loci including genes previously associated with aging, cancer, cardiovascular diseases and Alzheimer’s disease [rs1051730(CHRNA3), rs1317286 (CHRNA3/5), rs429358 (APOE), rs55730499 (LPA), rs1556516 (CDKN2BAS, CDKN2A/2B), rs28926173 (MC2R) and rs11065979(ATXN2)].5,54,55 Cholinergic receptor, nicotinic, alpha (CHRNA5-CHRNA3-CHRNB4) gene locus is located in chromosome15q25 region, and the gene codes for ligand-gated ion channels involved in neurotransmission. This locus is also implicated in smoking initiation and lung cancer.61 LPA encodes Lipoprotein(a) involved in cholesterol and triglyceride transport. Genetic variant in this LPA locus (6q26–27) are mainly implicated in type 2 diabetes, coronary heart disease and stroke.62,63 9p21 locus containing CDKN2BAS alias ANRIL, cyclin-dependent kinase inhibitors 2A/2B (CDKN2A/2B) is a GWAS hotspot associated with multiple complex diseases including cardiovascular disease, cancer, diabetes, dementia etc.64 The CDKN2A/2B are tumor-suppressor genes involved in the regulation of cell cycle, apoptosis, senescence and aging.65 MC2R codes for adrenocorticotropic hormone (ACTH) receptor involved in blood sugar regulation and immune system.66 Binding of ACTH to its receptors triggers production of glucocorticoid hormones (cortisol, corticosteroid) from the adrenal glands.
Longevity is a complex polygenic trait, and genetic studies are yet to establish its complete biology.6,14 Recent large scale efforts have yielded multiple loci and genes associated with longevity. While many of these gene variants have not shown consistent associations with longevity across studies, they play important functional roles, and are worth further study.
GENETICS OF FRAILTY
Initial genetic investigations of frailty like other complex traits targeted specific candidate genes and pathways. A major limitation in understanding genetics of frailty was the lack of a uniform definition for this condition. As discussed earlier, the 2 common methods used to define frailty are syndromic and cumulative deficit models of frailty. There has been a rapid growth in understanding the genetics of numerous traits in the past 2 decades following the completion of human genome project and the massive development of sequencing and genotyping technologies, bringing down cost and speed drastically. GWAS in complex traits like hypertension, coronary heart disease, Alzheimer’s disease, osteoarthritis, depressive symptoms and diabetes mellitus has revealed a multitude of significant genetic variants in multiple pathways.67 Many of these traits also have been associated with frailty in epidemiological studies.68 These observations suggest the possibility of shared etiologies and genetic risks of frailty with these diseases.
Candidate gene studies.
Candidate gene studies in frailty were built on observations of expression studies of frailty and aging, which pointed towards pathways associated with dysregulation in protein levels in inflammatory and endocrine pathways.69,70 Higher expression of inflammatory cytokines such as C-reactive protein (CRP), IL-6 and tumor necrosis factor-alpha (TNF–α) have been associated with frailty defined by both syndromic and cumulative deficit approaches.71,72 A metanalysis involving 32 cross sectional studies (n = 23,800) and 3 longitudinal studies (n = 3402) found serum IL-6 and CRP to be associated with frailty at cross-section but the 3 longitudinal studies showed that these inflammatory markers did not predict incident frailty.73 In other studies, lower expression of insulin-like growth factor 1(IGF-1) and dehydroepiandrosterone sulfate (DHEA-S) are associated with frailty pointing towards endocrine dysregulation as a possible mechanism.74
The primary focus of candidate gene studies in regard with frailty has been on inflammation, focusing on SNPs in cytokine genes (Table 1). In the English Longitudinal Study of Ageing, promoter variant rs1800629 in the proinflammatory cytokine gene TNF was associated with frailty using the syndromic frailty definition.75 While in the same cohort, rs360722 in the proinflammatory IL18 cytokine gene and 2 SNPs (rs4679868 and rs9852519) in the IL12 gene were associated with frailty using the cumulative deficit score.76 While none of these variants survived Bonferroni correction in both analyses, IL18 is still a promising gene with respect to understanding the biology of frailty. A higher IL-18 level is implicated in poorer physical performance and worse activities of daily living in older adults.77 C-reactive protein (CRP) is most consistent cytokine associated with frailty in expression studies. TT genotype of SNP rs1205(1846 C>T) in the 3’UTR region of CRP gene as well as higher CRP expression was associated with frailty in 3778 Australian community dwelling older adults.78 Interestingly the T allele of this SNP was associated with decrease in the expression of CRP. Another study involving 1723 elderly participants in the Rugao Longevity and Ageing Study study in China failed to replicate the association of this SNP with frailty.79 IL-6 expression is associated with frailty, aging and other age associated traits such as cardiovascular diseases, Alzheimer’s disease, type 2 diabetes and functional decline.80–82 But at genetic level, none of the studies have shown strong association of genetic variants in the IL6 gene region with frailty.83 Another important anti-inflammatory cytokine IL10 promoter polymorphism rs1800871 and rs1800896 was associated with the syndromic definition of frailty in 984 participants (368 nonfrail/309 prefrail/307 frail) at the allelic and haplotypic level involving these 2 SNPs.84
Table 1.
Genes which were shown to be associated with frailty in Candidate gene association studies
| Gene/SNPs | Ancestry/cohort | Age | Participants Frailty model | Pathway | Study | |
|---|---|---|---|---|---|---|
| TNF rs1800629 | Caucasian (ELSA) | ≥50 years | n = 3160 | Syndromic frailty | Immune response | Mekli et al., 201675 |
| IL18 rs360722 | Caucasian (ELSA) | ≥50 years | n = 3160 | Cumulative frailty Index (30–60 Variables) | Immune response | Mekli et al., 201576 |
| IL12 rs4679868 rs9852519 | Caucasian (ELSA) | ≥50 years | n = 3160 | Cumulative frailty Index (30–60 Variables) | Immune response | Mekli et al., 201576 |
| LRP1 rs1799986 | Caucasian (ELSA) | ≥50 years | n = 3160 | cumulative frailty Index (30–60 Variables) | Cholesterol transport | Mekli et al., 201576 |
| SELP rs6131 | Caucasian (ELSA) | ≥50 years | n = 3160 | Cumulative frailty Index (30–60 Variables) | cell adhesion | Mekli et al., 201576 |
| CRP rs360722 | Caucasian (HIMS/ Men only) | ≥65 years | n = 3778 | Syndromic frailty | Immune response | Almeida et.al., 201278 |
| IL10 rs1800871 rs1800896 | Mexican (SADEM) | ≥60 years | n = 984 | Syndromic frailty | Immune response | Cedillo, T.et al., 201984 |
| KL rs1207568 | Chinese (PLAD) | ≥90 years | n = 632 | Syndromic frailty | FGF23-mediated signal transduction, phosphate and calcium homeostasis, Aging | Hao et al., 201887 |
| APOE rs429358 rs7412 | Caucasian (HELIAD) | ≥65 years | n = 1234 | Syndromic frailty | Vesicle-mediated transport and Statin Pathway, Aging | Mourtzi et al., 201992 |
| NFIB rs518054 | Azhkenazi Jewish (LonGenity) | ≥65 years | n = 637 | Syndromic frailty | Cell differentiation, stem cell maintenance | Sathyan et al., 2018101 |
| MTR rs10925235 rs4659725 | Caucasian (WHAS/women only) | ≥70 years | n = 349 | Syndromic frailty | homocysteine and folate metabolism | Ho et al., 201185 |
| CASP8 rs3769827 rs2037815 | Caucasian (WHAS/women only) | ≥70 years | n =349 | Syndromic frailty | Apoptotic signaling | Ho et al., 201185 |
| FN1 rs7567647 | Caucasian (WHAS/women only) | ≥70 years | n = 349 | Syndromic frailty | cell adhesion, growth, migration, and differentiation. | Ho et al., 201185 |
ELSA, English longitudinal study of ageing; HIMS, health in men study; SADEM, Study on Aging and Dementia in Mexico; PLAD, Project of Longevity and Aging in Dujiangyan; HELIAD, Hellenic Longitudinal Investigation of Ageing and Diet; WHAS, Women’s Health and Aging Studies.
In one of the candidate gene studies in frailty, 1354 SNPs in 134 candidate genes having roles in physiological processes including apoptosis, cellular homeostasis and senescence were genotyped in 349 participants in the Womens Health and Aging Study. The syndromic definition of frailty was employed. No SNP survived multiple testing corrections in this study.85
Higher Klotho concentrations have been shown to be protective against frailty phenotype as well as being associated with lower likelihood of exhaustion,86 which is one of the components of the CHS frailty phenotype.17 An association of rs1207568 (G-395A), a variant in the promoter region of Klotho gene, with frailty among 632 oldest-old participants (mean age = 93.5 ± 3.2 years) was reported.87
APOE coding for Apolipoprotein E is a pleiotropic gene involved in physiological functions ranging from lipoprotein metabolism, inflammation, and oxidative stress as well as CNS physiology. Apolipoprotein E (APOE) ε4, ε3 and ε2 alleles are the most studied genotypes with regards to various adverse outcomes such as Alzheimer’s disease, stroke, hypertension, and cardiovascular diseases. The ε4 allele acts as risk allele for Alzheimer’s disease while ε3 is considered neutral and ε2 allele has been shown to have a cognitive protective effect.88,89 The role of APOE genotype with frailty has been confusing with mixed results in previous reports.90–92 In the first extensive study looking into relationship between APOE genotype and frailty, no association between APOE genotype and frailty (independent of the frailty definition adopted) was seen in 1452 older adults aged 70 years and above.90 The authors concluded that APOE was not a ‘frailty gene.’ This observation was repeated in a GWAS carried out in the UK biobank; the APOE ε4 variants were not associated with frailty in 164,610 participants aged 60–70 years.91 On the other hand, all the major longevity associated GWAS have pinpointed the APOE locus making it the most replicated longevity associated gene in humans.46,93 Interestingly, a recent study carried out in 1234 participants (65–99 years) in the Hellenic Longitudinal Investigation of Ageing and Diet study (HELIAD) showed that APOE ε4 allele carriers have over 2-fold higher odds of frailty (syndromic definition) compared to noncarriers.92 One possible reason for the inconsistent results might be differences in age range of participants in these studies. A decrease in the frequency of ε4 homozygotes (ε4ε4) with aging has been reported; from 2.7% in participants aged 60 years or less to 0.8% for those aged 85 years and above.94 The low prevalence of ε4 allele with age might be because it is a risk factor for multiple complex diseases besides Alzheimer’s disease, shortening survival of ε4 allele carriers in the population.95
Candidate gene and pathway-based studies have had limited success in deciphering the genetics of frailty. Genetic studies based on gene expression changes have inconsistent results with some showing association with frailty or provided negative results.96 These expression studies are inconclusive as changes in the protein levels might be related to the physiologic deterioration seen with aging, and not necessarily causal for frailty. Frailty being a highly polygenic trait, candidate gene studies might be too conservative to understand the underlying biology unless we have more robust predictors of frailty at the expression level.
We implemented a hypothesis-based strategy in a recent study. We took advantage of the fact that frailty is closely associated with age associated complex diseases including cardiovascular disease, diabetes and dementia.97–99 GWASs have successfully identified a large number of SNPs in diverse loci across genome associated with these complex diseases.100 Interestingly there are also few GWAS hotspot loci associated with multiple complex diseases. Based on the association of frailty with multiple complex diseases, we screened all common variants present in 9p21–23 region, a GWAS hotspot associated with multiple phenotypes including cardiovascular disease, diabetes, glaucoma, AD, aneurysms (intracranial and abdominal aortic) as well as cancers.64 We discovered a novel association of SNP rs518054 located in the enhancer region of NFIB gene at allelic and haplotypic level with the syndromic frailty phenotype.101 NFIB codes for transcription factor involved in cell differentiation, have antiapoptotic effect as well as involved in stem cell maintenance in adult tissues and act as epigenetic regulator.101
Such strategies are pivotal in developing hypotheses keeping in mind the shared etiology between different traits as well as pleiotropic effect of SNPs and gene. Growing evidence from GWASs also suggest that variants in the noncoding region might play a role through short range(promoter,3’UTR) as well as long range (enhancer) interaction with the gene. Even the changes at the expressional level of proteins as well as RNA might be regulated not only by variants which are located in genes as well as promoter and 3’UTR but also through epigenetic changes and variants present in the enhancer regions involved in long range interaction with promoter. Genetic studies in frailty have focused mainly on the common variants (MAF > 0.02) in the genome. The missing heritability might be also due to the rare variants across the genome, which has to be targeted by in-depth sequencing. Another major drawback has been again inconsistencies in frailty definitions across studies.
Genome Wide Association Studies (GWAS).
Only a few GWAS have been done to discover the genetic antecedents of frailty (Table 2).91,102 In a GWAS on frailty that included 8539 participants from United States (discovery cohort: Health and retirement study) and 5251 participants from United Kingdom (replication cohort: English Longitudinal Study of Ageing), SNP rs6765037 located in the intergenic region in Chr3q21.3 was associated with the cumulative deficit frailty index in the discovery cohort.102 This SNP was located in the 5’ region of (Kelch repeat and BTB Domain containing 12) gene. However, the finding could not be replicated in the replication cohort. Second top SNP rs7134291 in this study located in the GRIN2B gene showed suggestive association in both discovery (P = 1.81 × 10−6) and replication (P = 0.034) cohort.102 GRIN2B codes for glutamate binding NR2B subunit, and is expressed in human cerebral cortex. SNPs in GRIN2B gene have been associated with developmental delay103 and intellectual disabilities.104
Table 2.
Top genes and pathways associated in Genome wide association studies published in regard with frailty.
| Genomewide association studies | ||||||
|---|---|---|---|---|---|---|
| Study | Ancestry/cohort | Age | Participants | Frailty model | Top genes | Highlighted pathway |
| Atkins et al., 201991 | European (UK biobank) | 60–70 years | n = 164,610 | Cumulative frailty Index (49 Variables) | SYT14, LRPPRC, MYOSLID-AS1, RBM6, STAG1 NLGN1, HTT, LOC105379109, HLA-DQB1, HLA-DRB1, HLA-DQA1, PDE10A, FOXP2, HR CSMD3, EXD3, ANK3, NCAM1, LOC105369842, MVK, INO80, TMOD3, PAFAH1B1, KRT17P3, PHB, PIK3C3 | Synapse maintenance pathways |
| Mekli et al„ 2018102 | European (Discovery: HRS/Replication ELSA) | ≥65 years (Discovery)/≥50 years (Replication) | n = 13790 | Cumulative frailty Index (45 Variables) | GRIN2B, KBTBD12 | synaptic plasticity, Neuropathic pain signaling, GPCR-Mediated Nutrient Sensing |
| Livshits el al„ 2018110 | Europeans (TwinsUK, female only) | 17– 93 years | n = 3626 | Cumulative frailty Index (33 Variables) | No significant association | – |
HRS, Health and retirement study; ELSA, English longitudinal study of ageing.
A recent study in the UK Biobank involving 164,610 participants aged 60–70 years is the largest GWAS to date carried out for frailty.91 Frailty was defined by the cumulative frailty index. In this GWAS, 26 independent genetic variants at 24 loci were associated with frailty. Most of these loci were associated with cardiovascular disease, body mass index, smoking and personality traits like depression and neuroticism. Most frailty index associated variants were located in genes involved in neuronal functions. Novel loci associated exclusively with frailty include loci harboring CSMD3 (CUB and Sushi multiple domains 3), ANK3 (Ankyrin 3) and TMOD3 (Tropomodulin 3), which are involved in dendritic development, neurotransmission and cognitive function respectively.91 Pathway analysis showed that the synapse maintenance pathway was associated with frailty.91This finding might point towards possible cognitive determinants of frailty or shared biology with cognition.105,106 Various studies have shown frailty as such as well as main components of the frailty construct (e.g. slow gait speed) is related to cognitive complaints and dementia.107–109 Further studies need to be carried out to examine possible shared biology between frailty and cognitive syndromes.91 Loci implicated in longevity in previous studies including APOE, TERT, FOXO3A and CDKN2BAS/CDKN2A/CDKN2B were not associated with frailty in this GWAS. Heritability of frailty index using SNPs in this study was estimated to be 14%.91 One minor shortcoming of this study is the narrow age range (60–70 years) of participants, which may have resulted in missing out deteriorating frailty after age 70. The third GWAS in frailty involving 3626 participants from the NIHR BRC Twins UK BioResource consisting of female twins failed to show any variant to be associated with cumulative frailty index.110 Inclusion of only genetically related female twin participants in this GWAS might have led to negative results.110
More large scale approaches similar to the GWAS in the UK biobank is needed to provide a better picture of the underlying genetics of frailty.91 Frailty may be formed by the culmination of more pathways than assumed before.17 Individual components of the cumulative frailty index consist of complex diseases ranging from cardiovascular diseases, stroke, cancer, diabetes to chronic lung disease in addition to signs and symptoms of diseases. There is a possibility of different biological pathways getting activated in frailty, leading to a mosaic phenotype, which is difficult to capture at the population level. Frailty status can be an outcome of various primary drivers. The same score on the cumulative frailty score in different individuals could be achieved by different components like diabetes, cardiovascular diseases or neurological changes. Each of these traits possesses its own unique genetic components with certain amount of shared genetic components evidenced by the presence pleiotropic effects of SNPs and genes.111 Interestingly, the largest GWAS in the UK biobank too showed association of loci previously implicated in diverse traits.91 These phenotypic complexities along with missing heritability might be a barrier to understand the underlying biology of frailty as GWAS focus only on common variants. Rare variants as well as epigenetic changes might play an important role in frailty. At present, all GWAS in frailty were carried out in Caucasian populations in the United Kingdom and United States.91,110 Life expectancy also varies with different ethnicities as well as geographical location.112 This point towards the possibility of unique genetic as well as epigenetic signatures for age associated traits like frailty in diverse environments.
Most of the GWAS to date have used cumulative index for defining frailty.91,102,110 There is a need for GWAS to be carried out using syndromic definition of frailty in large cohorts like the UK biobank. A comparative analysis of pathways as well as genes using multiple definitions of frailty may discover unique pathways and genes associated with the frailty concept independent of the definitions used. More large scale efforts are needed in deciphering the genetics of frailty.
Other approaches.
Other genetic approaches like Mendelian randomization have been adopted to understand causal risk factors and shared genetic basis of other traits with frailty. Mendelian randomization approaches work on the principle that genetic variants that have robust effects or associations with modifiable exposures or risks that alter disease occurrence or outcome through modulation of specific exposure only. Genetic variants thus act as instrumental variable (IV) proxy for the exposure which might include biomarkers (e.g. HDL, LDL, triglyceride, and cholesterol).113 Basically, this approach looks for the impact of genetically determined risk factors on the outcome thus making it free from confounder effects. This approach has shown success in understanding risk factors associated with frailty. Instrumental variables created using genetic variants to instrument ‘life-long lowering of low-density lipoprotein cholesterol (LDL-C)’ showed association of genetically predicted life-long lowering of LDL-C with decreased frailty in 378,161 participants in the UK biobank.114 Two IVs were used in this study to predict ‘life-long lowering of low-density lipoprotein cholesterol (LDL-C),’ and both were associated with frailty. The larger IV set consists of 274 independent or untagged SNPs associated (P < 5.0 × 10–8) with LDL-C concentrations in GWAS.115 The£ second one is a more conservative smaller IV set consisting of only 50 SNPs associated with LDL-C but without strong association with any other lipids.114 In the Rugao Longevity and Ageing Study from China, Mendelian randomization approach was implemented by using single SNP (rs662799) in the APOA5 gene as IV for predicting triglyceride levels. But genetically predicted triglyceride level was not associated with frailty phenotype as well as longevity in this study.116
Recent large GWAS have shown that most complex traits including age associated traits are highly polygenic and influenced by genetic variants in multiple region across the genome.117 Individual level genetic predisposition to a trait is captured by polygenic score (PGS), which is based on the effect size of SNPs derived from trait specific largest GWAS. These approaches have successfully found PGSs for multiple risk factors to be associated with various outcomes including aging.118 Polygenic score for higher education has been associated with parental longevity.118 In the Health and Retirement Study, higher PGSs for educational achievement and well-being were found to be associated for younger subjective age; whereas higher PGSs for neuroticism, body mass index, waist circumference, and depressive symptoms were associated with higher subjective age of the participant.119 Subjective age in this study was defined as age felt by the individual relative to their chronological age and was computed as proportional discrepancy score by subtracting chronological age from felt age and then dividing that by chronological age.120 Frailty is influenced by multiple risk factors.121 It will be of interest to look for the relationship between the genetics of these risk factors represented as polygenic scores with frailty. For example, neuroticism, a personality trait characterized with emotional instability and negative effect, is assumed to be a risk factor for frailty.122 A recent study has found higher PGS for neuroticism to be significantly associated with in UK biobank and Swedish twin registry.123 Also, higher PGS for neuroticism predicted frailty in these large cohorts.123 Similarly low education level is associated with increasing the odds of being frail in the elderly population compared to high education levels.124,125 In the Health and Retirement Study, PGS for education attainment in 7064 participants was associated with frailty defined using both cumulative frailty index as well as the syndromic model of frailty.126 The association was weaker with advancing age, and was most evident till 75 years.126 The effect of PGS of educational attainment was absent after 80 years.126
In a large GWAS on frailty carried out in the UK biobank, the authors calculated PGS for 35 traits including only the most robust SNPs for each trait identified in previous GWAS studies.91 Interestingly PGS for educational attainment, BMI, waist to hip ratio, parental age, grip strength were associated with frailty index scores.91 Higher BMI and WHR PGS were associated with higher levels of frailty pointing towards effect of genetic predisposition of obesity on frailty.91 The PGSs for higher education and parent survival had protective effects on frailty levels.91
INFLUENCE OF LONGEVITY ON FRAILTY
Both aging and frailty are complex phenotypes with shared etiologies that may influence each other at the genetic level. At the phenotypic level, frailty increases with age, and age itself is a risk factor for frailty. Succumbing to frailty increases the rate of biological aging with an attendant increase in the incidence of comorbidities that eventually lead to death. The concept of longevity focuses on increase in lifespan to extremity like centenarians and super centenarians.
Though a clear overlap is seen with aging and frailty at the phenotypic level, the biological overlap is less clear-cut given the divergent etiologies associated with aging and frailty. There are 2 assumptions or hypothesis to be tested to understand genetics of frailty with respect to aging/longevity.
Are genetic risk factors associated with aging also a risk factor for frailty?
Do genetic factors associated with longevity have a protective effect against frailty?
For the first assumption, frailty genetics work the same as with other complex diseases (e.g., diabetes, cardiovascular diseases) with age acting as a risk factor though specific independent genetic risk also exist with diseases. There are few large genetic studies in frailty with most extant studies being candidate gene-based focusing on common variants in inflammatory and hormonal pathways. These studies have identified frailty related pathways shared with complex diseases as well as aging. In a recent large GWAS, many traditional aging related genetic variants such as APOE, 9p21.3, TERT and FOXO3A were not associated with frailty.91 While candidate gene studies have shown contradictory results with APOE ε4 associated with frailty in some but not all studies.90,92 Our study found a novel association of frailty with a SNP near 9p21.3 locus. Previously implicated SNPs with aging, cardiovascular diseases and diabetes in this locus were not associated with frailty.101 Larger genetic studies will be needed to delineate the frailty associated biological pathways and to understand the shared etiology between aging and other complex diseases.
The second assumption is that longevity promoting genes and genetic variants will have a protective effect against frailty. Longevity genes are involved in compensatory mechanisms that limit or repair the age associated damages (Fig 2).1 Studies have shown that individuals with exceptional longevity as well as their offspring possess resilience towards a wide range of complex disorders such as cardiovascular disease and Alzheimer’s disease. They also lead longer and healthier lives compared to their counterparts without personal or parental history of exceptional longevity.39,40,127–129 Offspring of centenarians have lower prevalence of heart disease, hypertension and diabetes as well as better self-rated health compared to age matched controls.130,131 A protective role of parental longevity in physical function has also been reported.132,133 The apparent health and lifespan advantages posed by individuals with longer parental longevity may be mediated by genetic factors.39 In the Louisiana Health Aging study, offspring of long-lived parent (at least one parent was age 90 or older) had 31% lower frailty index compared to offspring of short-lived parents (Both parent dead before age 76).134 Yearly acceleration in frailty index was higher in offspring of short-lived parents (2.7%) compared to offspring of long-lived parent (2.0%). Heritability of the frailty index was estimated to 0.39, and possible genetic predisposition for frailty was assumed.134 In another large study including 29,905 participants the same trend was observed with parental longevity having more protective effect in male offspring compared to female offspring. This is in line with previous observations that the impact of genetic factors on longevity is more pronounced in males compared to females.135 GWASs as well as candidate gene studies carried out in frailty have failed to establish strong genetic relationship between frailty and longevity.
Fig 2.

Different models for interaction of longevity and frailty. (a) In usual survivor longevity associated protection conferred by Geroprotector, including longevity factors (green) is less and accumulation of insults (red) leads to decrease in healthspan as well as lead to disabilities, frailty, and comorbidities finally leading to death. (b) In long-lived healthy individual have more Geroprotectors acting throughout life preventing insults as well as increasing stretch of healthspan as well as life span. Frailty and other comorbidities are confined to small time span. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)”
CONCLUSIONS AND FUTURE PERSPECTIVES
Frailty and longevity are complex phenotypes with multifactorial origins. The genetic components of longevity and normal aging have not been completely elucidated. Understanding the relationship between longevity and frailty will be furthered when we find unique genetic signatures for longevity and frailty. Large GWASs have found genetic variants associated with longevity, but the low number of studies that have replicated initial observations raises questions about the reliability and consistency of the initial findings. The complexity or uniqueness of extreme longevity is very difficult to be explained completely with few genetic variants discovered till date. The complexity of the frailty as well as missing heritability has made it difficult to determine its underlying genetics. The frailty phenotype is complex compared to other age associated complex diseases. But recent large frailty GWAS studies like those done in the UK biobank give us hope that we are moving in right direction. In the future, in-depth next generation sequencing strategy via whole genome or exome sequencing as well epigenetic studies might provide a more nuanced picture of frailty genetics. It is still early days for the frailty genetics field though leads from aging and longevity genetic studies are promising.
ACKNOWLEDGMENTS
Authors were supported by grant from National Institute on Aging (R01AG044829).
Abbreviations:
- ADL
Activity of Daily Living
- ANRIL
antisense non-coding RNA in the INK4 locus
- APOE
Apolipoprotein E
- CDKN2A/2B
cyclin-dependent kinase inhibitors 2A/2B
- CRP
C-reactive protein
- GWAS
Genome wide association study
- IL-6
Interleukin-6
- LDL-C
low-density lipoprotein cholesterol
- MMSE
Mini Mental State Examination
- RIR
Robustness Index Ratio
- RNA
Ribonucleic acid
- SNP
Single nucleotide polymorphism
- USP42
Ubiquitin carboxyl-terminal hydrolase 42
- 3′UTR
Three prime Untranslated Region
Footnotes
Conflicts of Interest: All authors have read the journal’s policy on conflicts of interests. All authors have read the journal’s authorship agreement. The authors have no conflict of interest to report.
REFERENCES
- 1.Johnson FB, Sinclair DA, Guarente L. Molecular biology of aging. Cell 1999;96:291–302. [DOI] [PubMed] [Google Scholar]
- 2.Lowsky DJ, Olshansky SJ, Bhattacharya J, Goldman DP. Heterogeneity in healthy aging. J Gerontol A Biol Sci Med Sci 2013;69:640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell 2013;153:1194–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy BK, Berger SL, Brunet A, et al. Geroscience: linking aging to chronic disease. Cell 2014;159:709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pilling LC, Kuo C-L, Sicinski K, et al. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging (Albany N Y) 2017;9:2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melzer D, Pilling LC, Ferrucci L. The genetics of human ageing. Nat Rev Genet 2019:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Berg N, Rodríguez-Girondo M, van Dijk IK, et al. Longevity defined as top 10% survivors and beyond is transmitted as a quantitative genetic trait. Nat Commun 2019;10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herskind AM, McGue M, Holm NV, Sörensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet 1996;97:319–23. [DOI] [PubMed] [Google Scholar]
- 9.Partridge L, Deelen J, Slagboom PE. Facing up to the global challenges of ageing. Nature 2018;561:45–56. [DOI] [PubMed] [Google Scholar]
- 10.Sebastiani P, Perls TT. The genetics of extreme longevity: lessons from the new England centenarian study. Front Genet 2012;3:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruby JG, Wright KM, Rand KA, et al. Estimates of the heritability of human longevity are substantially inflated due to assortative mating. Genetics 2018;210:1109–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplanis J, Gordon A, Shor T, et al. Quantitative analysis of population-scale family trees with millions of relatives. Science 2018;360:171–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks-Wilson AR. Genetics of healthy aging and longevity. Hum Genet 2013;132:1323–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shadyab AH, LaCroix AZ. Genetic factors associated with longevity: a review of recent findings. Ageing Res Rev 2015;19:1–7. [DOI] [PubMed] [Google Scholar]
- 15.Fried LP, Ferrucci L, Darer J, Williamson JD, Anderson G. Untangling the concepts of disability, frailty, and comorbidity: implications for improved targeting and care. J Gerontol A Biol Sci Med Sci 2004;59:M255–63. [DOI] [PubMed] [Google Scholar]
- 16.Fried LP, Darer J, Walston J. Frailty Geriatric Medicine. Springer; 2003:1067–76. [Google Scholar]
- 17.Fried LP, Tangen CM, Walston J, et al. Frailty in older adults evidence for a phenotype. J Gerontol 2001;56:M146–57. [DOI] [PubMed] [Google Scholar]
- 18.Levers MJ, Estabrooks CA, Ross Kerr JC. Factors contributing to frailty: literature review. J Adv Nursing 2006;56:282–91. [DOI] [PubMed] [Google Scholar]
- 19.Gobbens RJ, van Assen MA, Luijkx KG, Schols JM. Testing an integral conceptual model of frailty. J Adv Nursing 2012;68:2047–60. [DOI] [PubMed] [Google Scholar]
- 20.Dent E, Kowal P, Hoogendijk EO. Frailty measurement in research and clinical practice: a review. Eur J Intern Med 2016;31:3–10. [DOI] [PubMed] [Google Scholar]
- 21.Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. Lancet 2013;381:752–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitnitski AB, Song X, Rockwood K. The estimation of relative fitness and frailty in community-dwelling older adults using self-report data. J Gerontol A Biol Sci Med Sci 2004;59:M627–32. [DOI] [PubMed] [Google Scholar]
- 23.Rockwood K, Mogilner A, Mitnitski A. Changes with age in the distribution of a frailty index. Mech Ageing Dev 2004;125:517–9. [DOI] [PubMed] [Google Scholar]
- 24.Rockwood K, Mitnitski A. Frailty in relation to the accumulation of deficits. J Gerontol A Biol Sci Med Sci 2007;62:722–7. [DOI] [PubMed] [Google Scholar]
- 25.Makizako H, Shimada H, Doi T, Tsutsumimoto K, Suzuki T. Impact of physical frailty on disability in community-dwelling older adults: a prospective cohort study. BMJ open 2015;5:e008462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makary MA, Segev DL, Pronovost PJ, et al. Frailty as a predictor of surgical outcomes in older patients. J Am Coll Surg 2010;210:901–8. [DOI] [PubMed] [Google Scholar]
- 27.Kulminski AM, Ukraintseva SV, Kulminskaya IV, Arbeev KG, Land K, Yashin AI. Cumulative deficits better characterize susceptibility to death in elderly people than phenotypic frailty: lessons from the Cardiovascular Health Study. J Am Geriatr Soc 2008;56:898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitnitski AB, Mogilner AJ, Rockwood K. Accumulation of deficits as a proxy measure of aging. Sci World J 2001;1:323–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Searle SD, Mitnitski A, Gahbauer EA, Gill TM, Rockwood K. A standard procedure for creating a frailty index. BMC Geriatr 2008;8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young AC, Glaser K, Spector TD, Steves CJ. The identification of hereditary and environmental determinants of frailty in a cohort of UK twins. Twin Res Hum Genet 2016;19:600–9. [DOI] [PubMed] [Google Scholar]
- 31.Dato S, Montesanto A, Lagani V, Jeune B, Christensen K, Passarino G. Frailty phenotypes in the elderly based on cluster analysis: a longitudinal study of two Danish cohorts. Evidence for a genetic influence on frailty. Age 2012;34:571–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livshits G, Ni Lochlainn M, Malkin I, et al. Shared genetic influence on frailty and chronic widespread pain: a study from TwinsUK. Age Ageing 2018;47:119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murabito JM, Yuan R, Lunetta KL. The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long-lived individuals. J Gerontol A Biol Sci Med Sci 2012;67:470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giuliani C, Garagnani P, Franceschi C. Genetics of human longevity within an eco-evolutionary nature-nurture framework. Circul Res 2018;123:745–72. [DOI] [PubMed] [Google Scholar]
- 35.Poulain M, Herm A, Pes G. The Blue Zones: areas of exceptional longevity around the world. Vienna Yearbook Popul Res 2013;11:87–108. [Google Scholar]
- 36.McGue M, Vaupel JW, Holm N, Harvald B. Longevity is moderately heritable in a sample of Danish twins born 1870–1880. J Gerontol 1993;48:B237–44. [DOI] [PubMed] [Google Scholar]
- 37.Vaupel JW, Zhang Z, van Raalte AA. Life expectancy and disparity: an international comparison of life table data. BMJ open 2011;1:e000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timmers PR, Mounier N, Lall K, et al. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. eLife 2019;8:e39856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milman S, Barzilai N. Dissecting the mechanisms underlying unusually successful human health span and life span. Cold Spring Harb Perspect Med 2016;6:a025098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipton RB, Hirsch J, Katz MJ, et al. Exceptional parental longevity associated with lower risk of Alzheimer’s disease and memory decline. J Am Geriatr Soc 2010;58:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A 2008;105:13987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris BJ, Willcox DC, Donlon TA, Willcox BJ. FOXO3: a major gene for human longevity-a mini-review. Gerontology 2015;61:515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gurinovich A, Andersen SL, Puca A, Atzmon G, Barzilai N, Sebastiani P. Varying Effects of APOE Alleles on Extreme Longevity in European Ethnicities. J Gerontol A Biol Sci Med Sci 2019;74:S45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sebastiani P, Gurinovich A, Nygaard M, et al. APOE alleles and extreme human longevity. J Gerontol A Biol Sci Med Sci 2018;74:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schächter F, Faure-Delanef L, Guénot F, et al. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet 1994;6:29–32. [DOI] [PubMed] [Google Scholar]
- 46.Garatachea N, Emanuele E, Calero M, et al. ApoE gene and exceptional longevity: insights from three independent cohorts. Exp Gerontol 2014;53:16–23. [DOI] [PubMed] [Google Scholar]
- 47.Jian-Gang Z, Yong-Xing M, Chuan-Fu W, et al. Apolipoprotein E and longevity among Han Chinese population. Mech Ageing Dev 1998;104:159–67. [DOI] [PubMed] [Google Scholar]
- 48.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet 2007:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp Gerontol 2006;41:910–21. [DOI] [PubMed] [Google Scholar]
- 50.Broer L, Buchman AS, Deelen J, et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci 2014;70:110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Budovsky A, Craig T, Wang J, et al. LongevityMap: a database of human genetic variants associated with longevity. Trends Genet 2013;29:559–60. [DOI] [PubMed] [Google Scholar]
- 52.Daly AK, Day CP. Candidate gene case–control association studies: advantages and potential pitfalls. Br J Clin Pharmacol 2001;52:489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deelen J, Evans DS, Arking DE, et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat Commun 2019;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pilling LC, Atkins JL, Bowman K, et al. Human longevity is influenced by many genetic variants: evidence from 75,000 UK Biobank participants. Aging (Albany N Y) 2016;8:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joshi PK, Pirastu N, Kentistou KA, et al. Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity. Nat Commun 2017;8:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slagboom PE, van den Berg N, Deelen J. Phenome and genome based studies into human ageing and longevity: an overview. Biochim Biophys Acta Mol Basis Dis 2018;1864:2742–51. [DOI] [PubMed] [Google Scholar]
- 57.Fortney K, Dobriban E, Garagnani P, et al. Genome-wide scan informed by age-related disease identifies loci for exceptional human longevity. PLoS Genet 2015;11:e1005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sebastiani P, Gurinovich A, Bae H, et al. Four genome-wide association studies identify new extreme longevity variants. J Gerontol A Biol Sci Med Sci 2017;72:1453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hock AK, Vigneron AM, Vousden KH. Ubiquitin-specific peptidase 42 (USP42) functions to deubiquitylate histones and regulate transcriptional activity. J Biol Chem 2014;289:34862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng Y, Nie C, Min J, et al. Novel loci and pathways significantly associated with longevity. Sci Rep 2016;6:21243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25. 1. Nat Genet 2008;40:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mora S, Kamstrup PR, Rifai N, Nordestgaard BG, Buring JE, Ridker PM. Lipoprotein and risk of type 2 diabetes. Clin Chem 2010;56:1252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clarke R, Peden JF, Hopewell JC, et al. Genetic variants associated with Lp lipoprotein level and coronary disease. New Engl J Med 2009;361:2518–28. [DOI] [PubMed] [Google Scholar]
- 64.Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J 2011;25:444–8. [DOI] [PubMed] [Google Scholar]
- 65.Gu F, Pfeiffer R, Bhattacharjee S, et al. Common genetic variants in the 9p21 region and their associations with multiple tumours. Br J Cancer 2013;108:1378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meimaridou E, Hughes C, Kowalczyk J, Chan L, Clark A, Metherell L. ACTH resistance: genes and mechanisms Hormone Resistance and Hypersensitivity. Karger Publishers; 2013:57–66. [DOI] [PubMed] [Google Scholar]
- 67.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2013;42:D1001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiss CO. Frailty and chronic diseases in older adults. Clin Geriatr Med 2011;27:39–52. [DOI] [PubMed] [Google Scholar]
- 69.Walston J Frailty–the search for underlying causes. Science’s SAGE KE 2004;2004:4. [DOI] [PubMed] [Google Scholar]
- 70.Hubbard RE, Frailty Woodhouse KW. inflammation and the elderly. Biogerontology 2010;11:635–41. [DOI] [PubMed] [Google Scholar]
- 71.Walston J, McBurnie MA, Newman A, et al. Frailty and activation of the inflammation and coagulation systems with and without clinical comorbidities: results from the Cardiovascular Health Study. Arch Intern Med 2002;162:2333–41. [DOI] [PubMed] [Google Scholar]
- 72.Hubbard RE, O’Mahony MS, Savva GM, Calver BL, Woodhouse KW. Inflammation and frailty measures in older people. J Cell Mol Med 2009;13:3103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soysal P, Stubbs B, Lucato P, et al. Inflammation and frailty in the elderly: a systematic review and meta-analysis. Ageing Res Rev 2016;31:1–8. [DOI] [PubMed] [Google Scholar]
- 74.Leng SX, Cappola AR, Andersen RE, et al. Serum levels of insulin-like growth factor-I (IGF-I) and dehydroepiandrosterone sulfate (DHEA-S), and their relationships with serum interleukin-6, in the geriatric syndrome of frailty. Aging Clin Exp Res 2004;16:153–7. [DOI] [PubMed] [Google Scholar]
- 75.Mekli K, Nazroo JY, Marshall AD, Kumari M, Pendleton N. Proinflammatory genotype is associated with the frailty phenotype in the English Longitudinal Study of Ageing. Aging Clin Exp Res 2016;28:413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mekli K, Marshall A, Nazroo J, Vanhoutte B, Pendleton N. Genetic variant of Interleukin-18 gene is associated with the Frailty Index in the English Longitudinal Study of Ageing. Age Ageing 2015;44:938–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frayling TM, Rafiq S, Murray A, et al. An interleukin-18 polymorphism is associated with reduced serum concentrations and better physical functioning in older people. J Gerontol A Biol Sci Med Sci 2007;62:73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Almeida OP, Norman PE, van Bockxmeer FM, Hankey GJ, Flicker L. CRP 1846G>A polymorphism increases risk of frailty. Maturitas 2012;71:261–6. [DOI] [PubMed] [Google Scholar]
- 79.Liu Z-y, Wang Z-d, Li L-z, et al. Association of CRP gene polymorphisms with CRP levels, frailty and co-morbidity in an elderly Chinese population: results from RuLAS. Age Ageing 2016;45:360–5. [DOI] [PubMed] [Google Scholar]
- 80.Kritchevsky SB, Cesari M, Pahor M. Inflammatory markers and cardiovascular health in older adults. Cardiovasc Res 2005;66:265–75. [DOI] [PubMed] [Google Scholar]
- 81.Singh VK, Guthikonda P. Circulating cytokines in Alzheimer’s disease. J Psychiatr Res 1997;31:657–60. [DOI] [PubMed] [Google Scholar]
- 82.Cohen HJ, Pieper CF, Harris T, Rao KMK, Currie MS. The association of plasma IL-6 levels with functional disability in community-dwelling elderly. J Gerontol A Biol Sci Med Sci 1997;52:M201–8. [DOI] [PubMed] [Google Scholar]
- 83.Walston J, Arking D, Fallin D, et al. IL-6 gene variation is not associated with increased serum levels of IL-6, muscle, weakness, or frailty in older women. Exp Gerontol 2005;40:344–52. [DOI] [PubMed] [Google Scholar]
- 84.Juárez-Cedillo T, Vargas-Alarcón G, Martínez Rodríguez N, Juárez-Cedillo E, Fragoso JM, Escobedo-de-la-Peña J. Interleukin 10 gene polymorphisms and frailty syndrome in elderly Mexican people:(Sadem study). Mol Genet Genomic Med 2019;7:e918 10.1002/mgg3.918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ho Y-Y, Matteini AM, Beamer B, et al. Exploring biologically relevant pathways in frailty. J Gerontol A Biol Sci Med Sci 2011;66:975–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shardell M, Semba RD, Kalyani RR, et al. Plasma klotho and frailty in older adults: Findings from the InCHIANTI Study. J Gerontol A Biol Sci Med Sci 2017;74:1052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hao Q, Wang Y, Ding X, et al. G-395A polymorphism in the promoter region of the KLOTHO gene associates with frailty among the oldest-old. Sci Rep 2018;8:6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conejero-Goldberg C, Gomar J, Bobes-Bascaran T, et al. APOE2 enhances neuroprotection against Alzheimer’s disease through multiple molecular mechanisms. Mol Psychiatry 2014;19:1243. [DOI] [PubMed] [Google Scholar]
- 89.Raber J, Wong D, Yu G-Q, et al. Alzheimer’s disease: Apolipoprotein E and cognitive performance. Nature 2000;404:352. [DOI] [PubMed] [Google Scholar]
- 90.Rockwood K, Nassar B, Mitnitski A. Apolipoprotein E–polymorphism, frailty and mortality in older adults. J Cell Mol Med 2008;12:2754–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Atkins JL, Jylhava J, Pedersen N, et al. A Genome-Wide Association Study of the Frailty Index Highlights Synaptic Pathways in Aging. medRxiv 2019:19007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mourtzi N, Ntanasi E, Yannakoulia M, et al. Apolipoprotein ε4 allele is associated with frailty syndrome: results from the hellenic longitudinal investigation of ageing and diet study. Age Ageing 2019;48:917–21. [DOI] [PubMed] [Google Scholar]
- 93.Ryu S, Atzmon G, Barzilai N, Raghavachari N, Suh Y. Genetic landscape of APOE in human longevity revealed by high-throughput sequencing. Mech Ageing Dev 2016;155:7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McKay GJ, Silvestri G, Chakravarthy U, et al. Variations in apolipoprotein E frequency with age in a pooled analysis of a large group of older people. Am J Epidemiol 2011;173:1357–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ang LS, Cruz RP, Hendel A, Granville DJ. Apolipoprotein E, an important player in longevity and age-related diseases. Exp Gerontol 2008;43:615–22. [DOI] [PubMed] [Google Scholar]
- 96.Walston J, Arking D, Fallin D, et al. IL-6 gene variation is not associated with increased serum levels of IL-6, muscle, weakness, or frailty in older women. Exp Gerontol 2005;40:344–52. [DOI] [PubMed] [Google Scholar]
- 97.Newman AB, Gottdiener JS, McBurnie MA, et al. Associations of subclinical cardiovascular disease with frailty. J Gerontol A Biol Sci Med Sci 2001;56:M158–66. [DOI] [PubMed] [Google Scholar]
- 98.Perkisas S, Vandewoude M. Where frailty meets diabetes. Diabetes Metab Res Rev 2016;32:261–7. [DOI] [PubMed] [Google Scholar]
- 99.Buchman AS, Schneider JA, Leurgans S, Bennett DA. Physical frailty in older persons is associated with Alzheimer disease pathology. Neurology 2008;71:499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 2005;6:95. [DOI] [PubMed] [Google Scholar]
- 101.Sathyan S, Barzilai N, Atzmon G, Milman S, Ayers E, Verghese J. Genetic insights into frailty: Association of 9p21–23 locus with frailty. Frontiers in medicine 2018;5:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mekli K, Stevens A, Marshall AD, et al. Frailty Index associates with GRIN2B in two representative samples from the United States and the United Kingdom. PLoS One 2018;13:e0207824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morisada N, Ioroi T, Taniguchi-Ikeda M, et al. A 12p13 GRIN2B deletion is associated with developmental delay and macrocephaly. Hum Genome Var 2016;3:16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Endele S, Rosenberger G, Geider K, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet 2010;42:1021. [DOI] [PubMed] [Google Scholar]
- 105.Gale C, Ritchie SJ, Starr JM, Deary IJ. Physical frailty and decline in general and specific cognitive abilities: the Lothian Birth Cohort 1936. J Epidemiol Community Health 2020;74:108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Robertson DA, Savva GM, Kenny RA. Frailty and cognitive impairment—a review of the evidence and causal mechanisms. Ageing Res Rev 2013;12:840–51. [DOI] [PubMed] [Google Scholar]
- 107.Sathyan S, Ayers E, Gao T, et al. Frailty and risk of incident motoric cognitive risk syndrome. J Alzheimer’s Dis 2019;71:S85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wallace LM, Theou O, Godin J, Andrew MK, Bennett DA, Rockwood K. Investigation of frailty as a moderator of the relationship between neuropathology and dementia in Alzheimer’s disease: a cross-sectional analysis of data from the Rush Memory and Aging Project. Lancet Neurol 2019;18:177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Verghese J, Lipton RB, Hall CB, Kuslansky G, Katz MJ, Buschke H. Abnormality of gait as a predictor of non-Alzheimer’s dementia. New Engl J Med 2002;347:1761–8. [DOI] [PubMed] [Google Scholar]
- 110.Livshits G, Malkin I, Bowyer RC, et al. Multi-OMICS analyses of frailty and chronic widespread musculoskeletal pain suggest involvement of shared neurological pathways. Pain 2018;159:2565–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sivakumaran S, Agakov F, Theodoratou E, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet 2011;89:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Organization WH. World health statistics 2016: monitoring health for the SDGs sustainable development goals. World Health Organization; 2016. [Google Scholar]
- 113.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 114.Wang Q, Wang Y, Lehto K, Pedersen NL, Williams DM, Hägg S. Genetically-predicted life-long lowering of low-density lipoprotein cholesterol is associated with decreased frailty: a Mendelian randomization study in UK biobank. EBioMedicine 2019;45:487–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Z, Burgess S, Wang Z, et al. Associations of triglyceride levels with longevity and frailty: a Mendelian randomization analysis. Sci Rep 2017;7:41579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marioni RE, Ritchie SJ, Joshi PK, et al. Genetic variants linked to education predict longevity. Proc Natl Acad Sci U S A 2016;113:13366–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stephan Y, Sutin AR, Kornadt A, Terracciano A. Polygenic scores for education, health, and personality as predictors of subjective age among older individuals of European ancestry: evidence from the Health and Retirement Study. Psychol Aging 2019;34:139. [DOI] [PubMed] [Google Scholar]
- 120.Brothers A, Miche M, Wahl H-W, Diehl M. Examination of associations among three distinct subjective aging constructs and their relevance for predicting developmental correlates. J Gerontol B Psychol Sci Soc Sci 2015;72:547–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Feng Z, Lugtenberg M, Franse C, et al. Risk factors and protective factors associated with incident or increase of frailty among community-dwelling older adults: A systematic review of longitudinal studies. PLoS One 2017;12:e0178383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gale CR, Mõttus R, Deary IJ, Cooper C, Sayer AA. Personality and risk of frailty: the English Longitudinal Study of Ageing. Ann Behav Med 2016;51:128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Danielsdottir HB, Jylhava J, Hagg S, et al. Neuroticism as a Predictor of Frailty in Old Age: a Genetically Informative Approach. Psychosom Med 2019;81:799–807. [DOI] [PubMed] [Google Scholar]
- 124.Hoogendijk EO, van Hout HP, Heymans MW, et al. Explaining the association between educational level and frailty in older adults: results from a 13-year longitudinal study in the Netherlands. Ann Epidemiol 2014;24:538–44, e532. [DOI] [PubMed] [Google Scholar]
- 125.Franse CB, van Grieken A, Qin L, Melis RJ, Rietjens JA, Raat H. Socioeconomic inequalities in frailty and frailty components among community-dwelling older citizens. PLoS One 2017;12:e0187946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huibregtse BM, Newell-Stamper BL, Domingue BW, Boardman JD. Genes related to education predict frailty among older adults in the United States. J Gerontol B Psychol Sci Soc Sci 2019. 10.1093/geronb/gbz092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gubbi S, Schwartz E, Crandall J, et al. Effect of Exceptional Parental Longevity and Lifestyle Factors on Prevalence of Cardiovascular Disease in Offspring. Am J Cardiol 2017;120:2170–5. 10.1016/j.amjcard.2017.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Florez H, Ma Y, Crandall JP, et al. Parental longevity and diabetes risk in the Diabetes Prevention Program. J Gerontol A Biol Sci Med Sci 2011;66:1211–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van den Berg N, Beekman M, Smith KR, Janssens A, Slagboom PE. Historical demography and longevity genetics: Back to the future. Ageing Res Rev 2017;38:28–39. [DOI] [PubMed] [Google Scholar]
- 130.Atzmon G, Schechter C, Greiner W, Davidson D, Rennert G, Barzilai N. Clinical phenotype of families with longevity. J Am Geriatr Soc 2004;52:274–7. [DOI] [PubMed] [Google Scholar]
- 131.Terry DF, Wilcox M, McCormick MA, Lawler E, Perls TT. Cardiovascular advantages among the offspring of centenarians. J Gerontol A Biol Sci Med Sci 2003;58:M425–31. [DOI] [PubMed] [Google Scholar]
- 132.Ayers E, Barzilai N, Crandall JP, Milman S, Verghese J. Association of exceptional parental longevity and physical function in aging. Age 2014;36:9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ayers E, Barzilai N, Crandall JP, Milman S, Verghese J. Association of family history of exceptional longevity with decline in physical function in aging. J Gerontol A Biol Sci Med Sci 2017:glx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim S, Welsh DA, Cherry KE, Myers L, Jazwinski SM. Association of healthy aging with parental longevity. Age 2013;35:1975–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Romero-Ortuno R Frailty Index in Europeans: Association with determinants of health. Geriatr Gerontol Int 2014;14:420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


