Abstract
Pemphigus foliaceus (PF) is an autoimmune blistering skin disease characterized by the presence of pathogenic autoantibodies against desmoglein 1, a component of intercellular desmosome junctions. PF occurs sporadically across the globe and is endemic in some Brazilian regions. Because PF is a B‐cell‐mediated disease, we aimed to study the impact of variants within genes encoding molecules involved in the different steps of B‐cell development and antibody production on the susceptibility of endemic PF. We analysed 3,336 single nucleotide polymorphisms (SNPs) from 167 candidate genes genotyped with Illumina microarray in a cohort of 227 PF patients and 193 controls. After quality control and exclusion of non‐informative and redundant SNPs, 607 variants in 149 genes remained in the logistic regression analysis, in which sex and ancestry were included as covariates. Our results revealed 10 SNPs within or nearby 11 genes that were associated with susceptibility to endemic PF (OR >1.56; p < 0.005): rs6657275*G (TGFB2); rs1818545*A (RAG1/RAG2/IFTAP);rs10781530*A (PAXX), rs10870140*G and rs10781522*A (TRAF2); rs535068*A (TNFRSF1B); rs324011*A (STAT6);rs6432018*C (YWHAQ); rs17149161*C (YWHAG); and rs2070729*C (IRF1). Interestingly, these SNPs have been previously associated with differential gene expression, mostly in peripheral blood, in publicly available databases. For the first time, we show that polymorphisms in genes involved in B‐cell development and antibody production confer differential susceptibility to endemic PF, and therefore are candidates for possible functional studies to understand immunoglobulin gene rearrangement and its impact on diseases.
Keywords: genetics, immunogenetics, immunoglobulin, immunology, pemphigus foliaceus
Pemphigus foliaceus (PF) is a B‐cell‐mediated autoimmune skin disease found sporadically across the globe but endemic and highly prevalent in Brazil. The analysis of 167 genes knowingly implicated in antibody production and B‐cell development revealed 10 single nucleotide polymorphisms within or nearby 11 genes that were associated with susceptibility to endemic PF. Our unprecedented study includes a comprehensive in silico analysis suggesting that regulation of molecules involved in B‐cell activation, immunoglobulin isotype switching and hypermutation may explain the genetic associations.
Abbreviations
- Add
additive model
- AICDA
activation‐induced cytidine deaminase gene
- AID
activation‐induced cytidine deaminase molecule
- AIMs
ancestry‐informative markers
- BMP
bone morphogenetic protein
- CIITA
class II major histocompatibility complex transactivator
- cpm
case per million
- CSR
class‐switch recombination
- DNA‐PKcs
DNA‐dependent protein kinase, catalytic subunit
- Dom
dominant model
- eQTL
expression quantitative trait loci
- HGDP‐CEPH
Human Genome Diversity Project – Centre d'Etude du Polymorphisme Humain
- HLA
human leucocyte antigen
- HSC
haematopoietic stem cells
- IFTAP
intraflagellar transport‐associated protein
- Ig
immunoglobulin
- IGH
immunoglobulin heavy locus
- IGK
immunoglobulin kappa locus
- IGL
immunoglobulin lambda locus
- IL
interleukin
- IRF1
interferon regulatory factor 1
- LD
linkage disequilibrium
- MAF
minor allele frequency
- NHEJ1
non‐homologous end‐joining factor 1 (XLF)
- OR
odds ratio
- PAXX
PAXX non‐homologous end‐joining factor
- PF
pemphigus foliaceus
- RAG1
recombination‐activating gene 1
- RAG2
recombination‐activating gene 2
- Rec
recessive model
- SHM
somatic hypermutation
- SNP
single nucleotide polymorphism
- sQTL
splicing quantitative trait loci
- STAT6
signal transducer and activator of transcription 6
- TGFB2
transforming growth factor‐beta 2
- TNF
tumour necrosis factor
- TNFRSF1B
TNF receptor superfamily member 1B
- TRAF2
TNF receptor‐associated factor 2
- XRCC4
X‐ray repair cross complementing 4
- XRCC5
X‐ray repair cross complementing 5 (Ku80)
- XRCC6
X‐ray repair cross complementing 6 (Ku70)
- YWHAG
tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein gamma
- YWHAQ
tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein theta
INTRODUCTION
Pemphigus foliaceus (PF) is a blistering skin disease characterized by epidermal cell detachment (acantholysis) in the upper layer of the epidermis. In PF, the loss of cell adhesion is a consequence of the presence of autoantibodies, mostly IgG1 and IgG4, against desmoglein 1 (DSG1), a desmosomal component of keratinocytes. 1 , 2 PF occurs sporadically across the globe, with an incidence of one case per million (cpm). 3 , 4 , 5 , 6 In Brazil, however, its incidence reaches 25–35 cpm in some endemic regions. 7 As a multifactorial disease, multiple genetic and environmental factors contribute to the risk of developing PF. The environmental factors that trigger the disease in the Brazilian endemic regions are not well established; however, they are possibly related to exposure to sunlight, mosquito bites, certain foods and poor living conditions. 8 In terms of susceptibility, several genetic variants have been identified as playing a role in the risk of developing PF, 9 including HLA (human leucocyte antigen) genes, 10 , 11 , 12 , 13 KIR (killer‐cell immunoglobulin‐like receptor) 14 , 15 , 16 genes and genes of the complement system, 17 , 18 , 19 among others. 20 , 21 , 22 , 23 , 24
Antibodies are immunoglobulin (Ig) molecules produced by B cells after a series of somatic rearrangements in immunoglobulin genes. Structurally, antibodies can be divided into the variable domain, responsible for antigen binding, and the constant domain, which determines their effector function. Ig is a homoheterodimer composed by two identical heavy chains and two identical light chains; the heavy chain is encoded by the immunoglobulin heavy locus (IGH), and the light chains are encoded by immunoglobulin lambda locus (IGL) or immunoglobulin kappa locus (IGK). 25 Each one of these genes is composed of multiple gene segments in a way that their germline configuration does not allow the transcription to a functional mRNA. To be transcribed, these genes first undergo a complex somatic DNA rearrangement process called V(D)J recombination during the initial steps of B‐cell development, 26 which results in a V(D)J exon that will encode the variable region. 26 , 27 , 28 Following antigen encountering, a process called somatic hypermutation allows positive selection of B cells that exhibit Ig with increased antigen‐binding affinity and results in a more effective immune response. 29 Finally, immunoglobulin genes undergo a process called class‐switch recombination (CSR) that changes the isotype expressed by the B cells, from IgM and IgD to IgA, IgG or IgE (Figure 1), which changes the Ig effector function. 30 All these processes involve molecules responsible for the recognition of target sequences, double‐strand cleavage, non‐homologous end joining, as well as nucleotide deamination, excision, and addition. Because PF is an antibody‐mediated disease, we hypothesized that germline variants in genes that affect the development of immunoglobulins are candidates for disease association. Here, we screened 3,336 variants in 167 genes directly involved in antibody production and observed that ten single nucleotide polymorphisms (SNPs) were associated with PF.
Figure 1.
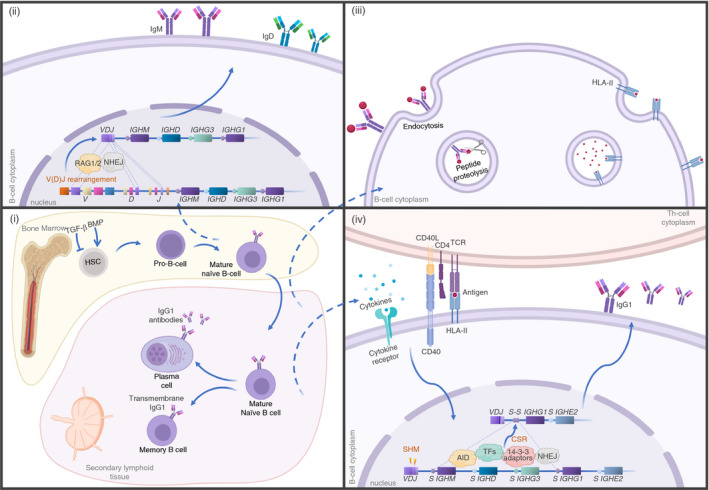
Stages of antibody production (i) In the bone marrow, haematopoietic stem cells (HSC) can give rise to common lymphoid progenitor when stimulated by bone morphogenetic protein (BMP) signalling and by the suppression of transforming growth factor‐beta (TGF‐β) stimulation. (ii) During B‐cell development, in the pro‐B‐cell stage, immunoglobulin heavy chain genes (IGH) undergo a somatic reassembly of their V, D and J segments through DNA cleavage by RAG1 and RAG2 (recombination‐activating proteins 1 and 2), followed by non‐homologous end joining (NHEJ) of the DNA. These steps result in a VDJ exon that will encode the variable region of Ig heavy chain and will be expressed with the adjacent constant gene segments, IGHM and IGHD (IgM and IgD heavy chain). If this rearrangement is successful, V and J gene segments of immunoglobulin light chains (kappa or lambda) are also rearranged (not shown), and complete IgM and IgD molecules are expressed. The cells are now called mature naïve B cells. (iii) In secondary lymphoid organs, immunoglobulin molecules on the surface of naïve B cells recognize and bind to their specific antigens, which are internalized by endocytosis and proteolyzed. (iv) The resulting peptides are presented in the context of HLA class II to cognate (with same antigen specificity) CD4+ T helper cells through their T‐cell receptor (TCR). Afterwards, T cells express CD40L and cytokines, which bind to specific receptors of B cells, promoting B‐cell activation, class‐switch recombination (CSR) and somatic hypermutation (SHM) of the immunoglobulin gene. CSR requires precise orchestration of signalling molecules, transcription factors and adaptor molecules (14‐3‐3) that recognized specific switch (S) regions of the DNA located upstream of each constant gene segment. 14‐3‐3 molecules are recognized by activation‐induced cytidine deaminase (AID) that promotes DNA cleavage, which is followed by NHEJ that links the rearranged VDJ exon to the selected constant IGH gene segment. As a result, this process changes the isotype expressed by the B cells from IgM and IgD to IgA, IgG or IgE. AID also plays a role in SHM by introducing single nucleotide substitutions in the V(D)J exon, which increases the diversity of immunoglobulins and allows positive selection of B cells with increased antigen‐binding affinity. These steps result in the transcription of the rearranged genes into immunoglobulin molecules that can be secreted by B cell, which are called antibodies, with different effector functions and higher binding affinity for a specific antigen. This figure was made with biorender (https://biorender.com/).
METHODS
Study population
DNA was isolated from peripheral blood of 227 unrelated endemic PF patients and 193 unrelated controls without a history of any autoimmune disease (Table 1). To minimize possible population stratification, we included only individuals of predominantly European ancestry. We excluded individuals self‐declared as non‐Euro‐descendants and those who reported a family history of miscegenation with non‐European. The study participants were contacted at Hospital Adventista do Pênfigo (Campo Grande, Mato Grosso do Sul), Hospital das Clínicas da Faculdade de Medicina da USP (Ribeirão Preto, São Paulo), Lar da Caridade – Hospital do Fogo Selvagem (Uberaba, Minas Gerais), Hospital de Clínicas da UFPR, Hospital de Dermatologia Sanitária São Roque and Hospital Santa Casa de Misericórdia (Curitiba, Paraná). The diagnosis was based on clinical characterization by specialized dermatologists, immunological tests, histopathology and immunohistochemistry of skin biopsies. All individuals voluntarily agreed to participate in this study and signed informed consent, according to the Declaration of Helsinki. This study was performed under Brazilian federal laws and approved by the Human Research Ethics Committee of the Federal University of Paraná under protocol number CAAE 02727412.4.0000.0096.
Table 1.
Characterization of patients and controls.
| Median age. Years old (range) | Sex | % of SSA | % of AMER | % of EUR | |
|---|---|---|---|---|---|
| Patients | 40.9 (6–83) | 53% female | 15 | 12 | 73 |
| Controls | 44.8 (11–86) | 52% female | 16 | 15 | 69 |
Percentages are the estimated proportion of sub‐Saharan African (SSA), Amerindian (AMER) and European (EUR) ancestry of the PF patients and control samples, based on the analysis of 71 ancestry‐informative markers and using the HGDP‐CEPH database 35 as reference.
Selection of candidate genes and genotyping
We selected 167 genes knowingly implicated in antibody production, including those encoding Ig (heavy, kappa and lambda chains). To select the candidate genes, we performed an extensive search in the literature databases (Google Scholar and PubMed) for review articles published in the last five years, using the terms 'V(D)J rearrangement', 'class‐switch recombination' (or 'CSR') and 'somatic hypermutation'. We comprehensively searched all the references cited by the retrieved articles. The full list of candidate genes is given in Table S1. Genotyping was performed by SNP microarrays using InfiniumTM CoreExome‐24 v1.1 BeadChip (Illumina, San Diego, USA).
Population structure analysis
Even though we carefully matched patients and controls for ancestry, the Brazilian population is admixed. To account for the possibility of population structure, we included another level of rigour by analysing a panel of 71 previously validated 31 , 32 , 33 , 34 ancestry‐informative markers (AIMs) (Table S2). The allelic frequencies of these SNPs differ across the major continental population groups (FST >0.25, δ > 0.40), and the genotypes of HGDP‐CEPH (Human Genome Diversity Project – Centre d'Etude du Polymorphisme Humain) populations are publicly available. 35 Pairwise FST was calculated using Arlequin v3.5.2 software, 36 and δ was considered the difference between the frequencies of pairs of populations. We compared the study population to the HGDP‐CEPH populations most closely related to the three major ancestral groups of the Brazilians: 37 sub‐Saharan Africans (n = 120) – Biaka Pygmy, Mbuti Pygmy, Mandenka, Yoruba, San, Bantu; Amerindians (n = 83) – Surui, Karitiana, Pima, Piapoco and Curripaco; and Europeans (n = 118) – French, French Basque, North Italian, Orcadian, Sardinian, Tuscan. For estimation of individual and populational ancestry, we used the software STRUCTURE v.2.2 38 , 39 , 40 with a run length of 100 000 burn‐in and 500 000 Markov chain Monte Carlo (MCMC) replications, the admixture model and independent allele frequency model.
Association analysis
We used PLINK v1.9 41 for all manipulation of SNP data. We extracted a total of 3,336 SNPs located within 3 Kbp upstream and downstream of each one of the 167 genes. From the total SNPs initially retrieved, we removed variants as follows: (a) whose genotypes deviated from the Hardy–Weinberg equilibrium in controls (p < 0.05); (b) or in strong linkage disequilibrium (LD) with any other variant (r 2 ≥ 0.80). We established MAF ≥0.20 (minor allele frequency) to reach at least 80% power with a one‐sided type I error rate α = 0.05 to detect small to moderate effect sizes (0.38 < d < 0.50) 42 , 43 . We established the significance threshold as p < 0.005. After quality control and removal of redundant or non‐informative SNPs, a total of 607 variants in 149 genes remained for the association analysis (Table S3). We performed logistic regression for additive, dominant and recessive models using sex and two principal components as covariates.
In silico analysis
We used the online tool HaploReg 44 to evaluate whether the associated SNPs could be implicated in structural and regulatory effects. To search for eQTL (expression quantitative trait loci) for variants that were associated with PF or in LD with them (r 2 > 0.8), we used the tool Qtlizer, 45 which compiles information from several databases. 44 , 54 We obtained splicing quantitative trait locus (sQTL) data from GTEx Portal 46 and searched the transcript variants of the affected genes in Ensembl. 55 We assessed LD between variants in the database LDlink 56 using European populations (CEU, TSI, FIN, GBR, IBS) of the 1000 Genomes Project. 57
RESULTS
Lack of population stratification in patients and controls
Patients and controls were previously classified as predominantly Euro‐descendants based on phenotypic characteristics and detailed assessment of family history. Here, our analysis using 71 AIMs showed that patients and controls were, in fact, homogeneous regarding ancestry (p = 0.14). The proportions of sub‐Saharan African, Amerindian and European compounds were 0.15, 0.12 and 0.73 for controls and 0.16, 0.15 and 0.69 for patients, respectively (Table 1, Figure 2).
Figure 2.
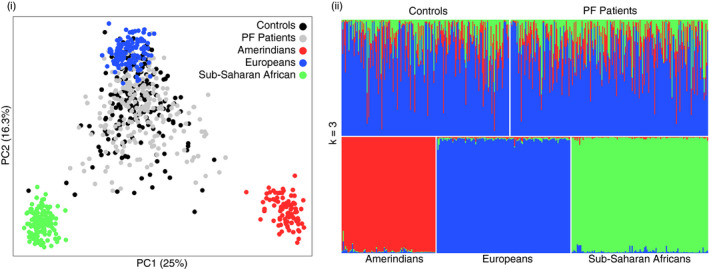
Lack of population structure in the study population. (i) Principal component analysis and (ii) bar plot of inferred ancestry proportions performed with 71 ancestry‐informative markers comparing PF patients and control samples with HGDP‐CEPH samples from three regional populations: sub‐Saharan Africans (n = 120), Amerindians (n = 83) and Europeans (n = 118) 35 .
Genetic associations
We found 10 SNPs within 11 genes, or in their vicinity, related to B‐cell development and antibody production associated with differential susceptibility to endemic PF either in the dominant (Dom), in the recessive (Rec) or in the additive (Add) models (Table 2). All variants associated with endemic PF were located in intronic or intergenic regions. One SNP is within a gene implicated in B‐cell development, two are in genes involved in V(D)J rearrangement, and seven are close to or within genes that participate in antibody class‐switch recombination.
Table 2.
Variants associated with increased risk to endemic pemphigus foliaceus.
| Association analyses | Predicted eQTL of the associated SNPs and their proxy | |||||
|---|---|---|---|---|---|---|
|
SNP Genomic position* |
Location | Model |
OR (95% CI) p‐value |
Affected gene | Cell, tissue or organ |
Effect p‐value |
| B‐cell modulation | ||||||
|
rs6657275*G Chr1:218596461 |
Intron of TGFB2 | Rec |
2.26 (1.33‐3.84) p = 2.6x10−3 |
TGFB2 | Lung, testis, brain | eQTL<8.2x10−3 |
| TGFB2‐AS1 | Whole blood, skeletal muscle | eQTL<2.8x10−9 | ||||
| V(D)J rearrangement | ||||||
| rs1818545*A Chr11:36612090 | Intergenic between RAG1, RAG2 and IFTAP | Dom |
1.85 (1.22‐2.81) p = 3.6 x10−3 |
IFTAP | Brain | eQTL<7.1x10−3 |
|
rs10781530*A Chr9:139885948 |
921 bp 5’ of PAXX | Add |
1.58 (1.16‐2.15) p = 3.6x10−3 |
PAXX | Whole blood, lung, heart, brain, artery, adipose | eQTL<8.9x10−5 |
| Class‐switch recombination and somatic hypermutation | ||||||
|
rs10870140*G Chr9:139796419 |
Intron of TRAF2 | Rec |
1.76 (1.19‐2.61) p = 4.9x10−3 |
TRAF2 | Whole blood | eQTL<1.2x10−9 |
| PAXX | Brain | eQTL<6.5 x10−6 | ||||
|
rs10781522*A Chr9:139815053 |
Intron of TRAF2 | Add |
1.61 (1.22‐2.14) p = 9x10−3 |
TRAF2 | Whole blood, transverse colon, testis, lymphoblastoid cell, monocytes, skeletal muscle, skin, tibial nerve, dendritic cells | eQTL<3.5 x10−4 |
| Rec |
1.99 (1.30‐3.05) p = 1.5x10−3 |
PAXX | Blood, brain | eQTL<1.2 x10−3 | ||
|
rs535068*A Chr1:12189561 |
Intron of TNFRSF8 | Dom |
3.11 (1.46‐6.62) p = 3.2x10−3 |
TNFRSF1B | Whole blood, skin, brain | eQTL<3.1 x10−3 |
|
rs324011*A Chr12:57502182 |
Intron of STAT6 | Add |
1.56 (1.16‐2.10) p = 3.3x10−3 |
STAT6 | Whole blood, CD4+ lymphocytes, monocytes, brain, liver, colon sigmoid | eQTL<1.4 x10−4 |
| Dom |
1.94 (1.30‐2.90) p = 1.3x10−3 |
|||||
|
rs2070729*C Chr5:131819921 |
Intron of IRF1 | Add |
1.56 (1.17‐2.09) p = 2.8x10−3 |
IRF1 | Whole blood, heart, monocytes | eQTL<4.7 x10−4 |
| Rec |
2.02 (1.25‐3.27) p = 4.4x10−3 |
IRF1‐AS1 | Whole blood, thyroid, spleen, skin, tibial nerve, Skeletal muscle, lung, heart, oesophagus, colon, brain, artery, adipose | eQTL<1.1 x10−4 | ||
| IL13 | Tibial nerve | eQTL<4.2 x10−5 | ||||
|
rs6432018*C Chr2:9721896 |
2.2 kb 3’ of YWHAQ | Add |
1.69 (1.28‐2.24) p = 3x10−3 |
YWHAQ | Tibial artery | sQTL<1.3 x10−6 |
| Dom |
2.04 (1.32‐3.14) p = 1.2 x10−3 |
|||||
|
rs17149161*C Chr7:75978229 |
intron of YWHAG | Add |
1.63 (1.20‐2.21) p = 1.7 x10−3 |
YWHAG | Monocytes, adipose, lung | eQTL<1.6 x10−4 |
SNP: rs ID of the single nucleotide polymorphism, Chr: chromosome, OR: odds ratio, CI: confidence interval. eQTL: expression quantitative trait loci. *Genomic position according to GRCh37.p13 primary assembly. The frequency of the associated alleles in patients and controls is given in Table S3. The complete list of proxy SNPs is available in Table S5.
Functional annotation
We performed a comprehensive in silico analysis using several publicly available databases and online tools. We found that all variants associated with endemic PF, or their proxy SNPs (r 2 > 0.8), had been previously associated with variable gene expression levels in different tissues, mainly in peripheral blood (Table 2). 45 Our in silico analysis also showed that these variants are located in sites containing chromatin regulatory histone marks and may be related to the regulation of promoters and enhancers in T and B cells 44 (Table S4), as summarized below.
The intergenic variant rs1818545*A is in nearly absolute LD with nine other SNPs (r 2 ≈ 1; D’ ≈ 1), including rs7104753*C, which is located in a predicted regulatory region of RAG2 (recombination‐activating gene 2) (Table S4). We found three variants that have eQTL effects: the regulatory region variant rs10781530*A, located at 921 bp 5’ of the PAXX (PAXX non‐homologous end‐joining factor) gene, which is involved in non‐homologous end joining; and rs10870140*G and rs10781522*A, two intronic variants of TRAF2 (TNF receptor‐associated factor 2), which have cis‐eQTL effects on TRAF2 in whole blood. Another SNP associated with endemic PF was the variant rs2070729*C, located in an intron of the gene IRF1 (interferon regulatory factor 1). This variant has a cis‐eQTL effect on IRF1 and also a trans‐eQTL effect on the gene IL13 (interleukin 13). We also observed that rs6432018*C, an intronic variant in the gene YWHAQ (tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein,theta isoform), is in high LD (r2 > 0.95) with other 12 SNPs that have sQTL effects on YWHAQ (Table 2, Table S5, and Table S6).
DISCUSSION
Break of immune homeostasis and self‐tolerance in some autoimmune disorders are directly related to the production of autoantibodies by B cells that differentiate into plasma cells. 58 In our study, we focused on genetic variants within genes related to B‐cell development, activation and maturation, and also immunoglobulin gene rearrangement, class‐switch recombination and somatic hypermutation. We aimed to contribute to a better understanding of these mechanisms in the context of endemic PF.
Our analysis with 71 previously validated ancestral‐informative markers confirmed no ancestry bias between patients and controls from our study. Their conspicuous differences in genotypic frequencies among Europeans, Africans and Amerindians indicate that these markers constitute a robust set to assess the continental ancestry of the study population. Nevertheless, we used two principal components as covariates in our logistic regression model to correct for possible minor differences that could contribute to spurious associations.
We used the p‐value of 0.005 as a cut‐off to identify genetic associations, as suggested by others, 59 , 60 , 61 to achieve a low risk of false associations while not excluding possible real associations with smaller effects. Although the use of arbitrary cut‐offs always brings the risk of type I error, we have taken several precautions to reduce the chances of false discoveries: (a) we only analysed variants for which our sample size allowed statistical power of at least 80% to detect low and moderate effects (MAF ≥0.20); (b) we quantified the individual ancestry compound of each individual and adjusted our analysis using principal components as covariates even though our samples were carefully matched; and (c) we adjusted our analysis for sex and age. As we discuss in detail below, all our associations are provided with a plausible biological explanation, increasing the confidence of our results. Nevertheless, further replication in pemphigus or other autoimmune disease cohorts would be desirable to corroborate our findings.
We found 10 variants located within or in the vicinity of 11 genes related to B‐cell development (TGFB2), V(D)J recombination (RAG1, RAG2, PAXX), B‐cell activation, somatic hypermutation and class‐switch recombination (TNFRSF1B, TRAF2, STAT6, IL13, IRF1, YWHAG and YWHAQ). The allele rs6657275*G, associated with increased risk to endemic PF, is located in the third intron of TGFB2 (transforming growth factor 2) and has been previously associated with susceptibility to cerebral malaria. 62 This SNP is in strong LD with other 39 variants, several of which have been predicted to have eQTL effects in different tissues on the gene TGFB2 and its antisense long non‐coding RNA (lncRNA) gene TGFB2‐AS1. Some of the SNPs in LD with rs6657275 are possibly related to regulatory histone marks H3K4me3 and H3K4me1 in T and B cells (Table S4), which suggest that they may be involved in transcriptional regulation of TGFB2, and/or of the lncRNA genes TGFB2‐AS1 and TGFB2‐OT1, which physically overlap (antisense gene overlap) the TGFB2 gene. Interestingly, the lncRNA TGFB2‐AS1 has been associated with regulatory functions on TGF‐β (product of TGFB2 gene) and bone morphogenetic protein (BMP) signalling on keratinocytes. 63 The TGF‐β and BMP also regulate the differentiation of haematopoietic stem cells (HSC) to the myeloid lineage or the lymphoid lineage, respectively. 64 Additionally, TGF‐β is secreted by B cells and may regulate B‐cell proliferation. 65 Dysregulation of TGF‐β pathways is known to be implicated in antibody‐mediated autoimmune disorders. 64
Two SNPs associated with endemic PF are located in genes whose products participate in the DNA cleavage and joining phases of the V(D)J recombination process: rs1818545 (RAG2) and rs10781530 (PAXX), respectively. The allele rs1818545*A has been previously associated with radiation‐induced pneumonitis in lung cancer patients. 66 The impact of rs1818545 on RAG1 and RAG2 expression can hardly be detected by the approaches applied for the investigation of gene expression because these genes are only expressed in developing lymphocytes, 67 which were not included in the published studies. However, this variant and their proxy SNPs have cis‐eQTL effects on the IFTAP (intraflagellar transport‐associated protein) gene, whose expression is inversely correlated with RAG1 and RAG2 expression. 68 Moreover, rs12283331 is in absolute LD (r 2 = 1; D’ =1) with rs1818545 and is located in a predicted binding motif for the transcription factor Ik‐2 in primary haematopoietic stem cells. 44 Ik‐2 promotes RAG1 and RAG2 transcription and downregulates IFTAP expression. 69 RAG1 is responsible for recognizing specific conserved recombination signal sequences adjacent to gene segments V, D and J of immunoglobulins, responsible for the specificity of the double‐strand cleavage of the DNA. RAG2, on the other hand, is necessary for the catalytic activity of RAG1. 70 Both RAG1 and RAG2 are part of the RAG complex, which also mediates allelic exclusion of Ig, ensuring that each B cell expresses only one allele of the Ig genes. 71 Therefore, the association of rs1818545*A with increased risk to endemic PF could be related to the RAG1 and RAG2 role in the V(D)J regulation.
The variant rs10781530*A is located upstream of PAXX and is associated with a higher expression of this gene. PAXX molecules stabilize the enzymatic complex composed by XRCC4, XRCC6 (Ku70), XRCC5 (Ku80), DNA‐PKcs, DNA ligase 4 and NHEJ1, which is required for the non‐homologous end‐joining pathway in V(D)J recombination. 72 This complex repairs the double‐strand break in the first step of the rearrangement process. The two SNPs rs10870140 and rs10781522, located within intronic regions of the gene TRAF2 (73 Kbp of the gene PAXX), are associated with endemic PF and have trans‐eQTL effects on PAXX. Therefore, it is plausible that the associations observed for these SNPs are related to the DNA repair process during V(D)J rearrangement.
After antigen recognition, B‐cell activation may be mediated by T cells. The variant rs2070729, located in the intron 9 of the IRF1 gene, together with five other SNPs in strong LD with it, have eQTL effects on IRF1 and IL13 gene expression. IRF1 is a transcription factor that promotes the expression of CIITA (class II major histocompatibility complex transactivator) molecules, 73 a critical regulator of HLA class II expression in antigen‐presenting cells, including B cells. HLA class II molecules are necessary for antigen presentation to T cells, T‐cell activation and, consequently, activation of B cells with the same antigen specificity. 74 Interestingly, a combination of certain HLA‐DRB1 alleles with a promoter variant in CIITA was strongly associated with endemic PF (OR = 14.05, p < 10−6). 13
T‐cell activation also stimulates the production of TNF (tumour necrosis factor) by B cells, which activates the TNF receptor superfamily member 1B (TNFRSF1B or CD120b) in B cells. Activated TNFRSF1B interacts with TNF receptor associated factor 2 (TRAF2), triggering the secretion of IgM. 75 Furthermore, Ig class switch is also regulated by another receptor on the surface of B cells, TNF receptor superfamily member 8 (TNFRSF8 or CD30), which prevents the class switching in B cells that are non‐antigen‐specific. 76 Associated with differential endemic PF risk was the intronic SNP rs535068 located in TNFRSF8. This SNP and other ten in strong LD with it (r 2 > 0.8) are located in a regulatory region of TNFRSF8. This region, which appears to be relevant for the regulation of T‐ and B‐cell lineages, includes enhancer and promoter segments that contain epigenetic chromatin marks such as H3K4me1, H3K4me3, H3K27ac and H3K9ac (Table S4). The SNP rs535068 and six of the SNPs in strong LD with it also have trans‐eQTL effects on the gene TNFRSF1B in blood and skin. Two other SNPs associated with endemic PF, rs10870140 and rs10781522, were located in intronic regions of TRAF2, whose product also plays a critical role in B‐cell activation. These variants have not only cis‐eQTL effect on TRAF2 but are also associated with differential expression of the gene PAXX, as mentioned above. The interaction between CD40 receptor and TRAF2 is essential for CD40‐regulated class switch of Ig from IgM to IgG and B‐cell activation. 77 , 78 Defects in these pathways have been suggested to cause the generation of pathogenic autoantibodies. 76
The class switch to IgG and IgE in B cells activated by IL‐4 and IL‐13 is also mediated by the transcription factor STAT6 (signal transducer and activator of transcription 6). 79 , 80 STAT6 acts as a regulator of several genes in IL‐4‐stimulated B cells, 81 which includes the AICDA. 82 The AICDA gene encodes the AID molecule (activation‐induced cytidine deaminase), which plays a pivotal role during class‐switch recombination by generating a double‐strand break of the DNA. 83 Moreover, AID action produces point mutations during somatic hypermutation. 84 The intronic SNP rs324011 in STAT6, also associated with endemic PF, is in LD with six other SNPs (Table S5). All these seven variants have eQTL effects on STAT6 expression in whole blood and other tissues. Additionally, four of them are located within enhancer or promoter regions and are predicted to participate in epigenetic chromatin modifications in several tissues, including T and B cells. Therefore, in the context of immunoglobulin gene rearrangement, it is plausible to suggest that the association of rs324011*A with endemic PF may be related to differential expression of STAT6, which consequently affects the expression of AICDA.
The CSR process requires the precise recognition of the DNA cleavage sites on the switch (S) regions. 85 The adaptor molecules 14‐3‐3 recognize these regions and function as scaffolds of the CSR machinery, interacting and stabilizing AID and other molecules, such as PKA‐Ca (cAMP‐dependent protein kinase catalytic subunit alpha) and Ung (uracil‐DNA glycosylase). 86 SNPs within two genes encoding 14‐3‐3 molecules (YWHAG and YWHAQ) were associated with differential susceptibility to endemic PF. The variant rs17149161*C has an eQTL effect on YWHAG, and rs6432018*C has an sQTL effect on YWHAQ. sQTL are genetic variants that can change the splicing ratios of gene transcripts. 87 Interestingly, of the five transcript variants that have been described for YWHAQ, two of them do not encode protein due to alternative splicing (Table S6).
One limitation of our study is that some critical variants implicated in the disease may have been excluded due to three main reasons: i) some relevant variants may not have been genotyped in the microarray. A large number of immune‐related genes, including immunoglobulin genes and others related to B‐cell function, are overall poorly covered in microarray chips due to homology and structural variation that impose technical limitations for genotyping. ii) Other variants may have been removed from the analysis due to their low frequency in our cohort. iii) Some relevant genes for B‐cell development are unknown or were missed. Therefore, some variants affecting PF pathogenesis may not have been uncovered by our study. However, we were able to screen over 3,000 SNPs and select the 607 most informative to our association analysis, which constitutes an unprecedented and comprehensive analysis of B‐cell‐related variants in this autoimmune disease. Our observations provide evidence that variation in B‐cell‐related genes has a pivotal effect on PF risk. Besides, we presented a comprehensive in silico analysis suggesting that regulation of molecules involved in B‐cell activation, immunoglobulin isotype switching and hypermutation may explain the associations that we observed for endemic PF. Therefore, our results justify further in‐depth analysis of genes that were not well covered in our study, applying different technologies that allow high‐resolution characterization.
In summary, we carefully explored the variation in genes implicated in B‐cell development and function in the context of the autoimmune B‐cell‐mediated nature of endemic PF. The associations that we observed can be explained by possible effects on the regulation of expression levels of molecules involved in the complex process of B‐cell modulation, DNA sequence recognition, DNA cleavage and joining, and somatic hypermutation. For the first time, we show that polymorphisms in genes involved in autoantibody production might confer differential susceptibility to this disease. Therefore, we identified candidate genes for possible high‐resolution and functional studies to understand Ig production and its impact on the aetiology of endemic PF and other diseases.
FUNDING INFORMATION
This study was supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Paraná, PRONEX (Convênio 116/2018 – Protocolo 50530) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, finance code 001).
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHOR CONTRIBUTIONS
VCS, DM and DA designed the study. DA performed microarray genotyping. VCS, LMA and TDJF analysed the data. MLPE, DA and DM contributed with reagents. VCS and DA drafted the manuscript. All authors significantly contributed with ideas and critically reviewed this manuscript.
ETHICAL APPROVAL INFORMATION
All individuals voluntarily agreed to participate in this study and signed informed consent, according to the Declaration of Helsinki. This study was performed under Brazilian federal laws and approved by the Human Research Ethics Committee of the Federal University of Paraná under protocol number CAAE 02727412.4.0000.0096.
Supporting information
Table S1. List of candidate genes considered in the association analysis
Table S2. Single nucleotide polymorphism selected as ancestry informative.
Table S3. Single nucleotide polymorphism of the candidate genes selected analysed in the association study.
Table S4. Predicted regulatory effects according to Haploreg database of the associated variants and of those in strong linkage disequilibrium with them (r² ≥ 0.8).
Table S5. Variants associated with increased risk to endemic pemphigus foliaceus, their proxy SNPs and eQTL and sQTL effect of these variants.
Table S6. Transcript variants of genes with associated SNP with sQTL effect.
ACKNOWLEDGMENTS
We warmly thank all the individuals who voluntarily enrolled in this study. Special thanks to Hospital Adventista do Pênfigo for kindly opening their doors for our laboratory members and for treating pemphigus patients with so much care and respect. We also thank the staff of the Laboratório de Genética Molecular Humana ‐ UFPR for their support.
DATA AVAILABILITY STATEMENT
All data are available in the manuscript and supplementary files.
REFERENCES
- 1. Amagai M, Stanley JR. Desmoglein as a target in skin disease and beyond. Journal of Investigative Dermatology 2012; 132(3):776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kasperkiewicz M, Ellebrecht CT, Takahashi H, Yamagami J, Zillikens D, Payne AS, et al Pemphigus. Nat Rev Dis Prim. 2017; 3(1):17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bastuji‐Garin S, Souissi R, Blum L, Turki H, Nouira R, Jomaa B, et al Comparative epidemiology of pemphigus in tunisia and France: unusual incidence of pemphigus foliaceus in young tunisian women. J Invest Dermatol. 1995; 104(2):302–5. [DOI] [PubMed] [Google Scholar]
- 4. Marazza G, Pham HC, Schärer L, Pedrazzetti PP, Hunziker T, Trüeb RM, et al Incidence of bullous pemphigoid and pemphigus in Switzerland: A 2‐year prospective study. Br J Dermatol. 2009; 161(4):861–8. [DOI] [PubMed] [Google Scholar]
- 5. Joly P, Litrowski N. Pemphigus group (vulgaris, vegetans, foliaceus, herpetiformis, brasiliensis). Clin Dermatol. 2011; 29(4):432–6. [DOI] [PubMed] [Google Scholar]
- 6. Kumar K. Incidence of pemphigus in Thrissur district, south India. Indian J Dermatol Venereol Leprol. 2008; 74(4):349. [DOI] [PubMed] [Google Scholar]
- 7. Diaz LA, Sampaio SA, Rivitti EA, Martins CR, Cunha PR, Lombardi C, et al Endemic pemphigus foliaceus (Fogo Selvagem): II. Current and historic epidemiologic studies. J Invest Dermatol. 1989; 92:4–12. [DOI] [PubMed] [Google Scholar]
- 8. Lombardi C, Borges PC, Chaul A, Sampaio SA, Rivitti EA, Friedman H, et al Environmental risk factors in endemic pemphigus foliaceus (Fogo selvagem). “The Cooperative Group on Fogo Selvagem Research”. J Invest Dermatol. 1992; 98(6):847–50. [DOI] [PubMed] [Google Scholar]
- 9. Petzl‐Erler ML. Beyond the HLA polymorphism: A complex pattern of genetic susceptibility to pemphigus. Genet Mol Biol. 2020; 43(3):e20190369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pavoni DP, Roxo VMMS, Marquart Filho A, Petzl‐Erler Ml. Dissecting the associations of endemic pemphigus foliaceus (Fogo Selvagem) with HLA‐DRB1 alleles and genotypes. Genes Immun. 2003; 4(2):110–6. [DOI] [PubMed] [Google Scholar]
- 11. Petzl‐Erler ML, Santamaria J. Are HLA class II genes controlling susceptibility and resistance to Brazilian pemphigus foliaceus (fogo selvagem)? Tissue Antigens 1989; 33:408–14. [DOI] [PubMed] [Google Scholar]
- 12. Brochado MJF, Nascimento DF, Campos W, Deghaide NHS, Donadi EA, Roselino AM. Differential HLA class I and class II associations in pemphigus foliaceus and pemphigus vulgaris patients from a prevalent Southeastern Brazilian region. J Autoimmun. 2016; 72:19–24. [DOI] [PubMed] [Google Scholar]
- 13. Piovezan BZ, Petzl‐Erler ML. Both qualitative and quantitative genetic variation of MHC class II molecules may influence susceptibility to autoimmune diseases: the case of endemic pemphigus foliaceus. Hum Immunol. 2013; 74(9):1134–40. [DOI] [PubMed] [Google Scholar]
- 14. Augusto DG, Lobo‐Alves SC, Melo MF, Pereira NF, Petzl‐Erler ML. Activating KIR and HLA Bw4 ligands are associated to decreased susceptibility to pemphigus foliaceus, an autoimmune blistering skin disease. PLoS One 2012; 7(7):e39991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Augusto DG, O’Connor GM, Lobo‐Alves SC, Bass S, Martin MP, Carrington M, et al Pemphigus is associated with KIR3DL2 expression levels and provides evidence that KIR3DL2 may bind HLA‐A3 and A11 in vivo. Eur J Immunol. 2015; 45(7):2052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farias TDJ, Augusto DG, de Almeida RC, Malheiros D, Petzl‐Erler ML. Screening the full leucocyte receptor complex genomic region revealed associations with pemphigus that might be explained by gene regulation. Immunology 2019; 156(1):86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bumiller‐Bini V, Cipolla GA, de Almeida RC, Petzl‐Erler ML, Augusto DG, Boldt ABW. Sparking fire under the skin? Answers from the association of complement genes with pemphigus foliaceus. Front Immunol. 2018; 9:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salviano‐Silva A, Petzl‐Erler ML, Boldt ABW. CD59 polymorphisms are associated with gene expression and different sexual susceptibility to pemphigus foliaceus. Autoimmunity 2017; 6934:1–9. [DOI] [PubMed] [Google Scholar]
- 19. Oliveira LC, Kretzschmar GC, dos Santos ACM, Camargo CM, Nisihara RM, Farias TDJ, et al Complement receptor 1 (CR1, CD35) polymorphisms and soluble CR1: a proposed anti‐inflammatory role to quench the fire of “Fogo Selvagem” pemphigus foliaceus. Front Immunol. 2019; 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malheiros D, Petzl‐erler ML. Individual and epistatic effects of genetic polymorphisms of B‐cell co‐stimulatory molecules on susceptibility to pemphigus foliaceus. Genes Immun. 2009; 10(6):547–58. [DOI] [PubMed] [Google Scholar]
- 21. Cipolla GA, Park JK, de Oliveira LA, Lobo‐Alves SC, de Almeida RC, Farias TDJ, et al A 3′UTR polymorphism marks differential KLRG1 mRNA levels through disruption of a miR‐584‐5p binding site and associates with pemphigus foliaceus susceptibility. Biochim Biophys Acta – Gene Regul Mech. 2016; 1859(10):1306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lobo‐Alves SC, Augusto DG, Magalhães WCS, Tarazona‐Santos E, Lima‐Costa MF, Barreto ML, et al Long noncoding RNA polymorphisms influence susceptibility to endemic pemphigus foliaceus. Br J Dermatol. 2019; 181(2):324–31. [DOI] [PubMed] [Google Scholar]
- 23. Spadoni MB, Bumiller‐Bini V, Petzl‐Erler ML, Augusto DG, Boldt ABW. First glimpse of epigenetic effects on pemphigus foliaceus. J Invest Dermatol. 2020; 140:488–91. [DOI] [PubMed] [Google Scholar]
- 24. Bumiller‐Bini V, Cipolla GA, Spadoni MB, Augusto DG, Petzl‐Erler ML, Beltrame MH, et al Condemned or Not to Die? Gene polymorphisms associated with cell death in pemphigus foliaceus. Front Immunol. 2019; 10:2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schroeder HW, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010; 125(2):S41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol 2005; 86:43–112. [DOI] [PubMed] [Google Scholar]
- 27. Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006; 24(1):541–70. [DOI] [PubMed] [Google Scholar]
- 28. Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell 2002; 109(2):S45–55. [DOI] [PubMed] [Google Scholar]
- 29. Jacobs H, Bross L. Towards an understanding of somatic hypermutation. Curr Opin Immunol. 2001; 13(2):208–18. [DOI] [PubMed] [Google Scholar]
- 30. Honjo T, Alt F, eds. Immunoglobulin genes. Annu Rev Immunol. 1983;1(1):499–528. [DOI] [PubMed] [Google Scholar]
- 31. Soundararajan U, Yun L, Shi M, Kidd KK. Minimal SNP overlap among multiple panels of ancestry informative markers argues for more international collaboration. Forensic Science International: Genetics 2016; 23:25–32. [DOI] [PubMed] [Google Scholar]
- 32. Lao O, Van DK, Kersbergen P, De KP, Kayser M. Proportioning whole‐genome single‐nucleotide–polymorphism diversity for the identification of geographic population structure and genetic ancestry. Am J Hum Genet. 2006; 78(4):680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009; 30(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mychaleckyj JC, Havt A, Nayak U, Pinkerton R, Farber E, Concannon P, et al Genome‐wide analysis in Brazilians reveals highly differentiated native american genome regions. Mol Biol Evol. 2017; 34(3):msw249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cann HM. A human genome diversity cell line panel. Science 2002; 296(5566):261–2. [DOI] [PubMed] [Google Scholar]
- 36. Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010; 10(3):564–7. [DOI] [PubMed] [Google Scholar]
- 37. Salzano FM. Interethnic variability and admixture in Latin America – social implications. Rev Biol Trop. 2014; 1(2):405. [DOI] [PubMed] [Google Scholar]
- 38. Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007; 7(4):574–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 2003; 164(4):1567–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155(2):945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second‐generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015; 4(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gauderman WJ. Sample size requirements for matched case‐control studies of gene‐environment interaction. Stat Med. 2002; 21(1):35–50. [DOI] [PubMed] [Google Scholar]
- 43. Gauderman WJ. Sample size requirements for association studies of gene‐gene interaction. Am J Epidemiol. 2002; 155(5):478–84. [DOI] [PubMed] [Google Scholar]
- 44. Ward LD, Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012; 40(D1):930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Munz M, Wohlers I, Simon E, Reinberger T, Busch H, Schaefer AS, et al Qtlizer: comprehensive QTL annotation of GWAS results. bioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, et al A novel approach to high‐quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank. 2015; 13(5):311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grimaldi‐Bensouda L, Rossignol M, Koné‐Paut I, Krivitzky A, Lebrun‐Frenay C, Clet J, et al Risk of autoimmune diseases and human papilloma virus (HPV) vaccines: Six years of case‐referent surveillance. J Autoimmun. 2017; 79:84–90. [DOI] [PubMed] [Google Scholar]
- 48. Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al Genetics and beyond ‐ the transcriptome of human monocytes and disease susceptibility. PLoS One 2010; 5(5):e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garnier S, Truong V, Brocheton J, Zeller T, Rovital M, Wild PS, et al Genome‐wide haplotype analysis of cis expression quantitative trait loci in monocytes. PLoS Genet. 2013; 9(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Westra H‐J, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013; 45(10):1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, et al SCAN: SNP and copy number annotation. Bioinformatics 2010; 26(2):259–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Franzén O, Ermel R, Cohain A, Akers NK, Di Narzo A, Talukdar HA, et al Cardiometabolic risk loci share downstream cis‐ and trans‐gene regulation across tissues and diseases. Science 2016; 353(6301):827–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leslie R, O’Donnell CJ, Johnson AD. GRASP: analysis of genotype‐phenotype results from 1390 genome‐wide association studies and corresponding open access database. Bioinformatics 2014; 30(12):i185–i194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buniello A, Macarthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al The NHGRI‐EBI GWAS Catalog of published genome‐wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47(D1):D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez‐Jarreta J, et al Ensembl 2020. Nucleic Acids Res. 2019. Nov 6; Available from: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkz966/5613682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Machiela MJ, Chanock SJ. LDlink: a web‐based application for exploring population‐specific haplotype structure and linking correlated alleles of possible functional variants: Fig. 1. Bioinformatics. 2015;31:3555–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Genomes Project Consortium , Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al A global reference for human genetic variation. Nature 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tsubata T. B‐cell tolerance and autoimmunity. F1000Research. 2017;6:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Di Leo G, Sardanelli F. Statistical significance: p value, 0.05 threshold, and applications to radiomics—reasons for a conservative approach. Eur Radiol Exp. 2020; 4(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ioannidis JPA. The proposal to lower P value thresholds to .005. J Am Med Assoc. 2018; 319(14):1429–30. [DOI] [PubMed] [Google Scholar]
- 61. Benjamin DJ, Berger JO, Johannesson M, Nosek BA, Wagenmakers EJ, Berk R, et al Redefine statistical significance. Nat Hum Behav. 2018; 2(1):6–10. [DOI] [PubMed] [Google Scholar]
- 62. Sambo MR, Trovoada MJ, Benchimol C, Quinhentos V, Gonçalves L, Velosa R, et al Transforming growth factor beta 2 and heme oxygenase 1 genes are risk factors for the cerebral malaria syndrome in Angolan children. PLoS One 2010; 5(6):e11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Papoutsoglou P, Tsubakihara Y, Caja L, Morén A, Pallis P, Ameur A, et al The TGFB2‐AS1 lncRNA regulates TGF‐β signaling by modulating corepressor activity. Cell Rep. 2019; 28(12):3182–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Naka K, Hirao A. Regulation of hematopoiesis and hematological disease by TGF‐β family signaling molecules. Cold Spring Harb Perspect Biol. 2017; 9(9):a027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kehrl JH, Taylor A, Kim S‐J, Fauci AS. Transforming growth factor‐beta is a potent negative regulator of human lymphocytes. Ann N Y Acad Sci. 1991; 628:345–53. [DOI] [PubMed] [Google Scholar]
- 66. Zhao L, Pu X, Ye Y, Lu C, Chang JY, Wu X. Association between genetic variants in DNA double‐strand break repair pathways and risk of radiation therapy‐induced pneumonitis and esophagitis in non‐small cell lung cancer. Cancers (Basel). 2016; 8(2):17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oettinger M, Schatz D, Gorka C, Baltimore D. RAG‐1 and RAG‐2, adjacent genes that synergistically activate V(D)J recombination. Science 1990; 248(4962):1517–23. [DOI] [PubMed] [Google Scholar]
- 68. Laszkiewicz A, Sniezewski L, Kasztura M, Bzdzion L, Cebrat M, Kisielow P. Bidirectional activity of the NWC promoter is responsible for RAG‐2 transcription in non‐lymphoid cells. PLoS One 2012; 7(9):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Łaszkiewicz A, Bzdzion Ł, Kasztura M, Śnieżewski Ł, Janik S, Kisielow P, et al Ikaros and RAG‐2‐mediated antisense transcription are responsible for lymphocyte‐specific inactivation of NWC promoter. PLoS One 2014; 9(9):e106927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Akamatsu Y, Oettinger MA. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol [Internet]. 1998; 18(8):4670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vettermann C, Schlissel MS. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev. 2010; 237(1):22–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kumar V, Alt FW, Frock RL. PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1‐arrested progenitor B‐cell line. Proc Natl Acad Sci USA. 2016; 113(38):10619–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morris AC, Beresford GW, Mooney MR, Boss JM. Kinetics of a gamma interferon response: expression and assembly of CIITA promoter IV and inhibition by methylation. Mol Cell Biol. 2002; 22(13):4781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Roche PA, Furuta K. The ins and outs of MHC class II‐mediated antigen processing and presentation. Nat Rev Immunol. 2015; 15(4):203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hostager BS, Bishop GA. Role of TNF receptor‐associated factor 2 in the activation of IgM secretion by CD40 and CD120b. J Immunol. 2002; 168(7):3318–22. [DOI] [PubMed] [Google Scholar]
- 76. Cerutti A, Schaffer A, Shah S, Zan H, Liou HC, Goodwin RG, et al CD30 is a CD40‐inducible molecule that negatively regulates CD40‐ mediated immunoglobulin class switching in non‐antigen‐selected human B cells. Immunity 1998; 9(2):247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jabara HH, Laouini D, Tsitsikov E, Mizoguchi E, Bhan AK, Castigli E, et al The binding site for TRAF2 and TRAF3 but not for TRAF6 is essential for CD40‐mediated immunoglobulin class switching. Immunity 2002; 17(3):265–76. [DOI] [PubMed] [Google Scholar]
- 78. Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, et al GeneCards Version 3: the human gene integrator. Database. 2010; 2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Linehan LA, Warren WD, Thompson PA, Grusby MJ, Berton MT. STAT6 is required for IL‐4‐induced germline Ig gene transcription and switch recombination. J Immunol. 1998; 161(1):302–10. [PubMed] [Google Scholar]
- 80. Turqueti‐Neves A, Otte M, da Costa OP, Höpken UE, Lipp M, Buch T, et al B‐cell‐intrinsic STAT6 signaling controls germinal center formation. Eur J Immunol. 2014; 44(7):2130–8. [DOI] [PubMed] [Google Scholar]
- 81. Schroder AJ, Pavlidis P, Arimura A, Capece D, Rothman PB. Cutting edge: STAT6 serves as a positive and negative regulator of gene expression in IL‐4‐stimulated B lymphocytes. J Immunol. 2002; 168:996–1000. [DOI] [PubMed] [Google Scholar]
- 82. Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity 2013; 46(2):83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Staszewski O, Baker RE, Ucher AJ, Martier R, Stavnezer J, Guikema JEJ. Activation‐induced cytidine deaminase induces reproducible DNA breaks at many non‐Ig loci in activated B cells. Mol Cell. 2011; 41(2):232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, et al Two levels of protection for the B cell genome during somatic hypermutation. Nature 2008; 451(7180):841–5. [DOI] [PubMed] [Google Scholar]
- 85. Dunnick W, Hertz GZ, Scappino L, Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993; 21(3):365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Stavnezer J, Schrader CE. IgH chain class switch recombination: mechanism and regulation. J Immunol. 2014; 193(11):5370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Monlong J, Calvo M, Ferreira PG, Guigó R. Identification of genetic variants associated with alternative splicing using sQTLseekeR. Nat Commun. 2014; 5:4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of candidate genes considered in the association analysis
Table S2. Single nucleotide polymorphism selected as ancestry informative.
Table S3. Single nucleotide polymorphism of the candidate genes selected analysed in the association study.
Table S4. Predicted regulatory effects according to Haploreg database of the associated variants and of those in strong linkage disequilibrium with them (r² ≥ 0.8).
Table S5. Variants associated with increased risk to endemic pemphigus foliaceus, their proxy SNPs and eQTL and sQTL effect of these variants.
Table S6. Transcript variants of genes with associated SNP with sQTL effect.
Data Availability Statement
All data are available in the manuscript and supplementary files.


