Summary
Animal embryos are patterned by a handful of highly conserved inductive signals. Yet in most cases it is unknown which pattern features (i.e., spatial gradients or temporal dynamics) are required to support normal development. An ideal experiment to address this question would be to “paint” arbitrary synthetic signaling patterns on “blank canvas” embryos to dissect their requirements. Here we demonstrate exactly this capability by combining optogenetic control of Ras/Erk signaling with the genetic loss of the receptor tyrosine kinase-driven terminal signaling patterning in early Drosophila embryos. Blue light illumination at the embryonic termini for 90 min was sufficient to rescue normal development, generating viable larvae and fertile adults from an otherwise-lethal terminal signaling mutant. Optogenetic rescue was possible even using a simple, all-or-none light input that reduced the gradient of Erk activity and eliminated spatiotemporal differences in terminal gap gene expression. Systematically varying illumination parameters further revealed that at least three distinct developmental programs are triggered at different signaling thresholds, and that the morphogenetic movements of gastrulation are robust to a three-fold variation in the posterior pattern width. These results open the door to controlling tissue organization with simple optical stimuli, providing new tools to probe natural developmental processes, create synthetic tissues with defined organization, or directly correct the patterning errors that underlie developmental defects.
Introduction
During animal development, the embryo is patterned by gradients of protein activity that define cells’ positions along the body axes and within developing tissues [1]. In recent years, many developmental patterns have been characterized in precise quantitative detail in individual embryos [2-4]. Yet in nearly every case it remains unknown which features of a signaling patterns carry essential information: the instantaneous protein concentration, the area-under-the-curve, or the total duration of signaling above a threshold. The quantity of information contained in a single pattern also remains mysterious: how many distinct levels are read out by the genetic networks that serve as signal interpretation systems, and how long does it take to transfer this information?
To address these questions, we envisioned an idealized experiment to better define the information contained in a developmental pattern (Figure 1A) [5]. First, one might prepare mutant embryos in which a specific signaling pattern is completely eliminated. On this background one might then apply a synthetic signaling pattern, varying features such as its shape, intensity, or duration and monitoring the capability of each to rescue the developmental process. Although such an experiment has historically been intractable, we reasoned optogenetic control over cell signaling opens the door to exactly this capability. An appropriately-tailored light input could be used to produce any spatiotemporal signaling pattern, enabling a biologist to test for the minimal features required to support proper development, or allowing a bioengineer to apply non-natural stimuli to implement novel tissue architectures or morphogenetic programs [5-7].
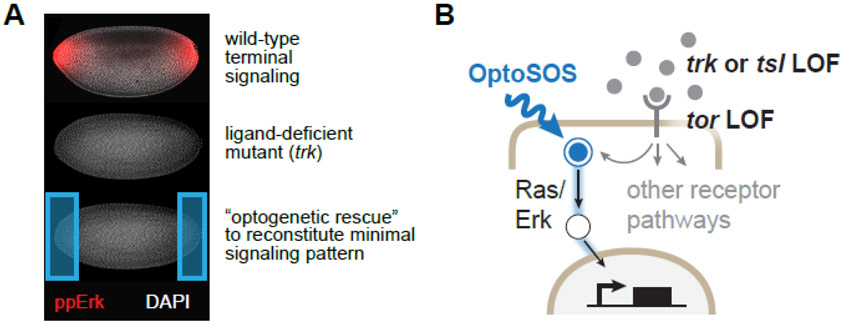
Figure 1. Painting developmental signaling patterns on a blank canvas.
(A) Upper: immunofluorescence (IF) for doubly phosphorylated Erk (ppErk; red) in a nuclear cycle 14 (NC14) embryo, exhibiting the characteristic terminal gradient. Middle: IF for ppErk in a trk1 mutant NC14 embryo, showing complete loss of terminal ppErk at the termini. Lower: Schematic of the proposed experiment, where light is applied on the trk mutant background to potentially restore Erk activity and function. All embryos in the figure are oriented with anterior to the left and ventral downward. (B) Because the light-activated OptoSOS system directly activates Ras/Erk pathway downstream of receptor tyrosine kinases, it can be functionally combined with the genetic loss of Tor, Tsl or Trk, the three receptor-level components normally active at the embryonic termini. See also Figure S1.
We thus set out to perform an optogenetic rescue of terminal signaling, the first pattern of receptor tyrosine kinase (RTK) activity during Drosophila embryogenesis [8]. Terminal signaling is orchestrated by localized activation of the RTK Torso (Tor) by its ligand Trunk (Trk) at the embryonic anterior and posterior poles (Figure 1B). Quantitative studies of terminal signaling in individual embryos have revealed a reproducible terminal-to-interior gradient that is dynamically established over a 2-hour window in early embryogenesis [9]. This gradient is essential: embryos from mothers lacking Tor, Trk, or the required co-factor Torso-like (Tsl) completely lack a terminal signaling gradient and are defective in a wide variety of anterior- and posterior-localized processes, including the formation of mouth parts and tail structures, the differentiation of many endoderm-derived tissues, and the ability to coordinate tissue movements during gastrulation [10, 11]. Yet the nature and quantity of information contained in the terminal pattern is still unclear. The naturally-observed gradient of Tor activity activates the two classic terminal gap genes Tll and Hkb in distinct but overlapping domains, supporting the notion that spatiotemporal variations in pathway activity play an important role [12-14]. On the other hand, seminal prior work demonstrated that many features of the terminal loss-of-function phenotype could be rescued by supplying rather crude sources of activity, for example by injection of tor RNA or constitutively-active Ras protein at the poles [15, 16]. The precise requirements for a rescuing terminal pattern thus remain to be defined.
Here, we report rescue of the full Drosophila life cycle from OptoSOS-trk embryos that completely lack receptor-level terminal signaling but whose Ras/Erk signaling can be controlled with light. Illuminated OptoSOS-trk embryos develop normal head and tail structures, gastrulate normally, hatch, metamorphose, mate and lay eggs. Full phenotypic rescue is possible despite the use of simple all-or-none light inputs that limit the graded information contained in the terminal pattern, for example eliminating expression differences in reporters of the terminal gap genes tll and hkb. We define the lower essential limits of terminal signaling, demonstrating that at least three distinct developmental switches are triggered at successively increasing illumination thresholds. Our study thus demonstrates that Ras activation by SOS is sufficient to recapitulate all the essential features of receptor tyrosine kinase signaling at the embryonic termini. It also suggests the spatial gradients of Erk activity normally observed at the termini are not required, at least in the presence of the embryo’s additional sources of anterior-posterior positional information. These data provide a first step towards defining the essential information contained in developmental signaling patterns and open the door to optically programming cell fates and tissue movements with high precision in developing tissues.
Results
Light-controlled terminal signaling rescues normal development
We first set out to establish a genetic background where light could be used as the sole source of Erk activity at the embryonic termini, so that its ability to rescue subsequent development could be assessed. Two attributes make terminal signaling an ideal system for optogenetic rescue. First, all three components of the Trk-Tor-Tsl receptor/ligand system are maternal-effect genes [10], so flies that are homozygous-null for any of the three genes develop normally, provided that the gene products are maternally deposited in the egg to produce the terminal pattern. Thus, in principle, one may be able to rescue the organism’s full life cycle by replacing this single developmental pattern with light. Second, we previously developed the OptoSOS optogenetic system for control over Ras/Erk signaling, a key downstream effector pathway of terminal signaling, in contexts ranging from cultured mammalian cells [17, 18] to the Drosophila embryo [19, 20]. In this system, a switch from darkness to light induces SOS membrane localization within seconds, followed by Erk activation and expression of Erk-dependent target genes (e.g. tll in the case of the early Drosophila embryo; see Ref. 19), whereas a switch to darkness triggers a rapid reversal of this process, returning Erk activity and gene expression to their baselines also on a timescale of minutes [17, 21, 22]. OptoSOS is ideal for attempting light-based rescue because it activates Ras downstream of receptor-level stimulation (Figure 1B), and can thus be combined with mutations targeting receptor-level signaling to place terminal Ras/Erk signaling solely under optogenetic control [23]. Indeed, in preliminary experiments comparing embryos harboring loss-of-function perturbations targeting receptor/ligand signaling (trk and tsl loss-of-function mutants and a Tor RNAi line; Figure S1; Figure 1A; Supplementary Methods), we found that OptoSOS-expressing embryos produced from trk1 mothers lack all endogenous terminal signaling activity [24], but when placed under uniform blue light these embryos exhibit phenotypes associated with strong gain-of-function terminal signaling [19]. We thus focused on these “OptoSOS-trk” embryos for subsequent experiments.
We next set out to determine whether applying light to OptoSOS-trk embryos would be sufficient to restore various embryonic structures that are dependent on terminal signaling, and if so, which features of the stimulus might prove to be essential. We began with a simple light stimulus: binary, all-or-none illumination of the anterior or posterior pole. We matched the light stimulus duration (90 min), spatial range (roughly 15% of the embryo’s length) and intensity level (one pulse every 30 sec) to roughly match the parameters observed for doubly-phosphorylated Erk during endogenous terminal signaling, which we quantified here (Figure S2) and in a prior study [4]. Importantly, our optogenetic stimulus eliminates both the complex temporal dynamics and spatial gradient of the endogenous terminal pattern. Yet even this simple all-or-none light stimulus, delivered to the anterior pole, was sufficient to restore head structures that were indistinguishable from those in wild-type embryos d>(Figure 2A; see Table S1 for number of embryos with rescued phenotypes). Similar results were obtained upon posterior illumination, which was sufficient to restore the formation of tail structures such as posterior spiracles as well as the 8th abdominal segment (Figure 2B).
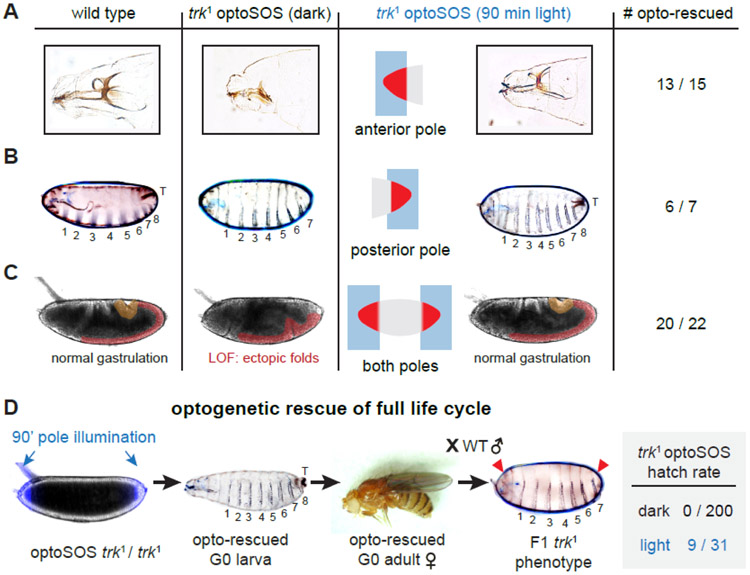
Figure 2. Light-controlled terminal signaling rescues normal development.
(A-B) Cuticle preparations from embryos that were illuminated for 90 min at the anterior-most 15% of the embryo (in A) or posterior-most 15% of the embryo (in B) with 0.6 sec pulses of saturating blue light delivered every 30 sec. Head structures, the 8th abdominal segment, and tail structures (marked “T”) are formed normally in wild-type embryos (left images) are truncated or absent in embryos lacking terminal signaling (middle images), but are rescued after 90 min of illumination at the appropriate pole (right images). (C) Still images from DIC time-lapse videos of gastrulation in wild-type embryos, OptoSOS-trk embryos without illumination and OptoSOS-trk embryos illuminated at both poles. Highlighted regions mark posterior midgut invagination (yellow) and germ band elongation (red). (D) Complete rescue of OptoSOS-trk animal development by 90 min illumination at both the anterior-most and posterior-most 15% of the embryo with 0.6 sec pulses of saturating blue light delivered every 30 sec. Embryos hatch, eclose, and mate. The embryos produced by female light-rescued flies exhibit the trk mutant phenotype (red arrows). All embryos in the figure are oriented with anterior to the left and ventral downward. See also Figure S2, Table S1 and Video S1.
To assess the rescue of other terminal signaling-dependent processes that are difficult to individually monitor, we applied similar all-or-none light patterns at both embryonic termini and visualized the remainder of their development by differential interference contrast (DIC) microscopy (Figure 2C). Approximately 30% of the embryos illuminated in this manner were able to gastrulate normally, complete the remainder of embryogenesis, and hatch from the imaging device (Video S1). We collected larvae that hatched on the microscope and maintained them in standard tubes, where they proceeded normally through each instar, pupated and produced normal adult flies (Figure 2D). Finally, we reasoned that optogenetically-rescued female adult flies produced in this manner should still be trk-null, so the embryos produced by these females should still harbor phenotypes consistent with the maternal loss of terminal signaling. Indeed, all embryos laid from light-rescued mothers failed to hatch, and cuticle preparations revealed the trk phenotype in all progeny (head defects; absence of the 8th abdominal segment and tail structures) (Figure 2D). Taken together, these data confirm the optogenetic rescue of terminal signaling in Drosophila embryogenesis. Simple synthetic signaling patterns, generated by local blue light illumination, were thus sufficient to overcome lethal defects in body segmentation, tissue morphogenesis, and cell differentiation to restore the entirety of the fly’s life cycle.
Optogenetic stimulation eliminates differences in terminal gap gene expression domains
Our optogenetic stimulation experiments relied on all-or-none light inputs, stimuli which we previously found to result in precise, subcellular spatial control over SOS membrane recruitment in the early Drosophila embryo [20]. However, many processes may still act to blur these precise inputs into a spatially-graded response (e.g., light scattering, diffusion of active components of the Ras/MAPK pathway within the syncytial embryo, or other gradients of gene expression along the anterior-posterior axis that might modulate the activity of the terminal signaling pathway). We thus set out to quantify the spatial distribution of Erk activity and downstream gene expression in response to the same all-or-none light stimulus used in our optogenetic rescue experiments. To circumvent the challenge of fixing and staining individual locally-illuminated embryos, we relied on live-cell fluorescent biosensors to measure Erk activity and gene expression with high spatiotemporal resolution.
To measure Erk activity, we turned to a recently-developed biosensor, miniCic, that translocates from the nucleus to cytosol upon phosphorylation by Erk in Drosophila (Figure S3A) [25]. We generated embryos that co-expressed miniCic-mCherry and the OptoSOS system (STAR Methods) and verified that this system could indeed be used in the early embryo by visualizing the endogenous terminal signaling gradient (Figure S3B). We then locally illuminated embryos and quantified nuclear miniCic as a function of position from the edge of our illumination pattern (Figure S3C-F). As a control, we quantified nuclear miniCic from the embryo’s poles along the endogenous terminal gradient. We fitted Hill curves to each embryo’s nuclear miniCic intensity as a function of position to measure the distance over which Erk was active as well as the steepness of its on-to-off switch (Figure S3G-H. We found that light could be used to trigger patterns on a shorter length-scale than the endogenous gradient: miniCic localization returned to baseline within 60 μm from the edge of the illuminated region, versus extending 120 μm from the termini in the endogenous pattern (Figure S3G). Light also resulted in a steeper on-to-off switch, measured by the distance over which miniCic localization switched from 10% to 90% of its baseline nuclear intensity (Figure S3H). Our approach likely over-estimates the sharpness of the endogenous pattern, as kinase biosensors are typically quite sensitive and can become saturated at sub-maximal levels of pathway activity [26], leading a shallow, high-amplitude gradient of Erk activity [4] to be clipped at the biosensor’s maximum value and thus appear to switch over a shorter range than the true activity gradient.
We next set out to characterize the spatial patterns of two Erk-dependent target genes, tll and hkb, that act to specify terminal cell fates and which are normally expressed in distinct domains. Prior studies revealed that tll is normally expressed over a broader range than hkb [20, 27, 28], a finding that is consistent with activation of tll by lower levels of active Erk [13, 19]. We generated embryos that expressed a fluorescent MCP protein and where either the tll and hkb upstream regulatory sequences drove expression of MS2-tagged mRNAs, in genetic backgrounds with normal terminal patterning or a variant of our optogenetic rescue system (OptoSOS-tsl) (Methods; Figure 3A; Video S2) [21]. Imaging the endogenous terminal pattern revealed distinct domains of tll and hkb transcriptional foci as expected, with tll expressed earlier (NC11 to early NC14) and over a broader domain, and hkb expressed primarily during NC13-14 and localized more tightly at the poles (Figure 3A; right panels; see Figure S3I for quantification over time). These distributions of RNA production were in good agreement with previously-measured distributions of total tll and hkb RNA [20].
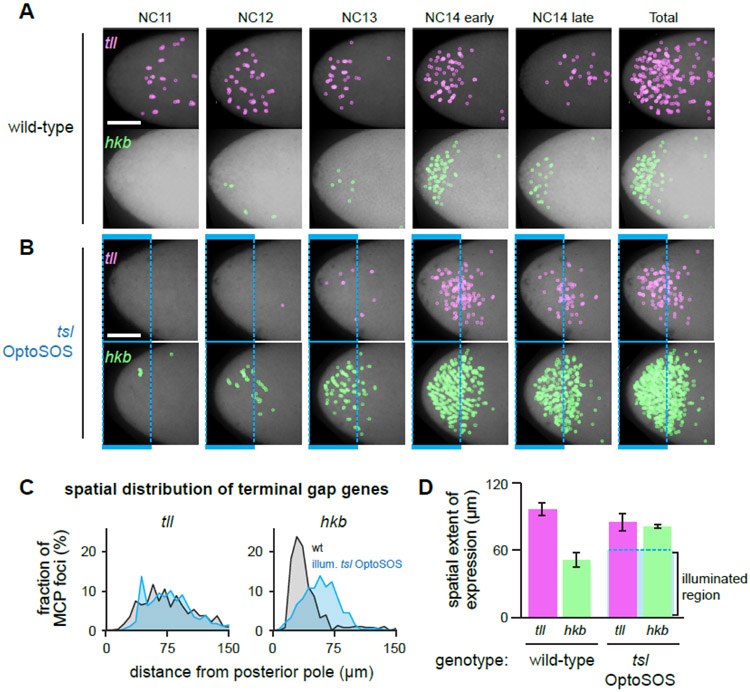
Figure 3. Light stimulation eliminates spatiotemporal differences in terminal gap gene expression.
(A-B) Images are shown of OptoSOS embryos (in A) and OptoSOS-tsl embryos (in B) expressing MCP-mCherry and harboring MS2 stem-loops driven by the tll or hkb upstream regulatory sequences (magenta and green, respectively). Embryos are oriented with posterior pole to the left. Images are maximum intensity projections across all z-frames and time points during the indicated nuclear cycles, with transcriptional foci marked with colored circles. In B, 0.6 sec pulses of saturating blue light were delivered every 30 sec to the shaded region. Scale bar: 50 μm. (C) Histogram showing the spatial distribution of transcriptional foci for tll (left panel) and hkb (right panel) for the endogenous gradient (embryos as in A; gray) and light stimulation (embryos as in B; blue). Each curve represents data pooled from at least three embryos. (D) The spatial extent of gene expression for tll and hkb was measured for the endogenous pattern (left bars) and light stimulation (right bars) for the same embryos quantified in C. Dotted blue box shows extent of illumination. Mean + S.E.M. is shown for at least three embryos. See also Figure S3, Table S2 and Video S2.
In contrast, stimulating OptoSOS-tsl embryos under the same all-or-none illumination conditions previously used for optogenetic rescue (0.6 sec pulses every 30 sec to the anterior-most and posterior-most 15% of individual embryos) produced a different result (Figure 3B). In this case, the expression domains for tll and hkb more closely matched one another in induction timing and spatial range. Both reporters exhibited transcriptional bursts in response to light that appeared between NC10-13, increasing in NC14 until gastrulation (Figure 3B; Figure S3J. The spatial distribution of gene expression was also similar across both reporters and resembled the broad distribution of the endogenous tll pattern (Figure 3C). We quantified the boundary of gene expression from the posterior pole in multiple light-stimulated embryos, which confirmed our observations and also revealed that terminal gene expression extended some tens of micrometers beyond the edge of the illumination pattern, just as had been observed for miniCic nuclear export (Figure 3D). No terminal gap gene expression was observed in control, dark-incubated OptoSOS-tsl embryos (Figure S3K-L).
Taken together, our data indicate that our all-or-none “rescue stimulus” also substantially reduces the amount of graded information contained within the terminal pattern. Most crucially, it eliminates major differences in the spatial domains and timing for reporters of tll and hkb, two target genes are thought to mediate the majority, if not the entirety, of terminal signaling. While some caution must be used in interpreting transcriptional reporters of regulatory regions, these reporters match the endogenous domains of tll and hkb expression and are activated only in response to OptoSOS stimulation, suggesting that at least Erk-dependent responses are intact and accurate. Importantly, quantification of Erk activity and transcriptional responses revealed that even our sharp, localized light stimulus is blurred tens of micrometers in the context of the embryo, suggesting that graded information is reduced but not perfectly eliminated by our optogenetic stimulus. Because the patterns of Erk activity and gene expression extend substantially further from the edge of the illumination pattern than the sharp boundaries of SOScat membrane recruitment [20], they likely do not represent light scattering, but rather reflect downstream intracellular processes such as signal propagation through the cytosolic MAP kinase cascade [29] or cytosolic flow during syncytial nuclear division cycles [30].
At least three levels of terminal signaling trigger distinct developmental programs
The rescue of all anterior and posterior tissue responses by a single all-or-none light pattern is consistent with two different models of terminal cell fate choice. First, Erk activity may be sensed by a single downstream program that triggers all terminal processes as pathway activation crosses a single threshold [16]. Alternatively, individual terminal processes may be rescued one by one as the signaling input crosses distinct fate-specific thresholds [13]. To distinguish the number of cell-fate switches and identify their thresholds, we set out to map terminal phenotypes in response to variations in the strength optogenetic stimulus (Figure 4; see Table S1 for number of embryos with rescued phenotypes). Optogenetic control is also ideally poised to further distinguish what feature of an input signal is sensed – its level, duration above a threshold, or the total dose (i.e., intensity * time) – and we indicate which is varied in each experiment that follows.
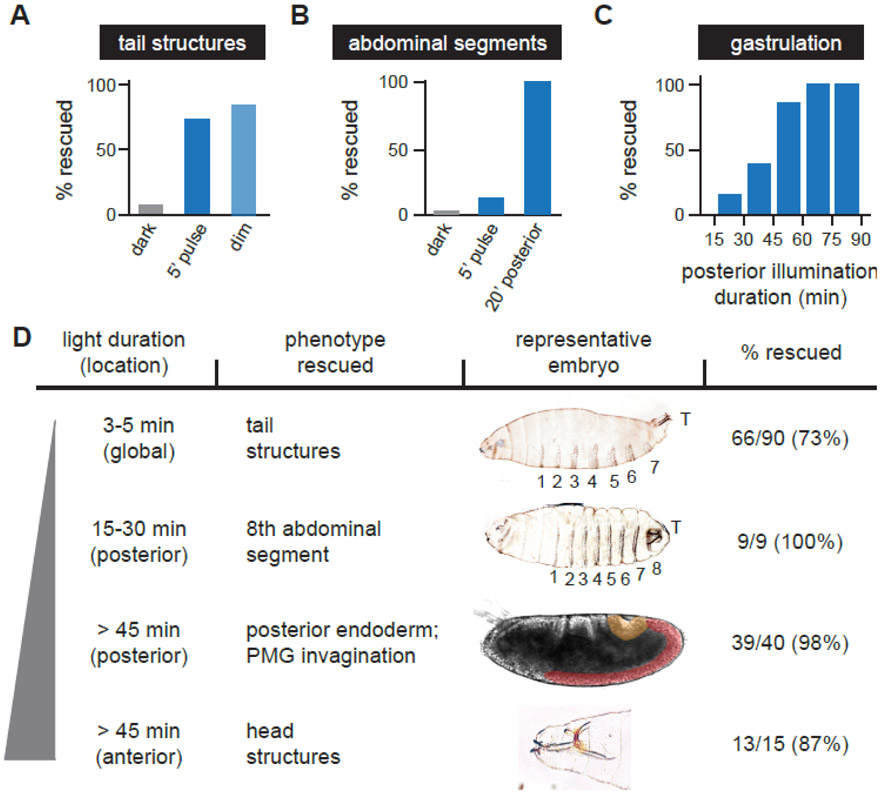
Figure 4: Three durations of terminal signaling trigger distinct developmental programs.
(A) The fraction of normal tail structures was quantified from embryos incubated in the dark, stimulated globally with a single 5-minute bolus of saturating blue light (“5’ pulse”), or 1 sec pulses of saturating blue light every 120 sec (“dim”; see Figure S1B for quantification of Erk activity at similar light doses). (B) The fraction of embryos with 8 abdominal segments was quantified from embryos incubated in the dark, subjected to a global 5-minute bolus of saturating blue light (“5’ pulse”) or illuminated for 20 min at the posterior-most 15% of the embryo with 0.6 sec pulses of saturating blue light every 30 sec. (C) Posterior tissue movements during gastrulation were scored by differential interference contrast (DIC) imaging of embryos illuminated at the posterior pole. (D) Developmental sequence of terminal phenotypes rescued in response to 0.6 sec pulses of saturating blue light delivered every 30 sec to the embryo’s anterior-most 15% (“anterior”) or posterior-most 15% (“posterior”), or in response to 1 sec pulses every 30 sec delivered to the entirety of the embryo (“global”). The stimulus duration, spatial position, developmental phenotype and a representative image are shown (OptoSOS-trk gastrulation and head structure images reproduced from Figure 2A and 2C). Embryos are oriented with anterior to the left and ventral downward. See also Figure S4 and Table S1.
We started with a brief light input – a single 5 min bolus of global, continuous illumination – reasoning that it would be much shorter than the 20-90 min periods of Erk activation that are typically triggered by RTK activation [31-34] and thus likely below the lower limit of detection by downstream phenotypic programs. Indeed, the 5 min pulse did not disrupt the development of a majority of OptoSOS embryos with wild-type terminal signaling, indicating that it was below the threshold for triggering substantial gain-of-function developmental defects (Figure S4A). However, we found that even this brief, uniform pulse of light was sufficient to restore tail structures in a majority of OptoSOS-trk embryos without altering other developmental programs (Figure 4A; Table S1). Tail structures were rescued even more efficiently by limiting the 5 min pulse to a narrower stimulation window of 90-150 min post fertilization (Figure S4B-C), presumably corresponding to a period in which terminal gap gene expression can be triggered most efficiently (Figure 3A-B).
We next tested whether tail formation could also be driven by weaker inputs delivered over a longer time period, and subjected embryos to 1 sec pulses delivered every 120 sec, a light intensity that results in less than 10% of the maximal Erk activity presented by the endogenous terminal gradient (Figure S2). Indeed, we found that equivalent rescue was obtained in response to either constant, low-intensity illumination or a brief, high-intensity pulse (Figure 4A). Together, these experiments reveal a set of remarkable requirements for a developmental cell fate choice: tail structure formation absolutely requires Ras/Erk signaling but is triggered at an extremely low total stimulus dose. Moreover, tail structures are rescued at the appropriate posterior position even by global illumination, a stimulus that does not contain any spatial information.
As we progressively increasing the duration of illumination at the anterior or posterior pole, using 0.6 sec pulses of saturating blue light every 30 sec, we observed that additional developmental processes were rescued in a well-defined sequence. The 8th abdominal segment was restored as the posterior light stimulus was increased to 20 min (Figure 4B), whereas normal gastrulation movements were only restored above 45 min of posterior illumination (Figure 4C). A similar 45-min pulse was also required at the anterior pole for the formation of head structures. We thus conclude that Ras/Erk activity is interpreted into at least three all-or-none developmental programs with duration thresholds spanning nearly an order of magnitude (5 min – 45 min), at a stimulus intensity that drives comparable Erk phosphorylation to the endogenous maximum terminal level (Figure 4D; Table S1). Our data are strongly diagnostic of a multiple-threshold model of terminal signal interpretation: we find that increasing the total duration of light stimulation triggers distinct developmental processes in a well-defined order. Furthermore, in at least two cases it appears that there is a correspondence between varying light intensity and duration, such that the phenotypic response would depend on the total light dose: tail formation (Figure 4A) and posterior midgut differentiation [13]. Importantly, the multiple-threshold model does not conflict with our prior observation of optogenetic rescue by a single, 45 min light stimulus. That is because mutant phenotypes appear to be restored in a cumulative fashion, so a given light stimulus rescues all developmental processes that are triggered at thresholds at or below this level.
Gastrulation movements are robust to variation in the spatial range of terminal patterning
The preceding experiments define the temporal requirements for terminal signaling, but what rules govern its permissible spatial parameters? We can again envision two extreme models. First, it is possible that only a highly restricted range of spatial pattern widths can support normal development, by balancing the proportion of cells committed to terminal and non-terminal fates. At the other extreme, many different spatial patterns could funnel into a proper developmental outcome [35], resembling the tolerance to variation in the Bicoid morphogen gradient as gene dosage is varied [36] or the Shh gradient in the neural tube of Gli3−/− mice [37].
To test these possibilities, we varied the spatial domain of terminal signaling at our standard illumination intensity (0.6 sec light pulses delivered every 30 sec) and monitored a model developmental response: tissue morphogenesis during gastrulation. Terminal signaling at the posterior pole drives formation of posterior midgut (PMG), which invaginates and moves across the embryo’s dorsal surface during germ band elongation (GBE). GBE is thought driven both by a combination of ‘pushing’ by cell intercalation at the ventral tissue (Figure 5A; red) and ‘pulling’ by invagination of the posterior endoderm itself (Figure 5A; yellow) [38, 39]. Embryos derived from trk-mutant mothers completely fail both PMG invagination and GBE, leading to buckling of the elongating tissue along the embryo’s ventral surface [10] (Figure S5A). Consistent with this requirement, we found that PMG invagination and GBE were absent in dark-incubated embryos as well as over 90% of embryos that were illuminated only at the anterior pole (Figure S5B).
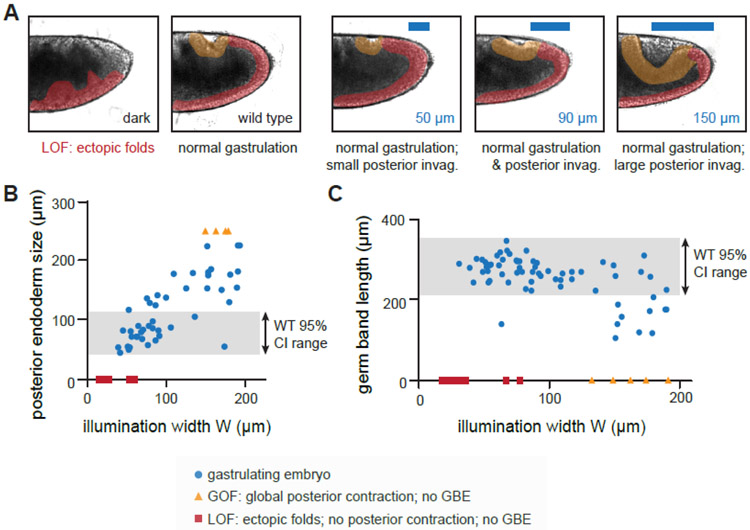
Figure 5: Tissue morphogenesis is robust to variations in terminal pattern width.
(A) Images of gastrulating wild-type embryos and OptoSOS-trk embryos stimulated with different illumination widths at the posterior pole with 90 minutes of 0.6 sec pulses of saturating blue light delivered every 30 sec. Yellow highlighted regions indicate posterior endoderm invagination, which expands as illumination width is increased. Red highlighted regions indicate the elongating germ band tissue, which buckles in loss-of-function (LOF) embryos. (B-C) Quantification of posterior endoderm perimeter (B) and germ band elongation length (C) as a function of the illumination width from the posterior pole. For some embryos (yellow triangles), posterior contraction was so large as to completely disrupt germ band elongation, a classic gain-of-function (GOF) phenotype. For others (red squares), no posterior contraction occurred, leading to loss-of-function (LOF) failure to extend a germ band at all. For both B-C, the shaded region indicates the normal wild-type size (mean +/− 95% confidence interval), quantified from 27 wild-type embryos. See also Figure S5 and Video S3.
We proceeded to systematically vary the width of posterior pattern and measured both the perimeter of the PMG and the maximum extent of GBE, comparing each to wild-type embryos as controls. We found that the size of the posterior invagination scaled linearly in proportion to the illumination width (Figure 5B), with illumination regions up to 150 μm inducing the formation of posterior invaginations more than twice the maximum observed in wild-type embryos (Figure 5A, right). Yet despite the different proportion of terminal vs non-terminal tissue, the mechanical processes of gastrulation were broadly unaffected, with PMG invagination and germ band elongation proceeding normally (Video S3). Quantitative analysis of the DIC videos indicated that germ band elongation was indistinguishable from wild-type controls as the light pattern was varied over a three-fold range, from 8–24% of the egg’s length (Figure 5C; Figure S5C). This result may partially explain the ease with which we obtained an optogenetic rescue even with imprecise illumination patterns. Furthermore, the ability to trigger morphogenetic movements at any spatial positions of interest will likely make OptoSOS-trk embryos a rich resource for informing and challenging models of tissue morphogenesis, along with other recent optogenetic tools for guiding tissue morphogenesis in vivo [40, 41].
Discussion
Here, we demonstrate that a developmental signaling pattern can be erased and replaced with a synthetic, patterned stimulus. Our approach relies on the tools of cellular optogenetics: unlike pharmacological or genetic perturbations, light can be applied and removed quickly, focused with high spatial precision, or shaped into arbitrary spatial patterns. We found that a simple all-or-none blue light stimulus, delivered to the embryonic termini, is sufficient to convert a lethal loss-of-function phenotype to rescue the full Drosophila life cycle: embryogenesis, larval development, pupation, adulthood and fecundity.
Our optogenetic rescue result provides two immediate insights into the interpretation of developmental RTK signaling. First, we find that recruiting the catalytic domain of SOS to the plasma membrane recapitulates all the essential developmental functions of Tor receptor tyrosine kinase signaling at the embryonic termini. This complete molecular sufficiency is non-obvious: we previously showed in mammalian cells that OptoSOS recruitment bypasses many intracellular pathways that are normally activated by RTKs (e.g. PI3K, Src, JNK, GSK3β) [17], some of which have been suggested to play roles in early Drosophila embryogenesis [42]. Nevertheless, our results are consistent with prior RNAseq data showing broad overlap between OptoSOS-stimulated and RTK-driven gene expression [22] and the observation that activating Ras pathway mutations are genetic suppressors of Tor partial loss-of-function alleles [43].
Second, our data suggest that the normally observed gradient of terminal signaling, resulting in spatially distinct domains of target gene expression, is not absolutely required for proper development. In support of this statement, our all-or-none rescue stimulus elicits a sharp boundary of OptoSOS membrane translocation [20], generates a steeper on-to-off switch in Erk activity than the endogenous terminal gradient (Figure S3), and substantially reduces differences in the expression kinetics and spatial distribution for reporters of the terminal gap genes tll and hkb (Figure 3). The sufficiency of even coarse terminal patterns has been long suggested by classic experiments in which the Torso receptor or constitutively-active Ras allele was injected at the termini of tor embryos, partially rescuing terminal processes [15, 16]; our data extends these early studies by quantifying the resulting patterns of gene expression and demonstrating the coarse input’s sufficiency for complete phenotypic rescue. However, even though a simple all-or-none pattern is enough to rescue, our data does not indicate that terminal signaling functions as a single all-or-none switch. Instead, we find that distinct developmental events are triggered at vastly different durations of Erk signaling, from rescue of tail structures with as little as 5 min of stimulation, to head structures and gastrulation movements only above 45 min. It is more likely that terminal processes operate as a series of switches with variable sensitivity, with stronger stimuli rescuing all phenotypes at or below that threshold.
How can such a simple stimulus pattern be reconciled with proper development? In wild-type embryos, terminal signaling triggers expression of the terminal gap genes Tll and Hkb in distinct domains, with Tll appearing earlier and extending further from the poles than Hkb [13]. Our optogenetic stimulus eliminates these differences, widening the expression domain of a hkb reporter to approximately match that of its tll counterpart. There is no reason to expect that this optogenetic scenario would prevent Tll and Hkb from playing their independent roles at the termini (e.g., Tll triggers posterior midgut invagination; Hkb represses Snail to block ventral furrow extension; Tll and Hkb each repress abdominal gap genes and specify endoderm cell fates) [28, 44-46]. On the contrary, we found no decrease in embryo viability when light activation was added to the endogenous terminal pattern, demonstrating robustness to the Erk dose at the termini [20]; and here, we further show that gastrulation movements are robust to variations in the spatial range of illumination (Figure 5). However, we would predict one important feature of the endogenous pattern to be entirely absent in light-rescued embryos: a terminal domain with high levels of Tll but low levels of Hkb [47, 48]. How light-rescued embryos compensate for loss of this “Tll AND NOT Hkb” signal, perhaps using other sources of anterior-posterior positional information, is an interesting question for future study [49]. This open question also reflects a broader challenge: we still lack a clear picture of the genetic circuits that decode developmental Erk signaling [50]. We expect that the approach we have taken here – combining controlled optogenetic stimulation with live-cell transcriptional imaging – could be applied to additional genes in the terminal response program to clearly define their signaling requirements in space and time.
It is also important to note that light-based rescue is far from perfect, with approximately 30% of embryos hatching after illumination. This loss in viability is likely to arise from both experimental and biological sources: challenges in reproducibly aligning embryos to the light pattern, leading to some error in the angle and extent of illumination at the termini; the procedure of mounting embryos in our imaging device for the entirety of embryogenesis; a loss of fitness from our simple all-or-none stimuli compared to the endogenous pattern; and the loss of parallel signaling pathways downstream of the Torso RTK that are bypassed by light-activated Ras. We anticipate that further advances in combining precise optical stimulation with non-invasive imaging will help to quantitatively determine how much each of these differences explains the increased lethality of our optogenetic stimulus relative to wild-type embryos.
There is considerable current interest in defining the rules that govern morphogenesis and patterning, both in vivo during embryo development and in engineered organoid-based systems. The optogenetic approaches defined here represent a first step toward the delivery of light-based programs to specific cells of interest within multicellular tissues. We find that even coarse synthetic signaling patterns can support normal tissue development and morphogenetic movements, suggesting that the tools of optogenetics and synthetic biology will likely be useful for generating developmental patterns that retain most or all of their essential functions [6, 51]. These capabilities could open the door to unprecedented control over developmental processes in both natural and synthetic multicellular systems.
Supplementary Material
Video S1: Complete rescue of embryogenesis in OptoSOS-trk embryo. Related to Figure 2.
Time-lapse DIC imaging of a trk1 optoSOS embryo without (top) and with (bottom) a 90-minute light stimulus of 0.6 sec pulses of saturating 450 nm light delivered every 30 sec at the anterior and posterior poles; this experimentally-delivered light input is shown in the video in blue. Embryo is oriented with anterior to the left and posterior downward.
Video S2: tll / mist transcription in the endogenous terminal gradient or after illumination. Related to Figure 3.
Time-lapse fluorescence imaging of tll (purple) or hkb (green) MS2/MCP bursts in either wild-type or OptoSOS-tsl embryos. The region of illumination is indicated by the distance from the pole to the blue line, and for all light stimuli, 0.6 sec pulses of saturating 450 nm light were delivered every 30 sec. Transcriptional foci were automatically detected as indicated by the green or purple circles, and the boundary of transcription is plotted as a vertical green or purple line. The elapsed time is indicated on each frame in hours:minutes. Embryos are oriented with posterior to the left.
Video S3: Gastrulation in an embryo exposed to a 150 μm-wide posterior light stimulus. Related to Figure 5.
Time-lapse DIC imaging of an optoSOS-trk embryo exposed to a wide posterior illumination pattern (lower panel) compared to a wild-type control (upper panel). For all light stimuli, 0.6 sec pulses of saturating 450 nm light were delivered every 30 sec. An abnormally large posterior invagination is followed by germ band elongation to an extent indistinguishable from a wild-type embryo. Embryos are oriented with anterior to the left and posterior downward.
Acknowledgements
H. E.J. was supported by the NIH Ruth Kirschstein fellowship F32GM119297. This work was also supported by NIH grant DP2EB024247 and NSF CAREER Award 1750663 (to J.E.T.), and NIH grant 5R01HD085870 (SYS). We also thank Dr. Gary Laevsky and the Molecular Biology Microscopy Core, which is a Nikon Center of Excellence, for microscopy support. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Footnotes
Declaration of Interests
The authors have no competing interests to declare.
References
- 1.Gurdon JB, and Bourillot PY (2001). Morphogen gradient interpretation. Nature 413, 797–803. [DOI] [PubMed] [Google Scholar]
- 2.Gregor T, Tank DW, Wieschaus EF, and Bialek W (2007). Probing the limits to positional information. Cell 130, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zagorski M, Tabata Y, Brandenberg N, Lutolf MP, Tkacik G, Bollenbach T, Briscoe J, and Kicheva A (2017). Decoding of position in the developing neural tube from antiparallel morphogen gradients. Science 356, 1379–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coppey M, Boettiger AN, Berezhkovskii AM, and Shvartsman SY (2008). Nuclear trapping shapes the terminal gradient in the Drosophila embryo. Current biology : CB 18, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson HE, and Toettcher JE (2018). Illuminating developmental biology with cellular optogenetics. Current opinion in biotechnology 52, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toda S, Blauch LR, Tang SKY, Morsut L, and Lim WA (2018). Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 361, 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kriegman S, Blackiston D, Levin M, and Bongard J (2020). A scalable pipeline for designing reconfigurable organisms. Proceedings of the National Academy of Sciences of the United States of America 117, 1853–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyal Y, Schupbach T, and Shvartsman SY (2018). A quantitative model of developmental RTK signaling. Developmental biology 442, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smits CM, and Shvartsman SY (2020). The design and logic of terminal patterning in Drosophila. Current topics in developmental biology 137, 193–217. [DOI] [PubMed] [Google Scholar]
- 10.Schupbach T, and Wieschaus E (1986). Maternal-effect mutations altering the anterior-posterior pattern of the Drosophila embryo. roux Arch Dev Biol 195, 302–317. [DOI] [PubMed] [Google Scholar]
- 11.Klingler M, Erdelyi M, Szabad J, and Nusslein-Volhard C (1988). Function of torso in determining the terminal anlagen of the Drosophila embryo. Nature 335, 275–277. [DOI] [PubMed] [Google Scholar]
- 12.Casanova J, and Struhl G (1989). Localized surface activity of torso, a receptor tyrosine kinase, specifies terminal body pattern in Drosophila. Genes & development 3, 2025–2038. [DOI] [PubMed] [Google Scholar]
- 13.Greenwood S, and Struhl G (1997). Different levels of Ras activity can specify distinct transcriptional and morphological consequences in early Drosophila embryos. Development 124, 4879–4886. [DOI] [PubMed] [Google Scholar]
- 14.Ghiglione C, Perrimon N, and Perkins LA (1999). Quantitative variations in the level of MAPK activity control patterning of the embryonic termini in Drosophila. Developmental biology 205, 181–193. [DOI] [PubMed] [Google Scholar]
- 15.Lu X, Chou TB, Williams NG, Roberts T, and Perrimon N (1993). Control of cell fate determination by p21ras/Ras1, an essential component of torso signaling in Drosophila. Genes & development 7, 621–632. [DOI] [PubMed] [Google Scholar]
- 16.Sprenger F, and Nusslein-Volhard C (1992). Torso receptor activity is regulated by a diffusible ligand produced at the extracellular terminal regions of the Drosophila egg. Cell 71, 987–1001. [DOI] [PubMed] [Google Scholar]
- 17.Toettcher JE, Weiner OD, and Lim WA (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bugaj LJ, Sabnis AJ, Mitchell A, Garbarino JE, Toettcher JE, Bivona TG, and Lim WA (2018). Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson HE, and Toettcher JE (2019). Signaling Dynamics Control Cell Fate in the Early Drosophila Embryo. Developmental cell 48, 361–370 e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson HE, Goyal Y, Pannucci NL, Schupbach T, Shvartsman SY, and Toettcher JE (2017). The Spatiotemporal Limits of Developmental Erk Signaling. Developmental cell 40, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keenan SE, Blythe SA, Marmion RA, Djabrayan NJ, Wieschaus EF, and Shvartsman SY (2020). Rapid Dynamics of Signal-Dependent Transcriptional Repression by Capicua. Developmental cell 52, 794–801 e794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson MZ, Ravindran PT, Lim WA, and Toettcher JE (2017). Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Molecular cell 67, 757–769 e755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goglia AG, Wilson MZ, Jena SG, Silbert J, Basta LP, Devenport D, and Toettcher JE (2020). A Live-Cell Screen for Altered Erk Dynamics Reveals Principles of Proliferative Control. Cell Syst 10, 240–253 e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm O, Sanchez Zini V, Kim Y, Casanova J, Shvartsman SY, and Wieschaus E (2012). Torso RTK controls Capicua degradation by changing its subcellular localization. Development 139, 3962–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno E, Valon L, Levillayer F, and Levayer R (2019). Competition for Space Induces Cell Elimination through Compaction-Driven ERK Downregulation. Current biology : CB 29, 23–34 e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillies TE, Pargett M, Minguet M, Davies AE, and Albeck JG (2017). Linear Integration of ERK Activity Predominates over Persistence Detection in Fra-1 Regulation. Cell Syst 5, 549–563 e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pignoni F, Baldarelli RM, Steingrimsson E, Diaz RJ, Patapoutian A, Merriam JR, and Lengyel JA (1990). The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell 62, 151–163. [DOI] [PubMed] [Google Scholar]
- 28.Bronner G, and Jackle H (1991). Control and function of terminal gap gene activity in the posterior pole region of the Drosophila embryo. Mech Dev 35, 205–211. [DOI] [PubMed] [Google Scholar]
- 29.Santos SD, Wollman R, Meyer T, and Ferrell JE Jr. (2012). Spatial positive feedback at the onset of mitosis. Cell 149, 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deneke VE, Puliafito A, Krueger D, Narla AV, De Simone A, Primo L, Vergassola M, De Renzis S, and Di Talia S (2019). Self-Organized Nuclear Positioning Synchronizes the Cell Cycle in Drosophila Embryos. Cell 777, 925–941 e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall CJ (1995). Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185. [DOI] [PubMed] [Google Scholar]
- 32.Lim B, Dsilva CJ, Levario TJ, Lu H, Schupbach T, Kevrekidis IG, and Shvartsman SY (2015). Dynamics of Inductive ERK Signaling in the Drosophila Embryo. Current biology : CB 25, 1784–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santos SD, Verveer PJ, and Bastiaens PI (2007). Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nature cell biology 9, 324–330. [DOI] [PubMed] [Google Scholar]
- 34.Murphy LO, Smith S, Chen RH, Fingar DC, and Blenis J (2002). Molecular interpretation of ERK signal duration by immediate early gene products. Nature cell biology 4, 556–564. [DOI] [PubMed] [Google Scholar]
- 35.Waddington CH (1959). Canalization of development and genetic assimilation of acquired characters. Nature 183, 1654–1655. [DOI] [PubMed] [Google Scholar]
- 36.Driever W, and Nusslein-Volhard C (1988). The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54, 95–104. [DOI] [PubMed] [Google Scholar]
- 37.Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, and Ribes V (2012). Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell 148, 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irvine KD, and Wieschaus E (1994). Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827–841. [DOI] [PubMed] [Google Scholar]
- 39.Collinet C, Rauzi M, Lenne PF, and Lecuit T (2015). Local and tissue-scale forces drive oriented junction growth during tissue extension. Nature cell biology 17, 1247–1258. [DOI] [PubMed] [Google Scholar]
- 40.Guglielmi G, Barry JD, Huber W, and De Renzis S (2015). An Optogenetic Method to Modulate Cell Contractility during Tissue Morphogenesis. Developmental cell 35, 646–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izquierdo E, Quinkler T, and De Renzis S (2018). Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nature communications 9, 2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pae J, Cinalli RM, Marzio A, Pagano M, and Lehmann R (2017). GCL and CUL3 Control the Switch between Cell Lineages by Mediating Localized Degradation of an RTK. Developmental cell 42, 130–142 e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuda L, Inoue YH, Yoo MA, Mizuno M, Hata M, Lim YM, Adachi-Yamada T, Ryo H, Masamune Y, and Nishida Y (1993). A protein kinase similar to MAP kinase activator acts downstream of the raf kinase in Drosophila. Cell 72, 407–414. [DOI] [PubMed] [Google Scholar]
- 44.Reuter R, and Leptin M (1994). Interacting functions of snail, twist and huckebein during the early development of germ layers in Drosophila. Development 120, 1137–1150. [DOI] [PubMed] [Google Scholar]
- 45.Weigel D, Jurgens G, Klingler M, and Jackle H (1990). Two gap genes mediate maternal terminal pattern information in Drosophila. Science 248, 495–498. [DOI] [PubMed] [Google Scholar]
- 46.Costa M, Wilson ET, and Wieschaus E (1994). A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell 76, 1075–1089. [DOI] [PubMed] [Google Scholar]
- 47.Kispert A, Herrmann BG, Leptin M, and Reuter R (1994). Homologs of the mouse Brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium, and Locusta. Genes & development 8, 2137–2150. [DOI] [PubMed] [Google Scholar]
- 48.Singer JB, Harbecke R, Kusch T, Reuter R, and Lengyel JA (1996). Drosophila brachyenteron regulates gene activity and morphogenesis in the gut. Development 122, 3707–3718. [DOI] [PubMed] [Google Scholar]
- 49.Wu LH, and Lengyel JA (1998). Role of caudal in hindgut specification and gastrulation suggests homology between Drosophila amnioproctodeal invagination and vertebrate blastopore. Development 125, 2433–2442. [DOI] [PubMed] [Google Scholar]
- 50.Patel AL, and Shvartsman SY (2018). Outstanding questions in developmental ERK signaling. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kicheva A, and Rivron NC (2017). Creating to understand - developmental biology meets engineering in Paris. Development 144, 733–736. [DOI] [PubMed] [Google Scholar]
- 52.Hunter C, and Wieschaus E (2000). Regulated expression of nullo is required for the formation of distinct apical and basal adherens junctions in the Drosophila blastoderm. The Journal of cell biology 150, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casanova J, Furriols M, McCormick CA, and Struhl G (1995). Similarities between trunk and spatzle, putative extracellular ligands specifying body pattern in Drosophila. Genes & development 9, 2539–2544. [DOI] [PubMed] [Google Scholar]
- 54.Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim HS, et al. (2015). The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 201, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savant-Bhonsale S, and Montell DJ (1993). torso-like encodes the localized determinant of Drosophila terminal pattern formation. Genes & development 7, 2548–2555. [DOI] [PubMed] [Google Scholar]
- 56.Fukaya T, Lim B, and Levine M (2016). Enhancer Control of Transcriptional Bursting. Cell 166, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ajuria L, Nieva C, Winkler C, Kuo D, Samper N, Andreu MJ, Helman A, Gonzalez-Crespo S, Paroush Z, Courey AJ, et al. (2011). Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development 138, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, and Levine M (2014). Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proceedings of the National Academy of Sciences of the United States of America 111, 10598–10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Groth AC, Fish M, Nusse R, and Calos MP (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bischof J, Maeda RK, Hediger M, Karch F, and Basler K (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proceedings of the National Academy of Sciences of the United States of America 104, 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson TK, Moore KA, Whisstock JC, and Warr CG (2017). Maternal Torso-Like Coordinates Tissue Folding During Drosophila Gastrulation. Genetics 206, 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Complete rescue of embryogenesis in OptoSOS-trk embryo. Related to Figure 2.
Time-lapse DIC imaging of a trk1 optoSOS embryo without (top) and with (bottom) a 90-minute light stimulus of 0.6 sec pulses of saturating 450 nm light delivered every 30 sec at the anterior and posterior poles; this experimentally-delivered light input is shown in the video in blue. Embryo is oriented with anterior to the left and posterior downward.
Video S2: tll / mist transcription in the endogenous terminal gradient or after illumination. Related to Figure 3.
Time-lapse fluorescence imaging of tll (purple) or hkb (green) MS2/MCP bursts in either wild-type or OptoSOS-tsl embryos. The region of illumination is indicated by the distance from the pole to the blue line, and for all light stimuli, 0.6 sec pulses of saturating 450 nm light were delivered every 30 sec. Transcriptional foci were automatically detected as indicated by the green or purple circles, and the boundary of transcription is plotted as a vertical green or purple line. The elapsed time is indicated on each frame in hours:minutes. Embryos are oriented with posterior to the left.
Video S3: Gastrulation in an embryo exposed to a 150 μm-wide posterior light stimulus. Related to Figure 5.
Time-lapse DIC imaging of an optoSOS-trk embryo exposed to a wide posterior illumination pattern (lower panel) compared to a wild-type control (upper panel). For all light stimuli, 0.6 sec pulses of saturating 450 nm light were delivered every 30 sec. An abnormally large posterior invagination is followed by germ band elongation to an extent indistinguishable from a wild-type embryo. Embryos are oriented with anterior to the left and posterior downward.