Abstract
Background
Children born extremely preterm are at heightened risk for intellectual and social impairment, including Autism Spectrum Disorder (ASD). There is increasing evidence for a key role of the placenta in prenatal developmental programming, suggesting that the placenta may, in part, contribute to origins of neurodevelopmental outcomes.
Methods
We examined associations between placental transcriptomic and epigenomic profiles and assessed their ability to predict intellectual and social impairment at age 10 years in 379 children from the Extremely Low Gestational Age Newborn (ELGAN) cohort. Assessment of intellectual ability (IQ) and social function was completed with the Differential Ability Scales-II and Social Responsiveness Scale (SRS), respectively. Examining IQ and SRS allows for studying ASD risk beyond the diagnostic criteria, as IQ and SRS are continuous measures strongly correlated with ASD. Genome-wide mRNA, CpG methylation and miRNA were assayeds with the Illumina Hiseq 2500, HTG EdgeSeq miRNA Whole Transcriptome Assay, and Illumina EPIC/850 K array, respectively. We conducted genome-wide differential analyses of placental mRNA, miRNA, and CpG methylation data. These molecular features were then integrated for a predictive analysis of IQ and SRS outcomes using kernel aggregation regression. We lastly examined associations between ASD and the multi-omic-predicted component of IQ and SRS.
Results
Genes with important roles in neurodevelopment and placental tissue organization were associated with intellectual and social impairment. Kernel aggregations of placental multi-omics strongly predicted intellectual and social function, explaining approximately 8% and 12% of variance in SRS and IQ scores via cross-validation, respectively. Predicted in-sample SRS and IQ showed significant positive and negative associations with ASD case–control status.
Limitations
The ELGAN cohort comprises children born pre-term, and generalization may be affected by unmeasured confounders associated with low gestational age. We conducted external validation of predictive models, though the sample size (N = 49) and the scope of the available out-sample placental dataset are limited. Further validation of the models is merited.
Conclusions
Aggregating information from biomarkers within and among molecular data types improves prediction of complex traits like social and intellectual ability in children born extremely preterm, suggesting that traits within the placenta-brain axis may be omnigenic.
Keywords: Prenatal neurodevelopmental programming, Social and cognitive impairment, Placental gene regulation, Epigenome-wide association, Differential expression analysis, Multi-omic aggregation
Background
Despite substantial research efforts to elucidate the etiology of neurodevelopmental impairment [1], little is known about transcriptomic and epigenomic factors influencing trajectories of neurodevelopment, such as those associated with preterm delivery [2]. Children born extremely preterm are at increased risk not only for intellectual impairment but also for Autism Spectrum Disorder (ASD) [3, 4], often accompanied by intellectual disability. In addition, preterm-born children have consistently been observed to manifest social difficulties (e.g., fewer prosocial behaviors) in childhood and adolecense that do not meet diagnostic criteria for ASD [5].
The placenta is posited as a critical determinant of both immediate and long-lasting neurodevelopmental outcomes in children [1]. The placenta is involved in hormone and neurotransmitter production and transfer of nutrients to the fetus, thus having direct influence on brain development. This intimate connection between the placenta and the brain is termed the placenta-brain axis [6, 7]. Epidemiological and animal studies have linked genomic and epigenomic alterations in the placenta with neurodevelopmental disorders and normal neurobehavioral development [8–10]. For example, the Markers of Autism Risk in Babies: Learning Early Signs (MARBLES) study has identified a differentially methylated region containing a putative fetal brain enhancer in placentas from children diagnosed with ASD (N = 24) compared to placentas from typically developing (N = 23) children [11]. The study of molecular interactions within and between the transcriptome and epigenome that represent the placenta-brain axis may advance our understanding of fetal mechanisms involved in aberrant neurodevelopment [6].
Most prior studies have investigated single molecular levels of the placenta transcriptome or epigenome, precluding analysis of possible interactions that could be linked to neurodevelopmental outcomes. Examining only a single molecular feature, or a single type of feature such as genotype even at a genome-wide scale can still result in much unexplained variation in phenotype due to potentially important interactions between multiple features [12, 13]. This observation is in line with Boyle et al.’s omnigenic model [14, 15], which proposes that gene regulatory networks are so highly interconnected that a large portion of the heritability of complex traits can be explained by effects on genes outside core pathways. Molecular integration to identify placental pathways related to fetal neurodevelopment in children has been largely unexplored but may prove to be insightful in associations with complex diseases [16].
We conducted a genome-wide analysis of DNA methylation (i.e., 5-methylcytosine), miRNA, and mRNA expression in the placenta, examining individual associations with social and intellectual impairment at 10 years of age in children from the Extremely Low Gestational Age Newborn (ELGAN) study [17]. We then combined the transcriptomic and epigenomic data to identify correlative networks of placental biomarkers predictive of social and intellectual impairment as continuous scales, thus allowing us to study neurodevelopmental difficulties beyond the ASD diagnostic categories [18]. To assess the convergent validity of our behavioral findings, we also examined the association of social and intellectual impairment in relation to ASD diagnoses [19]. This is among the first study of its kind to use multiple placental molecular signatures to predict intellectual and social impairment, which may inform a framework for predicting risk of adverse neurocognitive and neurobehavioral outcomes in young children.
Methods
ELGAN recruitment and study participants
From 2002 to 2004, women who gave birth at under 28 weeks gestation at one of 14 medical centers across five U.S. states enrolled in the ELGAN study [17]. The Institutional Review Board at each participating institution approved study procedures. Included were 379 of 889 children with both placental molecular data (CpG methylation, mRNA expression, and miRNA expression) and a 10-year neurodevelopment assessment.
Social and cognitive function and ASD at 10 years of age
Trained child psychologist examiners [5, 20] evaluated general cognitive ability (IQ) with the School-Age Differential Ability Scales-II (DAS-II) Verbal and Nonverbal Reasoning subscales [21]. The Social Responsiveness Scale (SRS) was used to assess severity of ASD-related social deficits in 5 subdomains: social awareness, social cognition, social communication, social motivation, and autistic mannerisms [22]. We used the gender-normed T-score (SRS-T; intended to correct gender differences observed in normative samples) as continuous measure of social deficit [23]. All participants were assessed for ASD [19]. Diagnostic assessment of ASD was conducted with three well-validated measures, administered sequentially. First, the Social Communication Questionnaire (SCQ) was administered to screen for potential ASD, using a score ≥ 11 to increase sensitivity relative to the standard criterion score of ≥ 15 [19, 24]. For children who screened positive on the SCQ criterion, we conducted the Autism Diagnostic Interview–Revised (ADI-R) with the primary caregiver [25]. All children who met ADI-R criteria for ASD, or who had a prior clinical diagnosis of ASD and/or exhibited symptoms of ASD during cognitive testing according to the site psychologist) were then assessed with the Autism Diagnostic Observation Schedule, Second Version (ADOS-2), which served as the criterion measure of ASD in this study [26]. All ADOS-2 administrations were independently scored by a second rater with autism diagnostic and ADOS-2 expertise. In cases of scoring disagreements, consensus was reached via discussion between raters. Item-by-item inter-rater agreement for the 14 ADOS-2 diagnostic algorithm scores was on average 0.93 (SD = 0.12). These developmental assessment procedures and all relevant test scores for ASD and intellectual function are reported in a prior publication [20].
Placental DNA and RNA extraction
After delivery, placentas were biopsied under sterile conditions. The ELGAN team collected a piece of the chorion, representing the fetal side of the placenta [27]. More specifically, placentas were placed in a sterilized basin and biopsied by pulling back the amnion to expose the chorion at the midpoint of the longest distance between the cord insertion and edge of the placental disk. A sample from the fetal side of the placenta was removed by applying traction to the chorion and underlying trophoblast tissue. The specimen was placed in a cryogenic vial and immersed in liquid nitrogen. Specimens were stored at − 80 °C for approximately 13–15 years until processed. For processing, a 0.2 g subsection of the placental tissue was cut from the frozen biopsy and washed with sterile 1 × phosphate-buffered saline to remove any remaining blood. Samples were homogenized using a lysis buffer, and the homogenate was separated into aliquots. This process was detailed in a prior publication [28]. Nucleic acids were extracted from the homogenate using AllPrep DNA/RNA/miRNA Universal kit (Qiagen, Germany). The quantity and quality of DNA and RNA were analyzed using the NanoDrop 1000 spectrophotometer and its integrity verified by the Agilent 2100 BioAnalyzer. As previously described [29], RNA quality was determined using LabChip (Perkin Elmer) instrument to generate RNA integrity numbers (RIN), which ranged from 1 to 3, and DV200 values, which were in acceptable range for placenta tissue [30].
Epigenome-wide placental DNA methylation
Extracted DNA sequences were bisulfate-converted using the EZ DNA methylation kit (Zymo Research, Irvine, CA) and followed by quantification using the Infinium MethylationEPIC BeadChip (Illumina, San Diego, CA), which measures CpG loci at a single nucleotide resolution, as previously described [27, 28, 31, 32]. Quality control and normalization were performed resulting in 856,832 CpG probes from downstream analysis, with methylation represented as the average methylation level at a single CpG site (β value) [28, 33–35]. DNA methylation data was imported into R for pre-processing using the minfi package [33]. Quality control was performed at the sample level, excluding samples that failed and technical duplicates; 411 samples were retained for subsequent analyses. Functional normalization was performed with a preliminary step of normal-exponential out-of band (noob) correction method [36] for background subtraction and dye normalization, followed by the typical functional normalization method with the top two principal components of the control matrix [34, 37]. Quality control was performed on individual probes by computing a detection P value and excluded 806 (0.09%) probes with non-significant detection (P > 0.01) for 5% or more of the samples. A total of 856,832 CpG sites were included in the final analyses. Lastly, the ComBat function was used from the sva package to adjust for batch effects from sample plate [38]. The data were visualized using density distributions at all processing steps. Each probe measured the average methylation level at a single CpG site. Methylation levels were calculated and expressed as β values (β = intensity of the methylated allele (M))/(intensity of the unmethylated allele (U) + intensity of the methylated allele (M) + 100). βvalues were logit transformed to M values for statistical analyses [39].
Genome-wide placental mRNA and miRNA expression
mRNA expression was determined using the Illumina QuantSeq 3′ mRNA-Seq Library Prep Kit, a method with high strand specificity. mRNA-sequencing libraries were pooled and sequenced (single-end 50 bp) on one lane of the Illumina Hiseq 2500. mRNA were quantified through pseudo-alignment with Salmon v.14.0 [40] mapped to the GENCODE Release 31 (GRCh37) reference transcriptome. miRNA expression profiles were assessed using the HTG EdgeSeq miRNA Whole Transcriptome Assay (HTG Molecular Diagnostics, Tucson, AZ). miRNA were aligned to probe sequences and quantified using the HTG EdgeSeq System [41]. Genes and miRNAs with less than 5 counts and variance less than 0.5 for each sample were filtered [42], resulting in 11,224 genes and 2047 miRNAs for downstream analysis. Distributional differences between lanes were first upper-quartile normalized [43]. Unwanted technical and biological variation (e.g. cell-type heterogeneity) was then estimated using RUVSeq [44], where we empirically defined transcripts not associated with outcomes of interest as negative control housekeeping probes [45]. One dimension of unwanted variation was removed from the variance-stabilized transformation of the gene expression data using the limma package [46, 47].
Statistical analysis
All code and functions used in the statistical analysis can be found at https://github.com/bhattacharya-a-bt/multiomics_ELGAN.
Correlative analyses between SRS, IQ, and ASD
Associations among SRS scores, IQ and ASD were assessed using Pearson correlations with estimated 95% confidence intervals, and the difference in distributions of SRS and IQ across ASD case–control was assessed using Wilcoxon rank-sum tests. Associations between demographic variables (race, sex, maternal age, number of gestational days, maternal smoking status, placental inflammation, birth weight Z-score and mother’s insurance) with SRS and IQ were determined using multivariable regression, assessing the significance of regression parameters using Wald tests of significance and adjusting for multiple testing with the Benjamini–Hochberg procedure [48].
Genome-wide molecular associations with SRS and IQ
Once associations between SRS and IQ and ASD were confirmed, we utilized continuous SRS and IQ measures as the main outcomes of interest. Associations between mRNA expression or miRNA expression with SRS and IQ were estimated through a negative binomial linear model using DESeq2 [46]. Epigenome-wide associations (EWAS) of CpG methylation sites with outcomes were assessed using robust linear regression with test statistic modification through an empirical Bayes procedure [47], described previously [28]. Both the differential mRNA and miRNA expression and EWAS models controlled for the following covariates: race, age, sex, number of gestational age days, birth weight Z-score, acute inflammation of the placental chorion, and education level of the mother. As in previous analyses, EWAS models also controlled for five surrogate variables to account for cell-type heterogeneity [28, 38]. Multiple testing was adjusted for using the Benjamini–Hochberg procedure. Sensitivity analyses of test power across various effect sizes (fold change for RNA-seq and miRNA-seq expression) and parameters (mean and dispersion of expression) were conducted for differential gene expression analysis [49], showing sufficient power to detect effect sizes of differential expression (Additional file 2: Supplemental Results—Figure S1A-B). Likewise, we found sufficient power to detect differentially methylated sites at large effect sizes (Additional file 2: Supplemental Results—Figure S1C) using the framework from Mansell et al.[50].
To examine placental cell type variability, we extended the differential mRNA expression analysis to consider cell-type specific proportions. We applied unmix, a reference-based deconvolution method [46], to estimate cell-type proportions using reference single cell RNA-seq expression profiles for extravillous trophoblasts, cytotrophoblasts, syncytiotrophoblasts, and mesenchymal stromal cells, derived from fetal placental tissue at 24 weeks of gestation [51]. Here, we refer to the mRNA expression data from ELGAN as the bulk signal, as it represents the mRNA expression from the bulk tissue that includes gene expression signal from all contributing cell (i.e. different trophoblasts, endothelial cells, epithelial cells, etc.). This algorithm estimates the contribution to the bulk mRNA signal from individual cell types on a sample-by-sample basis. We incorporated these cell-type proportions into the differential expression analysis by adding a covariate for cell-type proportions and an interaction term between gene expression and proportion. This cell-type interaction model subtly changes the interpretation of the main gene expression term, representing an estimate of the gene expression effect size on SRS or IQ when the bulk tissue contains 0% of the cell type. Thus, we can detect cell-type-specific differentially expressed genes by testing the interaction effect, which measures how the magnitude of the gene-to-outcome association differs in bulk tissue with 0% and 100% of the cell type in question [52] (details in Additional file 1: Supplemental Methods).
Placental multi-molecular prediction of SRS and IQ
We next assessed how well an aggregate of one or more of the molecular datasets (CpG methylation, mRNA expression, and miRNA expression) predicted continuous SRS and IQ scores. The analytical scheme is summarized in Fig. 1, using 379 samples with data for all three molecular datasets (DNA methylation, miRNA, and mRNA). Briefly, we first adjusted the outcome variables and molecular datasets for above noted demographic and clinical covariates using limma [53] to account for associations between the outcomes and these coviarates in the eventual predictive models. Next, to model the covariance between samples within a single molecular profile, we aggregated the molecular datasets with thousands of biomarkers each into a molecular kernel matrix. A molecular kernel matrix represents the inter-sample similarities in a given molecular profile (Additional file 1: Supplemental Methods). A linear or non-linear kernel aggregation may aid in prediction of complex traits by capturing non-additive effects [54–56], which represents a sizable portion of phenotypic variation [57, 58]. Using all individual, pairwise, and triplet-wise combinations of molecular kernel matrices, we fitted predictive models of SRS and IQ based on linear mixed modeling [56] or kernel regression least squares (KRLS) [59] and assessed predictive performance with McNemar’s adjusted R2 via Monte Carlo cross validation [60]. We also optimized predictive models for the number of included biomarkers per molecular profile with feature selection in the training sets. Extensive model details, as well as alternative models considered, are detailed in Additional file 1: Supplemental Methods.
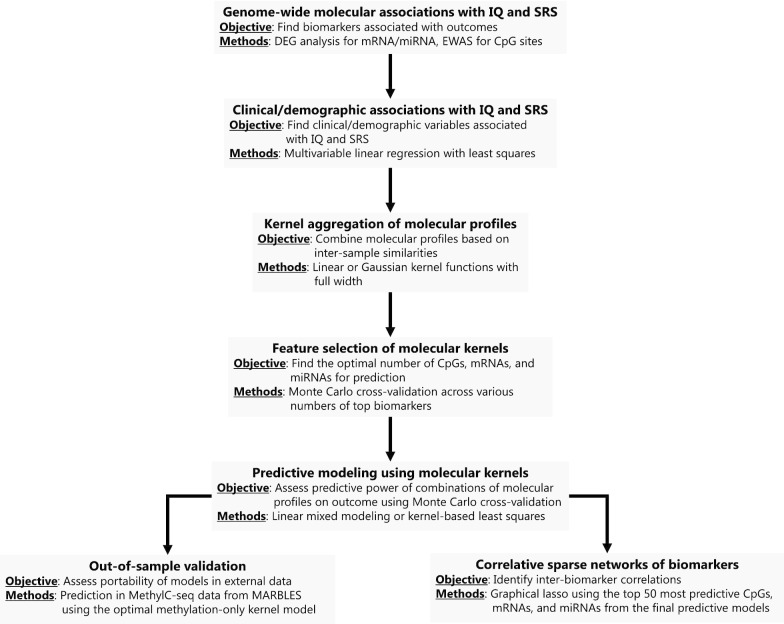
Fig. 1.
Scheme for kernel aggregation and prediction models. (1) Design matrices for CpG sites, mRNAs, and miRNAs are aggregated to form a linear or Gaussian kernel matrix that measures the similarity of samples. (2) Clinical variables are regressed out of the outcomes IQ and SRS and from the omic kernels to limit influence from these variables. (3) Using 50-fold Monte Carlo cross-validation on 75–25% training-test splits, we train prediction models with the kernel matrices for IQ and SRS in the training set and predict in the test sets. Prediction is assessed in every fold with adjusted and averaged for an overall prediction metric
Validation in external dataset
Lack of studies that consider placental mRNA, CpG methylation and miRNA data with long-term child neurodevelopment limit the ability to extablish external validation. We obtained one external placental CpG methylation dataset from the MARBLES cohort [11]. To assess out-of-sample performance of kernel models for methylation, we downloaded MethylC-seq data for 47 placenta samples, 24 of which were identified as ASD cases (NCBI Gene Expression Omnibus accession numbers GSE67615) [11]. β-values for DNA methylation were extracted from BED files and transformed into M-values with an offset of 1 [39], and used the best methylation-only predictive model to predict SRS and IQ in these 47 samples, as detailed in Additional file 1: Supplemental Methods.
Correlative networks and gene ontology enrichment analysis
In the final KRLS predictive models for both IQ and SRS including all three molecular profiles, we extracted the top 50 most predictive (largest point-wise effect sizes) CpGs, miRNAs, and mRNAs of SRS and IQ. A sparse correlative network was inferred among these biomarkers that links them based on the strength of correlative signals using graphical lasso in qgraph [61, 62]. We then conducted biological process and molecular function gene ontology over-representation analysis of genes identified in these correlative networks using WebGestalt [63].
Results
Social impairment (SRS) and cognition (IQ) are associated with ASD
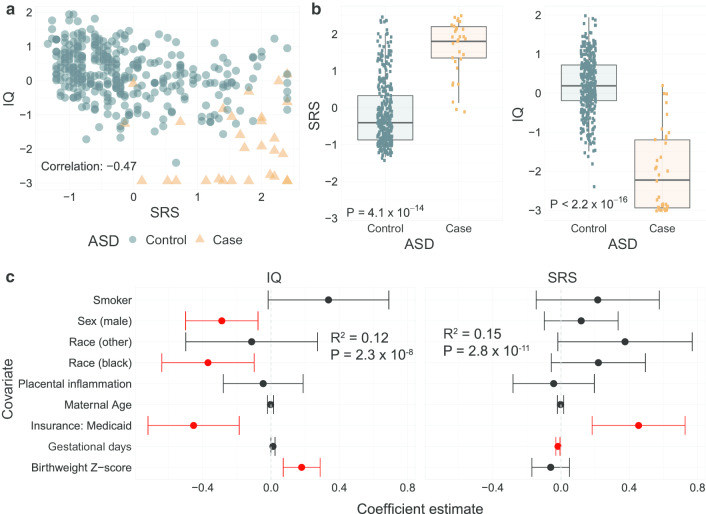
Although the sample is enriched for ASD cases (N = 35 cases, 9.3% of the sample) relative to non-preterm cohorts, there is still a relatively low case–control ratio for a genome-wide study of this sample size (descriptive statistics for relevant covariates in Table 1). Therefore, we considered continuous measures of SRS and IQ at age 10 for both associative and predictive analyses. Using continuous variables for SRS and IQ allow us to to study complexities beyond the ASD diagnostic categories [16, 18, 19]. Figure 2a, b shows the relationship between SRS, IQ, and ASD. SRS and IQ are negatively correlated [Pearson ρ = − 0.47, 95%CI (− 0.55, − 0.39)]. The mean SRS is significantly higher in ASD cases compared to controls [mean difference of 1.74, 95%CI (1.41, 2.07)]. Mean IQ is significantly lower in ASD cases versus controls [mean difference of − 2.23, 95%CI (− 2.46, − 1.96)]. We also measured associations between demographic characteristics with SRS and IQ using multivariable regression (Fig. 2c). Male sex is associated with lower IQ, while public health insurance is associated with both lower IQ and increased social impairment. Demographic variables included in the multivariable regression explain approximately 12% and 15% of the total variance explained in IQ and SRS, as measured by adjusted R2, with a summary of regression parameters in Table 2. Based on the associations identified here and the value of inclusion of continuous measures, subsequent transcriptomic and epigenomic analyses control for demographic covariates.
Table 1.
Descriptive statistics for demographic and clinical covariates
| Continuous variable | Mean, SD, median |
|---|---|
| Maternal age (years) | 29.6, 6.61, 29.5 |
| Gestational age (days) | 182.5, 9.17, 184.0 |
| Birthweight Z-score | 0, 1, 0.05 |
| Categorical variable | Number (proportion) |
|---|---|
| ASD | |
| Case | 35 (9.3%) |
| Control | 344 (90.7%) |
| Race | |
| White | 233 (61.5%) |
| Black | 112 (29.5%) |
| Other | 34 (9.0%) |
| Sex of baby | |
| Female | 180 (47.5%) |
| Male | 199 (52.5%) |
| Mother’s smoking status | |
| Non-smoker | 340 (89.7%) |
| Smoker | 39 (10.3%) |
| Mother’s insurance status | |
| Private | 251 (66.2%) |
| Medicaid | 128 (33.8%) |
| Placental chorion inflammation | |
| Not inflamed | 252 (66.5%) |
| Inflamed | 127 (33.6%) |
Fig. 2.
Associations between SRS, IQ, and ASD and with clinical variables. a Scatter plot of SRS (X-axis) and IQ (Y-axis) colored by ASD case (orange) and control (blue) status. b Boxplots of SRS and IQ across ASD case–control status. P value from a two-sample Mann–Whitney test is provided. c Caterpillar plot of multivariable linear regression parameters of IQ and SRS using clinical variables. Points give the regression parameter estimates with error bars showing the 95% FDR-adjusted confidence intervals [48]. The null value of 0 is provided for reference with the dotted line
Table 2.
Summary of multivariable regression models of SRS and IQ in relation to clinical covariates (self-reported race, sex, maternal age, smoking status, insurance level of the mother, gestational age, birthweight Z-score, and inflammation of the placental chorion)
| Parameter | SRS | IQ | ||
|---|---|---|---|---|
| Estimate (SE) | FDR-adjusted P value (Raw P value) |
Estimate (SE) | FDR-adjusted P value (Raw P value) |
|
| Race | ||||
| Black | 0.219 (0.13) | 0.165 (0.091) | − 0.369 (0.13) | 0.012 (0.004) |
| Other | 0.375 (0.19) | 0.087 (0.043) | − 0.113 (0.18) | 0.684 (0.533) |
| Sex | ||||
| Male | 0.119 (0.10) | 0.342 (0.243) | − 0.288 (0.10) | 0.012 (0.004) |
| Maternal age | − 0.002 (0.01) | 0.800 (0.800) | − 0.003 (0.01) | 0.792 (0.748) |
| Smoking status | ||||
| Yes | 0.215 (0.17) | 0.334 (0.204) | 0.337 (0.17) | 0.087 (0.043) |
| Mother’s insurance | ||||
| Medicaid | 0.454 (0.13) | 0.002 (0.001) | − 0.453 (0.13) | 0.003 (0.001) |
| Gestational age | − 0.017 (0.01) | 0.012 (0.002) | 0.012 (0.01) | 0.087 (0.043) |
| Birthweight Z-score | − 0.060 (0.05) | 0.342 (0.247) | 0.179 (0.05) | 0.003 (0.001) |
| Placental inflammation | − 0.042 (0.11) | 0.793 (0.705) | − 0.046 (0.11) | 0.793 (0.677) |
Genome-wide associations of mRNA, miRNA, and CpGs with SRS and IQ
Genome-wide association tests between each of the individual placental molecular datasets (e.g. the placental mRNA data, the CpG methylation, or the miRNA datasets) in relation to SRS and IQ (see "Methods") identified two genes with mRNA expression significantly associated with SRS at FDR-adjusted P < 0.01, namely Hdc Homolog, Cell Cycle Regulator (HECA), and LIM Domain Only 4 (LMO4). We did not find CpG sites or miRNAs associated with SRS (Table 3). Associations between IQ and the mRNA expression, at FDR-adjusted P < 0.01, were observed at four genes, namely Ras-Related Protein Rab-5A (RAB5A), Transmembrane Protein 167A (TMEM167A), Signal Transducer and Activator of Transcription 2 (STAT2), ITPRIP Like 2 (ITPRIPL2). One CpG site, cg09418354, located in the gene Carbohydrate Sulfotransferase 11 (CHST11) displayed an association with IQ, and no miRNAs were associated with IQ (Table 3). Manhattan plots (Additional file 2: Supplemental Results—Figure S2) show the strength of associations of all biomarkers by genomic position. No mRNAs, CpG sites, or miRNAs were significantly associated with both SRS and IQ. Summary statistics for these associations are provided in Additional file 2: Supplemental Results: Table S1.
Table 3.
Summary of genome-wide associations of molecular profiles with SRS and IQ at FDR-adjusted
| Biomarker | Effect size | FDR-adjusted value |
|---|---|---|
| SRS | ||
| mRNA expression | ||
| HECA | 0.571 | 0.001 |
| LMO4 | 0.467 | 0.001 |
| IQ | ||
| Biomarker | ||
| mRNA expression | ||
| RAB5A | − 0.516 | 0.002 |
| TMEM167A | − 0.632 | 0.004 |
| ITPRIPL2 | − 0.557 | 0.004 |
| STAT2 | − 0.584 | 0.004 |
| CpG methylation site | ||
| cg09418354 (within CHST11) | − 0.005 | 0.002 |
We also considered differential mRNA expression analysis specific to four key distinct cell-types that comprise the placenta: extravillous trophoblasts, cytotrophobalsts, syncytiotrophoblasts, and mesenchymal stromal cells [64]. Importantly, we did not detect any significant associations between placental cell-type proportions and the differentially expressed genes in the bulk tissue. Incorporating estimated cell-type proportions into an interaction-based differential mRNA expression model revealed no cell-type-specific differentially expressed genes at FDR-adjusted P < 0.01. To examine any cell-specific trends, at FDR-adjusted P < 0.05, we detected two SRS-associated stromal cell-specific differentially expression genes and two IQ-associated syncytiotrophoblast-specific differentially expression genes (Additional file 2: Supplemental Results—Table S2), all not detected without the interaction model. These SRS-associated genes include Bromodomain Containing 2 (BRD2), associated with fetal metabolic programming of newborns [65]. Furthermore, we detected a syncytiotrophoblasts-specific association between IQ and ATPase Plasma Membrane Ca2 + Transporting 1 (ATP2B1), a gene whose polymorphic variants have been shown to have associations with preeclampsia [66, 67].
Kernel regression shows predictive utility in aggregating multiple molecular datasets
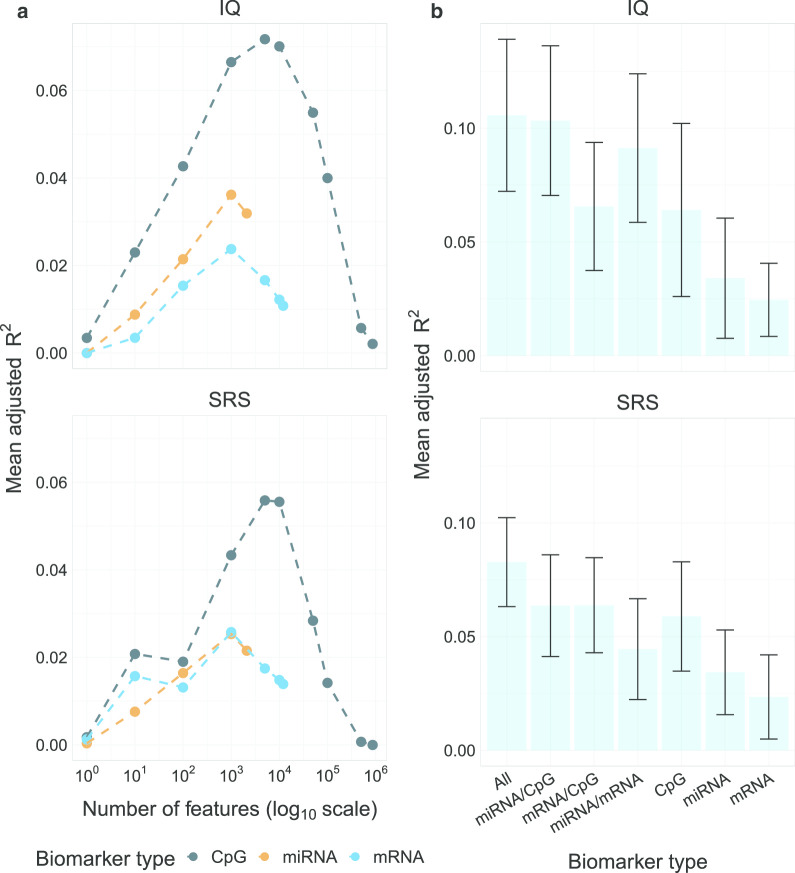
Because the genome-wide association analyses revealed few mRNAs, CpG sites or miRNAs that were associated with SRS or IQ with large effect sizes, we next assessed the impact of aggregating these molecular datasets on prediction of SRS and IQ. This was done to account for the considerable number of biomarkers that have moderate effect sizes on outcome. To find the most parsimonious model with the greatest predictive performance, we first selected the optimal number of biomarkers per molecular profile from the training set for each outcome that gave the largest mean adjusted R2 in predictive models with only one of the three molecular datasets (see Additional file 1: Supplemental Methods). Figure 3a shows the relationship between the number of biomarkers from the mRNA expression, CpG level, miRNA expression datasets and their predictive performance. In general, predictive performance steadily increased as the number of biomarker features increased until reaching a tipping point where predictive performance decreased (Fig. 3a). Overall, for CpG methylation, the top (lowest P values of association) 5000 CpG features showed the greatest predictive performance, and for the mRNA and miRNA expression datasets, the top 1000 features showed the greatest predictive performance.
Fig. 3.
In-sample predictive performance of kernel models. a Adjusted mean (Y-axis) of best kernel models over various numbers of the top biomarkers (X-axis) in the CpG (dark blue), miRNA (orange), and mRNA (light blue) omics over 50 Monte Carlo folds. The X-axis scale is logarithmic. b Bar plots of adjusted mean (Y-axis) for optimally tuned kernel predictive models using all combinations of omics (X-axis) over 50 Monte Carlo folds. The error bar gives a spread of one standard deviation around the mean adjusted
Using the fully-tuned 7000 biomarkers (5000 for CpG methylation and 1000 for both mRNA and miRNA expression) per molecular dataset with feature selection carried out in the training set, we trained predictive models (both linear and Gaussian kernel models) using all individual, pair-wise, and triplet-based combinations of the three molecular datasets. Figure 3b shows that whereas the mRNA had the lowest predicted performance to both IQ (R2 = 0.025) and SRS (R2 = 0.025), aggregating the mRNA expression, CpG methylation and miRNA expression datasets tends to increase the predictive performance. Specifically, in relation to both outcomes (SRS and IQ), the model using all three integrated datasets shows the greatest predictive performance (mean adjusted R2 = 0.11 in relation to IQ and R2 = 0.08 in relation to SRS).
Correlative networks of placental biomarkers
To gain further understanding of the associations among the identified mRNA, CpG and miRNA biomarkers in the context of IQ and SRS, we extracted (n = 50) mRNA, CpGs, and miRNAs with the largest effect sizes on IQ and SRS in the kernel regression models and inferred sparse correlative networks using the graphical lasso [61, 62] (see "Methods"). In the networks (Additional file 2: Supplemental Results—Figure S3), each molecular dataset clusters by itself, with minimal nodes extending between molecular datasets, and more correlation observed between miRNAs and CpG methylation versus mRNAs. These networks point to genes that have been shown in literature to play important roles in neuronal development and in placental function. For example, SMARCA2 (SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily A, Member 2) and DDX59 has been implicated in development disorders of the brain, such as Nicolaides-Baraitser syndrome and epilepsy, and other developmental disorders, such as dysfunctional central nervous system development and orofaciodigital syndrome [68–73]. Furthermore, ARL5B (ADP Ribosylation Factor Like GTPase 5B) and MPP5 (Membrane Palmitoylated Protein 5) have been associated with decidua and trophoblasts functions within the placenta [74–76]. Furthermore, at FDR-adjusted P < 0.05, over-representation analysis revealed gene enrichments for membrane organization processes (endomembrane system and membrane organization) for the IQ-associated gene set and nucleic acid and enzyme binding processes (RNA binding, ubiquitin protein ligase binding, heterocyclic compound binding, etc.) for the SRS-associated gene set (Additional file 2: Supplemental Results—Table S3).
Validation of in-sample and out-sample SRS and IQ prediction with ASD case and control
To contextualize our predictions, we tested whether the predicted SRS and IQ scores generated by our kernel models are associated with ASD case–control status; these predicted SRS and IQ scores represent the portion of the observed SRS and IQ values that our models can predict from placental genomic features. We used the optimal 7000 biomarker features identified with a tenfold cross-validation process, splitting samples into 10 hold-out sets and using the remaining samples as a training set to predict SRS and IQ for all 379 samples. After accounting for covariates, the predicted SRS and IQ values from the biomarker data were well-correlated with the observed clinical SRS and IQ values, explaining approximately 8% (approximate Spearman ρ =0.29, cross-validation R2 P value P = 7.5 × 10−9) and 12% (Spearman ρ = 0.35, P = 3.6 × 10−12) of the variance in the observed SRS and IQ variables, respectively. This shows that biomarkers in the placenta can explain considerable amount of the variance in SRS and IQ at 10 years of age.
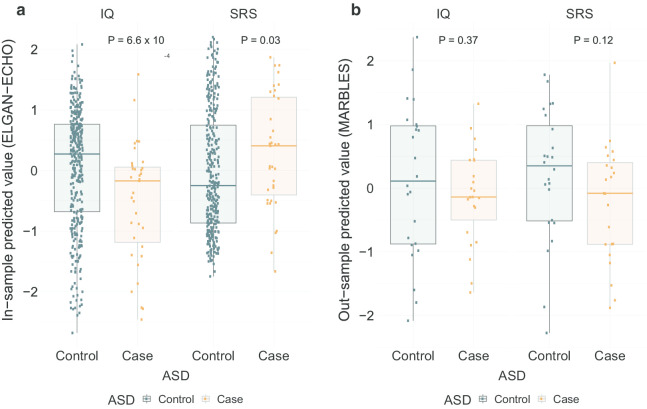
Lastly, we assessed associations between molecularly-predicted SRS and IQ values and ASD case–control status. In ELGAN, we found strong associations between the predicted SRS and IQ with ASD case and controls, mean difference of − 0.56 (test statistic W = 8121, P = 6.6 × 10−4) for IQ, and mean difference of 0.33 (W = 4717, P = 0.03) for SRS (Fig. 4a). Because of the lack of an available external dataset with all three molecular data (mRNA, CpG methylation, and miRNA) and IQ, SRS and ASD data, we assessed the out-of-sample predictive performance of the CpG methylation-only models using MethylC-seq data from the MARBLES cohort (GEO GSE67615) [11]. We computed predicted IQ and SRS values for 47 placental samples (24 cases of ASD) and assessed differences in mean predicted IQ and SRS across ASD case and control groups. The direction of the association is similar to our data for IQ, yet the differences in mean-predicted IQ (− 0.22, P = 0.37) and SRS (− 0.42, P = 0.12) across ASD groups in MARBLES are not significant (Fig. 4b). This external validation provides some evidence of the portability of our models and merits further future validation of these models, as more placental multi-omic datasets are collected.
Fig. 4.
Association of ASD case/control status with predicted SRS and IQ. a Box-plots of in-sample predicted IQ (left) and SRS (right) over ASD case/control in ELGAN over tenfold cross-validation. b Box-plots of out-sample predicted IQ (left) and SRS (right) over ASD case/control in MARBLES external validation dataset. -values presented as from a Mann–Whitney test of differences across the ASD case/control groups
Discussion
We evaluated the predictive capability of three types of molecular biomarker data, namely transcriptomic (mRNA), and epigenomic (miRNA expression, CpG methylation), in the placenta on cognitive and social impairment in relation to ASD at 10 years of age. The molecular biomarker data highlight that genes that play important roles in placental functioning (ARL5B and MPP5) and neurodevelopment (SMARCA2, DDX59, MPP5) were associated with or predictive of SRS and IQ. The multi-omic predictions of SRS and IQ are strong and explain up to 8% and 12% of the variance in the observed SRS and IQ variables in tenfold cross-validation, respectively. External validation of our models is inconclusive, however, and merits further investigation to minimize uncertainty in our findings, as mentioned in the limitations section. This study supports the utility of aggregating information from biomarkers within and among molecular datasets to improve prediction of complex neurodevelopmental outcomes like social and intellectual ability, suggesting that traits on the placenta-brain axis may be omnigenic.
Several genes with known ties to neurodevelopmental disorders distinguished individuals with and without intellectual or social impairments. For example, LMO4 (associated with social impairment) is a protein encoding gene with a broad spectrum of expression in human tissues and involved in multiple developmental pathways, including neurogenesis. Among its many roles, LMO4 promotes the acquisition of cortical neuronal identities by forming a complex with the protein neurogenin 2 (NGN2) and subsequently activating NGN2-dependent gene expression [77]. Humans with deletions in LMO4 display intellectual disabilities and occasionally autism [78]. Furthermore, this gene has also been associated with modulation of fear [79] and cue-reward leaning [80], which could result in perceptions and behaviors seen in association with social impairment. LMO4 also influence the growth factor-β (TGF-β) cytokine pathway, which plays important roles in mammalian development [81]. LMO4 and HECA has been identified in pathways and processes related to neural development via commonly regulated targets of Forkhead box protein P2 (FOXP2) and miR-3666; the LMO4 gene shows asymmetric expression in the embryonic brain possibly due to repression by FOXP2 and hence plays important roles in cortical patterning [82].
Among the biomarkers associated with IQ, we found RAB5A, a protein coding gene belonging to a family of small GTPases, involved in a variety of cellular processes including intracellular membrane trafficking. In the placental syncytiotrophoblasts, a specialized epithelial structure that interfaces the placenta and maternal blood, RAB5A has shown involvement in vesicular trafficking which could affect the syncytiotrophoblast function of transporting nutrients necessary for fetal development [83]. RAB5A can also affect the regulation of genes with roles in cell proliferation [84]. In terms of cognitive outcomes, RAB5A and the RAB family play critical roles in synaptic function [85, 86] and dendritic branching [87]. Finally, genetic variants of RAB5A have been associated with ASD [88]. Other relevant genes are STAT2 and CHST11. STAT2 is a well-known essential and specific positive effector of type I interferons (IFNs) signaling [89], and placental type I IFNs is an important immune modulator, including modulation of viral infection in the mother and fetus [90]. STAT2 was identified as one differentially expressed genes in ASD and co-morbidities that overlap with innate immunity pathways [91]. Also, with important roles in immune regulation, the genetic variation and methylation of the CHST11 gene, for which we found a methylated CpG site associated with intellectual impairment, has been linked to neurodevelopmental disorders [92, 93].
In our correlative gene network analysis, we detected a group of inter-related genes associated with IQ enriched for membrane organization processes. This enrichment may point to a previously established link: endothelial cell membrane dysfunction leads to deficient nutrient exchange and has a lasting impact on neurodevelopment [94]. For example, one of these genes is DDX59; in our differential expression analysis, DDX59 shows a nominally significant negative association with IQ. A recent study has shown that DDX59 is upregulated in syncytiotrophoblasts in severe preeclampsia patients compared to controls [95]. Another example is MPP5, expressed in the placenta, brain, nervous system and other tissues, which is essential for cell polarity, fate and survival. In the placenta, MPP5 seem to have significant roles: promotion of embryo-decidual adhesion [75], differentiation of extravillous trophoblasts [76], and gene expression levels from chorionic villus have been associated with severe early-onset preeclampsia [96]. In terms of neurodevelopment, both animal and human studies show that MPP5 has been found to be essential in neurogenesis [97, 98]; in a murine model Mpp5 depletion led to microcephaly, decreased cerebellar volume and cortical thickness, while humans with de novo variants of MPP5 suffer from global developmental delays with language regression and behavioral changes [97]. In our differential expression analysis, we found that MPP5 has a nominal negative association with IQ. Lastly, we also estimated that SMARCA2 has a strong predictive effect on and a nominally positive association with IQ; previous literature shows that epigenomic effects on and genetic dysfunction of SMARCA2 plays a role in development of Nicolaides-Baraitser syndrome, a developmental disorder categorized by intellectual disability and seizures [68, 69]. It is worth noting that these results are ultimately correlational in nature, and a causal interpretation should be avoided. Future research using in vitro and in vivo studies could elucidate the mechanistic influences of placental expression of these genes on the brain.
Not only did our cell-type-specific differential expression analysis show that the differentially expressed genes we detected in the bulk placenta were minimally affected by cell-type heterogeneity, we detected genes whose cell-type-specific expression has large associations with IQ and SRS. For instance, we detected a syncytiotrophoblast-specific association between IQ and ATP2B1, a gene that has been implicated in preeclampsia [66, 67], an in utero condition that is partially mediated by dysfunctional syncytiotrophoblasts [99] and has negative impacts with childhood neurodevelopment [100, 101]. In addition, ATP2B1 encodes PMCA1, a plasma membrane calcium ion pump, shown to have reduced activity in fetal-facing syncytiotrophoblast basal plasma membranes in patients with preeclampsia compared to controls [102–105]. This cell-type-specific analysis underscores the importance of not only accounting for cell-type-heterogeneity in bioinformatics analyses of the placenta but estimating cell-type-specific associations through deconvolution or single cell assays.
Comparing the individual molecular datasets, DNA methylation effects showed the strongest prediction of both SRS and IQ impairment. There is strong evidence suggesting inverse correlation between DNA methylation of the first intron or promoter region and gene expression across tissues and species [106]. We found that many of the CpG loci with the largest effect sizes on SRS and IQ identified in our analysis are located in genes with DNase hyperactivity or active regulatory elements for the placenta [107, 108], suggesting that these loci likely play regulatory functions. Experimental studies have demonstrated regions of the genome in which DNA methylation is causally important for gene regulation and those in which it is effectively silent [109]. We found that aggregating biomarkers within and among molecular datasets improves prediction of social and cognitive impairment. Specifially, this observation suggests new possibilities to the discovery of candidate genes in the placenta that convey neurodevelopmental risk, improving the understanding of the placenta-brain axis. Recent work in transcriptome-wide association studies (TWAS) are a promising tool that aggregates genetics and transcriptomics to identify candidate trait-associated genes [110, 111]. Incorporating information from regulatory biomarkers, like transcription factors and miRNAs, into TWAS increases study power to generate hypotheses about regulation [112, 113]. Given our observations in this analysis and the number of the integrated molecular datasets, we believe that the ELGAN study can be used to train predictive models for placental transcriptomics from genetics, enriched for regulatory elements [113]. These transcriptomic models can then be applied to genome-wide association study cohorts to study the regulation of gene-trait associations in the placenta.
Limitations
When interpreting the results of this study, some factors should be considered. Extremely preterm birth is strongly associated with increased risk for neurodevelopmental disorders [19]. This association may lead to bias in estimated associations between the molecular biomarkers and outcomes, mainly when unmeasured confounders are linked to both pre-term birth and autism [114]. Ideally, an external dataset with both multiomic data and pre-term birth phenotypes could be used to examine associations between molecular profiles and pre-term birth to investigate the degree to which collider bias affects associations between molecular profiles and SRS and IQ [114, 115]. Without this assessment of collider bias, results from our predictive models may not generalizable to term cohorts. Still, to our knowledge the ELGAN cohort is among the largest available placental repositories with both multiple molecular datasets and long-term neurodevelopmental assessment of the children.
Second, as the placenta is comprised of several heterogeneous cell types, cell type-specific molecular patterns in the placenta should be taken into consideration when interpreting these findings. We did consider deconvolution of our tissue samples using mRNA expression. Due to a dearth of placental cell-type-specific expression references, we opted for a reference-based deconvolution of four key cell types. These four cell types, however, do not fully represent the cellular compexity of the placenta. As these reference expression profiles become available, a more comprehensive analysis with reference-based deconvolution may reveal more cell-type-specific expression and methylation patterns that are specific to diverse populations of trophoblasts, stromal cells, endothelial cells, and pericytes. There are also considerations made about the degradation of RNA in the placental specimens over time. As the placental tissue was stored for ~ 15 years, we had to impose strict pre-filtering of genes whose expression have low counts and dispersions [42], resulting in a reduction of the analyzable transcriptome.
Lastly, to test the reproducibility and robustness of our kernel models, further out-of-sample validation is required, using datasets with larger sample sizes and similar molecular datasets. Though in-sample predictive performance is strong, platform differences between the ELGAN training set (assayed with the EPIC BeadChip) and the validation set (assayed with MethylC-seq) may lead to loss of predictive power. As our optimal models trained in ELGAN all aggregated the DNA methylation, miRNA, and mRNA datasets, the dearth of data for the placenta, in the context of social and intellectual impairment, makes out-of-sample validation of the full model especially challenging. In spite of these limitations, these data support the association between molecular features within the fetal placenta and social and cognitive outcomes in children that merits future investigation.
Conclusions
Our analysis underscores the importance of synthesizing data representing various levels of biological organization to understand distinct transriptomic and epigenomic underpinnings of complex developmental deficits, like intellectual and social impairment. This study provides novel evidence for the omnigenicity of the placenta-brain axis in the context of social and intellectual impairment.
Supplementary information
Additional file 1. Supplemental methods.
Additional file 2. Supplemental results.
Acknowledgements
We would like to thank the study participants of the ELGAN-ECHO study. We would also like to thank Michael Love and Lana Garmire for helpful discussion during the research process.
Abbreviations
- ASD
Autism Spectrum Disorder
- DAS-II
Differential Ability Scales-II
- ELGAN
Extremely Low Gestational Age Newborn
- EWAS
Epigenome-wide association study
- IQ
Intellectual ability
- KRLS
Kernel regression least squares
- MARBLES
Markers of Autism Risk in Babies-Learning Early Signs
- SRS
Social Responsiveness Scale
- SRS-T
SRS gender-normed T-score
Authors' contributions
HSP, RMJ, LS, KCKK, CJM, TMO, and RCF conceived and designed the study. HSP and AB analyzed the data. HSP, AB and RCF interpreted the data. HSP and AB drafted the work and all authors revised. All authors have approved the submitted version and are accountable for their own contributions. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the National Institutes of Health (NIH), specifically the National Institute of Neurological Disorders and Stroke (U01NS040069; R01NS040069), the Office of the NIH Director (UG3OD023348), the National Institute of Environmental Health Sciences (T32-ES007018), the National Institute of Nursing Research (K23NR017898; R01NR019245), and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD092374; R03HD101413 ).
Availability of data and materials
mRNA and miRNA expression data from the ELGAN study is available from the NCBI Gene Expression Omnibus GSE154829. CpG methylation data is freely available and can be requested from H.S.P or R.C.F. For validation, we used MethylC-seq data from the MARBLES study available at GSE67615. Single cell RNA-seq data used for reference-based deconvolution is available at GSE89497. Supplemental code and results are provided on Github: https://github.com/bhattacharya-a-bt/multiomics_ELGAN. Any questions about data availability can be directed to H.P.S.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of the University of North Carolina at Chapel Hill. All participants consented to the study as per IRB protocol.
Consent for publication
Not applicable.
Competing interests
The authors have no competing financial interests to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hudson P. Santos and Arjun Bhattacharya have co-first authorship
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13229-020-00402-w.
References
- 1.Hodyl NA, Aboustate N, Bianco-Miotto T, Roberts CT, Clifton VL, Stark MJ. Child neurodevelopmental outcomes following preterm and term birth: what can the placenta tell us? Placenta. 2017;57:79–86. doi: 10.1016/j.placenta.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Hu WF, Chahrour MH, Walsh CA. The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal S, Rao SC, Bulsara MK, Patole SK. Prevalence of autism spectrum disorder in preterm infants: a meta-analysis. Pediatrics. 2018;142:e20180134. doi: 10.1542/peds.2018-0134. [DOI] [PubMed] [Google Scholar]
- 4.Xie S, Heuvelman H, Magnusson C, Rai D, Lyall K, Newschaffer CJ, et al. Prevalence of autism spectrum disorders with and without intellectual disability by gestational age at birth in the Stockholm Youth Cohort: a register linkage study. Paediatr Perinat Epidemiol. 2017;31:586–594. doi: 10.1111/ppe.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korzeniewski SJ, Joseph RM, Kim SH, Allred EN, O’shea TM, Leviton A, et al. Social responsiveness scale assessment of the preterm behavioral phenotype in 10-year-olds born extremely preterm. J Dev Behav Pediatr. 2017;38:697–705. doi: 10.1097/DBP.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenfeld CS. The placenta-brain-axis. J Neurosci Res. 2020 doi: 10.1002/jnr.24603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shallie PD, Naicker T. The placenta as a window to the brain: a review on the role of placental markers in prenatal programming of neurodevelopment. Int J Dev Neurosci. 2019;73:41–49. doi: 10.1016/j.ijdevneu.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld CS. Placental serotonin signaling, pregnancy outcomes, and regulation of fetal brain development. Biol Reprod. 2020;102:532–538. doi: 10.1093/biolre/ioz204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meakin CJ, Martin EM, Santos HP, Mokrova I, Kuban K, O’Shea TM, et al. Placental CpG methylation of HPA-axis genes is associated with cognitive impairment at age 10 among children born extremely preterm. Horm Behav. 2018;101:29–35. doi: 10.1016/j.yhbeh.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paquette AG, Houseman EA, Green BB, Lesseur C, Armstrong DA, Lester B, et al. Regions of variable DNA methylation in human placenta associated with newborn neurobehavior. Epigenetics. 2016;11:603–613. doi: 10.1080/15592294.2016.1195534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroeder DI, Schmidt RJ, Crary-Dooley FK, Walker CK, Ozonoff S, Tancredi DJ, et al. Placental methylome analysis from a prospective autism study. Mol Autism. 2016;7:51. doi: 10.1186/s13229-016-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G, et al. Psychiatric genomics: an update and an agenda. Am J Psychiatry. 2018;175:15. doi: 10.1176/appi.ajp.2017.17030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Li YI, Pritchard JK. Trans effects on gene expression can drive omnigenic inheritance. Cell. 2019;177:1022–1034. doi: 10.1016/j.cell.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;14:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Shea TM, Allred EN, Dammann O, Hirtz D, Kuban KCK, Paneth N, et al. The ELGAN study of the brain and related disorders in extremely low gestational age newborns. Early Hum Dev. 2009;85:719–725. doi: 10.1016/j.earlhumdev.2009.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torske T, Nærland T, Bettella F, Bjella T, Malt E, Høyland AL, et al. Autism spectrum disorder polygenic scores are associated with every day executive function in children admitted for clinical assessment. Autism Res. 2020;13:207–220. doi: 10.1002/aur.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph RM, O’Shea TM, Allred EN, Heeren T, Hirtz D, Paneth N, et al. Prevalence and associated features of autism spectrum disorder in extremely low gestational age newborns at age 10 years. Autism Res. 2017;10:224–232. doi: 10.1002/aur.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joseph RM, O’Shea TM, Allred EN, Heeren T, Hirtz D, Jara H, et al. Neurocognitive and academic outcomes at age 10 years of extremely preterm newborns. Pediatrics. 2016;137:e20154343. doi: 10.1542/peds.2015-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott CD. Differential ability scales. 2. San Antonio: Harcourt Assessment; 2007. [Google Scholar]
- 22.Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, et al. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord. 2003;33:427–433. doi: 10.1023/A:1025014929212. [DOI] [PubMed] [Google Scholar]
- 23.Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P. Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry. 2010;167:1349–1356. doi: 10.1176/appi.ajp.2010.09101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutter M, Bailey A, Lord C. The social communication questionnare. Los Angeles: Western Psychological Services; 2003. [Google Scholar]
- 25.Risi S, Lord C, Gotham K, Corsello C, Chrysler C, Szatmari P, et al. Combining information from multiple sources in the diagnosis of autism spectrum disorders. J Am Acad Child Adolesc Psychiatry. 2006;45:1094–1103. doi: 10.1097/01.chi.0000227880.42780.0e. [DOI] [PubMed] [Google Scholar]
- 26.Lord C, Rutter M, DiLavore P, Risi S, Gotham K, Bishop S. Autism diagnostic observation schedule. Los Angeles: Western Psychological Services; 2012. [Google Scholar]
- 27.Addo KA, Bulka C, Dhingra R, Santos HP, Jr, Smeester L, et al. Acetaminophen use during pregnancy and DNA methylation in the placenta of the extremely low gestational age newborn (ELGAN) cohort. Environ Epigenetics. 2019;5:dvz010. doi: 10.1093/eep/dvz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos HP, Bhattacharya A, Martin EM, Addo K, Psioda M, Smeester L, et al. Epigenome-wide DNA methylation in placentas from preterm infants: association with maternal socioeconomic status. Epigenetics. 2019;14:751–765. doi: 10.1080/15592294.2019.1614743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eaves L, Phookphan P, Rager J, Bangma J, Santos HP, Smeester L, et al. A role for microRNAs in the epigenetic control of sexually dimorphic gene expression in the human placenta. Epigenomics. 2020;12:1543–1558. doi: 10.2217/epi-2020-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fajardy I, Moitrot E, Vambergue A, Vandersippe-Millot M, Deruelle P, Rousseaux J. Time course analysis of RNA stability in human placenta. BMC Mol Biol. 2009;10:21. doi: 10.1186/1471-2199-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulka CM, Dammann O, Santos HP, VanderVeen DK, Smeester L, Fichorova R, et al. Placental CpG methylation of inflammation, angiogenic, and neurotrophic genes and retinopathy of prematurity. Investig Ophthalmol Vis Sci. 2019;60:2888–2894. doi: 10.1167/iovs.18-26466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark J, Martin E, Bulka CM, Smeester L, Santos HP, O’Shea TM, et al. Associations between placental CpG methylation of metastable epialleles and childhood body mass index across ages one, two and ten in the Extremely Low Gestational Age Newborns (ELGAN) cohort. Epigenetics. 2019;14:1102–1111. doi: 10.1080/15592294.2019.1633865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fortin J-P, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 36.Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fortin JP, Triche TJ, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–560. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:e161. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Z, Wang L, Desai K, Cogswell J, Stern M, Lawson B, et al. Reliable gene expression profiling from small and hematoxylin and eosin–stained clinical formalin-fixed, paraffin-embedded specimens using the HTG EdgeSeq Platform. J Mol Diagnostics. 2019;21:796–807. doi: 10.1016/j.jmoldx.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gagnon-Bartsch JA, Speed TP. Using control genes to correct for unwanted variation in microarray data. Biostatistics. 2012;13:539–552. doi: 10.1093/biostatistics/kxr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat. 2016;10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 49.Zhao S, Li CI, Guo Y, Sheng Q, Shyr Y. RnaSeqSampleSize: real data based sample size estimation for RNA sequencing. BMC Bioinformatics. 2018;19:191. doi: 10.1186/s12859-018-2191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. Guidance for DNA methylation studies: Statistical insights from the Illumina EPIC array. BMC Genomics. 2019;20:1–15. doi: 10.1186/s12864-019-5761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Fan X, Wang R, Lu X, Dang YL, Wang H, et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018;28:819–832. doi: 10.1038/s41422-018-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.André G, Westra H-J, Arends D, Esko T, Peters MJ, Schurmann C, et al. Cell specific eQTL analysis without sorting cells. PLoS Genet. 2015;11:e1005223. doi: 10.1371/journal.pgen.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morota G, Gianola D. Kernel-based whole-genome prediction of complex traits: a review. Front Genet. 2014;5:363. doi: 10.3389/fgene.2014.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu B, Song N, Shen R, Arora A, Machiela MJ, Song L, et al. Integrating clinical and multiple omics data for prognostic assessment across human cancers. Sci Rep. 2017;7:16954. doi: 10.1038/s41598-017-17031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Endelman JB. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome. 2011;4:250–255. doi: 10.3835/plantgenome2011.08.0024. [DOI] [Google Scholar]
- 57.Zhu W, Hu B, Becker C, Doğan ES, Berendzen KW, Weigel D, et al. Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol. 2017;18:1–16. doi: 10.1186/s13059-017-1281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng Y, Amador C, Xia C, Marioni R, Sproul D, Walker RM, et al. Parent of origin genetic effects on methylation in humans are common and influence complex trait variation. Nat Commun. 2019;10:1–13. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hainmueller J, Hazlett C. Kernel Regularized least squares: reducing misspecification bias with a flexible and interpretable machine learning approach. Polit Anal. 2014;22:143–168. doi: 10.1093/pan/mpt019. [DOI] [Google Scholar]
- 60.Xu QS, Liang YZ. Monte Carlo cross validation. Chemom Intell Lab Syst. 2001;56:1–11. doi: 10.1016/S0169-7439(00)00122-2. [DOI] [Google Scholar]
- 61.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9:432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Epskamp S, Cramer AOJ, Waldorp LJ, Schmittmann VD, Borsboom D. Qgraph: network visualizations of relationships in psychometric data. J Stat Softw. 2012;48:1–18. doi: 10.18637/jss.v048.i04. [DOI] [Google Scholar]
- 63.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:199–205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang X, Jia L, Qian Z, Jia Y, Chen X, Xu X, et al. Diversity in human placental microvascular endothelial cells and macrovascular endothelial cells. Cytokine. 2018;111:287–294. doi: 10.1016/j.cyto.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Houde AA, Ruchat SM, Allard C, Baillargeon JP, St-Pierre J, Perron P, et al. LRP1B, BRD2 and CACNA1D: New candidate genes in fetal metabolic programming of newborns exposed to maternal hyperglycemia. Epigenomics. 2015;7:1111–1122. doi: 10.2217/epi.15.72. [DOI] [PubMed] [Google Scholar]
- 66.Wan JP, Wang H, Li CZ, Zhao H, You L, Shi DH, et al. The common single-nucleotide polymorphism rs2681472 is associated with early-onset preeclampsia in Northern Han Chinese women. Reprod Sci. 2014;21:1423–1427. doi: 10.1177/1933719114527354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun X-M, Yang M, Jiang C-X. Association of ATP2B1 gene polymorphism with incidence of eclampsia. Eur Rev Med Pharmacol Sci. 2019;23:10609–10616. doi: 10.26355/eurrev_201912_19756. [DOI] [PubMed] [Google Scholar]
- 68.Chater-Diehl E, Ejaz R, Cytrynbaum C, Siu MT, Turinsky A, Choufani S, et al. New insights into DNA methylation signatures: SMARCA2 variants in Nicolaides–Baraitser syndrome. BMC Med Genomics. 2019;12:105. doi: 10.1186/s12920-019-0555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang S, Hughes E, Lascelles K, Simpson MA, Pal DK, Marini C, et al. New SMARCA2 mutation in a patient with Nicolaides–Baraitser syndrome and myoclonic astatic epilepsy. Am J Med Genet Part A. 2017;173:195–199. doi: 10.1002/ajmg.a.37935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K, et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet. 2009;18:2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- 71.Shamseldin HE, Rajab A, Alhashem A, Shaheen R, Al-Shidi T, Alamro R, et al. Mutations in DDX59 implicate RNA helicase in the pathogenesis of orofaciodigital syndrome. Am J Hum Genet. 2013;93:555–560. doi: 10.1016/j.ajhg.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paine I, Posey JE, Grochowski CM, Jhangiani SN, Rosenheck S, Kleyner R, et al. Paralog studies augment gene discovery: DDX and DHX genes. Am J Hum Genet. 2019;105:302–316. doi: 10.1016/j.ajhg.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salpietro V, Efthymiou S, Manole A, Maurya B, Wiethoff S, Ashokkumar B, et al. A loss-of-function homozygous mutation in DDX59 implicates a conserved DEAD-box RNA helicase in nervous system development and function. Hum Mutat. 2018;39:187–192. doi: 10.1002/humu.23368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Løset M, Mundal SB, Johnson MP, Fenstad MH, Freed KA, Lian IA, et al. A transcriptional profile of the decidua in preeclampsia. Am J Obstet Gynecol. 2011;204:84.e1–83.e27. doi: 10.1016/j.ajog.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paidas MJ, Krikun G, Huang SJ, Jones R, Romano M, Annunziato J, et al. A genomic and proteomic investigation of the impact of preimplantation factor on human decidual cells. Am J Obstet Gynecol. 2010;202:459.e1–459.e8. doi: 10.1016/j.ajog.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies J, Pollheimer J, Yong HEJ, Kokkinos MI, Kalionis B, Knöfler M, et al. Epithelial-mesenchymal transition during extravillous trophoblast differentiation. Cell Adhes Migr. 2016;10:310–321. doi: 10.1080/19336918.2016.1170258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Asprer JST, Lee B, Wu CS, Vadakkan T, Dickinson ME, Lu HC, et al. LMO4 functions as a co-activator of neurogenin 2 in the developing cortex. Development. 2011;138:2823–2832. doi: 10.1242/dev.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang L, Qin Z, Ricke KM, Cruz SA, Stewart AFR, Chen HH. Hyperactivated PTP1B phosphatase in parvalbumin neurons alters anterior cingulate inhibitory circuits and induces autism-like behaviors. Nat Commun. 2020;11:1017. doi: 10.1038/s41467-020-14813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maiya R, Kharazia V, Lasek AW, Heberlein U. Lmo4 in the basolateral complex of the amygdala modulates fear learning. PLoS ONE. 2012;7:34559. doi: 10.1371/journal.pone.0034559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maiya R, Mangieri RA, Morrisett RA, Heberlein U, Messing RO. A selective role for Lmo4 in cue–reward learning. J Neurosci. 2015;35:9638–9647. doi: 10.1523/JNEUROSCI.1740-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Z, Lam KS, Wang N, Xu X, Cortes M, Andersen B. LMO4 can interact with Smad proteins and modulate transforming growth factor-β signaling in epithelial cells. Oncogene. 2006;25:2920–2930. doi: 10.1038/sj.onc.1209318. [DOI] [PubMed] [Google Scholar]
- 82.Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, et al. FOXP2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7:1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vandré DD, Ackerman WE, IV, Tewari A, Kniss DA, Robinson JM. A placental sub-proteome: the apical plasma membrane of the syncytiotrophoblast. Placenta. 2012;33:207–213. doi: 10.1016/j.placenta.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, et al. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–456. doi: 10.1016/S0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 85.Mignogna ML, D’Adamo P. Critical importance of RAB proteins for synaptic function. Small GTPases. 2018;9:145–157. doi: 10.1080/21541248.2016.1277001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Binotti B, Jahn R, Chua J. Functions of Rab proteins at presynaptic sites. Cells. 2016;5:7. doi: 10.3390/cells5010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moya-Alvarado G, Gonzalez A, Stuardo N, Bronfman FC. Brain-derived neurotrophic factor (BDNF) regulates Rab5-positive early endosomes in hippocampal neurons to induce dendritic branching. Front Cell Neurosci. 2018;12:493. doi: 10.3389/fncel.2018.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek GA, Samocha KE, Cicek EA, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arimoto KI, Löchte S, Stoner SA, Burkart C, Zhang Y, Miyauchi S, et al. STAT2 is an essential adaptor in USP18-mediated suppression of type i interferon signaling. Nat Struct Mol Biol. 2017;24:279–289. doi: 10.1038/nsmb.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Racicot K, Aldo P, El-Guindy A, Kwon J-Y, Romero R, Mor G. Cutting edge: fetal/placental type I IFN can affect maternal survival and fetal viral load during viral infection. J Immunol. 2017;198:3029–3032. doi: 10.4049/jimmunol.1601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nazeen S, Palmer NP, Berger B, Kohane IS. Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities. Genome Biol. 2016;17:228. doi: 10.1186/s13059-016-1084-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsetsos F, Padmanabhuni SS, Alexander J, Karagiannidis I, Tsifintaris M, Topaloudi A, et al. Meta-analysis of tourette syndrome and attention deficit hyperactivity disorder provides support for a shared genetic basis. Front Neurosci. 2016;10:340. doi: 10.3389/fnins.2016.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richetto J, Massart R, Weber-Stadlbauer U, Szyf M, Riva MA, Meyer U. Genome-wide DNA methylation changes in a mouse model of infection-mediated neurodevelopmental disorders. Biol Psychiatry. 2017;81:265–276. doi: 10.1016/j.biopsych.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 94.Bronson SL, Bale TL. The placenta as a mediator of stress effects on neurodevelopmental reprogramming. Neuropsychopharmacology. 2016;41:207–218. doi: 10.1038/npp.2015.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gormley M, Ona K, Kapidzic M, Garrido-Gomez T, Zdravkovic T, Fisher SJ. Preeclampsia: novel insights from global RNA profiling of trophoblast subpopulations. Am J Obstet Gynecol. 2017;217:200.e1–200.e17. doi: 10.1016/j.ajog.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 96.Nevalainen J, Skarp S, Savolainen ER, Ryynänen M, Järvenpää J. Intrauterine growth restriction and placental gene expression in severe preeclampsia, comparing early-onset and late-onset forms. J Perinat Med. 2017;45:869–877. doi: 10.1515/jpm-2016-0406. [DOI] [PubMed] [Google Scholar]
- 97.Sterling N, Duncan AR, Park R, Koolen DA, Shi J, Cho S-H, et al. De novo variants in MPP5 cause global developmental delay and behavioral changes. Hum Mol Genet. 2020; ddaa224. [DOI] [PMC free article] [PubMed]
- 98.Park JY, Hughes LJ, Moon UY, Park R, Kim SB, Tran K, et al. The apical complex protein Pals1 is required to maintain cerebellar progenitor cells in a proliferative state. Development. 2016;143:133–146. doi: 10.1242/dev.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Redman CWG, Staff AC. Preeclampsia, biomarkers, syncytiotrophoblast stress, and placental capacity. Am J Obstet Gynecol. 2015;213:S9.e1–S9.e4. doi: 10.1016/j.ajog.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 100.Gumusoglu SB, Chilukuri ASS, Santillan DA, Santillan MK, Stevens HE. Neurodevelopmental outcomes of prenatal preeclampsia exposure. Trends Neurosci. 2020;43:253–268. doi: 10.1016/j.tins.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lodygensky GA, Seghier ML, Warfield SK, Tolsa CB, Sizonenko S, Lazeyras F, et al. Intrauterine growth restriction affects the preterm infant’s hippocampus. Pediatr Res. 2008;63:438–443. doi: 10.1203/PDR.0b013e318165c005. [DOI] [PubMed] [Google Scholar]
- 102.Nardulli G, Proverbio F, Limongi FG, Marín R, Proverbio T. Preeclampsia and calcium adenosine triphosphatase activity of red blood cell ghosts. Am J Obstet Gynecol. 1994;171:1361–1365. doi: 10.1016/0002-9378(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 103.Casart Y, Proverbio T, Marín R, Proverbio F. Comparative study of the calcium adenosine triphosphatase of basal membranes of human placental trophoblasts from normotensive and preeclamptic pregnant women. Gynecol Obstet Invest. 2001;51:28–31. doi: 10.1159/000052886. [DOI] [PubMed] [Google Scholar]
- 104.Carrera F, Casart YC, Proverbio T, Proverbio F, Marín R. Preeclampsia and calcium-ATPase activity of plasma membranes from human myometrium and placental trophoblast. Hypertens Pregnancy. 2003;22:295–304. doi: 10.1081/PRG-120024033. [DOI] [PubMed] [Google Scholar]
- 105.Carreiras MM, Proverbio T, Proverbio F, Marín R. Preeclampsia and Calcium-ATPase activity of red cell ghosts from neonatal and maternal blood. Hypertens Pregnancy Hypertens Pregnancy. 2002;21:97–107. doi: 10.1081/PRG-120004027. [DOI] [PubMed] [Google Scholar]
- 106.Anastasiadi D, Esteve-Codina A, Piferrer F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin. 2018;11:37. doi: 10.1186/s13072-018-0205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–29. [DOI] [PMC free article] [PubMed]
- 108.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lea AJ, Vockley CM, Johnston RA, Del Carpio CA, Barreiro LB, Reddy TE, et al. Genome-wide quantification of the effects of DNA methylation on human gene regulation. Elife. 2018;7:e37513. doi: 10.7554/eLife.37513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang W, Voloudakis G, Rajagopal VM, Readhead B, Dudley JT, Schadt EE, et al. Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits. Nat Commun. 2019;10:3834. doi: 10.1038/s41467-019-11874-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bhattacharya A, Li Y, Love MI. MOSTWAS: Multi-omic strategies for transcriptome-wide association studies. bioRxiv. 2020; 10.1101/2020.04.17.047225. [DOI] [PMC free article] [PubMed]
- 114.Paternoster L, Tilling K, Davey SG. Genetic epidemiology and Mendelian randomization for informing disease therapeutics: Conceptual and methodological challenges. PLOS Genet. 2017;13:e1006944. doi: 10.1371/journal.pgen.1006944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bhattacharya A, García-Closas M, Olshan AF, Perou CM, Troester MA, Love MI. A framework for transcriptome-wide association studies in breast cancer in diverse study populations. Genome Biol. 2020;21:42. doi: 10.1186/s13059-020-1942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplemental methods.
Additional file 2. Supplemental results.
Data Availability Statement
mRNA and miRNA expression data from the ELGAN study is available from the NCBI Gene Expression Omnibus GSE154829. CpG methylation data is freely available and can be requested from H.S.P or R.C.F. For validation, we used MethylC-seq data from the MARBLES study available at GSE67615. Single cell RNA-seq data used for reference-based deconvolution is available at GSE89497. Supplemental code and results are provided on Github: https://github.com/bhattacharya-a-bt/multiomics_ELGAN. Any questions about data availability can be directed to H.P.S.