Abstract
Background
Cardiac fibroblasts (CF) have a central role in the ventricular remodeling process associated with different types of fibrosis. Recent studies have shown that fibroblasts do not respond homogeneously to heart injury. Due to the limited set of bona fide fibroblast markers, a proper characterization of fibroblast population heterogeneity in response to cardiac damage is still missing. The purpose of this study was to define the CF heterogeneity during ventricular remodeling and the underlying mechanisms that regulate their function.
Methods
Collagen1α1-GFP+ CF were characterized after myocardial infarction (MI) by single-cell and bulk RNA-seq, ATAC-seq and functional assays. Swine and patient samples were studied using bulk RNA-seq.
Results
We identified and characterized a unique CF subpopulation that emerges after MI in mice. These activated fibroblasts exhibit a clear pro-fibrotic signature, express high levels of Collagen Triple Helix Repeat Containing 1 (Cthrc1) and localize into the scar. Non-canonical TGF-β signaling and different transcription factors including SOX9 are important regulators mediating their response to cardiac injury. Moreover, the absence of CTHRC1 results in pronounced lethality due to ventricular rupture. Finally, a population of CF with a similar transcriptome was identified in a swine model of MI and in heart tissue from patients with MI and dilated cardiomyopathy.
Conclusions
We report CF heterogeneity, their dynamics during the course of MI and redefine the CF that respond to cardiac injury and participate in myocardial remodeling. Our study identifies Cthrc1 as a novel regulator of the healing scar process, and as a target for future translational studies.
Keywords: Myocardial infarction, cardiac fibroblasts, TGF-β1, Single-cell RNA-seq, CTHRC1
INTRODUCTION
Cardiac fibroblasts (CF) represent only 10% of the total number of cells in the myocardium; however, they play a critical role in the structural and mechanical maintenance of the heart 1, 2. After myocardial infarction (MI), CF become activated, orchestrating a fibrotic response that leads to the generation of a collagen scar, which prevents cardiac rupture 3, 4. The absence of reliable markers has traditionally hampered our understanding of the specific role of CF in cardiac homeostasis. However, several studies using lineage-tracing reporter strains have allowed to track the origin and roles of fibroblasts showing that the fibroblast response to cardiac injury is rather heterogeneous (reviewed in 3). Importantly, these studies revealed that different CF subtypes may play different roles during the healing process that follows MI 5–7. In this context, Fu et al. have recently described a new subpopulation of activated CF, the matrifibrocytes, which support the mature scar and are characterized by the expression of extracellular matrix (ECM) and tendon genes 8.
This highlights the need for a better understanding of CF heterogeneity and its impact on processes that mediate repair of the ischemic myocardium. The development of single-cell RNA-sequencing (scRNA-seq) represents an ideal tool to address this knowledge gap. Two recent publications have revealed the presence of three subpopulations of CF in pathological conditions at single-cell resolution using a SORT-seq protocol 7, 9. In accordance with previous studies, both manuscripts show that activated CF are characterized by the expression of Postn. In contrast, Kretzschmar et al. described a total of eleven subpopulations of CF after pooling samples from neonatal, adult and pathological hearts 10. However, all these studies have a poor representation of CF because of the low number of collected cells (63 CF/185 cardiac cells in 7, 935 cardiac cells in 9, and 243 CF in 10). In contrast, Skelly et al. performed an unbiased analysis of 10,519 cardiac cells distinguishing four subpopulations of CF but only during homeostasis 11. More recently, Farbehi et al. described eight CF subpopulations within a total of eleven that emerged from unbiased clustering of 16,787 Pdgfrα+ cardiac interstitial cells isolated from healthy, 3 and 7 days post-infarcted hearts 12. However, no scRNA-seq study to date has focused exclusively on CF, their anatomic location and their dynamics during the course of MI.
Here, we use scRNA-seq to identify a new subpopulation of CF that mediates ventricular remodeling associated with different cardiac fibrotic processes. This subpopulation is characterized by a specific transcriptomic signature that includes Collagen Triple Helix Repeat Containing 1 (Cthrc1), an essential molecule involved in the synthesis and deposition of the fibrotic scar 13, 14. Moreover, we describe and characterize the role of some putative regulatory transcription factors (TF) and validate the non-canonical TGF-β signaling pathway as mediator of this specific signature. Further, using a knockout model for genetic ablation of Cthrc1, we provide evidence for the essential role of this specific fibroblast population during cardiac healing. Finally, we provide evidence of the conservation of this CTHRC1+ CF subpopulation in a preclinical swine model of myocardial ischemia and in patients with different types of cardiac fibrosis.
METHODS
All data, analytic methods and study materials are available from the corresponding author upon request.
Animal models
All animal requisitions, housing, treatments and procedures were performed according to all state and institutional laws, guidelines and regulations. All studies were approved by the Ethics Committee for Animal Research at the University of Navarra and the Government of Navarra. Studies conducted at Maine Medical Center Research Institute were approved by its IACUC and were compliant with The Guide.
Human samples
The study protocol was approved by the Medical Ethics Committee and informed consent was obtained from all patients. For RNA studies, human heart samples were kindly provided by Professor S. Janssens. For histological analysis, human heart samples were kindly provided by the Maine Medical Center BioBank.
Statistical Analysis
In vitro scratch and morphometric assay experiments statistical significance was analyzed using a non-parametric one-way analysis of variance with a Kruskal-Wallis post-hoc test. For the Kaplan-Meier survival curve, log-rank Mantel-Cox test was used to determine statistical difference between the survival curves of the two groups of animals. Quantification of Collagen deposition statistical significance was analyzed by an unpaired t-test.
Statistical analysis to obtain the significance when comparing Z-score distributions was done with a two-sided Wilcoxon rank sum test. Differential expression analysis for normalized values was performed using negative binomial generalized linear models.
A Two-ways ANOVA was used to determine statistical significance between the relative qPCR values in different anatomical regions for POSTN, COL1α1 and CTHRC1 in pig samples. For the comparison of the echocardiography, resonance and infarct size between reduced and preserved pigs a two-tailed t-test with a Mann-Whitney post-hoc test was used.
Data resources
All single-cell RNA-seq, bulk RNA-seq, and ATAC-seq data are available in a SuperSeries at NBCI’s Sequence Read Archive database under accession number GSE132146.
Detailed methods are provided in the Supplemental Materials, including Methods, Supplemental Tables I-VI, Supplemental Figures I-XVI, Supplementary Movie I and Supplementary File I.
RESULTS
Characterization of cardiac fibroblast heterogeneity
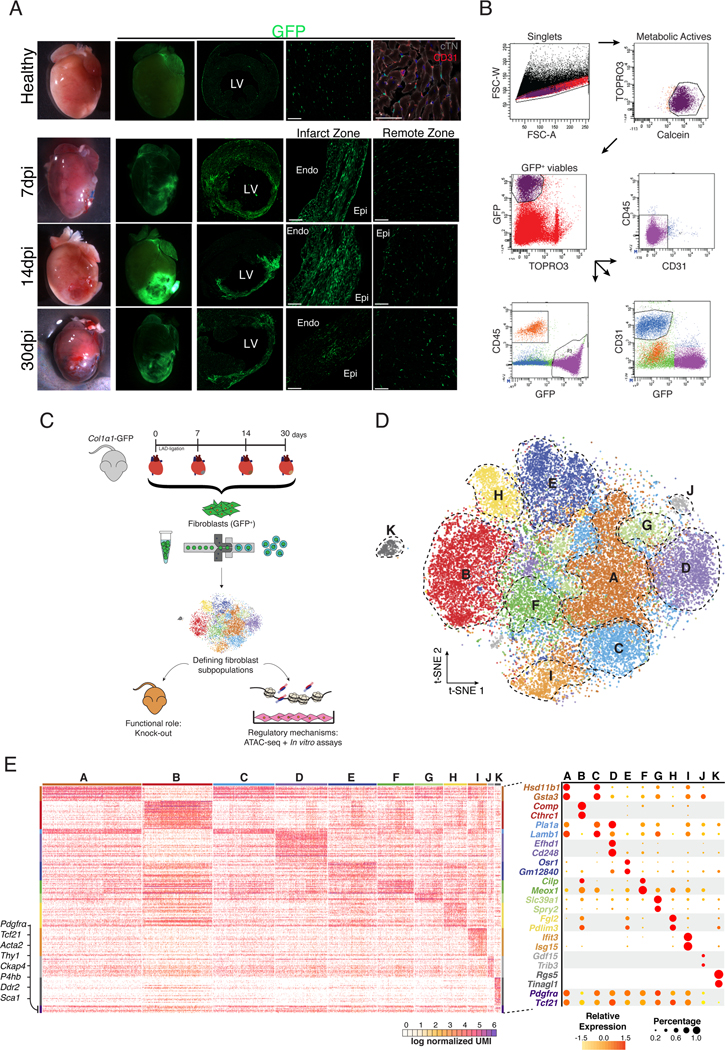
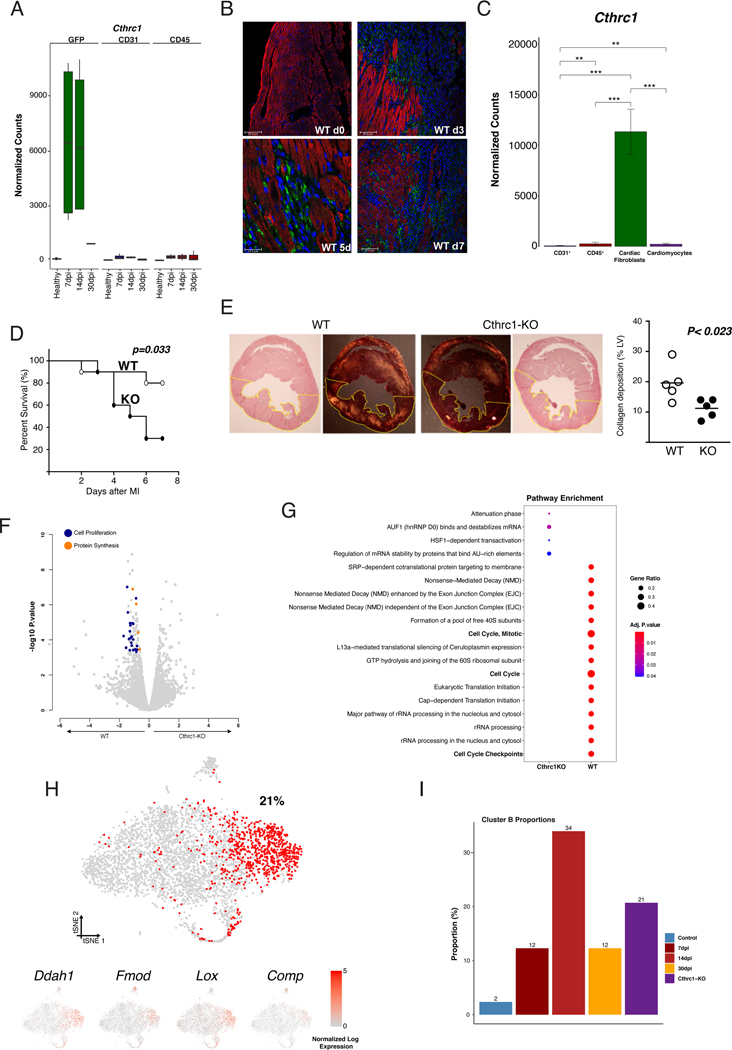
To follow the dynamics of the global CF population during MI, we used the Col1α1-GFP reporter mice. This strain has shown to homogeneously label fibroblasts from different origins 15 during homeostasis and in different cardiac fibrosis models 1, 6, 7, 16. Histological analysis of healthy and infarcted myocardium at different time-points showed changes in the amount and distribution pattern of GFP+ cells around the injured site (Figure 1A). In order to validate this model, we isolated putative CF (GFP+/CD31−/CD45−), endothelial (GFP−/CD31+/CD45−) and bone marrow-derived cells (GFP−/CD31−/CD45+) from healthy myocardium along with 7, 14, and 30 days post-infarction (dpi) (Figure 1B). No GFP+ cells were found in the CD31+ and CD45+ cell fractions at any time point by FACS or immunohistochemistry (Figure 1B, Figure IA in the Supplement). Additionally, no transcriptional markers for CF were found in these populations of cells. The transcriptional profiles of putative CF and tail/dermal fibroblasts (GFP+/CD31−/CD45−) were shown to be closely similar (Figure IB, IC and ID in the Supplement). An increased percentage and total number of GFP+ cells was observed in the infarct (IZ) and border zones (BZ) after MI (Figure IIB in the Supplement). Col1α1-GFP+/CD31−/CD45− cells expressed a set of surface membrane markers and a transcriptomic profile consistent with classical CF (Figure IIA and IIC in the Supplement) and mEFSK4+/CD31−/CD45− CF 1, 17 (Figure IID and IIE in the Supplement). Taken together, these results indicate that cardiac GFP+ cells are bona fide fibroblasts, validating the Col1α1-GFP model to study CF biology during cardiac repair.
Figure 1: CF are a heterogeneous population of cells.
(A) Overview of healthy hearts and infarcted ones at 7, 14 and 30 dpi using bright field and fluorescence microscopy. Overview and detail of transverse sections showing GFP+ cell distribution in the free wall of the LV in healthy and 7, 14 and 30 dpi in the infarct and remote zones. Detail of GFP+ cardiac interstitial cells. Cardiac Troponin-I+ (cTN) cardiomyocytes (grey), CD31+ endothelial cells (red), nuclei (DAPI, blue). (B) Representative gating for isolation of GFP+/CD31−/CD45− CIC as performed in healthy and at 7, 14 and 30 dpi hearts. From left to right: gating for singlets, metabolically active (Calcein+/TOPRO3−), viable cells (GFP+/TOPRO3−), and GFP+ cells that are negative for CD45 and CD31. (C) Schematic representation of the experimental design. Col1α1-GFP mice were subjected to MI and GFP+ CF were isolated at different time-points (healthy, 7, 14 and 30 dpi). These cells were used for scRNA-seq. The functional role of CF was analyzed with a mouse knock-out model and their regulatory mechanisms with ATAC-seq and in vitro assays to validate the hypothesis generated. (D) t-distributed stochastic neighbor embedding (t-SNE) plot of the global CF population (29,176 cells) comprising the four different times. Plot is color coded by clusters (A-K) identified through unsupervised analysis. Dashed lines delimit clusters. (E) Heatmap showing log normalized UMIs for scRNA-seq analyses cells. Top and side bars indicate clusters. Traditional marker genes for CF are indicated (left). Dot plot of expression and specificity of top markers (two) for identified clusters. Dot size represents percentage of cells per cluster expressing the given marker and color represents the relative expression. CIC = cardiac interstitial cells; Endo = endocardium; Epi = epicardium; LV = left ventricle. Scale bars: 100 μm (37 μm in top right picture in A).
To characterize the fibroblast response after MI at single-cell resolution, we profiled the single-cell transcriptomes of GFP+ CF (Figures 1C). Transcriptomes from each time point (7,079 cells in healthy; 10,448 cells at 7dpi; 8,337 cells at 14dpi; and 6,805 cells at 30dpi) were subjected to quality control filtering and merged into a single data set of 29,176 individual cells using a canonical correlation approach (Figure III in the Supplement). Unsupervised clustering of the global dataset revealed eleven clusters of GFP+ cells (Figure 1D). Ten of these clusters (A-J) represent different fibroblast subpopulations, characterized by high levels of fibroblast associated molecules, while cluster K comprises a population with high expression of classical pericyte markers 11 (Figure 1E, Figure IVA and IVB in the Supplement). Clusters B, D, I, and J showed specific expression profiles, representing potential functional CF subgroups, while the remaining clusters (A, C, E, F, G, and H) showed less specific transcriptomic identities, likely reflecting intermediate CF types within a larger population (Figure 1E, Figure IVC in the Supplement). We also compared our sorted GFP+ CF with CF identified in a recent study at scRNA-seq resolution (Figure V in the Supplement) and found that all GFP+ CF fall within previously identified CF 12.
Cluster B represents a subpopulation of activated periostin+ fibroblasts localized in the infarcted myocardium
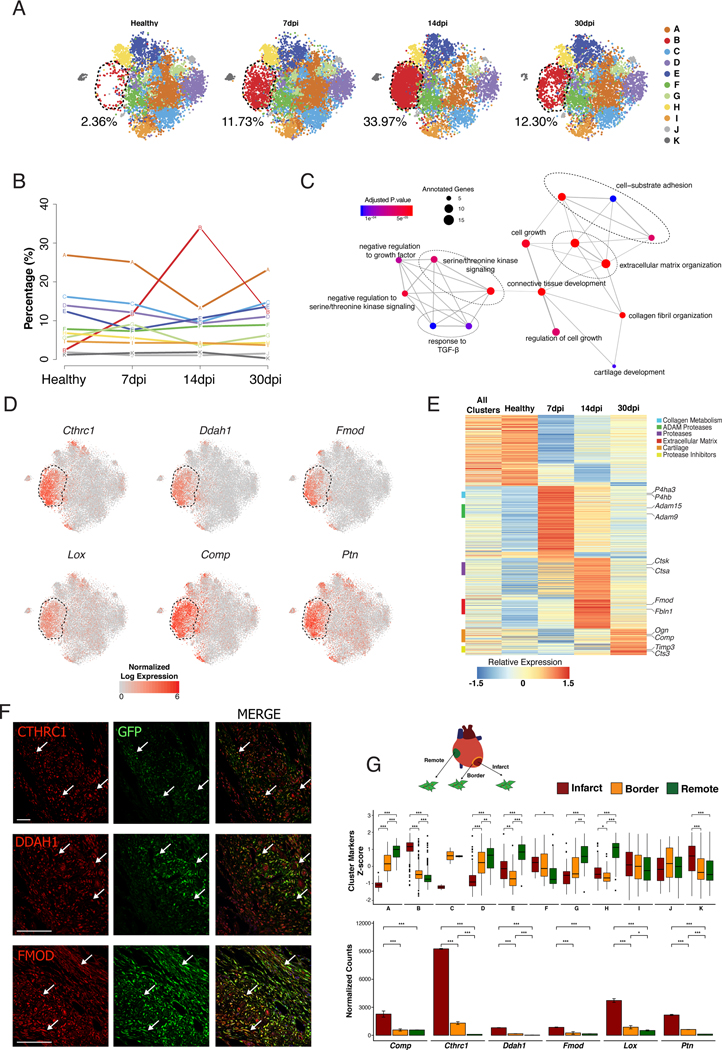
To explore the potential role of the different clusters during MI, we analyzed changes in the percentage of cells in each cluster. Clusters F and H-K remained constant at the different time-points (Figure VIA in the Supplement), while cluster B manifested significant changes after MI. Only 2.3% of the total GFP+ cells belonged to cluster B in healthy myocardium and the percentage rose sharply after MI (12% at 7dpi and 34% at 14dpi) followed by a decrease at 30dpi (12%) (Figure 2A and 2B, Figure VIA in the Supplement). Cluster B’s transcriptomic signature was significantly enriched in pathways and gene ontology (GO) terms related to ECM organization, cell proliferation and cell-substrate adhesion, in comparison with other clusters (Figure 2C, Figure IVD in the Supplement). This was reflected by high and specific expression of structural molecules, including Fmod and Comp, and by enzymes involved in collagen metabolism including Ddah1, Lox and Ptn 18, 19. Additionally, Collagen triple helix repeat containing 1 (Cthrc1), a gene linked to vascular remodeling and fibrotic processes 20–24, was identified as a top marker of cluster B (Figure 2D, Figure VIB in the Supplement). Changes in the pro-repair expression signature of cluster B fibroblasts were observed at 7, 14 and 30 dpi and were consistent with potential functions involved in specific phases of the repair process (Figure 2E, Figure VIC in the Supplement). The highly specific expression of ECM related genes in cluster B suggests a profound association of this population with the formation of the fibrotic scar.
Figure 2: Dynamics of CF heterogeneity reveals a unique subpopulation that responds to MI.
(A) t-SNE plot showing CF heterogeneity at different time-points. Number indicates percentage of cells in cluster B. (B) Representation showing the dynamics of the percentages of cells by cluster along infarction. Line color-coded by clusters (A-K). (C) Network representation of enriched pathways based on cluster B markers. Dot sizes represent number of cluster B markers annotated for each pathway and color scale statistical significance for each function. (D) t-SNE representation of normalized expression of Cthrc1, Ddah1, Lox, Comp, Fmod and Ptn comprising the four time-points. Dashed lines delimit cluster B. (E) Scaled gene expression heatmap showing transcriptional dynamics of cluster B along time-points. (F) Representative immunohistochemistries of Col1α1-GFP infarcted hearts at 14dpi showing the spatial location of CTHRC1, DDAH1, or FMOD (red) in the infarct zone. GFP+ (green), Nuclei (DAPI, blue). Co-localizations in yellow (arrows). Scale bars: 100 μm. (G) Experimental design: CF (GFP+/CD31−/CD45−) from remote, border and infarct zones were sorted for bulk RNA-seq (above). Boxplot showing the Z-score distribution of cluster markers in the three zones at 7dpi, Wilcoxon signed-rank test, n=2 (middle). Bar plot showing normalized expression values for top cluster B markers (below), linear model differential expression, n=2. Data, mean ± SD. Scale bars: 100 μm. * p≤0.05, ** p≤0.01, *** p≤0.001.
Immunohistochemistry analysis of three specific cluster B markers (Cthrc1, Ddah1, Fmod) revealed that these cells were almost exclusively located in the IZ and BZ at 7, 14 and 30 dpi (Figure 2F, Figure VII in the Supplement). In agreement, zonal RNA-seq profiling of the infarcted heart at 7dpi showed a clear enrichment in the cluster B signature in the infarct area compared to the remaining clusters (Figures 2G). Taken together, our results identify cluster B as a defined subpopulation of CF that (i) emerges in response to MI, (ii) presents a strong and dynamic pro-fibrotic signature and (iii) localizes to the damaged tissue.
To analyze the origin of cluster B, both RNA velocity and latent time analysis were performed. Using RNA velocity 25, we observed a dynamic movement among all clusters of CF in healthy heart (Figure VIII in the Supplement). At 7dpi, the previously identified transitions were clearly reduced, and a specific transition from cluster F to cluster B was observed. This transition was prominent at 14dpi, suggesting that cluster B cells were likely originating from cluster F. At 30dpi most of the identified transitions were reduced, including F to B, resembling the dynamics between subpopulations observed in healthy myocardium (Figure VIIIA in the Supplement). Using latent time analysis 26, cluster B cells grouped in regions with the highest latent time values. In addition, the expression of Cthrc1 was almost exclusive to cluster B, while other marker genes for quiescent (as Cd90/Thy1 or Pdgfrα/Cd140a) or activated CF (Periostin (Postn) or Acta2/αSMA) were expressed in earlier stages of activation (Figure VIIIB in the Supplement). This pattern suggests that cluster B fibroblasts corresponds to the final activation stage of a subset of Postn+ CF generated in response to MI. In this process, expression of Cthrc1 seems to be specific for the final stages of activation and associated with the healing process through scar formation.
To determine the differences between cluster B cells and other subpopulations of activated CF 4, 8, we compared the transcriptomic profile of CF that (co)express the specific markers Postn (activated CF), Acta2 (matrifibrocites), and/or Cthrc1 (cluster B fibroblasts). Depending on the combined expression of these three markers, different subsets of activated CF appeared (Figure IXA in the Supplement). Again, Cthrc1+ CF emerged as a subset of Postn+ CF, but with enrichment in GO terms related to ECM assembly and organization, as well as collagen fibril organization (Figure IXB in the Supplement). In agreement with these results and our scRNA-seq analysis, different subpopulations of activated CF were detected at 7dpi by immunohistochemistry (Figure IXC in the Supplement). In addition, a small number of CTHRC1+ CF were identified at 60dpi (Figure IXD in the Supplement).
Finally, to determine whether cluster B can be identified in other models of cardiac fibrosis, we examine the presence of CTHRC1+ CF in a model of chronic cardiac fibrosis generated by infusion of angiotensin-II for 28 days 5. As shown in Figure X in the Supplement CTHRC1+ CF were detected two days after the beginning of treatment with angiotensin-II (dpt), with an increase at 7dpt. At 14dpt, this population decreased and was no longer detected 28 days after infusion of angiotensin-II.
Collectively, these results suggest that cluster B cells represent a specific subpopulation of activated Postn+ CF with a major role in reparative cardiac fibrotic processes. Hence, we termed these cells Reparative Cardiac Fibroblasts (RCF).
Characterization of the molecular regulation of the RCF response
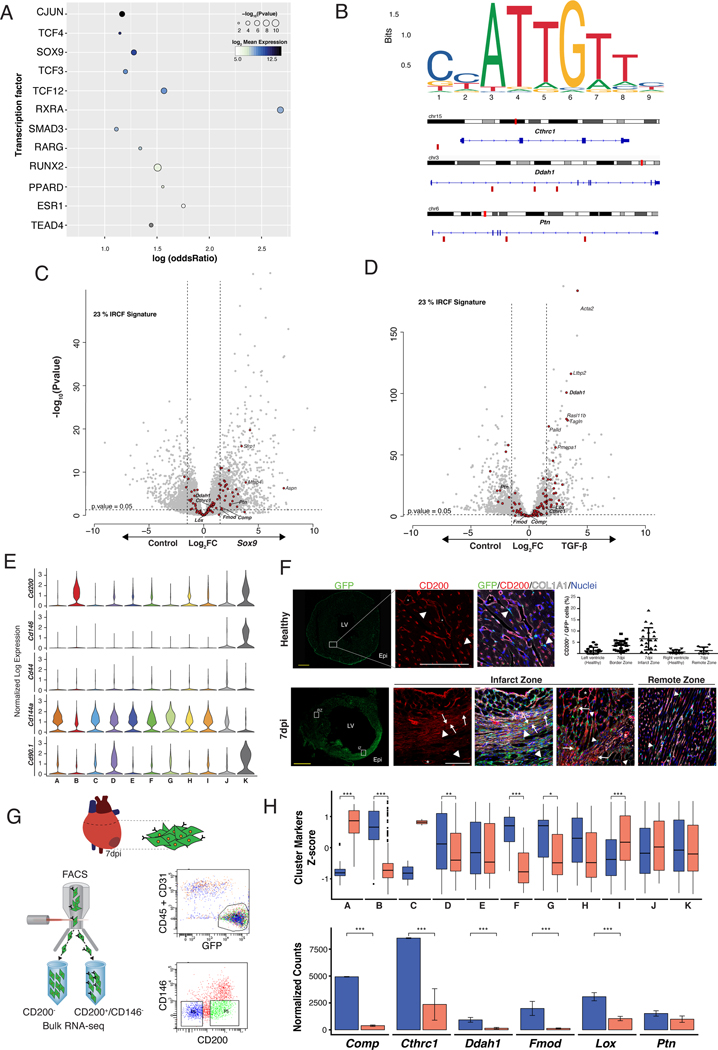
In order to identify the molecular regulation underpinning the generation of RCF, we undertook different approaches. First, we leveraged publicly available ChIP-seq datasets to identify TF whose binding patterns are enriched in the vicinity of RCF genes. This approach identified several TF, such as SOX9 and SMAD3 (Figure 3A). Some of the top marker genes of RCF showed binding motifs for SOX9 (Figure 3B). In addition, Sox9 overexpression in cultured CF induced 23% of the RCF signature (28 genes, FC>1.5 p-value <0.05) (Figure 3C), similar to the response observed after incubation of CF with TGF-β (33 genes, FC>1.5 p-value<0.05), a classical regulator of CF activation 27, 28 (Figure 3D). These results suggested the potential of SOX9 to promote an RCF phenotype and are consistent with previous studies describing CF activation programs 28, 29. However, Sox9 overexpression only partially explained the transcriptional signature of RCF.
Figure 3: CD200+/CD146− CF provides the most specific characterization of RCF.
(A) TF target gene enrichments. Dot size represents enrichment p-value of TF and color represents log2 transformed expression in CD200+ bulk RNA-seq. (B) SOX9 DNA binding motif sequence logo and its location in some of the top markers for cluster B. Small red rectangles below the gene sequence show confirmed SOX9 binding sites, as determined by JASPAR. (C and D) Volcano plots showing differential gene expression of in vitro grown CF overexpressing Sox9 (left) or treated with TGF-β (right). Genes with Log Fold Change of ±1.5 and p-value <0.05 were considered differentially expressed. Red dots represent RCF markers. (E) Violin plots showing single-cell normalized expression of selected surface markers in the pooled CF population (healthy, 7, 14 and 30 dpi). (F) Transverse sections of Col1α1-GFP hearts at 7dpi. Scale bars: 1 mm. Immunofluorescence analysis of GFP+ (green), CD200+ (red), COL1α1 (grey) and DAPI/nuclei (blue) in healthy LV, and IZ (left) and RZ (right) at 7dpi. Co-localization of GFP+ and CD200+ in yellow (arrows) in IZ (and co-localize with COL1α1 in light yellow). Arrowheads indicate GFP+/CD200− cells and asterisks indicate GFP−/CD200+. Scale bars: 100 μm. Quantification of GFP+/CD200+ cells in healthy and at 7dpi (right, top). (G) Gating strategy for isolation of GFP+/CD31−/CD45−/CD200+/CD146− (CD200+) and GFP+/CD31−/CD45−/CD200−/CD146− (CD200−) CIC at 7dpi. (H) Boxplot representation of Z-score distributions for cluster markers in CD200+ and CD200− CF (above), Wilcoxon signed-rank test, n=2. Normalized expression bar plots (mean ± SD) for top RCF markers in both subpopulations (below), linear model differential expression, n=2. Data, mean ± SD. * p≤0.05, ** p≤0.01, *** p≤0.001. IZ = infarct zone; RZ = remote zone. Other of abbreviations as in Figure 1.
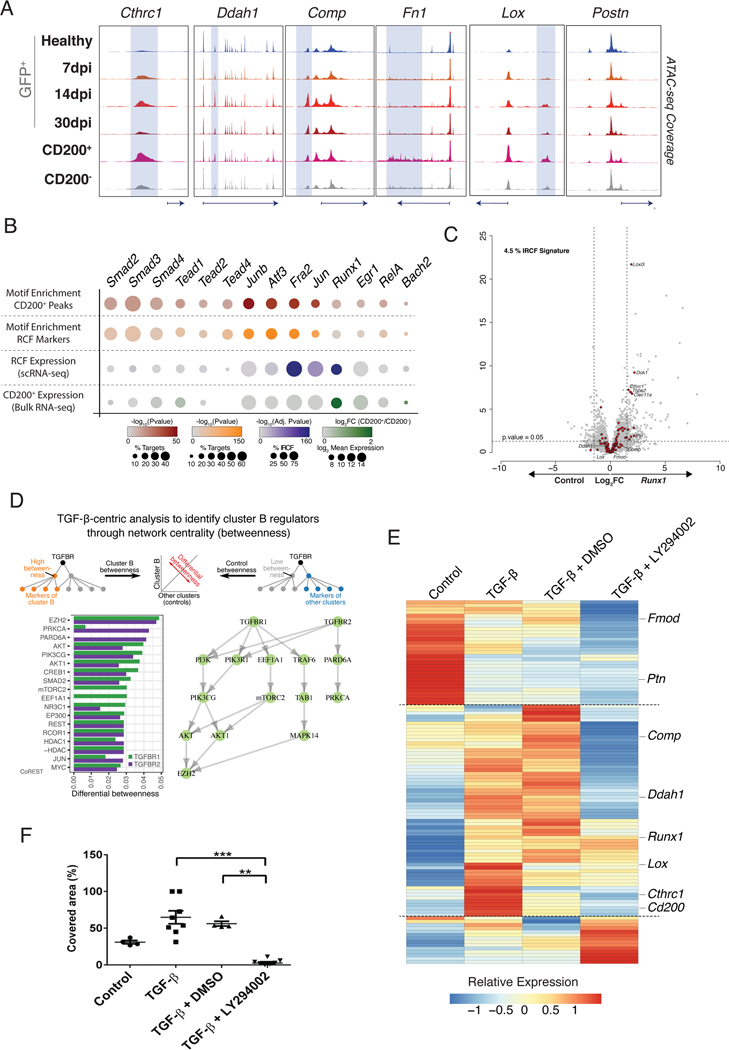
To further identify TF involved in RCF generation, we aimed to profile chromatin accessibility (ATAC-seq) at 7dpi. We examined the expression patterns of membrane surface markers in our single-cell dataset to identify a specific combination that would allow the isolation of RCF. None of the classical markers of fibroblast such as Thy1 (Cd90), Pdgfrα (Cd140a) or Pgp1 (Cd44) 17, 30 were specific for RCF (Figure 3E). Only Ox-2/Cd200 was expressed in 62.8% of cluster B cells and in 50% of cluster K cells, which comprise RCF and pericytes respectively. The percentages of Cd200 were substantially lower in other clusters (Figure XIA in the Supplement). Additionally, CD200+/GFP+ cells co-localized with COL1α1 protein in the center of the scar (Figure 3F, Figure XIC in the Supplement). We next profiled FACS-sorted CD200− and CD200+ fibroblasts at 7dpi after negative selection for CD146 (Mcam) to discard pericytes (Figure 3G). CD200+/CD146− CF showed a significant similarity with the aggregated expression signature of cluster B cells (Figure XIB in the Supplement) and significantly higher levels of cluster B, F, G and H markers in comparison with CD200− CF (Figure 3H). Using the single-cell data from 7dpi, 37% of Cd200 cells were assigned to RCF while other clusters showed smaller proportions (Figure XID in the Supplement). In conclusion, no combination of surface markers allowed for the unequivocal selection of cluster B CF, but the expression of Cd200+/Cd146- provides the most specific characterization of RCF. Based on these results, we performed the chromatin accessibility patterns (ATAC-seq) on CD200+/CD146−, CD200−/CD146− at 7dpi, as well as on GFP+ CF population at different time-points (Figure 4A). The highest chromatin accessibility for RCF specific loci was observed in CD200+/CD146− CF. Furthermore, a motif enrichment analysis identified several TF such as RUNX1, SMAD, AP-1(JUN) and TEAD as candidate regulators of RCF (Figure 4B). With the exception of Runx1, most of the single-cell expression patterns of the identified TF showed no specificity for RCF (Figure XIIA in the Supplement). Although Runx1 has been reported to play a role in CF activation 31 overexpression of Runx1 in cultured CF was unable to induce the RCF specific transcription signature (Figure 4C). As a result, we considered that the analysis and the characterization of the TF role should be extended to compare the ATAC-seq fingerprint between CD200+/CD146− and CD200−/CD146− (Figure XIIB in the Supplement). Based on motifs detected in peaks from both populations and in the percentage of increased or decreased accessibility, TF were grouped in three different sets: enriched in CD200+ peaks but not differentially expressed (Set 1); enriched in CD200− peaks but not differentially expressed (Set 2); and enriched in CD200− peaks and differentially expressed (Set 3). No TF enriched in CD200+ peaks and differentially expressed were found (Figure XIIC in the Supplement). To characterize TF in Set 1 and 3, we used peak-gene mapping to conduct a Gene-Set Analysis per TF. These analyses revealed specific TF in Set 1 (ATF3, JUN or ZNF93) and in Set 3 (RUNX1, WT1 or KLF5) (Figure XIID in the Supplement). Together, these results represent an initial approach for the characterization of the TF involved in RCF generation.
Figure 4: Molecular regulation of RCF identity.
(A) Genome browser snapshots showing accessibility profiles of representative loci in global CF population at different time-points and in CD200+ and CD200− CF subpopulations at 7dpi. Shadowed areas mark distal regulatory elements displaying increased accessibility in CD200+ subpopulation. (B) Dot plot representing motif enrichment and expression specificity of potential TF mediating RCF response (cluster B/CD200+ CF). First row, analysis of CD200+ specific accessible distal regulatory elements (+/− 1.5 Kb from TSS). Second row, analysis of distal regulatory elements found within RCF specific loci. Third row, expression values of TF in scRNA-seq. Fourth row, expression values for TF in CD200+ bulk RNA-seq. (C) Volcano plot showing differential gene expression of in vitro grown CF overexpressing Runx1. Genes with Log Fold Change of ±1.5 and p-value <0.05 were considered differentially expressed. Red dots represent RCF markers. (D) TGF-β network centrality analysis revealed non-canonical PI3K-Akt pathway related with RCF markers. (E) Heatmap showing relative expression of RCF markers in non-treated (Control), TGF-β, TGF-β + Vehicle (DMSO) and TGF-β + LY294002 cultured CF. Linear model differential expression, n=4 per group. (F) Quantification of area covered by cultured CF after 23 hours of treatment in wound healing experiment. One-way analysis of variance, Kruskal-Wallis post-hoc test, n=4–8. ** p≤0.01, *** p≤0.001.
As an additional effort to characterize molecular regulation of RCF, we built a network spanning protein signaling and gene-regulatory interactions using our single-cell expression dataset based on the distances between cluster markers and signaling receptors. This analysis highlighted the non-canonical TGF-β1/PI3K-Akt pathway over the canonical one as the main driver of RCF gene expression (Figure 4D). To validate this, we stimulated in vitro CF with TGF-β in the presence or absence of LY294002, a known PI3K-Akt inhibitor 32. This inhibitor abolished the transcriptomic signature of RCF induced by TGF-β (Figure 4E, Figure XIIIA and XIIIB in the Supplement) as well as the ability of fibroblast to migrate and proliferate (Figure 4F, Movie I in the Supplement). These results emphasize the relevance of the non-canonical TGF-β1/PI3K-Akt pathway in controlling RCF identity and function.
In order to explore in depth the role of TGF-β, we cultured adult CF in the presence of small molecules that specifically inhibit different elements of the TGF-β1 signaling pathway (Table II in the Supplement, Figure XIIIC in the Supplement). The inhibition of specific elements of the canonical (SMAD2/3) 33, 34 and non-canonical (AKT and p38) 33, 35–38 TGF-β1 signaling pathways reduced the expression of both ECM (Col1α1, Col3α1, Lox) and CF activation (Cthrc1, Ddah1, Postn, Acta2) related genes (Figure XIIID in the Supplement). However, PD98059, a specific inhibitor of ERK 33, 34, 38, did not affect any of the genes (Figure XIIID in the Supplement). These results indicate that activation of the pro-fibrotic profile and the phenotypic transformation of RCF are regulated by a balance between SMAD2/3 and AKT/p38, but not via ERK. Remarkably, SB-431542, a specific inhibitor of TGF-β receptor 1/ALK5 33, 38, 39, selectively reduced the RCF markers together with Acta2 and Col1α1, but not Postn and Col3α1. This highlights the complexity that underlies the regulation of CF activation.
CTHRC1 mediated RCF activity is essential for the formation of the healing scar
As Cthrc1 is exclusively expressed in CF but not in other cardiac cell populations such as CD45, CD31 or cardiomyocytes (CM) (Figure 5A, 5B, and 5C, Figure XIVB and XIVC in the Supplement), we decided to analyze the functional role of RCF in mice with genetic ablation of Cthrc1 (Cthrc1-KO) (Figure XIVA in the Supplement). Cthrc1-KO mice appear normal both during development and adulthood. No changes were observed in size or weight, and more importantly, no cardiac phenotype was detected 22. However, after MI, KO mice showed a dramatic decrease in survival (80% WT versus 30% KO) due to ventricular rupture (Figure 5D). This was associated with a significant decrease in collagen deposition in the free wall of the left ventricle of KO infarcted hearts (50%) (Figure 5E). This phenotype was not associated with differences in the transcriptomic profile of other cardiac cell populations between WT and KO mice, suggesting that CTHRC1 had no pleiotropic effects (Figure XIVC in the Supplement). In contrast, we found significant differences between KO and WT CF in GO terms related to cell division, proliferation, and protein synthesis upon MI (Figure 5F and 5G). Cultured KO CF from healthy mice stimulated with TGF-β1 showed a decrease in GO terms related to angiogenesis, muscle contraction and vasculature development when compared to WT CF, indicating a reduced capacity to respond to TGF-β1 (Figure XV in the Supplement).
Figure 5: CTHRC1 is an essential effector of RCF for the healing repair process.
(A) Normalized expression bar plots (mean ± SD) of Cthrc1 in CF (GFP+/CD31−/CD45−), endothelial (CD31+), and bone marrow-derived cells (CD45+) at different time-points. (B) Localization of CTHRC1 (green) in the left ventricle of healthy wild type (WT) mice, and at 3, 5 and 7 dpi. Cardiac Troponin-I+ (cTN) cardiomyocytes (red), Nuclei (DAPI, blue). (C) Normalized expression bar plots (mean ± SD) of Cthrc1 in endothelial (CD31+), bone marrow-derived cells (CD45+), CF (mEFSK4+/CD31−/CD45−) and cardiomyocytes from WT hearts at 5dpi, linear model with normalized counts. (D) Kaplan-Meier survival curves after MI in WT and Cthrc1 knockout (KO) mice. Log-rank Mantel-Cox test, n=10 per group. (E) Representative images of collagen deposition in left ventricle of WT (left) and KO (right) mice. Quantification of collagen deposition in the LV in both genotypes at 3dpi (WT, open circle; KO, closed circle), unpaired t-test (right). (F) Volcano plots showing differential gene expression between WT and KO CF at 5dpi. (G) Dot plot comparison of enriched pathways in the bulk RNA-seq analysis between Cthrc1-KO (left column) and WT (right column) at 7dpi. (H) t-SNE representation of 4,189 CF from one KO heart at 7dpi. Red dots represent RCF-like fibroblasts, and the number the percentage of them in relation to the total isolated CF. t-SNE representation for RCF markers (below). (I) Proportion of cluster B CF in each of the datasets. ** p≤0.01, *** p≤0.001. KO = knockout; WT = wild type.
scRNA-seq analysis on Cthrc1-KO hearts at 7dpi (4,189 cells) did not show a reduction of RCF but rather an increased percentage of RCF-like cells in comparison with WT mice (21% versus 14%) (Figure 5H and 5I). RCF-like CF and other subpopulations of activated CF such as POSTN+ were found in the IZ and BZ of Cthrc1-KO infarcted mice, indicating that the ventricular rupture phenotype is derived from the absence of CTHRC1 (Figure XIVD and XIVE in the Supplement). Accordingly, deletion of Cthrc1 was associated with downregulation of genes related to ECM organization, collagen biosynthesis and TGF-β mediated regulation of the ECM in CF in comparison with WT mice (Figure XIVF in the Supplement). Globally, these results suggested that RCF orchestrate cardiac repair via secretion of CTHRC1 which affects the deposition and synthesis of ECM molecules and promotes the proliferation of CF.
An RCF-like expression signature is detected in a pre-clinical model of MI and in human cardiac fibrotic tissue
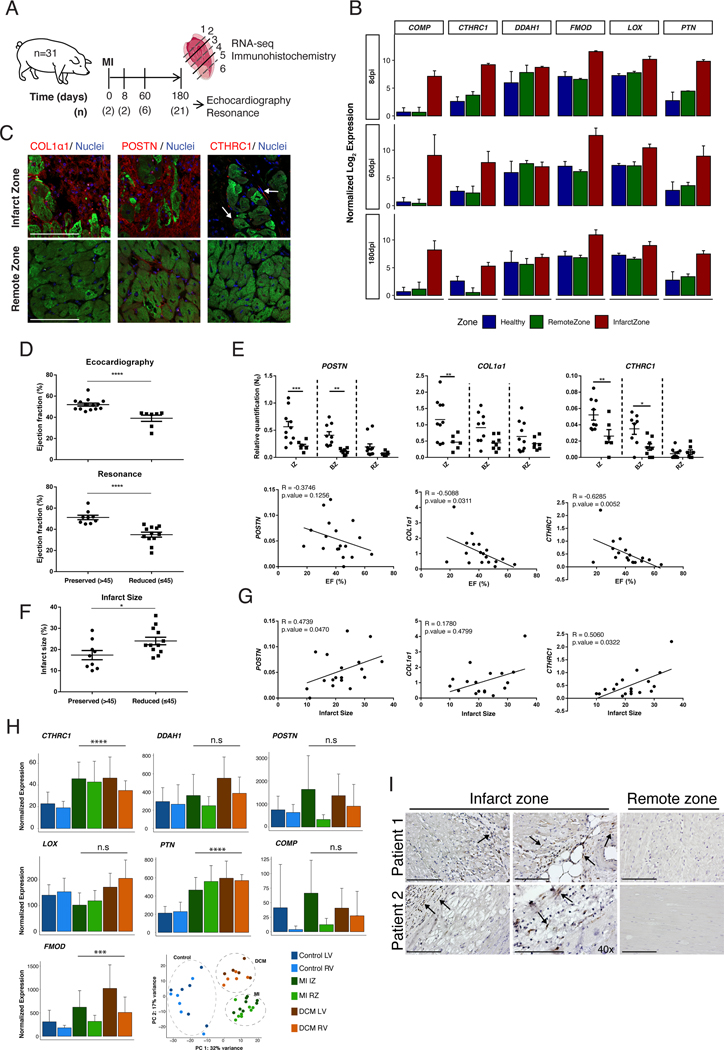
To assess the translational potential of our findings, we examined whether the RCF signature can be detected in a preclinical pig model of MI (Figure 6A). This was done by comparing the global transcriptome of representative biopsies from IZ and RZ of infarcted pig hearts at 8, 60 and 180 dpi (Figure 6B). Despite the lack of cell type resolution, we were able to detect the induction of 33%, 42% and 32% of the RCF signature at 8, 60 and 180 dpi, respectively, in the IZ but not in RZ (Figure XVIA in the Supplement). Consistent with murine data, CTHRC1+ cells localized to the IZ but not to the RZ, in the same region where deposits of COL1α1 and POSTN were found (Figure 6C). CTHRC1 expression was lower at 180dpi in comparison with 8 and 60 dpi (Figure 6B) with no CTHRC1+ cells detected at 180dpi by immunohistochemistry (data not shown). At 8dpi, we identified three different subpopulations of activated CF, CTHRC1+, αSMA+ and αSMA+/CTHRC1+ in the IZ and BZ, with a decrease at 60dpi, corroborating the heterogeneity of activated CF observed in mice (Figure XVIB in the Supplement). These results indicated that an RCF-like population also appears after MI in our pre-clinical swine model with similar dynamics as in mice.
Figure 6: RCF markers expression correlates with cardiac function in infarcted pigs and is conserved in humans.
(A) Twenty-nine pigs were submitted to Ischemia/Reperfusion surgery and 2 animals were used as controls. Cardiac function was determined in 21 pigs after 6 months using echocardiography and MRI. (B) Normalized expression bar plots (mean ± SD) for a zonal transcriptomic profiling at 8dpi (above) (n=2), 60dpi (middle) (n=6) and 180dpi (below) (n=9). Normalized expression bar plots (mean ± SD) for top RCF markers for healthy (blue) and at 8, 60 and 180 dpi in RZ (green) and IZ (red). (C) Immunohistochemistry of COL1α1, POSTN and CTHRC1 in IZ and RZ at 8dpi. (D) Distribution of pigs with reduced (≤45%, circles) (n=9) or preserved (>45%, squares) (n=12) ejection fraction (EF) at 180dpi by echocardiography or MRI. Two-tailed t-test with a Mann-Whitney post-hoc test. (E) Comparison of POSTN, COL1a1 and CTHRC1 expression levels in different anatomical regions between infarcted pigs with reduced or preserved EF. Two-ways ANOVA, n=8–10 (above). Correlation between EF and expression level of POSTN, COL1a1 and CTHRC1 in IZ. Spearman correlation coefficient (below). (F) Distribution of pigs with reduced (≤45%, circles) or preserved (>45%, squares) infarct area at 180dpi by MRI. Two-tailed t-test with a Mann-Whitney post-hoc test, n=21. (G) Correlations between infarct area and expression level of POSTN, COL1a1 and CTHRC1 in IZ. Spearman correlation coefficient. (H) Normalized expression bar plots for top RCF markers (CTHRC1, DDAH1, POSTN, FMOD, LOX, PTN and COMP) in human samples from healthy (LV and RV) (n=6), infarcted (IZ and RZ) (n=8), and dilated cardiomyopathy (DCM) (LV and RV) (n=5). Likelihood ratio test patients. PCA scatter plot of human samples subjected to transcriptomic profiling. (I) Representative images of the immunohistochemistry analysis of CTHRC1+ CF (in brown, arrows) performed on sections of the infarct and the remote zones obtained from the LV of two different patients that suffered MI. BZ = border zone; IZ = infarct zone; LV = left ventricle; RV = right ventricle; RZ = remote zone. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001. Scale bars: 100 μm.
To assess the relation between the expression of CTHRC1 and cardiac function, we compared the level of expression of CTHRC1 in the IZ between animals with mid-range, preserved (>45%) and reduced (≤45%) ejection fraction (EF) 40 at 180dpi (n=12 versus n=9, respectively) (Figure 6D). Despite the critical role of CTHRC1 in the early stages after MI, we found that at later stages (180dpi) the level of CTHRC1 (BZ, IZ) was significantly higher in the group with cardiac dysfunction (Figure 6E). Consequently, a negative correlation between the EF and the expression of CTHRC1, COL1α1 and POSTN in the IZ was found (Figure 6E), which was only statistically significant for CTHRC1 and COL1α1. Additionally, a positive and significant correlation between the infarct area and expression levels were observed for CTHRC1 and POSTN but not for COL1α1 (Figure 6F and 6G). Taken together, our results suggest that the upregulation of CTHRC1 could be beneficial immediately after MI, whereas increased expression at later time-points may reflect incomplete repair with progressive fibrosis.
Finally, the transcriptomic signature of RCF was partially found in biopsies obtained from the ischemic zone (IZ) of patients with ischemic cardiomyopathy and in biopsies from both right and left ventricle of patients with dilated cardiomyopathy (DCM). Several genes of the RCF transcriptomic signature (CTHRC1, PTM, FMOD) were significantly overexpressed in all the pathological conditions included in this study compared to controls as shown in the barplots from Figure 6H. Moreover, CTHRC1+ CF were found in the IZ but not in RZ of infarcted hearts from patients (Figure 6I). These findings highlight the potential role of CTHRC1+ RCF in orchestrating the cardiac repair process in patients suffering cardiac fibrosis.
DISCUSSION
As the main cellular component responsible for cardiac healing after MI, CF are an attractive therapeutic target. However, the lack of appropriate markers, the cellular heterogeneity and the limited understanding of the molecular mechanisms underlying their activation has precluded the development of successful therapies targeting fibrotic remodeling 4. In this study, we characterize at the single-cell level the heterogeneity of CF using Col1α1-GFP mice. We identify a specific subpopulation of CF defined by the expression of a unique transcriptional signature responsible for the healing response after cardiac injury. Among the top markers, we identified Cthrc1, a crucial molecule involved in the ventricular remodeling process. In a recent study, a similar role for Cthrc1+ fibroblasts has been described in fibrotic lungs 41. Furthermore, we also identified RCF in a large swine model and in patients with cardiac diseases that lead to myocardial fibrosis.
Non-myocyte cardiac cell heterogeneity has been recently characterized with a particular focus on CF 7–10, 12. Most of these studies focus on the early phase after cardiac injury 12 or assess other pathological conditions 7, 9, 10. In the study by Kretzschmar and colleagues 10, the transcriptome of 282 CF at 14dpi was studied, identifying eleven clusters one of which was defined as activated CF (FstlI+). This subpopulation had similar properties as RCF reported in this study but both subpopulations showed different transcriptomic signatures. Fu et al. recently described a new subpopulation of activated CF, the matrifibrocytes, characterized by the expression of ECM and tendon genes 8. Despite expression of several RCF marker genes in the matrifibrocytes, it was not possible to compare both subpopulations at the single-cell level. Moreover, RCF have a role during the healing scar, in contrast to matrifibrocytes with their role in supporting the mature scar at later stages. Although markers associated with RCF have been previously described in other CF subpopulations 7–9, 12, our transcriptomic, histological and functional results clearly indicate that RCF are distinct from Postn+, Acta2+ or matrifibrocytes. An important difference between our work and previous studies is that we focused exclusively on the CF population, unlike recently reported scRNA-seq studies in models of MI 9, 10, 12. This distinction together with the carefully description of the anatomic location and the temporal dynamics of RCF after MI make for a better characterization of CF heterogeneity and a refined definition of new subpopulations.
Understanding the regulation of RCF will be essential for the identification of therapeutic strategies. On the one hand, our transcriptomic studies together with our binding site analyses indicate a potential role for different TF in the generation of RCF. We validated the role of SOX9, consistent with recent publications 28, 42. Moreover, our ATAC-seq data revealed TF that could be involved in the regulation of RCF gene markers and additionally identify other TF involved in the regulation of CD200+ cells through chromatin accessibility.
Our in silico approaches emphasize the role of the TGF-β signaling pathway as a master regulator of CF activation and the development of cardiac fibrosis 43, 44. TGF-β signaling is known to promote myofibroblast formation and ECM production 28, 29, 45–48. Interestingly, our results demonstrate that both canonical and non-canonical TGF-β signaling pathways play a significant contribution in the activation of RCF, with a predominant role for the PI3K-Akt pathway in generating RCF. This pathway upregulates Cthrc1, the key marker gene for RCF, and the migratory and proliferative capacities of CF. These results suggest that the development of specific inhibitors of the PI3K-Akt pathway could define new targets for modulating cardiac fibrosis, although further studies of the downstream mechanisms are required.
CTHRC1 is a secreted protein that participates in collagen matrix synthesis through TGF-β signaling 21, 22, 49. Moreover, the expression of Cthrc1 has been recently identified as a potential marker for activated CF in the heart 7, 9, 12 and lungs 41. The longer follow-up in our study and the evaluation of Cthrc1 deletion in the context of MI may explain why it was not previously identified as a key player in the cardiac repair process. Consistent with other studies where key components of activated CF were ablated 7, 10, the deletion of Cthrc1 was associated with increased mortality due to ventricular rupture. In our case, RCF were still detected in the myocardium of KO mice, together with other subpopulations of activated CF. Moreover, our in silico data indicated that RCF originate from activated Postn+ CF with a specific role in the secretion and deposition of collagen in the healing scar. These results suggest that the ventricular rupture described by others could be an effect of the deletion of CF that give rise to RCF 36, 50, 51. Taken together, these data indicate that Cthrc1 may be an effector of RCF, promoting the healing process, rather than an inducer of the activation of this population.
Our results in a pre-clinical model of MI and in myocardial biopsy specimens of patients with ischemic or dilated cardiomyopathy provide additional translational relevance to our findings. Similar to the observations in mice, expression of CTHRC1 was upregulated in the fibrotic areas with identification of CTHRC1+ CF in these zones and not in the healthy myocardium. Also, the pattern of expression after MI in pigs showed a significant increase in the level of CTHRC1 in the early stages after MI, followed by a reduction of its expression levels in the chronic phase. Of note, the presence of more CTHRC1+ CF or higher levels of CTHRC1 expression, particularly at later time-points, was associated with worse cardiac function which would seem counterintuitive as CTHRC1 expression is associated with cardiac healing. These findings suggest that both responses depend upon ECM deposition by the CTHRC1+ CF lineage, such that ECM contributes to acute cardiac repair and prevents rupture, whereas sustained ECM deposition at later stages may reflect pathological remodeling. Although additional studies are needed, our findings suggest this molecule might be considered as a potential candidate to study “adverse remodeling” versus “myocardial recovery” in heart failure, based on three properties of the molecule: (i) the dynamic transcriptome of CTHRC1; (ii) its essential role on ECM during the early healing response; and (iii) its potentially negative effect on cardiac function during later stages of adverse ventricular remodeling 52.
In conclusion, our study, including the largest number of CF interrogated by scRNA-seq, clearly demonstrates the remarkable heterogeneity of CF after MI and defines a specific subpopulation, RCF, characterized by the expression of Cthrc1. Insights into molecular regulation and biological function distinctly support the role of Cthrc1+ CF in the early healing process after MI. These novel findings should facilitate the search for a more personalized treatment through control of the cardiac fibrotic process in patients.
Study limitations
We recognize the limitations of our study. First, we are aware that the current analysis does not provide complete mechanistic insight because regulation based on post-translational modifications of TF was not considered. Second, with the apparent complexity underlying the regulation of CF, we cannot rule out that other molecules or subpopulations of CF could have an essential role in the ventricular remodeling process. Third, future studies should address whether deletion of RCF using Cre-inducible DTR transgenic mouse models instead of CTHRC1 null mice results in a different outcome 53.
Competency in medical knowledge
Unraveling CF heterogeneity for understanding different cardiac fibrotic processes is critical. In the case of MI, Cthrc1+ CF are responsible for a proper healing response to cardiac injury. However, its role in chronic models requires further studies.
Translational Outlook
Further investigations are needed to determine the potential role of CTHRC1 during MI related to cardiac repair and its mechanism of regulation to identify the therapeutic windows that may allow controlling the size of the fibrotic scar.
Supplementary Material
Clinical Perspective.
What is New?
Our study identifies a new subpopulation of CF, Reparative Cardiac Fibroblasts (RCF), characterized by a distinct transcriptional profile, including Cthrc1, with a major role in the fibrotic healing response after MI.
Experiments performed in a mouse model deficient in Cthrc1 reveal an increase in mortality after MI and a decrease in the fibrotic response.
We identified CTHRC1+ CF in a swine model of MI and in patients with MI and dilated cardiomyopathy, supporting the conservation of the RCF subpopulation in humans.
What are the clinical implications?
Understanding the role of CF after MI is highly relevant to identify new therapeutic targets for controlling the adverse remodeling after MI.
The present study provides a unique resource for the cardiovascular field by characterizing the subpopulation of CF responsible for cardiac repair after cardiac fibrosis and identifying molecular mechanisms implicated in the generation of this subpopulation.
The conservation of this population in swine and human and the relation between Cthrc1 expression and cardiac function after MI suggest that this molecule might be a potential target in patients suffering from MI.
Acknowledgements
The authors thank D.A. Brenner and T. Kisseleva (University of California San Diego, UCSD) for the gift of Col1α1-GFP mice. The authors are grateful to S. Sarvide for her comments in the preparation of this manuscript. In addition, this study was also enabled in part by the MMC BioBank, a core facility of Maine Medical Center Research Institute.
Sources of funding
This work was supported by funds from the ISCIII and FEDER funds (PI16/00129, CPII15/00017), Red TERCEL RETIC RD16/0011/0005 and MINECO (Program RETOS Cardiomesh), ERANET II (Nanoreheart), and H2020 Program BRAVE. A.R.V. is supported by FSE/Ministerio de Economía, Industria y Competitividad - Agencia Estatal de Investigación/ IJCI-2016-30254, and the Spanish Ministerio de Ciencia, Innovación y Universidades (RTI2018–095410-BI00). N.F. is supported by a fellowship from the European Molecular Biology Organization (EMBO ALTF 241-2017). C.B. is supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant (EU Horizon 2020 research and innovation program, grant agreement n° 679146). This work was supported in part, by the National Heart, Lung, and Blood Institute of the National Institute of Health under grants R01 HL136560 and HL146504 (V.L. and S.R.). This work also utilized services of the Physiology Core, Histopathology Core, Confocal Microscopy Core, and Mouse Transgenic Core which are supported by NIH/NIGMS P30GM106391 and P20GM121301.
Non-standard Abbreviations and Acronyms:
- MI
Myocardial infarction
- CF
Cardiac Fibroblasts
- dpi
days post-infarction
- RCF
Reparative Cardiac Fibroblasts
- dpt
days post beginning of treatment
- IZ
Infarct Zone
- BZ
Border Zone
- RZ
Remote Zone
Footnotes
Disclosures: None
Conflict of interest: None declared.
REFERENCES
- 1.Pinto AR, Ilinykh A, Ivey MJ, Kuwabara JT, D’Antoni ML, Debuque R, Chandran A, Wang L, Arora K, Rosenthal NA, et al. Revisiting Cardiac Cellular Composition. Circ Res. 2016;118:400–9. doi: 10.1161/CIRCRESAHA.115.307778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frangogiannis NG. Pathophysiology of Myocardial Infarction. Compr Physiol. 2015;5:1841–75. doi: 10.1002/cphy.c150006 [DOI] [PubMed] [Google Scholar]
- 3.Shinde AV, Frangogiannis NG. Mechanisms of Fibroblast Activation in the Remodeling Myocardium. Curr Pathobiol Rep. 2017;5:145–152. doi: 10.1007/s40139-017-0132-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tallquist MD, Molkentin JD. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol. 2017;14:484–491. doi: 10.1038/nrcardio.2017.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruiz-Villalba A, Simon AM, Pogontke C, Castillo MI, Abizanda G, Pelacho B, Sanchez-Dominguez R, Segovia JC, Prosper F, Perez-Pomares JM. Interacting resident epicardium-derived fibroblasts and recruited bone marrow cells form myocardial infarction scar. J Am Coll Cardiol. 2015;65:2057–66. doi: 10.1016/j.jacc.2015.03.520 [DOI] [PubMed] [Google Scholar]
- 6.Moore-Morris T, Guimaraes-Camboa N, Banerjee I, Zambon AC, Kisseleva T, Velayoudon A, Stallcup WB, Gu Y, Dalton ND, Cedenilla M, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest. 2014;124:2921–34. doi: 10.1172/JCI74783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, SC JL, Aronow BJ, Tallquist MD, et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun. 2016;7:12260. doi: 10.1038/ncomms12260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC, et al. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest. 2018;128:2127–2143. doi: 10.1172/JCI98215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A, van Rooij E. Single-Cell Sequencing of the Healthy and Diseased Heart Reveals Cytoskeleton-Associated Protein 4 as a New Modulator of Fibroblasts Activation. Circulation. 2018;138:166–180. doi: 10.1161/CIRCULATIONAHA.117.030742 [DOI] [PubMed] [Google Scholar]
- 10.Kretzschmar K, Post Y, Bannier-Helaouet M, Mattiotti A, Drost J, Basak O, Li VSW, van den Born M, Gunst QD, Versteeg D, et al. Profiling proliferative cells and their progeny in damaged murine hearts. Proc Natl Acad Sci U S A. 2018;115:E12245–E12254. doi: 10.1073/pnas.1805829115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR. Single-Cell Transcriptional Profiling Reveals Cellular Diversity and Intercommunication in the Mouse Heart. Cell Rep. 2018;22:600–610. doi: 10.1016/j.celrep.2017.12.072 [DOI] [PubMed] [Google Scholar]
- 12.Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, Ho JW, Nordon RE, Harvey RP. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife. 2019;8:e43882. doi: 10.7554/eLife.43882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyagay P, Heroult M, Wang Q, Lehnert W, Belden J, Liaw L, Friesel RE, Lindner V. Collagen triple helix repeat containing 1, a novel secreted protein in injured and diseased arteries, inhibits collagen expression and promotes cell migration. Circ Res. 2005;96:261–8. doi: 10.1161/01.RES.0000154262.07264.12 [DOI] [PubMed] [Google Scholar]
- 14.Jin YR, Stohn JP, Wang Q, Nagano K, Baron R, Bouxsein ML, Rosen CJ, Adarichev VA, Lindner V. Inhibition of osteoclast differentiation and collagen antibody-induced arthritis by CTHRC1. Bone. 2017;97:153–167. doi: 10.1016/j.bone.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yata Y, Scanga A, Gillan A, Yang L, Reif S, Breindl M, Brenner DA, Rippe RA. DNase I-hypersensitive sites enhance alpha1(I) collagen gene expression in hepatic stellate cells. Hepatology. 2003;37:267–76. doi: 10.1053/jhep.2003.50067 [DOI] [PubMed] [Google Scholar]
- 16.Ivey MJ, Kuwabara JT, Pai JT, Moore RE, Sun Z, Tallquist MD. Resident fibroblast expansion during cardiac growth and remodeling. J Mol Cell Cardiol. 2018;114:161–174. doi: 10.1016/j.yjmcc.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivey MJ, Tallquist MD. Defining the Cardiac Fibroblast. Circ J. 2016;80:2269–2276. doi: 10.1253/circj.CJ-16-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn MA, Trafford AW. Aging and the cardiac collagen matrix: Novel mediators of fibrotic remodelling. J Mol Cell Cardiol. 2016;93:175–85. doi: 10.1016/j.yjmcc.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez A, Schelbert EB, Diez J, Butler J. Myocardial Interstitial Fibrosis in Heart Failure: Biological and Translational Perspectives. J Am Coll Cardiol. 2018;71:1696–1706. doi: 10.1016/j.jacc.2018.02.021 [DOI] [PubMed] [Google Scholar]
- 20.Binks AP, Beyer M, Miller R, LeClair RJ. Cthrc1 lowers pulmonary collagen associated with bleomycin‐induced fibrosis and protects lung function. Physiological Reports. 2017;5:e13115. doi: 10.14814/phy2.13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeClair RJ, Durmus T, Wang Q, Pyagay P, Terzic A, Lindner V. Cthrc1 is a novel inhibitor of transforming growth factor-beta signaling and neointimal lesion formation. Circ Res. 2007;100:826–33. doi: 10.1161/01.RES.0000260806.99307.72 [DOI] [PubMed] [Google Scholar]
- 22.Stohn JP, Perreault NG, Wang Q, Liaw L, Lindner V. Cthrc1, a novel circulating hormone regulating metabolism. PLoS One. 2012;7:e47142. doi: 10.1371/journal.pone.0047142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Lee M, Yu G, Lee H, Han X, Kim D. CTHRC1 activates pro-tumorigenic signaling pathways in hepatocellular carcinoma. Oncotarget. 2017;8:105238. doi: 10.18632/oncotarget.22164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu G, Fan W, Wang F, Lu H, Xing X, Zhang R, Jiang P. CTHRC1 as a novel biomarker in the diagnosis of cervical squamous cell carcinoma. Int J Clin Exp Pathol. 2018;11:847–854. [PMC free article] [PubMed] [Google Scholar]
- 25.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nature Biotechnology. 2020:1–7. doi: 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- 27.Hu HH, Chen DQ, Wang YN, Feng YL, Cao G, Vaziri ND, Zhao YY. New insights into TGF-beta/Smad signaling in tissue fibrosis. Chem Biol Interact. 2018;292:76–83. doi: 10.1016/j.cbi.2018.07.008 [DOI] [PubMed] [Google Scholar]
- 28.Lacraz GPA, Junker JP, Gladka MM, Molenaar B, Scholman KT, Vigil-Garcia M, Versteeg D, de Ruiter H, Vermunt MW, Creyghton MP, et al. Tomo-Seq Identifies SOX9 as a Key Regulator of Cardiac Fibrosis During Ischemic Injury. Circulation. 2017;136:1396–1409. doi: 10.1161/CIRCULATIONAHA.117.027832 [DOI] [PubMed] [Google Scholar]
- 29.Khalil H, Kanisicak O, Prasad V, Correll RN, Fu X, Schips T, Vagnozzi RJ, Liu R, Huynh T, Lee SJ, et al. Fibroblast-specific TGF-beta-Smad2/3 signaling underlies cardiac fibrosis. J Clin Invest. 2017;127:3770–3783. doi: 10.1172/JCI94753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huebener P, Abou-Khamis T, Zymek P, Bujak M, Ying X, Chatila K, Haudek S, Thakker G, Frangogiannis NG. CD44 is critically involved in infarct healing by regulating the inflammatory and fibrotic response. J Immunol. 2008;180:2625–2633. doi: 10.4049/jimmunol.180.4.2625 [DOI] [PubMed] [Google Scholar]
- 31.Kim W, Barron DA, San Martin R, Chan KS, Tran LL, Yang F, Ressler SJ, Rowley DR. RUNX1 is essential for mesenchymal stem cell proliferation and myofibroblast differentiation. Proc Natl Acad Sci U S A. 2014;111:16389–94. doi: 10.1073/pnas.1407097111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montiel-Duarte C, Cordeu L, Agirre X, Roman-Gomez J, Jimenez-Velasco A, Jose-Eneriz ES, Garate L, Andreu EJ, Calasanz MJ, Heiniger A, et al. Resistance to Imatinib Mesylate-induced apoptosis in acute lymphoblastic leukemia is associated with PTEN downregulation due to promoter hypermethylation. Leuk Res. 2008;32:709–16. doi: S0145–2126(07)00344-X [pii] 10.1016/j.leukres.2007.09.005 [DOI] [PubMed] [Google Scholar]
- 33.Vivar R, Humeres C, Ayala P, Olmedo I, Catalán M, García L, Lavandero S, Díaz-Araya G. TGF-β1 prevents simulated ischemia/reperfusion-induced cardiac fibroblast apoptosis by activation of both canonical and non-canonical signaling pathways. Biochim Biophys Acta. 2013;1832:754–762. doi: 10.1016/j.bbadis.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 34.Voloshenyuk TG, Landesman ES, Khoutorova E, Hart AD, Gardner JD. Induction of cardiac fibroblast lysyl oxidase by TGF-β1 requires PI3K/Akt, Smad3, and MAPK signaling. Cytokine. 2011;55:90–97. doi: 10.1016/j.cyto.2011.03.024 [DOI] [PubMed] [Google Scholar]
- 35.Ellis IR, Jones SJ, Lindsay Y, Ohe G, Schor AM, Schor SL, Leslie NR. Migration Stimulating Factor (MSF) promotes fibroblast migration by inhibiting AKT. Cell. Signal. 2010;22:1655–1659. doi: 10.1016/j.cellsig.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 36.Molkentin JD, Bugg D, Ghearing N, Dorn LE, Kim P, Sargent MA, Gunaje J, Otsu K, Davis J. Fibroblast-Specific Genetic Manipulation of p38 Mitogen-Activated Protein Kinase In Vivo Reveals Its Central Regulatory Role in Fibrosis. Circulation. 2017;136:549–561. doi: 10.1161/CIRCULATIONAHA.116.026238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papakrivopoulou J, Lindahl GE, Bishop JE, Laurent GJ. Differential roles of extracellular signal-regulated kinase 1/2 and p38MAPK in mechanical load-induced procollagen α1 (I) gene expression in cardiac fibroblasts. Cardiovasc Res. 2004;61:736–744. doi: 10.1016/j.cardiores.2003.12.018 [DOI] [PubMed] [Google Scholar]
- 38.Hu J, Wang X, Wei S-M, Tang Y-H, Zhou Q, Huang C-X. Activin A stimulates the proliferation and differentiation of cardiac fibroblasts via the ERK1/2 and p38-MAPK pathways. European journal of pharmacology. 2016;789:319–327. doi: 10.1016/j.ejphar.2016.07.053 [DOI] [PubMed] [Google Scholar]
- 39.Bollong MJ, Yang B, Vergani N, Beyer BA, Chin EN, Zambaldo C, Wang D, Chatterjee AK, Lairson LL, Schultz PG. Small molecule-mediated inhibition of myofibroblast transdifferentiation for the treatment of fibrosis. Proceedings of the National Academy of Sciences. 2017;114:4679–4684. doi: 10.1073/pnas.1702750114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JG, Coats AJ, Falk V, Gonzalez-Juanatey JR, Harjola V-P, Jankowska EA. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. European heart journal. 2016;37:2129–2200. doi: 10.1093/eurheartj/ehw128 [DOI] [PubMed] [Google Scholar]
- 41.Tsukui T, Sun K-H, Wetter JB, Wilson-Kanamori JR, Hazelwood LA, Henderson NC, Adams TS, Schupp JC, Poli SD, Rosas IO. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Comm. 2020;11:1–16. doi: 10.1038/s41467-020-15647-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scharf GM, Kilian K, Cordero J, Wang Y, Grund A, Hofmann M, Froese N, Wang X, Kispert A, Kist R. Inactivation of Sox9 in fibroblasts reduces cardiac fibrosis and inflammation. JCI insight. 2019;4doi: 10.1172/jci.insight.126721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu G, Ma C, Yang H, Zhang PY. Transforming growth factor beta and its role in heart disease. Exp Ther Med. 2017;13:2123–2128. doi: 10.3892/etm.2017.4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobaczewski M, Chen W, Frangogiannis NG. Transforming growth factor (TGF)-beta signaling in cardiac remodeling. J Mol Cell Cardiol. 2011;51:600–6. doi: 10.1016/j.yjmcc.2010.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frangogiannis NG, Ren G, Dewald O, Zymek P, Haudek S, Koerting A, Winkelmann K, Michael LH, Lawler J, Entman ML. Critical role of endogenous thrombospondin-1 in preventing expansion of healing myocardial infarcts. Circulation. 2005;111:2935–42. doi: 10.1161/CIRCULATIONAHA.104.510354 [DOI] [PubMed] [Google Scholar]
- 46.Lal H, Ahmad F, Zhou J, Yu JE, Vagnozzi RJ, Guo Y, Yu D, Tsai EJ, Woodgett J, Gao E, et al. Cardiac fibroblast glycogen synthase kinase-3beta regulates ventricular remodeling and dysfunction in ischemic heart. Circulation. 2014;130:419–30. doi: 10.1161/CIRCULATIONAHA.113.008364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bujak M, Ren G, Kweon HJ, Dobaczewski M, Reddy A, Taffet G, Wang XF, Frangogiannis NG. Essential role of Smad3 in infarct healing and in the pathogenesis of cardiac remodeling. Circulation. 2007;116:2127–38. doi: 10.1161/CIRCULATIONAHA.107.704197 [DOI] [PubMed] [Google Scholar]
- 48.Campa CC, Silva RL, Margaria JP, Pirali T, Mattos MS, Kraemer LR, Reis DC, Grosa G, Copperi F, Dalmarco EM. Inhalation of the prodrug PI3K inhibitor CL27c improves lung function in asthma and fibrosis. Nat Comm. 2018;9:1–16. doi: 10.1038/s41467-018-07698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Cao J, Li M, Yu Y, Yang Y, Xiao X, Wu Z, Wang L, Tu Y, Chen H. Collagen triple helix repeat containing-1 inhibits transforming growth factor-b1-induced collagen type I expression in keloid. Br J Dermatol. 2011;164:1030–1036. doi: 10.1111/j.1365-2133.2011.10215.x [DOI] [PubMed] [Google Scholar]
- 50.Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, Lorts A, Brunskill EW, Dorn GW 2nd, Conway SJ, et al. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res. 2007;101:313–21. doi: 10.1161/CIRCRESAHA.107.149047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaur H, Takefuji M, Ngai CY, Carvalho J, Bayer J, Wietelmann A, Poetsch A, Hoelper S, Conway SJ, Mollmann H, et al. Targeted Ablation of Periostin-Expressing Activated Fibroblasts Prevents Adverse Cardiac Remodeling in Mice. Circ Res. 2016;118:1906–17. doi: 10.1161/CIRCRESAHA.116.308643 [DOI] [PubMed] [Google Scholar]
- 52.Kim GH, Uriel N, Burkhoff D. Reverse remodelling and myocardial recovery in heart failure. Nature Reviews Cardiology. 2018;15:83. doi: 10.1038/nrcardio.2017.139 [DOI] [PubMed] [Google Scholar]
- 53.Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–26. doi: 10.1038/nmeth762 [DOI] [PubMed] [Google Scholar]
- 54.Döring H. The isolated perfused heart according to Langendorff technique-function-application. Physiol Bohemoslov. 1990;39:481. [PubMed] [Google Scholar]
- 55.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–9. doi: 10.1126/science.1247651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169:750–765 e17. doi: 10.1016/j.cell.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bagnoli JW, Ziegenhain C, Janjic A, Wange LE, Vieth B, Parekh S, Geuder J, Hellmann I, Enard W. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat Commun. 2018;9:2937. doi: 10.1038/s41467-018-05347-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–9. doi: 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–203. doi: 10.1038/ng.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stohn JP, Wang Q, Siviski ME, Kennedy K, Jin YR, Kacer D, DeMambro V, Liaw L, Vary CP, Rosen CJ. C thrc1 controls adipose tissue formation, body composition, and physical activity. Obesity. 2015;23:1633–1642. doi: 10.1002/oby.21144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruiz-Villalba A, van Pelt-Verkuil E, Gunst QD, Ruijter JM, van den Hoff MJ. Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR). Biomol Detect Quantif. 2017;14:7–18. doi: 10.1016/j.bdq.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruijter JM, Ruiz Villalba A, Hellemans J, Untergasser A, van den Hoff MJ. Removal of between-run variation in a multi-plate qPCR experiment. Biomol Detect Quantif. 2015;5:10–4. doi: 10.1016/j.bdq.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruiz-Villalba A, Mattiotti A, Gunst QD, Cano-Ballesteros S, van den Hoff MJ, Ruijter JM. Reference genes for gene expression studies in the mouse heart. Sci Rep. 2017;7:24. doi: 10.1038/s41598-017-00043-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nygard AB, Jorgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol. 2007;8:67. doi: 10.1186/1471-2199-8-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356doi: 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li W, Cerise JE, Yang Y, Han H. Application of t-SNE to human genetic data. J Bioinform Comput Biol. 2017;15:1750017. doi: 10.1142/S0219720017500172 [DOI] [PubMed] [Google Scholar]
- 70.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Team RC. R: a language and environment for statistical computing. R Foundation for Statistical Computing website; 2019. [Google Scholar]
- 72.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic acids research. 2019;47:e47–e47. doi: 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferreira JA. The Benjamini-Hochberg method in the case of discrete test statistics. Int J Biostat. 2007;3:Article 11. doi: 10.2202/1557-4679.1065 [DOI] [PubMed] [Google Scholar]
- 78.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang R, Grishagin I, Wang Y, Zhao T, Greene J, Obenauer JC, Ngan D, Nguyen D-T, Guha R, Jadhav A. The NCATS BioPlanet–an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front Pharmacol. 2019;10:445. doi: 10.3389/fphar.2019.00445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, Ma’ayan A. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016;2016doi: 10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–D386. doi: 10.1093/nar/gkx1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Perfetto L, Briganti L, Calderone A, Cerquone Perpetuini A, Iannuccelli M, Langone F, Licata L, Marinkovic M, Mattioni A, Pavlidou T, et al. SIGNOR: a database of causal relationships between biological entities. Nucleic Acids Res. 2016;44:D548–54. doi: 10.1093/nar/gkv1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- 88.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell iden [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: Clustering, classification and density estimation using gaussian finite mixture models. R J. 2016;8:289–317. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.