Summary
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported at the end of 2019 in China. By the end of February 2020, the virus has spread worldwide through continuous human-to-human transmission via contact and droplet infection, demonstrating the ease with which emerging viruses disperse globally through the mass transport system. Here, we summarize our knowledge of other coronaviruses that have infected humans in comparison with SARS-CoV-2.
Keywords: SARS-CoV-2, human coronaviruses, intermediate host, reservoir
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, provisionally named 2019-nCoV) emerged in Wuhan, Hubei Province, China at the end of 2019 as the seventh human coronavirus (HCoV) (1) and spread throughout the world. SARS-CoV-2 causes coronavirus disease 2019 (COVID-19), which can be asymptomatic or cause fever, cough, shortness of breath, acute respiratory distress syndrome (ARDS), pneumonia, and death (2).
As of February 27, 2020, the WHO reported that more than 82,000 cases of COVID-19 have been identified in 46 countries/regions of Asia, South and North America, Europe, Africa, and Oceania (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/). In particular, South Korea, Italy, Japan, Iran, and Singapore, together with China, have each reported more than 100 cases of COVID-19.
Most of the recent COVID-19 patients in Japan claim not to have visited China or to have had close contact with COVID-19 patients, indicating that SARS-CoV-2 might have invaded the Japanese population. The National Institute of Infectious Disease (NIID) and local institutes of public health in Japan are using nested RT-PCR and/or real-time RT-PCR for diagnosis, and the NIID together with Japanese companies has initiated the development of a rapid diagnosis kit (based on immunochromatography). The development of vaccines and antivirals against SARS-CoV-2 are also planned in Japan as well as other countries. Furthermore, basic research on SARS-CoV-2 is underway to understand its behavior in natural and intermediate hosts and humans.
SARS-CoV-2, which belongs to lineage B of Betacoronavirus within the order Nidovirales and family Coronaviridae, possesses a single positive-strand RNA (~30 kb) as its genome. It encodes at least 10 structural (S, M, E, and N) and nonstructural proteins. The spike (S) protein protrudes from the virion surface and binds to human angiotensin converting enzyme 2 (ACE2) via the receptor binding domain (RBD) of the S protein to allow the virus to gain cell entry (3). Given this function, the S protein is a promising target for the development of a vaccine and monoclonal antibody therapy. The 3D structure of the S protein and its binding mode to human ACE2 has been described (4), supporting vaccine development and analysis of monoclonal antibodies. The nucleoprotein (N) associates with the viral genome within the virus particle and is thought to be a good target for an immunochromatography-based rapid diagnosis kit. Both the matrix (M) and the envelope (E) protein are important for virus particle formation at the ER-Golgi intermediate compartment. However, the precise roles of these proteins are unknown because SARS-CoV-2 has just emerged. Therefore, we need to understand the virus and the disease through both clinical observations and laboratory experiments.
The alphacoronaviruses HCoV-NL63 and -229E and the betacoronaviruses HCoV-OC43 and -HKU1 are widely known as human pathogens for the common cold, whereas the betacoronaviruses severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome (MERS)-CoV caused outbreaks with high case fatality rates (10% and 30%, respectively) in humans in 2002-2003 and 2014-2015, respectively (5). Large scale virus surveillance in wild and domestic animals revealed that diverse CoVs related to HCoV-NL63, -229E, -OC43, and -HKU1 can be detected in bats and rodents, whereas CoVs closely related to HCoV-229E and -OC43 are found in camels and cattle (5), suggesting that bats and rodents are reservoirs of HCoVs, whereas camels and cattle are intermediate hosts (Figure 1). Similarly, bats are a reservoir of SARS- and MERS-CoV, whereas civets or camels play a direct role in transmission to humans as intermediate hosts for SARS- and MERS-CoV (Figure 1).
Figure 1.
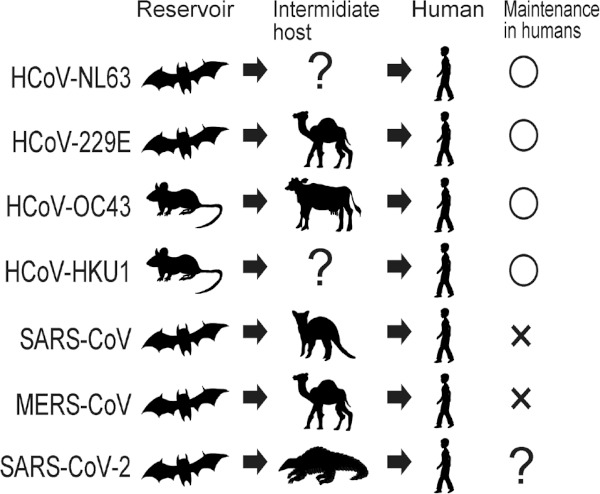
The putative reservoirs and intermediate hosts of seven CoV that have infected humans. The reservoirs and intermediate hosts of each Cov are listed. HCoV-NL63, -229E, -OC43, and -HKU1 cause human infections every year, whereas SARS- and MERS-CoV were not maintained in the human population until now.
Although all six CoVs cause mild-to-severe disease in humans, the low virulent HCoV-NL63, -229E, -OC43, and -HKU1 viruses continue to transmit between humans, whereas the relatively highly virulent SARS- and MERS-CoV are not maintained in the human population; SARS-CoV has disappeared from the human population and MERS-CoV infection has been limited at the interface between humans and camels. We do not know what determines whether a novel coronavirus that emerges from animals will subsequently become established in the human population. Based on our scientific knowledge of other human coronaviruses, researchers immediately sought a reservoir and an intermediate host for SARS-CoV-2 and found coronaviruses genetically similar to SARS-CoV-2 in bats and pangolins (3,6). Genetic analysis showed that the viruses detected in pangolins are relatively similar to SARS-CoV-2 compared with those detected in bats, indicating that bats and pangolins likely serve as a reservoir and an intermediate host, respectively. However, further analysis is warranted to definitively ascertain which animals are reservoirs and intermediate hosts of these viruses.
It may be too early to discuss what will happen after the outbreak; we cannot predict whether SARS-CoV-2 will disappear from the human population like SARS-CoV or will continue to transmit between humans like the four common cold-related CoVs. Since the case fatality rate of COVID-19 is currently 2-3%, which is lower than those of SARS- and MERS-CoV but higher than those of the four common cold-related CoVs, we speculate that SARS-CoV-2 will be maintained in humans and remain the causative agent of COVID-19 after the outbreak. However, if we can contain SARS-CoV-2, it should disappear from the human population; although this possibility is rapidly diminishing due to the fact that the virus has already spread worldwide.
Acknowledgements
We thank Susan Watson for scientific editing and Aya Ogawa for drawing illustrations.
References
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020; 382:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020; 395:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018; 100:163-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lam TTY, Shum MHH, Zhu HC, Tong YG, Ni XB, Liao YS, Wei W, Cheung WYM, Li WJ, Li LF, Leung GM, Holmes EC, Hu YL, Guan Y. Identification of 2019- nCoV related coronaviruses in Malayan pangolins in southern China. Preprint on bioRxiv. 2020; https://doi.org/10.1101/2020.02.13.945485. [Google Scholar]


