Abstract
Background
Accumulating evidences have shown that circular RNAs (circRNAs) play important roles in regulating the pathogenesis of cancer. However, the role of circRNAs in gastric cancer (GC) remains largely unclear.
Methods
In this study, we identified a novel upregulated circRNA, hsa_circ_0001829, in chemically induced malignant transformed human gastric epithelial cells using RNA-seq. Subsequent qRT-PCR and ISH assays were performed to detect the expression level of hsa_circ_0001829 in GC cell lines and tissues. Functional roles of hsa_circ_0001829 in GC were then explored by loss- and gain-of- function assays. Bioinformatic prediction and luciferase assay were used to investigate potential mechanisms of hsa_circ_0001829. Finally, the mice xenograft and metastasis models were constructed to assess the function of hsa_circ_0001829 in vivo.
Results
We found that hsa_circ_0001829 was significantly upregulated in GC tissues and cell lines. Loss- and gain-of- function assays showed that hsa_circ_0001829 promotes GC cells proliferation, migration and invasion, and the affected cell cycle progression and apoptosis rates may account for the effect of hsa_circ_0001829 on GC proliferation. In addition, bioinformatic prediction and luciferase assay showed that hsa_circ_0001829 acts as a molecular sponge for miR-155-5p and that SMAD2 was a target gene of miR-155-5p; moreover, hsa_circ_0001829 sponges miR-155-5p to regulate SMAD2 expression and hsa_circ_0001829 promotes GC progression through the miR-155-5p–SMAD2 pathway. Finally, suppression of hsa_circ_0001829 expression inhibited tumor growth and aggressiveness in vivo.
Conclusion
Taken together, our findings firstly demonstrated a novel oncogenic role of hsa_circ_0001829 in GC progression through miR-155-5p–SMAD2 axis, and our study may offer novel biomarkers and therapeutic targets for GC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13046-020-01790-w.
Keywords: Gastric cancer, Progression, hsa_circ_0001829, miR-155-5p, SMAD2
Background
As the fifth most common malignancies, gastric cancer (GC) is the third leading cause of cancer-associated deaths worldwide and continues to be a major threat to human health [1]. Despite of great improvement in diagnosis and treatments for GC, the prognosis of patients with GC remains poor, with a 5-year overall survival below 40% in most countries, mainly because a large majority of diagnostic cases are at advanced-stage with extensive metastasis and high recurrence [2]. GC is a multifactorial disease and a number of risk factors play roles in GC etiology, among which environmental carcinogens have been considered as one of the most important factors [3]. N-nitroso compounds (NOCs), a class of powerful carcinogens, are ubiquitous in environment and dietary sources, and exhibit genotoxicity and carcinogenesis through DNA damage [4, 5]. As a kind of NOCs, N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) is a well-known chemical mutagen and carcinogen that can induce genotoxic stress and lead to the occurrence of tumors, especially GC [6].
In recent years, with the advances in molecular biology, noncoding RNAs (ncRNAs) have emerged as key molecular players in many pathological conditions, especially cancers [7, 8]. Increasing studies have revealed the role of ncRNAs in carcinogenesis induced by environmental carcinogens, and demonstrated that alterations in ncRNAs expression are associated with exposure to chemical carcinogens [9, 10]. In the study by Yang et al., malignant-transformed human gastric epithelial cells (GES-1-T cells) induced by MNNG were established to explore the molecular mechanism of MNNG-induced gastric carcinogenic processes in humans [11]; moreover, Yang et al. and we respectively showed that microRNA (miRNA) miR-21 and long noncoding RNA LOC101927497 were aberrantly expressed and involved in malignant progression of MNNG-induced GC [11, 12].
As a new type of endogenous ncRNAs, circular RNAs (circRNAs) are characterized by a covalently closed loop without 5′ caps and 3′ poly (A) tails [13–15]. Due to their circular structures, circRNAs are unusually stable. Previous studies have revealed their high conservation, cell or tissue-specific expression and abundant presence in plasma and exosomes [16–18]. Furthermore, emerging lines of studies have revealed that circRNAs play important roles in physiological and pathological conditions including malignant tumors. Hence, circRNAs have great potential as diagnosis or prognosis biomarkers and therapeutic targets for cancers. Recently, it has been reported that circRNAs harbor abundant conserved miRNA response elements (MREs) and serve as miRNA sponge to regulate gene expression, which is involved in tumor initiation and progression [19]. Although our previous studies have demonstrated that hsa_circ_006100 and hsa_circ_0000592 were involved in the regulation of MNNG induced gastric tumorigenesis [20, 21], the role of circRNAs in the occurrence and development of GC induced by environmental carcinogens has been poorly elucidated and more studies are needed.
In present study, we analyzed the expression profile of circRNAs in GES-1-T and non-malignant control GES-1-N cells using high-throughput RNA sequencing (RNA-seq), and identified a novel circRNA hsa_circ_0001829, which was significantly upregulated in GC cell lines and tissues. Loss- and gain-of- function assays showed that hsa_circ_0001829 promotes GC cells proliferation, migration and invasion. Mechanistically, hsa_circ_0001829 functions as a miR-155-5p sponge to upregulate SMAD2 expression and consequently facilitates GC progression. Our findings provide a further insight into mechanism of carcinogen-related GC tumorigenesis and reveal a novel candidate for GC diagnosis/prognosis and therapy.
Materials and methods
Cell culture and tissue specimens
GES-1-T cells and control GES-1-N cells were constructed in the previous study [11], and were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Human GC cell lines (MKN-28, SGC-7901, MGC-803, BGC-823) were cultured in RPMI 1640 medium (HyClone, USA) supplemented with 10% FBS and 1% penicillin-streptomycin. All cells were incubated in a humidified atmosphere containing 5% CO2 at 37 °C.
A human tissue microarray of 98 cases of GC patients which contained 83 paired GC samples (Cat No. HStmA180Su15) was purchased from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). 20 paired of GC and adjacent samples were obtained from patients underwent surgery at the Fifth Affiliated Hospital of Guangzhou Medical University. All samples and clinical data were collected with patients’ consents. All experiments were approved by the Ethics Committee of the Fifth Affiliated Hospital of Guangzhou Medical University.
High-throughput sequencing and data analysis
High-throughput sequencing was performed and data was analyzed as we previously described [12, 20, 21]. CircRNAs were subjected to further analysis including annotation, sequence prediction, and the calculated expression levels.
RNA extraction, nuclear-cytoplasmic fractionation and quantitative reverse transcription-polymerase chain reaction(qRT-PCR)
RNA extraction, nuclear-cytoplasmic fractionation and qRT-PCR assay were performed as previously described [20].GAPDH or U6 acted as internal standards. The 2-ΔΔCt method was used to analyze relative expression levels. The specific primers were listed in Supplementary Table 1.
RNA in situ hybridization (ISH)
After dewaxing and hybridization, the tissue microarray was dealt with Proteinase K, fix in 4% paraformaldehyde and hybridized with digoxigenin-labeled hsa_circ_0001829 probes at 55 °C overnight and subsequently incubated overnight at 4 °C with anti-digoxin monoclonal antibody (Roche, Mannheim, Germany). After staining with Strept-Avidin-Biotin-Peroxidase Complex, the tissue microarray was observed and images were captured under a microscope. The analysis software Image-pro 6.0 (Media Cybernetics, Inc., Rockville, MD, USA) was applied to acquire the Integrated Optical Density (IOD) for evaluating the expression level of has_circ_0001829 in GC tissues.
Actinomycin D and RNase R treatment
Actinomycin D (10 μg/ml) or DMSO was added to the culture medium to evaluate the stability of circRNA and its linear isoform. For RNase R treatment, total RNA was incubated for 30 min at 37 °C with or without 3 U/μg RNase R (Epicentre, Madison, WI, USA). After treatment with actinomycin D and RNase R, the expression levels of SLC45A4 and hsa_circ_0001829 were measured by qRT-PCR.
Transfection
The overexpression vector was constructed with PCR product of hsa_circ_0001829 based on pcDNA3.1. MiRNA mimics/inhibitors and siRNAs were synthesized by GenePharma (Shanghai, China). The siRNAs were designed base on the circRNA backspliced region. For transient transfection, GC cells were cultured and transfected with these reagents using Lipofectamine™ 2000 (Invitrogen, USA) according to the manufacturer’s instructions. To stably knockdown hsa_circ_0001829, sh-circRNA-1# and sh-circRNA-2# lentivirus vectors were constructed and the lentiviruses were packaged and purified by GenePharma (Shanghai, China). Stable transfection procedures with lentivirus vectors were performed according to manufacturer’s instructions.
Fluorescence in situ hybridization analysis (FISH)
Cy3-labeled hsa_circ_0001829 specific probe was designed for RNA FISH, and FAM-labeled hsa_circ_0001829 specific probe and CY5-labeled miR-155-5p probe were designed and used for co-localized RNA FISH. Cells attached to slides were immobilized and then digested with protease K. After dehydration with 70, 85 and 100% alcohol, hybridization was performed at 42 °C overnight in dark. The slides were then washed with 50% formamide/2 × SSC preheated to 43 °C, 0.1% NP- 40/2 × SSC preheated to 37 °C, and DAPI staining solution at room temperature. Images were captured under a laser confocal microscope (Leica, Mannheim, Germany).
Cell counting Kit-8 (CCK-8) assay
Cell proliferation was assayed by CCK-8 (Dojindo, Tokyo) according to the manufacturer’s instructions. A total of 3000 cells were plated in each well of a 96-well plate. Then, on the indicated day, 10 μl of CCK-8 solution was added to each well. Following 1 h of incubation at 37 °C, the absorbance of each well at 450 nM was measured by Synergy 2 microplate reader (BioTek, Winooski, VT, USA).
5-Ethynyl-2′-deoxyuridine (EdU) assay
EdU assays were carried out using a Cell-light EdU DNA Cell Proliferation Kit (RiboBio, Guangzhou, China) according to the manufacturer’s protocol. Cells (1 × 104) were seeded in each well of a 96-well plate. After incubation with 50 μM EdU for 2 h, the cells were fixed in 4% paraformaldehyde and stained with Apollo Dye Solution. The Hoechst 33342 was used to stain the nucleic acids. Then the EdU-positive cells were photographed and IPP software was used to analyze the resultant data.
Colony formation assay
GC cells were seeded into 6-well plate at a density of 500 cells per well and cultured routinely for two weeks. When a clone is visible in the dish, the culture was terminated. Then the colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Cell colonies were counted and analyzed.
Transwell invasion assay
Transfected cells were placed into the upper chamber (Corning Costar, 8.0 μm pore size), which was coated with Matrigel (BD, New Jersey, USA). A total of 500 μL medium supplemented with 20% FBS was added into the lower chamber. After incubation for 24 h, the non-invaded cells on the upper side of the chamber were removed with a cotton swab, while invaded cells were fixed in 4% paraformaldehyde and stained with 0.1% crystal violet solution. The stained cells were analyzed.
Scratch wounding assay
Transfected cells were seeded into 6-well plates and cultured overnight at 37 °C. When the cells were fully confluent, a uniform straight scratch was made in the center of the well using a sterile 200 μl pipet tip. Images were obtained as a baseline after washing three times with PBS. Subsequently, fresh medium contained 2% FBS was added. After 24 h, images of the same location were acquired.
Flow cytometric analysis
Flow cytometric analysis was performed as previously described [12]. Cell cycle and apoptosis were determined on a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Dual-luciferase reporter assay
293-T cells were seeded into 24-well plates. Cells at 60–70% confluence were co-transfected with wild-type or mutated hsa_circ_0001829 3′ UTR or SMAD2 3′ UTR reporter plasmids, and with miR-155-5p mimics or negative controls using the Lipofectamine™2000 as transfection reagent. After 24 h incubation, luciferase assays were conducted using the Dual Luciferase Reporter Assay System (Promega, WI, USA) to detect firefly and Renilla luciferase activities. The ratio of firefly fluorescence and Renilla fluorescence was calculated as the relative luciferase activity.
Western blot
The GES-1-T and MKN-28 cells were lysed with RIPA lysis buffer (Beyotime, China), and protein was harvested and quantified by bicinchoninic acid (BCA) analysis (Beyotime, China). Protein extractions were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Merck Millipore, Billerica, MA, USA). After 2 h blocking with 5% bovine serum albumin, the membranes were incubated with primary anti-GAPDH antibody (1:2000), anti SMAD2 antibody (1:1000) (Abcam, Cambridge, UK) overnight at 4 °C. The membranes were then incubated with a secondary antibody. After washes, signals were detected and analyzed using an Odyssey Imaging System (LI-COR, Lincoln, NE, USA).
Animal experiments
Four-week-old female NOG mice were used for animal experiments. For tumor growth studies, GES-1-T cells (5 × 106) stably transfected with vectors were injected into the flanks of NOG mice (n = 5 per group). The tumor volumes and weights were measured at the indicated time points and the tumor tissues were then harvested for immunostaining analysis. For metastasis studies, GES-1-T cells (5 × 106) stably transfected with vectors were injected from tail vein of NOG mice (n = 5 per group). The mice lung and liver were carefully examined for tumor metastasis 4 weeks later.
Immunohistochemistry (IHC)
Fresh tumor tissues from the NOG mice were fixed in 4% formalin, embedded in paraffin and sections (4 μm) were prepared. Then, immunohistochemistry was performed using the primary antibodies against MMP2, PCNA and SMAD2 from Abcam. The complex was visualized with DAB complex, and the nuclei were counterstained with haematoxylin. All sections were scored by the semi-quantitative H-score approach.
Statistical analysis
All experiments were repeated at least three times. Statistical analysis was performed using SPSS 13.0 or the Prism statistical software package. Differences between the different groups were evaluated using the Student’s t-test or analysis of variance (ANOVA). Pearson’s correlation coefficient analysis was used to analyze correlations. All experimental data were presented as the mean ± S.D.. The differences were considered to be significant at p < 0.05.
Results
hsa_circ_0001829 is upregulated in GC cell lines and tissues, and is a stable circular RNA
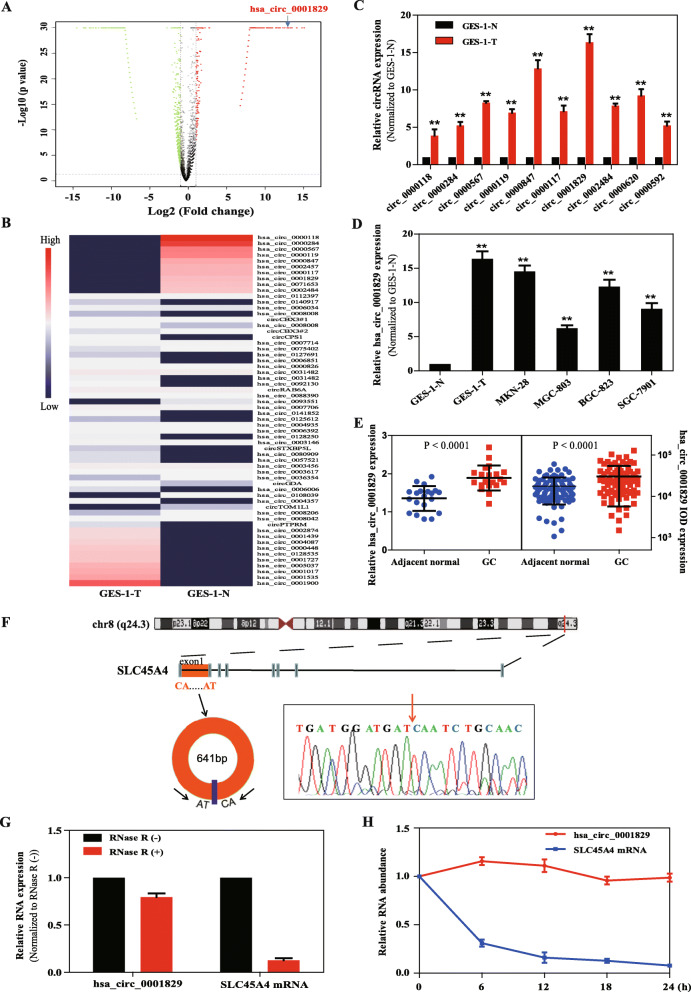
To investigate the potential role of circRNAs in MNNG-induced malignant transformation of GC cells, we analyzed the expression profile of circRNAs in GES-1-T and GES-1-N cells using ribosomal RNA depleted and RNase R treated RNA-seq, and found a total of 4650 differentially expressed circRNAs with a criterion of fold change (FC) more than 2.0 or less than 0.5 and P < 0.05, of which 1509 circRNAs were upregulated and 3141 circRNAs were downregulated in GES-1-T cells (Fig. 1a). The heat map expression of the top 10 upregulated circRNAs, the top 10 downregulated circRNAs and 40 randomly selected differentially expressed circRNAs were shown in Fig. 1b. For further analysis, we focused on the upregulated circRNAs and validated the induced expression of the top 10 upregulated circRNAs in GES-1-T cells by qRT-PCR assay. Consistent with the results of RNA-seq, the expressions of the 10 circRNAs were significantly increased in GES-1-T cells, with hsa_circ_0001829 showing the greatest increase (Fig. 1c). Furthermore, hsa_circ_0001829 exhibited upregulated expression in four GC cell lines (MKN-28, MGC-803, BGC-823 and SGC-7901) (Fig. 1d). Moreover, qRT-PCR analysis with 20 pairs of matched GC and adjacent tissue samples revealed that hsa_circ_0001829 expression was upregulated in GC tissues, and this result was further confirmed by ISH assay using tissue microarray in 83 GC samples (Fig. 1e).
Fig. 1.
Screening and expression of hsa_circ_0001829 in gastric cancer cells. a Volcano plot of the differentially expressed circRNAs in GES-1-T and GES-1-N cells (FC ≥ 2 or FC ≤ 0.5 and P < 0.05). Significantly upregulated circRNAs are indicated in red and downregulated circRNAs are indicated in green. The blue arrow indicates hsa_circ_0001829. b Heat map of the differentially expressed circRNAs in GES-1-T and GES-1-N cells. Red indicates a higher fold-change and blue indicates a lower fold-change. c Expression of the top 10 upregulated circRNAs in GES-1-T compared with GES-1-N cells was detected via qRT-PCR. d Expression of hsa_circ_0001829 in four gastric cell lines compared with GES-1-N cells. e Expression of hsa_circ_0001829 in 20 patients with GC (P < 0.0001) and in a tissue microarray of 83 GC paired patients (P < 0.0001). f The genomic loci of the SLC45A4 gene and hsa_circ_0001829. Red arrow indicates the back-splicing of hsa_circ_0001829 confirmed by Sanger sequencing. f qRT-PCR analysis of hsa_circ_0001829 and SLC45A4 mRNA after treatment with RNase R in GES-1-T cells. g The abundance of hsa_circ_0001829 and SLC45A4 mRNA was, respectively, tested by qRT-PCR in GES-1-T cells treated with Actinomycin D at the indicated time points. *p < 0.05, **p < 0.01
With a backspliced length of 641 bp, hsa_circ_0001829 is derived from exon 1 within solute carrier family 45 member 4 (SLC45A4) gene locus (Fig. 1f) and is located at chr8:142264087–142,264,728. Sanger sequencing of RT-PCR product confirmed the head-to-tail splicing of hsa_circ_0001829 (Fig. 1f). Then, the stability of hsa_circ_0001829 was evaluated. Compared with linear SLC45A4, hsa_circ_0001829 was resistant to digestion induced by RNase R exonuclease (Fig. 1g), indicating that hsa_circ_0001829 harbors a loop structure. Following treatment with Actinomycin D (an inhibitor of transcription), the half-life of hsa_circ_0001829 exceeded 24 h, while that of linear SLC45A4 displayed only about 4.3 h, revealing that hsa_circ_0001829 is highly stable (Fig. 1h).
These results demonstrated that hsa_circ_0001829 was upregulated in GC cell lines and tissues, and was a stable circular RNA.
-
2.
Silencing of hsa_circ_0001829 inhibits GC cells proliferation, migration and invasion in vitro
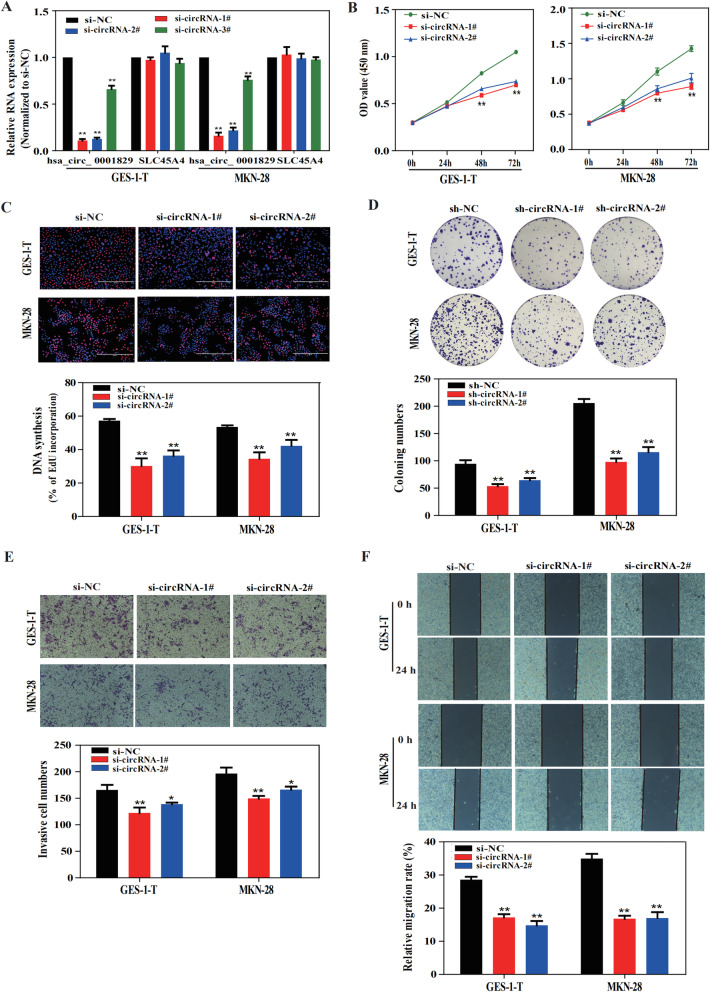
To investigate the biological function of hsa_circ_0001829, three small interfering RNAs (siRNAs) targeting the junction sites of hsa_circ_0001829 were designed to silence hsa_circ_0001829 expressions. These siRNAs significantly decreased hsa_circ_0002819 expression level with no effect on SLC45A4 linear isoform in both GES-1-T and MKN-28 cell lines (Fig. 2a). And we selected si-circRNA-1# and si-circRNA-2# for the subsequent experiments due to the relative higher interference efficiency. Following transfection of GES-1-T and MKN-28 cells with si-circRNA-1# and si-circRNA-2#, CCK-8 and EdU assays were employed to measure the cell proliferation. As shown, silencing of hsa_circ_0001829 markedly inhibited cell proliferation in both cell lines (Fig. 2b, c). Furthermore, colony-forming ability of these two cell lines stably transfected with sh-circRNA-1# and sh-circRNA-2# by lentiviral infection were significantly reduced after silencing of hsa_circ_0001829 (Fig. 2d). Additionally, knockdown of hsa_circ_0001829 impaired migration and invasion ability of GES-1-T and MKN-28 cells, as demonstrated by transwell and wound healing assays (Fig. 2e, f).
Fig. 2.
Silencing of hsa_circ_0001829 inhibits GC cells proliferation, migration and invasion in vitro. a The interference efficiency of three hsa_circ_0001829-targeting siRNAs on hsa_circ_0001829 and SLC45A4 mRNA level were examined by qRT-PCR. b-c CCK-8 and EdU assays of GES-1-T and MKN-28 cells transfected with control or siRNAs. d Colony formation assay of GES-1-T and MKN-28 cells stably transfected with control or shRNAs by lentiviral infection. e-f Transwell and wound healing assays of GES-1-T and MKN-28 cells transfected with control or siRNAs. *p < 0.05, ** p < 0.01
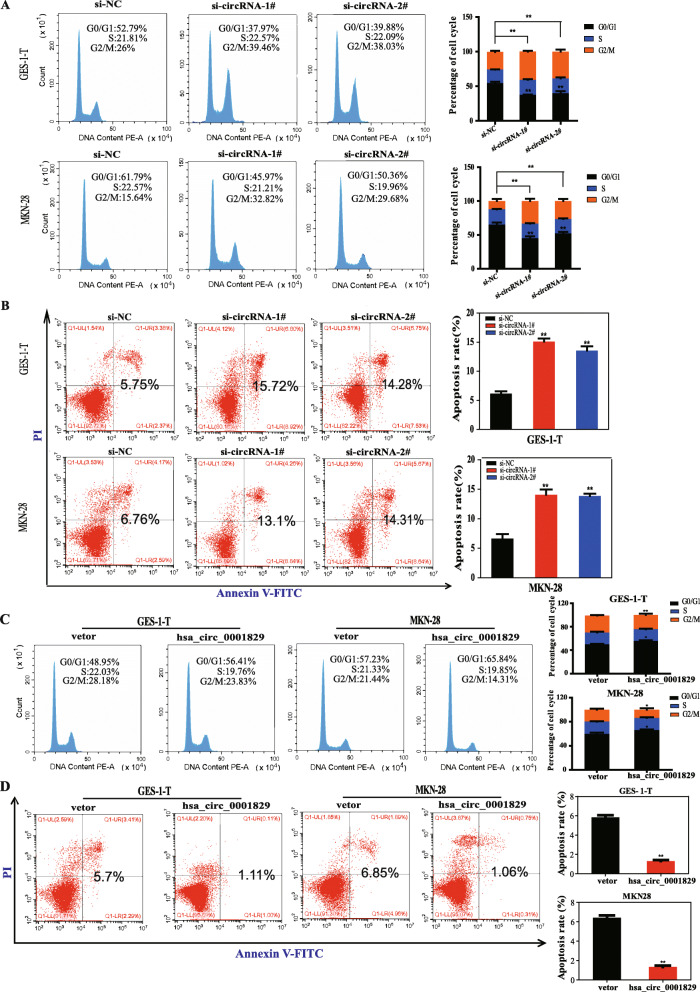
Then assays were conducted to evaluate whether hsa_circ_0001829 silencing had effect on the cell cycle and apoptosis. As shown in Fig. 4a, more cells were distributed in G2/M phase upon silencing hsa_circ_0001829, suggesting that knockdown of hsa_circ_0001829 induced cell cycle arrest at G2/M. Moreover, the apoptosis rates of GES-1-T and MKN-28 cells upon hsa_circ_0001829 knockdown were obviously increased (Fig. 4b). These results demonstrated that hsa_circ_0001829 silencing induced G2/M cell cycle arrest and increased apoptosis rates.
Fig. 4.
The changes of cell cycle and apoptosis rate of GC cells with silencing or overexpression of hsa_circ_0001829. a FCM analysis of the cell cycle in GES-1-T and MKN-28 cells transfected with control or siRNAs. b Apoptosis alteration of GES-1-T and MKN-28 cells after transfection with siRNAs. c FCM analysis of the cell cycle in GES-1-T and MKN-28 cells transfected with control vector or pcDNA-hsa_circ_0001829. d Apoptosis alter of GES-1-T and MKN-28 cells after transfection with control vector or pcDNA-hsa_circ_0001829. Data were presented as mean ± S.D., *p < 0.05, ** p < 0.01
Collectively, these results indicated that silencing of hsa_circ_0001829 inhibits GC cells proliferation, migration and invasion, and the decreased cell cycle progression and increased apoptosis may account for the inhibitory effect of hsa_circ_0001829 knockdown on GC proliferation.
-
3.
Overexpression of hsa_circ_0001829 promotes GC cells proliferation, migration and invasion in vitro
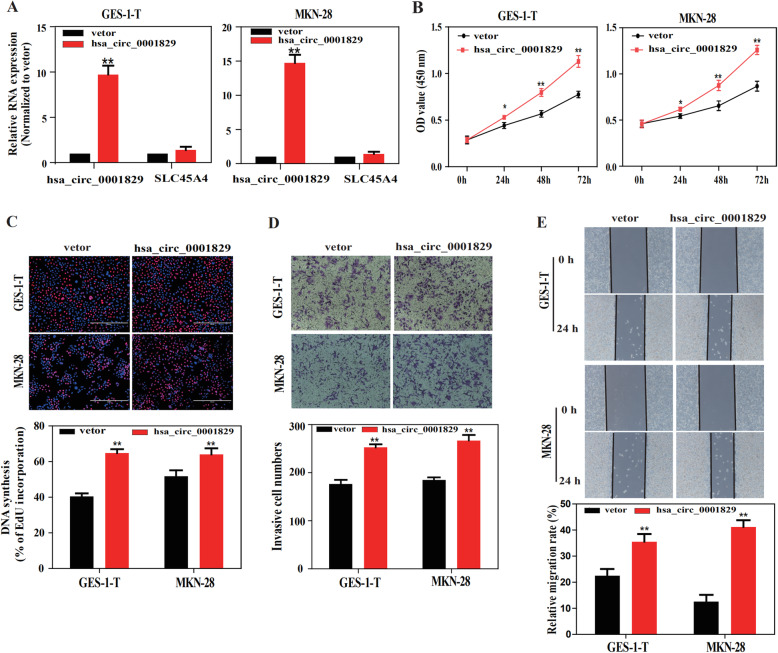
To further verify the functional role of hsa_circ_0001829, hsa_circ_0001829-overexpressing plasmid (pcDNA-circ_0001829) was constructed. The increase of hsa_circ_0001829 expressions induced by pcDNA-circ_0001829 was confirmed by qRT-PCR analysis, which showed approximately tenfold and fifteenfold overexpression in GES-1-T and MKN-28 cells, respectively; meanwhile, no significant change in SLC45A4 mRNA level was observed (Fig. 3a). Subsequent CCK-8 and EdU cell proliferation assays revealed that overexpression of hsa_circ_0001829 obviously increased the growth of both GES-1-T and MKN-28 cells (Fig. 3b, c). Additionally, the migration and invasion abilities of GES-1-T and MKN-28 cells were enhanced by transfection with hsa_circ_0001829 overexpression vector, as indicated by the results of transwell and wound healing assays (Fig. 3d, e).
Fig. 3.
Overexpression of hsa_circ_0001829 promotes GC cells proliferation, migration and invasion in vitro. a The overexpression efficiency of pcDNA-hsa_circ_0001829 on hsa_circ_0001829 and SLC45A4 mRNA level were measured by qRT-PCR. b CCK-8 assay of GES-1-T and MKN-28 cells transfected with control vector or pcDNA-hsa_circ_0001829. c EdU assay of GES-1-T and MKN-28 cells transfected with control vector or pcDNA-hsa_circ_0001829. Transwell d and wound healing e assays of GES-1-T and MKN-28 cells transfected with control vector or pcDNA-hsa_circ_0001829. *p < 0.05, **p < 0.01
Next, the effect of hsa_circ_0001829 overexpression on cell cycle and apoptosis rates of GES-1-T and MKN-28 cells were also analyzed. Contrary to the effect induced by hsa_circ_0001829 knockdown, a significant reduction of cells in the G2/M phase and an increase of cells in the G0/G1 phase were observed upon hsa_circ_0001829 overexpression (Fig. 4c). Furthermore, hsa_circ_0001829 overexpression reduced the apoptotic rates of GES-1-T and MKN28 cells (Fig. 4d). These results demonstrated that hsa_circ_0001829 overexpression stimulated cell cycle progression and decreased apoptosis rates.
Taken together, these data indicated that overexpression of hsa_circ_0001829 promotes GC cells proliferation, migration and invasion, and the promoting cell cycle progression and decreased apoptosis may account for the promotional effect of hsa_circ_0001829 overexpression on GC proliferation.
-
4.
Hsa_circ_0001829 acts as a molecular sponge for miR-155-5p
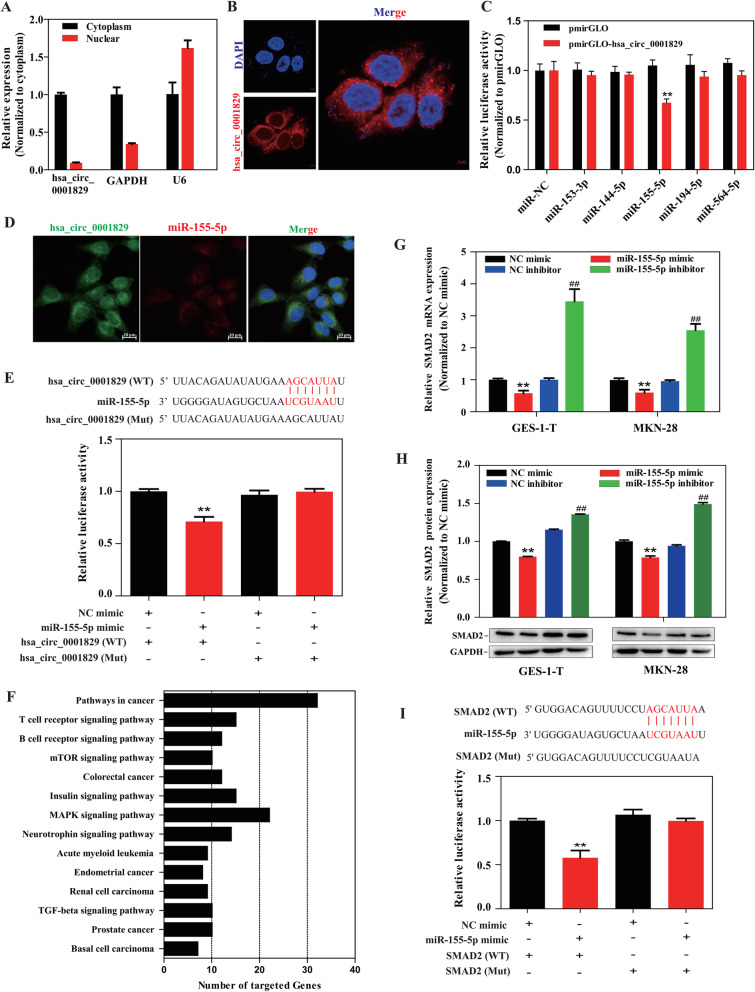
To explore the regulatory mechanism of hsa_circ_0001829 in GES-1-T cells, we first evaluated its subcellular distribution. The subcellular localization was first evaluated via qRT–PCR in nuclear or cytoplasmic fractions; as shown in Fig. 5a, hsa_circ_0001829 was mainly distributed in the cytoplasm of GES-1-T cells. Then FISH assay was employed, and the red fluorescent distribution also indicated that hsa_circ_0001829 was mainly distributed in the cytoplasm of GES-1-T cells (Fig. 5b).
Fig. 5.
hsa_circ_0001829 acts as a molecular sponge for miR-155-5p, and SMAD2 was validated as a target gene of miR-155-5p. a Cytoplasmic and nuclear distribution of hsa_circ_0001829 in GES-1-T cells was detected by qRT-PCR. b The subcellular localization of hsa_circ_0001829 in GES-1-T cells by FISH assay. The nuclei were stained with DAPI for blue color, and the cytoplasmic hsa_circ_0001829 was stained for red color. c Luciferase activity in GES-1-T cells co-transfected with five miRNA mimics and hsa_circ_0001829 wild-type luciferase reporter vector. d Dual RNA FISH for hsa_circ_0001829 (green) and miR-155-5p (red) was detected in GES-1-T cells, miR-155 was co-localized with hsa_circ_0001829 in cytoplasm, with nuclei staining with DAPI (blue). Scale bars: 20 μm. e (Up) The binding sites of wild type or mutant hsa_circ_0001829 with miR-155-5p. (Down) Dual luciferase reporter assays demonstrated that hsa_circ_0001829 could sponge miR-155-5p. f KEGG pathway analysis of the 556 predicted target genes. g The effects of miR-155-5p mimics and inhibitors on the expression of SMAD2 mRNA detected by qRT-PCR. *vs NC mimic group, #vs NC inhibitor group. h The effects of miR-155-5p mimics and inhibitors on the protein expression level of SMAD2 by western blot. *vs NC mimic group, #vs NC inhibitor group. i (Up) The binding sites of wild type or mutant SMAD2 3′-UTR with miR-155-5p. (Down) Dual luciferase reporter assays demonstrated that SMAD2 is a direct target of miR-155-5p. *p < 0.05, **p < 0.01, # p < 0.05, ## p < 0.01
Given that circRNAs in the cytoplasm have been widely reported to serve as miRNA sponges, we subsequently explored whether hsa_circ_0001829 could act as a miRNA sponge. TargetScan [22] and miRanda [23] were applied to predict the potential absorbing miRNAs of hsa_circ_0001829. Then, 13 overlapped miRNAs of the two databases were found to potentially interact with hsa_circ_0001829. Among the 13 candidate miRNAs, we mainly focused on five miRNAs (miR-153-3p, miR-144-5p, miR-194-5p, miR-155-5p and miR-564-5p), which have been reported to be closely related to cancer development [24–28]. For further investigation, dual luciferase reporter assays were performed and demonstrated that relative luciferase activity was reduced by more than 30% when cells were co-transfected with hsa_circ_0001829 wild-type vector and miR-155-5p mimics, whereas co-transfection with the other four miRNAs mimics induced non-significant differences, suggesting that hsa_circ_0001829 interacts directly with miR-155-5p (Fig. 5c). Consistently, the merged images obtained from FISH detection showed that hsa_circ_0001829 and miR-155-5p were co-localized mainly in the cytoplasm of GES-1-T cells (Fig. 5d). To further confirm the direct binding of miR-155-5p and hsa_circ_0001829, luciferase reporter plasmids with a wild type (WT) of hsa_circ_0001829 sequence and a mutant (Mut) hsa_circ_0001829 sequence in the binding sites of miR-155-5p were generated. We found that co-transfection of hsa_circ_0001829 (WT) and miR-155-5p mimic significantly reduced the luciferase activity, while co-transfection of hsa_circ_0001829 (Mut) and miR-155-5p mimic had no effect on luciferase activity (Fig. 5e).
These results indicated that hsa_circ_0001829 directly binds to miR-155-5p, acting as a sponge for miR-155-5p.
-
5.
SMAD2 was validated as a target gene of miR-155-5p
Having investigated the interaction between hsa_circ_0001829 and miR-155-5p, we next used TargetScan to predict the target genes of miR-155-5p and 556 target genes were obtained. Then KEGG pathway analysis by David [29] was applied to predict the involvement pathways of 556 genes, and 14 significantly enriched pathways were acquired (Fig. 5f). Importantly, “pathway in the cancer” ranked first, and SMAD2 as one of genes in this pathway was further investigated since besides TargetScan prediction, SMAD2 also got strongly-evidenced interactions with miR-155-5p by miRTarBase [30], an experimentally validated microRNA-target interactions database, and several reports indicate a promotional effect of SMAD2 on GC progression [31–33].
We then overexpressed or silenced miR-155-5p expression and measured the expression levels of SMAD2. Results showed that transfection of miR-155-5p mimics decreased the expression of SMAD2 mRNA, while transfection of miR-155-5p inhibitors increased SMAD2 mRNA expression in both GES-1-T and MKN-28 cells (Fig. 5g). We also detected SMAD2 protein expression in GES-1-T and MKN-28 cells after miR-155-5p knockdown or overexpression. MiR-155-5p overexpression notably decreased SMAD2 protein expression while miR-155-5p knockdown increased it (Fig. 5h). Thus, SMAD2 is a putative target gene of miR-155-5p. To confirm the interaction between miR-155-5p and SMAD2, we conducted dual luciferase reporter assays and found that cells co-transfected with the plasmid containing 3′-UTR-WT regions of SMAD2 (SMAD2 WT) and miR-155-5p mimic had significantly less luciferase activity than the controls, while mutation of the potential miR-155-5p binding sites in the SMAD2 3′-UTR (SMAD2 Mut) abolished this effect (Fig. 5i).
Collectively, these results indicated that miR-155-5p directly targets SMAD2.
-
6.
Hsa_circ_0001829 promotes GC progression via the miR-155-5p-SMAD2 pathway
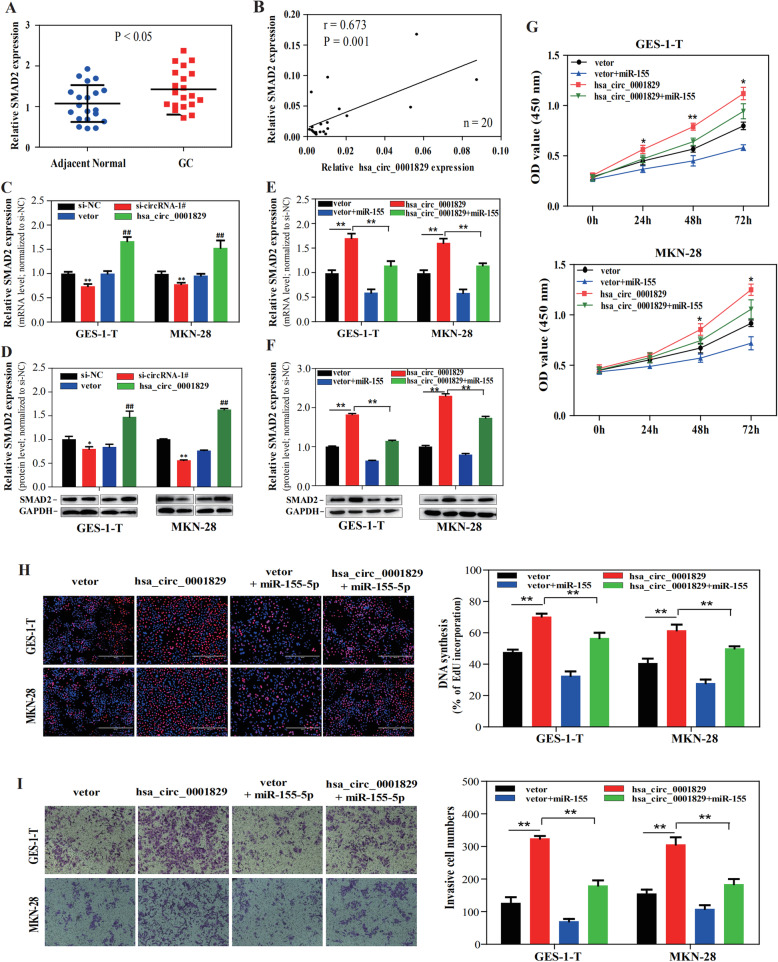
We further determined whether hsa_circ_0001829 regulates the expression of SMAD2 by sponging miR-155-5p. First, we tested the expression of SMAD2 mRNA in 20 pairs of matched GC and adjacent tissue samples. Compared with adjacent normal tissue, SMAD2 mRNA was highly expressed in cancer tissues (Fig. 6a). And we found that the level of hsa_circ_0001829 was positively correlated with the mRNA level of SMAD2 (Fig. 6b). Furthermore, hsa_circ_0001829 silencing decreased the RNA and protein expression level of SMAD2 and hsa_circ_0001829 overexpression significantly increased it, in both GES-1-T and MKN-28 cells (Fig. 6c, d). Next, GES-1-T and MKN-28 cells were co-transfected with miR-155-5p mimics and hsa_circ_0001829 overexpression vectors. The qRT-PCR assay showed that the promoting effect of hsa_circ_0001829 overexpression on SMAD2 expression was reversed by miR-155-5p mimics (Fig. 6e), which was also verified at the protein level by Western blot assays (Fig. 6f). These findings indicated that hsa_circ_0001829 may sponge miR-155-5p to regulate SMAD2 expression.
Fig. 6.
hsa_circ_0001829 promotes GC progression via the miR-155-5p-SMAD2 pathway. a SMAD2 expression in gastric cancer tissues compared with adjacent normal tissues (n = 20, P < 0.05). b Correlations between hsa_circ_0001829 and SMAD2 expression were found with Pearson′ correlation analysis in 20 paired GC tissues (P = 0.001). c-d The mRNA and protein expression level of SMAD2 in GES-1-T and MKN-28 cells with hsa_circ_0001829 knockdown or overexpression by qRT-PCR and western blot. * vs si-NC group, #vs vector group. e-i GES-1-T and MKN-28 cells were transfected with vector, miR-155-5p, hsa_circ_0001829, or miR-155-5p + hsa_circ_0001829. Then the relative mRNA and protein expression of SMAD2 was respectively measured by qRT-PCR analysis (e) and western blot assays (f), and the ability of cell proliferation and invasion was assessed by CCK-8 assay (g), EdU assay (h) and invasion assay (i). *p < 0.05, ** p < 0.01, # p < 0.05, ## p < 0.01
To determine whether hsa_circ_0001829 promotes GC progression by sponging miR-155-5p, rescue experiments were performed by co-transfecting hsa_circ_0001829 vectors and miR-155-5p mimics into GES-1-T or MKN-28 cells. The CCK-8 and EdU assay showed that miR-155-5p mimics effectively reversed the hsa_circ_0001829-induced promotion of proliferation in GES-1-T and MKN-28 cells (Fig. 6g, h). Likewise, transwell assay indicated that miR-155-5p mimics attenuated the ability of hsa_circ_0001829 to promote invasion of GES-1-T and MKN-28 cells (Fig. 6i). Taken together, these results demonstrated that hsa_circ_0001829 promotes GC progression through the miR-155-5p-SMAD2 pathway.
-
7.
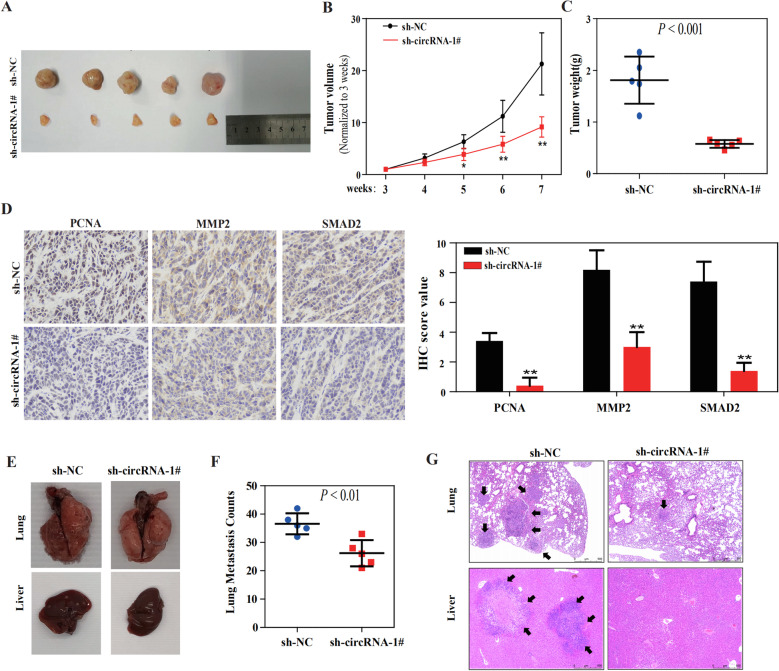
Suppression of hsa_circ_0001829 expression inhibits tumor growth and aggressiveness in vivo
To further clarify whether hsa_circ_0001829 influences tumor growth and aggressiveness in vivo, we performed tumor growth and metastasis experiments in NOG mice. For tumor growth studies, GES-1-T cells stably transfected with sh-circRNA-1# or negative control (sh-NC) were constructed and subcutaneously injected into the flank of NOG mice. Consistent with the observations in vitro, stably transfection of sh-circRNA-1# resulted in a significant decrease in the size and weight compared to those in the sh-NC group (Fig. 7a-c). IHC analysis demonstrated that the expression of PCNA and MMP2 was decreased by hsa_circ_000289 knockdown (Fig. 7d), which suggest that hsa_circ_0001829 knockdown inhibited the proliferative and invasion abilities of subcutaneous tumors. Also, the SMAD2 protein showed lower expression in the tumors of the sh-circRNA-1# group (Fig. 7d). For metastasis studies, GES-1-T cells stably transfected with sh-circRNA-1# or sh-NC were injected from tail vein of NOG mice. Notably, NOG mice in the sh-circRNA-1# group displayed less lung metastatic colonies than the sh-NC group (Fig. 7e-g). Moreover, metastatic foci in the liver could be observed in the sh-NC group, but not in the sh-circRNA-1# group (Fig. 7e, g).
Fig. 7.
Suppression of hsa_circ_0001829 expression inhibits tumor growth and aggressiveness in vivo. a-c) Schematic representation (a), in vivo growth curve (b), and weight at the end points (c) of xenograft tumors formed by subcutaneous injection of GES-1-T cells stably transfected with sh-NC or sh-circRNA-1# into the flanks of NOG mice (n = 5 for each group). d Representative images (left panel) and quantification (right panel) of immunohistochemical staining showing the expression of PCNA, MMP2 and SMAD2 within xenograft tumors from the sh-NC and sh-circRNA-1# groups. e-g Representative images (e), quantification (f) of lung metastatic colonization and H&E staining (g) of NOG mice treated with tail vein injection of GES-1-T cells stably transfected with sh-NC or sh-circRNA-1# (n = 5 for each group). *p < 0.05, ** p < 0.01
These results indicated that hsa_circ_0001829 knockdown inhibits tumor growth and aggressiveness of GC in vivo.
Discussion
Despite environmental carcinogen exposure is one of the most important causes of GC, the roles of circRNAs in environmental carcinogen-induced malignant transformation and their underlying mechanisms remain largely unknown. In this study, we applied RNA-seq analysis to screen differentially expressed circRNAs in MNNG induced malignant-transformed gastric epithelial cells, and then, we focused on the top 10 upregulated circRNAs and hsa_circ_0001829 was identified as the most highly upregulated circRNA through qRT-PCR assay, which was subsequently confirmed to be also upregulated in other four GC cell lines and GC tissues. Furthermore, results of loss- and gain-of-function assays showed that hsa_circ_0001829 promotes GC cells proliferation, migration and invasion, and the affected cell cycle progression and apoptosis rates may account for the effect of hsa_circ_0001829 on GC proliferation. Moreover, animal experiments revealed that hsa_circ_0001829 knockdown repressed tumor growth in vivo. These findings collectively indicated that hsa_circ_0001829 is upregulated in GC and promotes GC progression.
With the deepening of research on the role of circRNAs in tumors, numerous aberrantly expressed circRNAs have been unveiled in tumor tissues and cell lines, and they were considered as promising biomarkers for tumors in terms of their structural stability, specificity and high abundance [34, 35]. For example, Lu’s study demonstrated that circ-RanGAP1 was significantly upregulated in both GC tissues and exosomes from the plasma of GC patients and high circ-RanGAP1 expression was closely associated with an advanced TNM stage and worse survival, which suggest that circ-RanGAP1 might act as a potential prognostic biomarker [36]. Herein, we found that hsa_circ_0001829 was upregulated in GC tissues and cell lines, and functioned as a promoter of GC progression, implying that hsa_circ_0001829 might be a potent biomarker for diagnosis or prognosis. However, whether hsa_circ_0001829 is present in plasma samples or circulating exosomes requires further investigation.
Although our understanding of the functions of circRNAs is still nascent, increasing studies have shown that circRNAs can sequester microRNAs or proteins, modulate transcription, interfere with splicing, and even translate to produce polypeptides or proteins [37]. CircRNAs that act as miRNA sponges are predominantly in the cytoplasm, which occupy the same space of miRNAs, and these circRNAs generally function via a circRNA-miRNA-mRNA axis. For example, circRNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis [38]; circLARP4 inhibits cell proliferation and invasion of GC by sponging miR-424-5p and regulating LATS1 expression [39]. In our study, we showed that hsa_circ_0001829 was mainly located in the cytoplasm and miRNA-predicted analysis showed that hsa_circ_0001829 harbored sponging sites for miR-155-5p. We further confirmed that hsa_circ_0001829 was capable of binding to miR-155-5p by luciferase reporter assays. Therefore, our results indicated that hsa_circ_0001829 could bind to miR-155-5p and had the potential to play its oncogenic role by acting as a miRNA sponge. However, other functions of hsa_circ_0001829 still need to be elucidated in GC cells.
MiR-155-5p is a multifaceted regulator of cell proliferation, cell cycle, development, immunity and inflammation that plays pivotal roles in numerous cancers [40]. MiR-155-5p has been demonstrated to be elevated in breast and lung cancer and correlated with poor prognosis [41, 42]. However, the role of miR-155-5p in GC remains contradictory and largely unclear. Li and colleagues reported that miR-155-5p is one of the most downregulated miRNAs and may serve as a tumor suppressor in GC [43], whereas Qu′s group claimed that the levels of miR-155-5p were obviously increased and miR-155-5p acts as an oncogene in GC [44]. In our study, the CCK-8 and EdU assays showed that miR-155-5p mimics effectively reversed the hsa_circ_0001829-mediated promotion of GC cell proliferation, and likewise, the transwell assay showed that miR-155-5p mimics attenuated the ability of hsa_circ_0001829 to promote invasion of GC cells. Therefore, our results support the notion that miR-155-5p plays a suppressing role in GC development.
SMAD2, as a receptor-regulated SMAD member, is the primary intracellular signaling mediator and transcription factor for transforming growth factor-β (TGFβ) family signaling. Zhang et al. reported that both the mRNA and protein levels of SMAD2 were elevated in MNNG-initiated GC rats [31], and Bruna et al. found that high TGFβ-SMAD activity is present in aggressive, highly proliferative gliomas [32]. The reports that the expression of p-SMAD2 is associated with malignant phenotype and poor prognosis in patients with advanced GC [33] and that high TGFβ-SMAD activity confers poor prognosis in patients with glioma [32] support the promotional effect of SMAD2 on GC progression. Moreover, TGFβ can induce epithelial-to-mesenchymal transition (EMT) through canonical SMAD2/3 pathways [45, 46] and the EMT induced by SMAD2 may account for the effect of SMAD2 on GC cells invasion. In our study, we indicated that miR-155-5p could directly bind to SMAD2 and negatively regulate SMAD2 expression by dual luciferase reporter assay and qPCR. In addition, SMAD2 mRNA was highly expressed in cancer tissues and was positively correlated with the level of hsa_circ_0001829. We also found that SMAD2 expression was positively regulated by hsa_circ_0001829, which was attenuated by miR-155-5p. These results indicated that hsa_circ_0001829 could regulate SMAD2 expression by sponging miR-155-5p, thereby promoting proliferation and invasion of GC cells.
Conclusions
Taken together, we identified a novel circRNA, hsa_circ_0001829 that is upregulated in GC cell lines and tissues. Furthermore, we demonstrated that hsa_circ_0001829 promotes the malignant behaviors of GC cells by sponging miR-155-5p to regulate SMAD2 expression. Our findings firstly identify the role of hsa_circ_0001829 in GC, which may offer an effective biomarker for diagnosis/prognosis and a promising target for therapy in GC.
Supplementary Information
Acknowledgements
The authors thank Boyun Shi (Department of Oncology, The Fifth Affiliated Hospital of Guangzhou Medical University) for experimental supports.
Authors′ contributions
QLN, ZJD, ML and WXY performed experiments, analyzed data and wrote the paper; YWL, HL, MZL and XZ performed some experiments and analyzed data; WXY, JSW and XKZ conceived, designed the experiments and revised the draft of manuscript. All authors read and approved the final manuscript.
Abbreviations
- GC
Gastric cancer
- circRNA
Circular RNA
- NOCs
N-nitroso compounds
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- ncRNA
Noncoding RNA
- GES-1-T
Malignant transformed human gastric epithelial cells
- MREs
miRNA response elements
- RNA-seq
High-throughput RNA sequencing
- qRT-PCR
Quantitative reverse transcription-polymerase chain reaction
- SLC45A4
Solute carrier family 45 member 4
- TGFβ
Transforming growth factor-β
Funding
This study was supported by the National Natural Science Foundation of China (81673206 and 31500142), the Science and Technology Program of Guangzhou (201902020001 and 201905010004), and the Project of Educational Commission of Guangdong Province (2019KTSCX142).
Availability of data and materials
All data and materials can be provided upon request.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Research Ethics Committee of the Fifth Affiliated Hospital of Guangzhou Medical University. Primary GC samples were from the Fifth Affiliated Hospital of Guangzhou Medical University with informed consent.
Consent for publication
The authors agree for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Q. Niu, Z. Dong, and M. Liang are co-first authors.
This article has been retracted. Please see the retraction notice for more detail: 10.1186/s13046-023-02673-6
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/26/2023
This article has been retracted. Please see the Retraction Notice for more detail: 10.1186/s13046-023-02673-6
Contributor Information
Wenxia Yao, Email: yaowenxia917@126.com.
Jinsheng Weng, Email: 1280789568@qq.com.
Xinke Zhou, Email: zxkstar@126.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385(9972):977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomark Prev. 2014;23(5):700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007;6(4):429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19(12):1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol. 2004;5(2):148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 7.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams BD, Anastasiadou E, Esteller M, He L, Slack FJ. The inescapable influence of noncoding RNAs in Cancer. Cancer Res. 2015;75(24):5206–5210. doi: 10.1158/0008-5472.CAN-15-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Li X, Xu Y, Yang T, Yang Q, Yang C, et al. Identification of a long non-coding RNA NR_026689 associated with lung carcinogenesis induced by NNK. Oncotarget. 2016;7(12):14486–14498. doi: 10.18632/oncotarget.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caiment F, Gaj S, Claessen S, Kleinjans J. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res. 2015;43(5):2525–2534. doi: 10.1093/nar/gkv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Q, Xu E, Dai J, Wu J, Zhang S, Peng B, et al. miR-21 regulates N-methyl-N-nitro-N'-nitrosoguanidine-induced gastric tumorigenesis by targeting FASLG and BTG2. Toxicol Lett. 2014;228(3):147–156. doi: 10.1016/j.toxlet.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Luo Y, Liang M, Yao W, Liu J, Niu Q, Chen J, et al. Functional role of lncRNA LOC101927497 in N-methyl-N'-nitro-N-nitrosoguanidine-induced malignantly transformed human gastric epithelial cells. Life Sci. 2018;193:93–103. doi: 10.1016/j.lfs.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in Cancer. Cell. 2019;176(4):869–881. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang M, Liu Z, Lin H, Shi B, Li M, Chen T, et al. High-throughput sequencing reveals circular RNA hsa_circ_0000592 as a novel player in the carcinogenesis of gastric carcinoma. Biosci Rep. 2019;39(6):BSR20181900. [DOI] [PMC free article] [PubMed]
- 21.Liang M, Huang G, Liu Z, Wang Q, Yu Z, Lin H, et al. Elevated levels of hsa_circ_006100 in gastric cancer promote cell growth and metastasis via miR-195/GPRC5A signalling. Cell Prolif. 2019;52(5):e12661. doi: 10.1111/cpr.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H, Tian T, Liu X, Xia M, Chen C, Mai L, et al. Upregulated circ_0005576 facilitates cervical cancer progression via the miR-153/KIF20A axis. Biomed Pharmacother. 2019;118:109311. doi: 10.1016/j.biopha.2019.109311. [DOI] [PubMed] [Google Scholar]
- 25.Song L, Peng L, Hua S, Li X, Ma L, Jie J, et al. miR-144-5p Enhances the Radiosensitivity of Non-Small-Cell Lung Cancer Cells via Targeting ATF2. BioMed Res Int. 2018;2018:1–10. [DOI] [PMC free article] [PubMed]
- 26.Wu S, Sun H, Wang Y, Yang X, Meng Q, Yang H, et al. MALAT1 rs664589 polymorphism inhibits binding to miR-194-5p, contributing to colorectal Cancer risk, growth, and metastasis. Cancer Res. 2019;79(20):5432–5441. doi: 10.1158/0008-5472.CAN-19-0773. [DOI] [PubMed] [Google Scholar]
- 27.Al-Haidari A, Algaber A, Madhi R, Syk I, Thorlacius H. MiR-155-5p controls colon cancer cell migration via post-transcriptional regulation of human antigen R (HuR) Cancer Lett. 2018;421:145–151. doi: 10.1016/j.canlet.2018.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Mutlu M, Saatci O, Ansari SA, Yurdusev E, Shehwana H, Konu O, et al. miR-564 acts as a dual inhibitor of PI3K and MAPK signaling networks and inhibits proliferation and invasion in breast cancer. Sci Rep. 2016;6:32541. doi: 10.1038/srep32541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Li Y. miR-148a downregulates the expression of transforming growth factor-beta2 and SMAD2 in gastric cancer. Int J Oncol. 2016;48(5):1877–1885. doi: 10.3892/ijo.2016.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, Arias A, et al. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11(2):147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 33.Shinto O, Yashiro M, Toyokawa T, Nishii T, Kaizaki R, Matsuzaki T, et al. Phosphorylated smad2 in advanced stage gastric carcinoma. BMC Cancer. 2010;10:652. doi: 10.1186/1471-2407-10-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 35.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 38.Xie F, Li Y, Wang M, Huang C, Tao D, Zheng F, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. 2018;17(1):144. doi: 10.1186/s12943-018-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nariman-Saleh-Fam Z, Saadatian Z, Daraei A, Mansoori Y, Bastami M, Tavakkoli-Bazzaz J. The intricate role of miR-155 in carcinogenesis: potential implications for esophageal cancer research. Biomark Med. 2019;13(2):147–159. doi: 10.2217/bmm-2018-0127. [DOI] [PubMed] [Google Scholar]
- 41.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33(6):679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Xie S, Liu M, Chen Z, Liu X, Wang L, et al. The clinical significance of downregulation of mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric cancer tumorigenesis. Int J Oncol. 2014;45(1):197–208. doi: 10.3892/ijo.2014.2415. [DOI] [PubMed] [Google Scholar]
- 44.Qu Y, Zhang H, Sun W, Han Y, Li S, Qu Y, et al. MicroRNA-155 promotes gastric cancer growth and invasion by negatively regulating transforming growth factor-beta receptor 2. Cancer Sci. 2018;109(3):618–628. doi: 10.1111/cas.13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, Feng XH. TGF-beta signaling in cell fate control and cancer. Curr Opin Cell Biol. 2019;61:56–63. doi: 10.1016/j.ceb.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Tsubakihara Y, Moustakas A. Epithelial-Mesenchymal Transition and Metastasis under the Control of Transforming Growth Factor beta. Int J Mol Sci. 2018;19(11):3672. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials can be provided upon request.